Abstract
The transcription factor, Hoxc8, is a member of the homeobox gene family that is vital for growth and differentiation. Previously, we identified 34 genes whose expression levels were changed at least 2-fold by forced expression of Hoxc8 in C57BL/6J mouse embryo fibroblast cells using a mouse 16,463-gene oligonucleotide microarray. In the present study, we used the combined power of microarray profiling, global Hoxc8 DNA-binding site analysis, and high-throughput chromatin immunoprecipitation assays to identify direct and biologically relevant targets of Hoxc8 in vivo. Here we show that 19 of the 34 responsive genes contain Hoxc8 consensus DNA-binding sequence(s) in their regulatory regions. Chromatin immunoprecipitation analysis indicated that Hoxc8-DNA interaction was detected in five of the 19 candidate genes. All of these five target genes have been implicated in oncogenesis, cell adhesion, proliferation, and apoptosis. Overall, the genes described here should aid in the understanding of global regulatory networks of Hox genes and to provide valuable insight into the molecular basis of Hoxc8 in development and carcinogenesis.
Keywords: cancer, chromatin immunoprecipitation, Hox protein binding sites, microarray
Homeobox proteins are transcription factors that regulate the coordinated expression of multiple genes involved in development, differentiation and malignant transformation. The homeobox is a 183-bp DNA sequence coding a 61-aa domain defined as the homeodomain that is responsible for recognizing and binding sequence-specific DNA motifs (1). Several homeobox gene families have been identified, HOX, EMX, PMX, and MSX, as well as many more divergent homeobox genes. Among these, the Hox genes are most intriguing for having a recognized regulatory network structure. The Hox gene family consists of 39 genes organized in four clusters on four different chromosomes. The individual clusters span ≈200-kb and contain 9–11 genes (reviewed in ref. 2). These genes are expressed in a tissue-specific, stage-related fashion and play pivotal roles in development and differentiation. In addition to their important function in embryonic development, both gain and loss of expression for a wide variety of Hox genes have been reported in malignancies of the brain, breast, colon, kidney, lung, ovary, prostate, skin, and uterus (reviewed in ref. 3). Loss of Hoxc6 expression can induce apoptosis in prostate cancer cells (4), and Hoxb13 is a repressor of hormone-activated androgen receptor inducing growth suppression of prostate cancer cells (5, 6). Hoxb7 is a key factor for a tumor-associated angiogenic switch shown to activate basic fibroblast growth factor that in turn promotes cellular proliferation (7). Specific roles for Hoxb3 and Hoxd3 in cell proliferation have been reported during hematopoesis and angiogenesis (8, 9). Additionally, Hoxa5 and Hoxa10 regulate P53 expression in human breast cancer cells (10, 11).
Hoxc8 is expressed in both the neural tube and somitic mesoderm in the prospective thorax, and is essential for mouse forelimb and skeletal development (12). Hoxc8-null mutant mice show neuromuscular defects in the forelimb and skeletal defects in the ribs and vertebrae of the thorax (13). Overexpression of a Hoxc8 transgene has been shown to cause cartilage defects with an accumulation of proliferating chondrocytes and reduced maturation in skeletal elements (14). Hoxc8 plays an essential role in cancer development. Expression of Hoxc8 correlates with higher Gleason grades in prostate tumors, and the overexpression of Hoxc8 can suppress androgen-dependent transcription in prostate cancer cells (15, 16). It is selectively activated in cervical cancer cells (17) and expressed only in esophageal squamous cell carcinoma tissue, but not in noncancerous mucosa (18).
It is important to study the direct functional role of the Hox genes in carcinogenesis and development. Recently, we identified 34 putative mouse Hoxc8 target genes using microarray analysis (19). In the present study, we combined the power of expression profiling, computer analysis of the putative Hoxc8-DNA recognition sequence, and high-throughput chromatin immunoprecipitation (ChIP) assays to identify Hoxc8 direct target genes. We identified five of the 34 genes as direct downstream targets for Hoxc8 in vivo. These are (i) the zinc finger protein regulator of apoptosis (Zac1), a tumor suppressor gene; (ii and iii) neural cell adhesion molecules (Ncam) and Cadherin 11 (Cdh11) supporting cell–cell and cell–matrix adhesion; (iv) pigment epithelium-derived factor (PEDF) playing an important role in angiogenesis; and (v) osteopontin (OPN) serving as a key regulator of carcinogenesis, metastasis, and skeletogenesis. These five genes are all involved in crucial biological processes such as morphogenesis, differentiation, and tumorigenesis. Our results substantially expand our knowledge of the Hoxc8 regulatory network as it pertains to development and neoplasia.
Results
Search for Hoxc8 Consensus Recognition Sequences in the 5′-Flanking Regions of Perturbed Genes.
In our previous microarray study, we identified 34 genes whose expression level was changed at least 2-fold by forced expression of Hoxc8 in C57BL/6J MEF cell lines (19). To identify genes more likely to be direct targets of Hoxc8 regulation, we aligned the 34-gene set with mouse genomic sequences to map their 5′ sequences. Because most of the known Hox binding sequences are located in the proximal promoter region, the area of the upstream region between −1 and −1,000 relative to the transcription start site was retrieved. We then used the transfac match program (20) to search for Hox consensus recognition sequences in the upstream regions of the 34-gene set. Nineteen genes were found to contain one or more Hox consensus sequences in their proximal upstream region. Besides these proximal upstream regions, we also searched the Hox consensus recognition sequences in the 10-kb upstream regions of these 34 candidate genes and found that the Hox protein core consensus binding elements TTAT, TAAT, and TTAC, were present in all sequences searched. Because of the widespread distribution of Hox consensus binding sites in the mouse genome, the presence of binding sites is not likely to be instructive in identifying direct versus indirect targets of Hoxc8 gene regulation. Therefore, to further identify the direct targets of Hoxc8, we used ChIP to detect binding of Hoxc8 to the predicted Hoxc8-binding sequences in the Hoxc8 overexpressing MEF cell lines.
Hoxc8 Binds to Five of the Regulated Genes in a DNA Sequence-Specific Manner.
To distinguish direct versus indirect targets of Hoxc8 regulation, high-throughput ChIP assays were adopted. We focused on 19 genes whose proximal promoters were found to contain one or more consensus Hox binding sites. The ChIP assays were performed on the predicted Hoxc8 DNA-binding sites in the promoter of these 19 genes. MEF cell lines transfected with pcDNA4-Hoxc8 or pcDNA4-control were treated with formaldehyde to crosslink proteins to DNA. After sonification, the cross-linked chromatin was immunoprecipitated with Hoxc8-specific antibody. The cross-linked protein was then uncoupled from DNA by de-crosslink and proteolysis. Finally, the immunoprecipitated DNA was analyzed by quantitative PCR to determine which potential DNA-binding sites were actually bound by Hoxc8 in vivo. The ChIP assays were considered positive if the predicted Hoxc8-binding sequence(s) was enriched 3-fold greater than a no antibody control in three independent experiments. PCR primers were designed to generate a PCR amplicon that contained the predicted Hoxc8-binding sites. It should be emphasized that all 19 TTAT-, TAAT-, and TTAC-positive genes were subjected to ChIP analysis, and only the five indicated genes (see below) were ChIP positive, indicating the direct binding specificity.
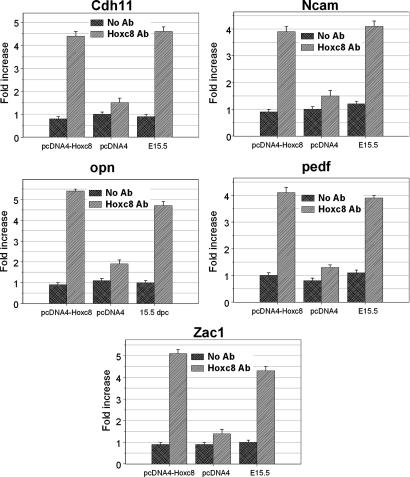
The ChIP results (Fig. 1) demonstrate that Hoxc8 protein interacts directly with predicted Hoxc8 binding sequences in Cdh11 (Cadherin 11); Ncam (Neural cell adhesion molecule); Opn (Osteopontin); pedf (pigment epithelium-derived factor) and Zac1 (the zinc finger protein regulator of apoptosis). Hoxc8 binding sequence(s) was enriched 5.5, 4.9, 6.0, 4.1, and 5.7 times in Cdh11, Ncam, Opn, pedf, and Zac1, respectively.
Fig. 1.
Hoxc8-DNA interaction detected by ChIP assay. MEF cells transfected with Hoxc8 expression plasmid (pcDNA4-Hoxc8) or pcDNA4 empty vector were used for ChIP assays. E15.5 mouse embryos were also used for ChIP assay. The assays were performed by using antibody recognizing Hoxc8 or no antibody as a negative control. Immunoprecipitated DNA was analyzed by quantitative PCR using Hoxc8 binding site-specific primers. Cdh11, Ncam, opn, pedf, and Zac1 were enriched >3-fold greater than no antibody negative control. The ChIP assays were repeated in three independent experiments with similar results.
Because MEF cell lines used in the above ChIP analysis were cultured from 15.5-days postcoitum C57BL/6J embryos, we wished to further detect whether Hoxc8 protein can bind to the promoter of these five genes at the same mouse embryonic stage. For this reason, embryonic day 15.5 (E15.5) mouse embryos were cross-linked with 1% formaldehyde diluted in serum-free DMEM for 15 min at room temperature. Quantitative PCR using the Hoxc8 antibody precipitated chromatin showed the predicted Hoxc8 binding sites were enriched 5.8, 3.4, 4.7, 3.6, and 4.3 times in Cdh11, Ncam, Opn, pedf, and Zac1, respectively, in the E15.5 ChIP assay (Fig. 1). The above results further verified that these five genes are authentic Hoxc8 target genes in vivo.
Hoxc8 Regulates the Level of pedf, Ncam, Zac1, Opn, and Cdh11 in MEF and HeLa Cells.
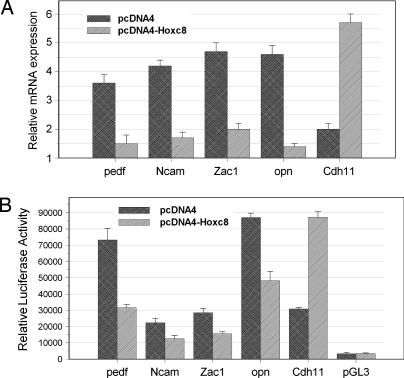
To determine whether the expression level of the five identified Hoxc8 direct target genes are regulated by Hoxc8 protein, quantitative RT-PCR assays were performed to detect their expression differences between the MEF cell lines transfected with pcDNA4-Hoxc8 or the pcDNA4 empty vector. As shown in Fig. 2A, pedf, Ncam, Zac1, and Opn were down-regulated 2.4, 2.5, 2.4, and 3.3-fold, respectively when Hoxc8 was overexpressed in MEF cell lines; in contrast, the expression of Cdh11 was increased 2.9-fold by Hoxc8 overexpression. Quantitative RT-PCR results are consistent with the previous microarray analysis (19). Comparison of activation of these five Hoxc8 target genes in the control and the Hoxc8 overexpressed NIH 3T3 cell lines demonstrated similar results with MEF cells (data not shown).
Fig. 2.
Effects of Hoxc8 on pedf, Ncam, Zac1, opn, and Cdh11 mRNA and promoter expressions in MEF and HeLa cells. (A) Quantitative RT-PCR analysis of pedf, Ncam, Zac1, opn, and Cdh11 mRNA expression in MEF cells transfected with Hoxc8 expression plasmid (pcDNA4-Hoxc8) or pcDNA4 empty vector. All results were normalized to the β-actin value. (B) Relative luciferase activities for different constructs. HeLa cells were plated at a density of five × 104 cells per well in 12-well plates for transfections. pedf, Ncam, Zac1, opn, and Cdh11 promoter constructs (0.3 μg) were cotransfected with 0.1 μg of Hoxc8 expression plasmid (pcDNA4-Hoxc8) or 0.1 μg of pcDNA4 empty vector separately. Transfected cells were harvested and lysed in 48 h after transfection. The luciferase activity was measured and normalized to the Renilla luciferase level as internal controls. Data are shown with the mean of ± SD of triplicates.
To gain further insight into the Hoxc8 regulation of pedf, Ncam, Zac1, Opn, and Cdh11 transcription, we have cloned an ≈1-kb DNA fragment of the promoter regions of each of these five genes into a pGL3-basic luciferase reporter vector to detect their responsiveness to Hoxc8. The five luciferase reporter constructs were cotransfected in HeLa cells with pcDNA4-Hoxc8 or pcDNA4 expression vector separately. The transfection of the five constructs resulted in a significant increase in luciferase activity compared with the pGL3-basic control vector, especially Opn, that showed a 26-fold induction (Fig. 2B). In addition, Hoxc8 has an obvious effect on pedf, Ncam, Zac1, Opn, and Cdh11 gene expression. As shown in Fig. 2B, an increase of Hoxc8 protein expression induces an ≈2.4-, 1.5-, 1.7-, and 1.9-fold decrease in the expression of pedf, Ncam, Zac1, and Opn, respectively, and a 2.8-fold increase in the expression of cdh11. Our luciferase results provide an additional line of evidence that Hoxc8 can regulate the expression of these five genes.
Target Gene Expression in Vivo.
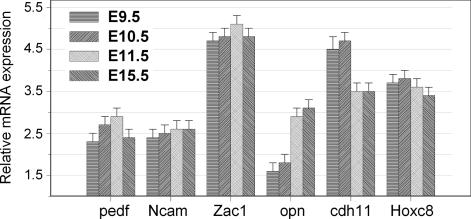
To determine whether the identified five Hoxc8 target genes are normally expressed in embryo stages that express Hoxc8, and therefore present an opportunity for regulation, we performed quantitative RT-PCR on these five genes for expression in the E9.5, E10.5, E11.5, and E15.5 stage embryos (Fig. 3). As shown in Fig. 3, Hoxc8, pedf, Ncam, Zac1, and Cdh11 express highly in all embryonic stages. Interestingly, Opn expresses at a low level in E9.5 and E10.5 stages, and then after E11.5 rises to relatively high levels. Opn mRNA levels increase ≈6-fold from E9.5 to E15.5 (Fig. 3).
Fig. 3.
pedf, Ncam, Zac1, opn, Cdh11 and Hoxc8 expression level at different mouse embryo stages. Quantitative RT-PCR analysis of pedf, Ncam, Zac1, opn, Cdh11, and Hoxc8 mRNA expression in E9.5, E10.5, E11.5, and E15.5 mouse embryo stages. The level of gene expression was normalized to β-actin. Results are representative of three independent experiments.
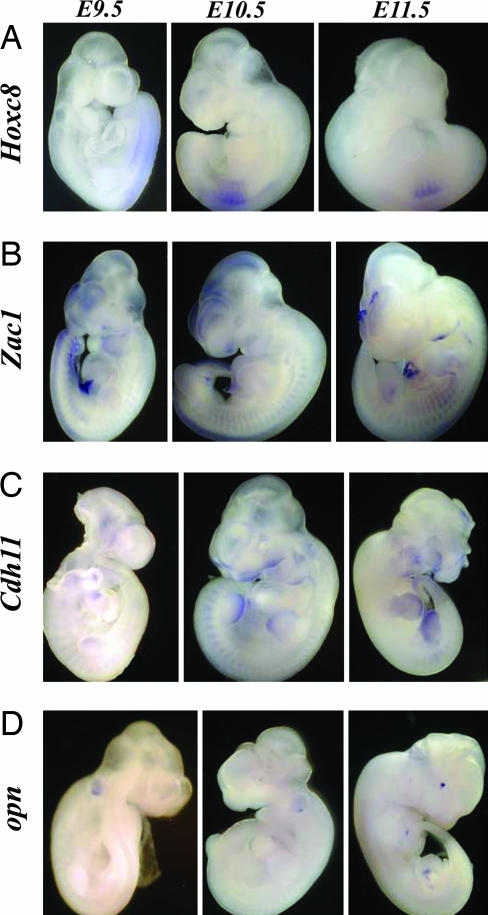
After quantifying variations in the RNA levels of these five candidate genes over developmental time, we performed whole-mount in situ hybridization (WISH) assays using E9.5, E10.5, and E11.5 stage embryos to detect whether Zac1, Opn, and Cdh11 have an in vivo expression pattern that overlaps with Hoxc8. Hoxc8 expression was detected in the paraxial mesoderm, somites, and neural tube (Fig. 4A). High levels of Zac1 expression was detected in brain, somites, neural tube, hindlimb, and forelimb (Fig. 4B). As shown in Fig. 4C, upon WISH staining, Cdh11 expression is clearly observed in branchial arches, somites, forelimb, and hindlimb (Fig. 4C). Opn expression could not be detected at E10.5 along the anterior/posterior axis; however, its expression was clearly detectable in the eye, forelimb, and hindlimb at E11.5 (Fig. 4D).
Fig. 4.
Hoxc8, Zac1, Cdh11, and opn expression pattern during mouse early embryogenesis. Whole-mount in situ hybridization of Hoxc8 (A), Zac1 (B), Cdh11 (C), opn (D) mRNA at E9.5, E10.5, and E11.5.
Discussion
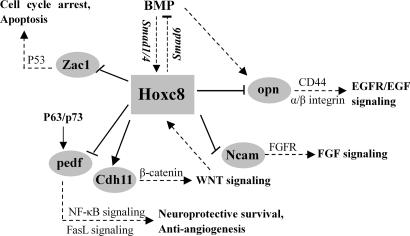
Hox genes encode transcription factors that play a vital role in embryonic development, cell fate determination, differentiation, and proliferation. Recent reports have demonstrated differences in Hox gene expression between normal and neoplastic tissues, but Hoxc8’s role in neoplasia remains elusive. To decipher the mechanisms by which Hox genes regulate development and neoplasia, it is necessary to characterize their upstream regulators and downstream target genes. We have used microarray and ChIP to validate direct targets of Hoxc8, namely, Ncam, Cdh11, Opn, pedf, and Zac1 as Hoxc8 direct target genes. Strikingly, the function of these five potential Hoxc8 target genes has been implicated in embryonic and cancer development. In fact, all of these five candidate genes are members of the classical FGF, BMP, WNT, p53, or NF-KB signaling pathways. The seminal regulatory roles of these five Hoxc8 target genes (Fig. 5) are described below.
Fig. 5.
The Hoxc8-regulated transcriptional network in vivo. Hoxc8 can regulate the cross-talk between Wnt, BMP, FGF signaling pathway by direct down-regulating Ncam, opn, pedf, and Zac1 and up-regulating Cdh11 in vivo. Direct pathways are indicated by solid lines and arrows, and indirect consequences of pathway activation and inactivation are indicated by dotted lines.
Cell Adhesion.
During embryonic development, cell–cell interactions mediated by cell adhesion molecules play key roles in the control of cell movement, aggregation, and migration, and also influence cell proliferation and differentiation. Hox proteins similarly are important developmental regulators that interact in multiple germ layers to coordinate pattern formation, cell division, cell migration, and differentiation. Increasing evidence functionally links Hox genes to cell adhesion molecules. Here, we identify three cell adhesion molecules, Ncam, Cdh11 and Opn, as Hoxc8 direct target genes.
Ncam.
Ncam is down-regulated 2.5-fold by Hoxc8 in MEF cells (Fig. 2A). NCAM is a Ca2+-independent Ig related cell–cell adhesion molecule that mediates homotypic and heterotypic cell–cell and cell–matrix adhesion (21). Ncam transcripts are first detected around day 8.5 in somites and neural tube, and it is thought to play an important role as a regulator of morphogenetic processes. The regional specificity of NCAM in the neural tube added further weight to the notion that Ncam may be a downstream target of Hox gene products (22). Hoxc6, which displays overlapping expression with Ncam along the body axis induces a positive regulatory effect on the promoter of Ncam. Hoxb9 induces Ncam transcription by binding to its promoter; however, cotransfection with Hoxb8 antagonizes transactivation (reviewed in ref. 23). Although Ncam has been extensively studied as a cell–cell adhesion molecule, recent reports highlight the role of Ncam in signal transduction. Loss of NCAM function during tumor progression affects cell–matrix adhesion through the loss of FGFR-induced, integrin-mediated cell–matrix adhesion (24).
Cdh11.
Cadherins modulate calcium ion-dependent cell–cell adhesion and are important in cell aggregation, migration, and cell sorting. Cadherin 11, or OB-cadherin, is a member of the cadherin family (25). Hoxc8 up-regulates Cdh11 expression 2.9-fold in MEF cells (Fig. 2A). WISH staining shows that cdh11 is highly expressed in the branchial arches, head mesenchyme, somites, and the distal portion of limb mesenchyme (Fig. 4B). The expression pattern of Cdh11 correlates with mesenchymal morphogenesis in the head, somite, and limb buds of early mouse embryos. Cdh11 is essential for the process of the segmentation of the paraxial mesoderm layer (26). In the trunk region of the Hoxc8 knockout mice, several skeletal segments were transformed into the likeness of more anterior ones (13). Cdh11, also localized to the cell membrane, can mediate the formation of a functional adherens junction complex, recruiting β-catenin to the membrane. The expression of Cdh11 does not dramatically alter anchorage-independent growth or cellular proliferation rate, but has been shown to produce significant changes in the invasive capacity of cancer cells (27).
OPN.
OPN is a secreted, integrin-binding glycophosphoprotein that has been linked to tumor progression and survival in several solid tumors, including head and neck cancers (28). Here, we identified Opn as a Hoxc8 direct target gene that is down-regulated 3.3-fold by Hoxc8 in MEF cells (Fig. 2). Previously, Shi et al. (29) showed that Hoxc8 binds directly to the Opn promoter region by gel-shift analysis assay. Our ChIP analysis supports and extends their findings. Upon WISH staining, Opn expresses in the eye and forelimb and hindlimb buds (Fig. 4D). Bone morphogenetic protein (BMP)/Smads signaling has been reported to induce Opn expression in a Hoxc8- regulated way (29). Currently, cumulative evidence suggests that OPN functions in the regulation of tumor metastasis and invasion. OPN is a ligand for the integrin and CD44 families of receptors, and this binding allows OPN to mediate adhesive cell–matrix interaction and to activate cellular second messengers in signal transduction pathways. It has been reported that the integrin-dependent induction of human mammary epithelial cells migration by OPN involves and depends on the activation of at least two different growth factor/receptor pathways (EGF ligands/EGFR and HGF/Met) and multiple different signal transduction pathways (30, 31).
Angiogenesis and Apoptosis.
Hox gene expression is linked to apoptosis and angiogenesis. Hoxa5 and Hoxa10 can regulate p53 expression in human breast cancer cell lines (10, 11). Induction of HOXA5 expression causes caspase 2- and caspase 8-mediated apoptotic cell death (32). Hoxb5 and Hoxb7 also have roles in angiogenesis (7).
pedf.
Here, we show that pedf is a direct target of Hoxc8 in that it was down-regulated 2.4-fold by Hoxc8 in MEF cells (Fig. 2A). PEDF, an angiogenesis inhibitor with neurotropic properties, balances angiogenesis in the eye and blocks tumor progression (33). However, PEDF is not simply an antiangiogenic factor: it has neuroprotective effects in the nervous system and an apoptotic effect in endothelial cells. PEDF interacts with the NF-κB pathway to stimulate the transcription of neuroprotective genes in the nervous system; and PEDF can generate anti-angiogenic and apoptosis signal by activating the Fas–Fas ligand (FasL) death cascade in endothelial and epithelial cells (34). Additionally, PEDF contributes as a natural angiogenesis inhibitor in two hormone-sensitive organs, the prostate and pancreas (35). pedf has also been reported to be a direct target of p63 or p73, p53 family member genes (36).
Zac1.
Zac1 is directly down-regulated 2.4-fold by Hoxc8 in MEF cell (Fig. 2A). Zac1 is a zinc finger transcription factor that elicits antiproliferative activity and is a potential tumor suppressor gene that exhibits its tumor suppressor activity characterized by induction of apoptosis and G1 arrest. Zac1 is the first gene besides p53 that concurrently induces apoptosis and cell cycle arrest (37). As a transcription factor, Zac1 shares a number of similar functions with p53 (38). During mouse embryogenesis, Zac1 was shown to have strong expression in brain areas; in addition, Zac1 expression was detected in the neural tube, somites, forelimbs, and hindlimbs (Fig. 4B). The Zac1 transcript is predominantly localized in developing chondrogenic tissue and suggested that Zac1 is a potential regulatory gene involved in chondrogenic differentiation and cartilage development (39, 40). Overexpression of a Hoxc8 transgenic mice has been reported to cartilage defects (14). Here, we identified Zac1 as a Hoxc8 target gene in vivo, so it is possible that Hoxc8 protein altered the chondrocyte differentiation by regulating the Zac1 expression in the early embryo stage.
Hox genes are master transcription factors, and they function at the top of a genetic hierarchy controlling pathway. Multiple cellular processes are regulated through the Hox network. Hoxc8 can regulate the cross-talk between Wnt, BMP, and FGF signaling pathways by direct targeting Ncam, Cdh11, Opn, pedf, and Zac1 in vivo (Fig. 5). Our results provide insights in understanding the roles of Hox genes in cancer and development.
Materials and Methods
Cell Culture.
Cells were cultured in DMEM (GIBCO/BRL) supplemented with 10% FCS (HyClone) and 100 μg/ml penicillin–streptomycin–glutamine (GIBCO/BRL). HeLa and NIH 3T3 cells were purchased from ATCC. pcDNA4-Hoxc8 and pcDNA4-control expressing mouse embryonic fibroblast cell lines have been described (19).
Plasmid Constructions.
The plasmid pcDNA4-Hoxc8 harboring mouse Hoxc8 has been described (19). The 1-kb promoters for the pedf, Ncam, Zac1, Opn, and Cdh11 were amplified by PCR from mouse genomic DNA and cloned into the SmaI and BglII sites of the pGL3-basic vector (Promega) to generate a luciferase reporter constructs: pedf-luc, Ncam-luc, Zac1-luc, Opn-luc, and Cdh11-luc.
Quantitative ChIP Assay.
The formaldehyde cross-link and ChIP assays of tissue culture cells were performed as described (19). We followed a protocol established by the Farnham laboratory for ChIP performed with mouse embryonic tissues (http://mcardle.oncology.wisc.edu/farnham/protocols/tissues.html). Quantitative PCR using ChIP samples was carried out by using a SYBR Green qPCR kit (New England Biolabs). The fold change in the specific binding was normalized to GAPDH and mock IP values. The primer sequences are available upon request.
RNA Isolation and Real-time PCR Quantification.
Total RNA was isolated with RNeasy miniprep columns plus RNase-free DNaseI set (Qiagen). The cDNA for real-time PCR was made by using the OmniScript kit (Qiagen). Real-time PCR was performed in triplicate by using Taqman probes and the ABI prism 7900 sequence detection system (Applied Biosystems). Samples were normalized to the β-actin values.
Transfection and Luciferase Assay.
HeLa cells (5 × 104 cells in 12-well plates) were transiently transfected by using Lipofectamine 2000 reagent (Invitrogen) with a total of 0.3 μg of luciferase reporter plasmid (pedf-luc, Ncam-luc, Zac1-luc, Opn-luc, and Cdh11-luc) and different expression plasmids as indicated. Luciferase activities were assayed 48 h after transfection by using the Dual Luciferase assay kit (Promega) according to the manufacturer’s directions. Luciferase values shown in Fig. 2B are representative of transfection experiments performed in triplicate in three independent experiments.
Whole-Mount in Situ Hybridization.
Embryos were collected from matings of FVB mice at E9.5, E10.5, and E 11.5. Embryos were staged by assigning noon of the day of vaginal plug as E0.5. In situ probe templates were gifts from M. Chu (zac1, ref. 39) and D. C. Page (Opn and Cdh11, ref. 41). Hoxc8 probe template was from our laboratory (42). DNA template was linearized and DIG labeled antisense probes were prepared by in vitro transcription with SP6, T3 or T7 polymerase (Roche). WISH using embryos fixed in 4% paraformaldehyde was performed according to standard procedures.
Acknowledgments
We thank Dr. M. Chu (Thomas Jefferson University, Philadelphia) for providing Zac1 WISH probe, Dr. D. C. Page (Massachusetts Institute of Technology, Boston) for the Opn and Cdh11 WISH probes, and Dr. Hailong Wang for providing statistical analysis. This work was supported by National Institutes of Health Grant GM09966.
Abbreviations
- ChIP
chromatin immunoprecipitation
- En
embryonic day n
- WISH
whole-mount in situ hybridization.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Gehring W. J., Hiromi Y. Annu. Rev. Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 2.Cillo C., Cantile M., Faiella A., Boncineli E. J. Cell Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- 3.Abate-Shen C. Nat. Rev. Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran S., Liu P., Young A. N., Yin-Goen Q., Lim S. D., Laycock N., Amin M. B., Carney J. K., Marshall F. F., Petros J. A., et al. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 5.Jung C. Y., Kim R. S., Zhang H. J., Lee S. J., Jeng M. H. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 6.Jung C. Y., Kim R. S., Lee S. J., Wang C., Jeng M. H. Cancer Res. 2004;64:3046–3051. doi: 10.1158/0008-5472.can-03-2614. [DOI] [PubMed] [Google Scholar]
- 7.Carè A., Felicetti F., Meccia E., Bottero L., Parenza M., Stoppacciaro A., Peschle C., Colombo M. P. Cancer Res. 2001;61:6532–6539. [PubMed] [Google Scholar]
- 8.Boudreau N., Andrews C., Srebrow A., Ravanpay A., Cheresh D. A. J. Cell Biol. 1997;139:257–264. doi: 10.1083/jcb.139.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers C., Charboneau A., Cheung I., Hanks D., Boudreau N. Am. J. Pathol. 2002;161:2099–2109. doi: 10.1016/S0002-9440(10)64488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman V., Martensen S. A., Reisman D., Evron E., Odenwald W. F., Jaffee E., Marks J., Sukumar S. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 11.Chu M. C., Selam F. B., Taylor H. S. Cancer Biol. Ther. 2004;3:568–572. doi: 10.4161/cbt.3.6.848. [DOI] [PubMed] [Google Scholar]
- 12.Shashikant C. S., Ruddle F. H. Proc. Natl. Acad. Sci. USA. 1996;93:12364–12369. doi: 10.1073/pnas.93.22.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouellic H. L., Lallemand Y., Brulet P. Cell. 1992;69:251–264. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- 14.Yueh Y. G., Gardner D. P., Kappen C. Proc. Natl. Acad. Sci. USA. 1998;95:9956–9961. doi: 10.1073/pnas.95.17.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller G. J., Miller H. L., Bokhoven A, Lambert J. R., Werahera P. N., Schirripa O., Lucia M. S., Nordeen S. K. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 16.Waltregny D., Alami Y., Clausse N., Leval J., Castronovo V. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 17.Alami Y., Castronovo V., Belotti D., Flagiello D., Clausse N. Biochem. Biophys. Res. Commun. 1999;257:738–745. doi: 10.1006/bbrc.1999.0516. [DOI] [PubMed] [Google Scholar]
- 18.Chen K. N., Gu Z. D., Ke Y., Li J. Y., Shi X. T., Xu G. W. Clin. Cancer Res. 2005;11:1044–1049. [PubMed] [Google Scholar]
- 19.Lei H. Y., Wang H. L., Juan A. H., Ruddle F. H. Proc. Natl. Acad. Sci. USA. 2005;102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Homischer K., Karas D., Kel A. E., Kel-Margoulis O. V., et al. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crossin K. L., Krushel L. A. Dev. Dyn. 2000;218:260–279. doi: 10.1002/(SICI)1097-0177(200006)218:2<260::AID-DVDY3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Bally-Cuif L., Goridis C., Santoni M.-J. Development (Cambridge, U.K.) 1993;117:543–552. doi: 10.1242/dev.117.2.543. [DOI] [PubMed] [Google Scholar]
- 23.Akin Z. N., Nazarali A. J. Cell Mol. Neurobiol. 2005;25:697–741. doi: 10.1007/s10571-005-3971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christofori G. EMBO J. 2003;22:2318–2323. doi: 10.1093/emboj/cdg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christofori G., Cavallaro U. Nat. Rev. Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 26.Kimura Y., Matsunami H., Inoue T., Shimamura K., Uchida N., Ueno T., Miyazaki T., Takeichi M. Dev. Biol. 1995;169:347–358. doi: 10.1006/dbio.1995.1149. [DOI] [PubMed] [Google Scholar]
- 27.Feltes C. M., Kudo A., Blaschuk O., Byers S. W. Cancer Res. 2002;62:6688–6697. [PubMed] [Google Scholar]
- 28.Rittling S. R., Chambers A. F. Br. J. Cancer. 2004;90:1877–1881. doi: 10.1038/sj.bjc.6601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi X., Yang X., Chen D., Chang Z., Cao X. J. Biol. Chem. 1999;274:13711–13717. doi: 10.1074/jbc.274.19.13711. [DOI] [PubMed] [Google Scholar]
- 30.Wai P. Y., Kuo P. C. J. Surg. Res. 2004;121:228–241. doi: 10.1016/j.jss.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 31.Tuck A. B., Hota C., Wilson S. M., Chambers A. F. Oncogene. 2003;22:1198–1205. doi: 10.1038/sj.onc.1206209. [DOI] [PubMed] [Google Scholar]
- 32.Chen H., Chung S., Sukumar S. Mol. Cell. Biol. 2004;24:924–935. doi: 10.1128/MCB.24.2.924-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tombran-Tink J., Barnstable C. J. Biochem. Biophys. Res. Commun. 2004;316:573–579. doi: 10.1016/j.bbrc.2004.02.076. [DOI] [PubMed] [Google Scholar]
- 34.Tombran-Tink J., Barnstable C. J. Nat. Rev. Neurosci. 2003;4:628–636. doi: 10.1038/nrn1176. [DOI] [PubMed] [Google Scholar]
- 35.Doll J. A., Stellmach V. M., Bouck N., Bergh A. R., Lee C., Abramson L. P., Cornwell M. L., Pins M. R., Borensztajn J., Crawford S. E. Nat. Med. 2003;9:774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki Y., Naishiro Y., Oshima Y., Imai K., Nakamura Y., Tokino T. Oncogene. 2005;24:5131–5136. doi: 10.1038/sj.onc.1208695. [DOI] [PubMed] [Google Scholar]
- 37.Spengler D., Villalaba M., Hoffmann A., Pantaloni C., Houssami S., Bockaert J., Journot L. EMBO J. 1997;16:2814–2825. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S. M., Schonthal A. H., Stallcup M. R. Oncogene. 2001;20:2134–2143. doi: 10.1038/sj.onc.1204298. [DOI] [PubMed] [Google Scholar]
- 39.Tsuda T., Markova D., Wang H., Evangelisti L., Pan T. C., Chu M. L. Dev. Dyn. 2004;229:340–348. doi: 10.1002/dvdy.10439. [DOI] [PubMed] [Google Scholar]
- 40.Valente T., Auladell C. Mech. Dev. 2001;108:207–211. doi: 10.1016/s0925-4773(01)00492-0. [DOI] [PubMed] [Google Scholar]
- 41.Menke D. B., Page D. C. Gene Expr. Patterns. 2002;2:359–367. doi: 10.1016/s1567-133x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 42.Belting H. G., Shashikant C. S., Ruddle F. H. J. Exp. Zool. 1998;282:196–222. [PubMed] [Google Scholar]