Abstract
Species radiations provide unique insights into evolutionary processes underlying species diversification and patterns of biodiversity. To compare plant diversification over a similar time period to the recent cichlid fish radiations, which are an order of magnitude faster than documented bird, arthropod, and plant radiations, we focus on the high-altitude flora of the Andes, which is the most species-rich of any tropical mountains. Because of the recent uplift of the northern Andes, the upland environments where much of this rich endemic flora is found have been available for colonization only since the late Pliocene or Pleistocene, 2–4 million years (Myr) ago. Using DNA sequence data we identify a monophyletic group within the genus Lupinus representing 81 species endemic to the Andes. The age of this clade is estimated to be 1.18–1.76 Myr, implying a diversification rate of 2.49–3.72 species per Myr. This exceeds previous estimates for plants, providing the most spectacular example of explosive plant species diversification documented to date. Furthermore, it suggests that the high cichlid diversification rates are not unique. Lack of key innovations associated with the Andean Lupinus clade suggests that diversification was driven by ecological opportunities afforded by the emergence of island-like habitats after Andean uplift. Data from other genera indicate that lupines are one of a set of similarly rapid Andean plant radiations, continental in scale and island-like in stimulus, suggesting that the high-elevation Andean flora provides a system that rivals other groups, including cichlids, for understanding rapid species diversification.
Keywords: leguminosae, Lupinus, phylogeny, species diversification
Studies of rapid episodes of species diversification or species radiations have provided a continuously rich source of new insights into the evolutionary processes underlying diversification and modern patterns of biodiversity for the last 150 years (1–6). A striking feature of species radiations is the discrepancy between the very high rates of species diversification reported for cichlid fish radiations in east African rift lakes and all bird, arthropod and plant radiations (2, 7, 8). This discrepancy has been attributed in part to the recency of the fish radiations (7). Accurate measurement of peak episodes of diversification embedded within older radiations requires fully sampled and resolved phylogenies (9). In the absence of such phylogenies, measurements of diversification rates for most radiations are less precise because they average out episodes of faster and slower speciation. Furthermore, these approaches measure net diversification rates, and the effects of extinction are not assessed. If speciation is concentrated in the early phases of radiations (3, 7, 10, 11), this could explain the discrepancy between the exceptional rates of species diversification reported for very recent [<2 million years (Myr)] lacustrine fish and other documented bird, arthropod, or plant radiations, which are generally older (>5 Myr) (7). Direct comparisons with the cichlid fish diversification rates have been lacking because few comparably recent radiations have been found in other groups.
To compare plant diversification over a similarly short time period to the very recent fish radiations, we examined patterns of species diversification in the high-altitude flora of the northern Andes, which is by far the most species-rich of any tropical mountain massif (12). It forms part of the tropical Andean biodiversity hotspot, which contains an estimated 45,000 plant species, 44% of which are endemic (13). Endemism is even higher, reaching 60%, in the high-altitude north Andean páramos (14). The cold upland habitats where much of this rich endemic flora is found today have been available for plant colonization only since the late Pliocene or early Pleistocene 2–4 Myr ago (15–18) after final uplift of the northern Andes (15). This recent uplift implies that at least some Andean plant groups must have diversified very recently and rapidly (16, 19). Despite this presumption, there are few reliable estimates of species diversification rates for Andean groups because of uncertainty about species numbers and the lack of robust, well resolved, and densely sampled phylogenies (20, 21).
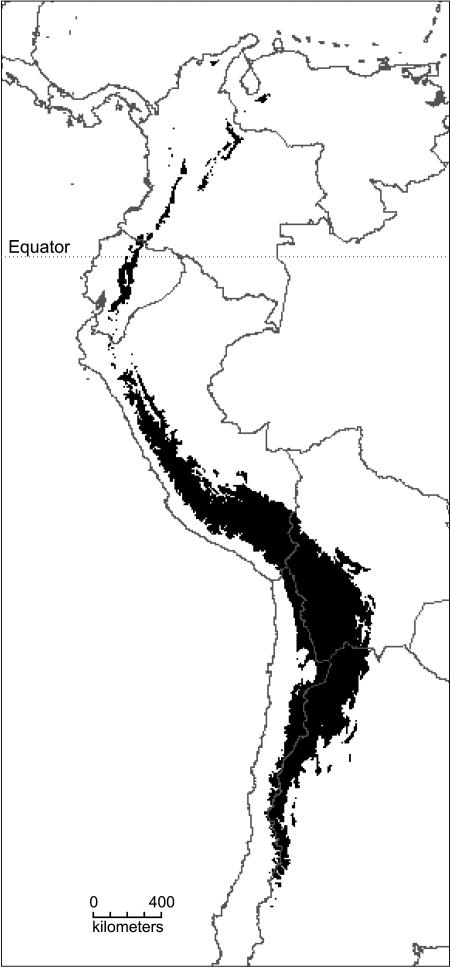
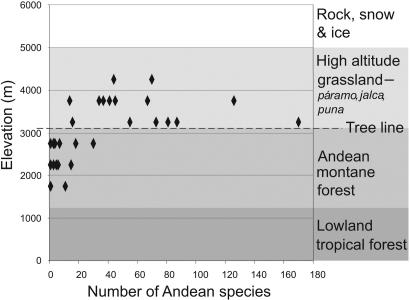
The prevalence of north and south temperate genera in the high-altitude Andean flora (12) supports the idea that colonization of the emerging high Andes was comparable to colonization of a newly formed island or island archipelago (16, 22) (Fig. 1). On recently formed islands and island-like formations such as lakes and mountains, rapid diversification has been attributed to ecological opportunities afforded by the availability of new habitats and absence of competition (1, 3, 7, 8, 10, 11, 23). Conversely, on continents, where ecological opportunity is harder to demonstrate, rapid episodes of diversification have generally been associated with key morphological or physiological innovations (4, 11). Numerous north temperate plant genera, such as Alnus, Draba, Lupinus, Quercus, Salix, Sambucus, Valeriana, and Viburnum, are postulated to have arrived in the Andes from North America after uplift of the northern Andes (17, 18). The altitudinal distribution of Andean species diversity for these genera shows that diversification is restricted largely to those growing at high elevations (Fig. 2). Although the monophyly of most of these endemic Andean species flocks has not been tested, this pattern supports the idea that island-like opportunities for diversification were available above the tree line in the high-altitude grassland zone and that plants that were preadapted to cooler conditions were most able to exploit such opportunities (Fig. 2).
Fig. 1.
The Andes as an island archipelago. This map shows the distribution of land over 3,000 m elevation, corresponding to the lower limit of the high-altitude grassland (páramo, jalca, and puna) vegetation zone. Although a few Andean Lupinus species occur at lower elevations, the genus is most diverse in the 3,000- to 4,000-m zone. Lupinus species are ubiquitous in all of the high-elevation “islands” from Venezuela and the Sierra Nevada Santa Marta in northern Colombia in the north to northwest Argentina in the south, and the geographical extent of the Andean Lupinus radiation (Fig. 3, k) closely matches the overall area of the Andean “archipelago” as delimited here.
Fig. 2.
Altitudinal variation in Andean species diversification. Data points represent plant genera with putative north temperate origins (see Methods). Diversification is concentrated above 3,000 m in the high-altitude grassland vegetation zone (Fig. 1).
The Genus Lupinus
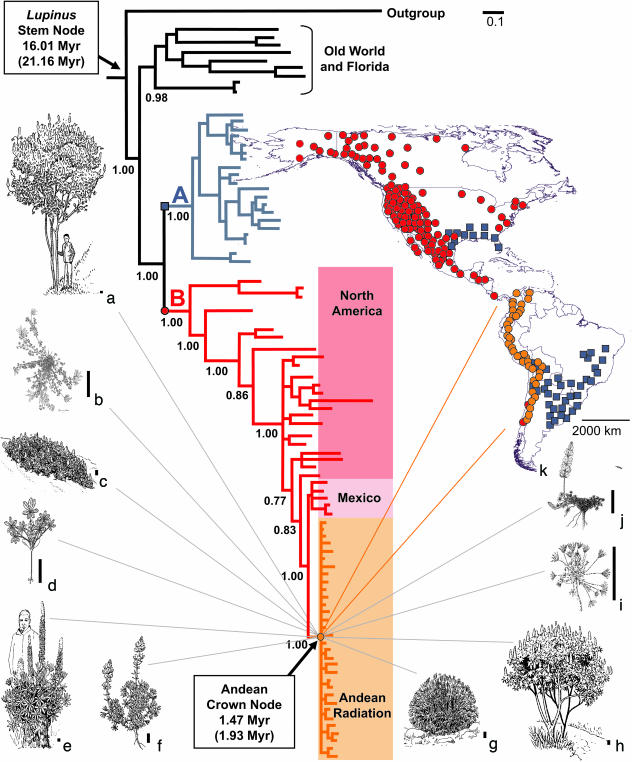
To gain insight into colonization of the recently formed high-elevation Andean habitats, we examined species diversification in the legume genus Lupinus, which comprises ≈275 species of annual herbs and herbaceous and woody perennials with an amphiatlantic distribution. The majority of Lupinus species occur in the New World, with two main centers of species diversity in western North America (≈100 species) and the Andes (≈85 species). Guided by previous estimates of Lupinus phylogeny based on internal transcribed spacer (ITS) sequences (24), we generated a densely sampled, well resolved, and robust phylogeny for Lupinus based on nuclear ITS and CYCLOIDEA gene sequences (Fig. 3). Our analysis shows that the Andean species flock is not monophyletic. Instead, Andean species are present in each of two well supported New World lineages. Nested within a large western New World group (Fig. 3, clade B) that spans North America, Mexico, and the Andes is a well supported clade representing ≈81 Andean species that are distributed from Venezuela to Argentina (Fig. 3, k). Four species distributed in the south-central Andes as far north as Lake Titicaca fall outside this clade. These species are placed in a predominantly lowland eastern New World lineage (Fig. 3, clade A). Species from both clades occur in sympatry in Bolivia and northern Argentina (Fig. 3, k). These distribution patterns suggest that there were at least two independent incursions of Lupinus into the Andes.
Fig. 3.
Phylogeny of Lupinus. Fifty percent majority rule Bayesian tree from analysis of the combined ITS/LEGCYCIA data sets. Posterior probabilities of major clades are shown below nodes; all nodes had posterior probabilities >0.5. This tree is congruent with the strict consensus derived from parsimony analysis. For clarity, taxon names have been omitted (see supporting information). Branch lengths are proportional to changes on the tree. A and B denote two well supported New World clades: A, eastern New World; B, western North America, Mexico, and the Andes. The map shows the distributions of these two New World clades to be largely allopatric, with limited overlap in the southern U.S. and the south-central Andes. The geographical extent of the main Andean radiation is shown in orange. Chilean accessions of Lupinus microcarpus group with their North American counterparts at the base of clade B, but the occurrence of L. microcarpus in central Chile (shown in red) is doubtfully native. Line drawings illustrate life forms encompassed by species in the Andean Lupinus radiation: a, treelet, Lupinus semperflorens; b, prostrate herb, Lupinus sp. nov; c, perennial woody shrublet, Lupinus smithianus; d, ephemeral annual herb, L. mollendoensis; e, giant stem rosette, Lupinus weberbaueri; f, woody perennial shrub, Lupinus sp. nov; g, acaulescent rosette, Lupinus nubigenus; h, perennial woody shrublet, Lupinus sp. nov; i, dwarf acaulescent rosette, Lupinus pulvinaris; j, prostrate herb, Lupinus prostratus. (Scale bars: 5 cm.)
Species belonging to the main Andean clade exhibit diverse life forms (Fig. 3, a–j) and ecologies. The majority of species grow in the 3,000- to 4,000-m high-elevation grassland (páramo, jalca, and puna) zone spanning wet eastern Andean ranges and drier inter-Andean valley and Pacific slopes (Fig. 1). A subset of prostrate (Fig. 3, b and j) and acaulescent (Fig. 3, e, g, and i) species are adapted to the extreme freeze–thaw conditions found at higher elevations between 4,000 and 5,200 m. Although predominantly a high-elevation group, a small number of mainly annual species adapted to lower elevation (<3,000 m) drier environments are included in the main Andean clade, including one ephemeral annual species, Lupinus mollendoensis, that grows in the coastal desert of southern Peru.
Results and Discussion
Independent dates derived from evolutionary rates analysis of DNA sequence data and paleobotanical evidence support the idea that the main Andean Lupinus radiation is very recent. The well supported monophyly of the large Andean Lupinus clade (parsimony bootstrap 92%, Bayesian posterior probability 1.0) and its placement as sister group to a clade of Mexican species suggest that diversification of this clade followed a single colonization from North America. The low sequence divergence and lack of resolution within the large Andean clade in comparison to the rest of the Lupinus phylogeny (Fig. 3) suggest rapid and recent diversification in the Andes. To test this, an evolutionary rates analysis of the combined ITS and LEGCYCIA sequence data sets was carried out. This analysis estimated the age of the Andean Lupinus radiation to be 1.47 ± 0.29 Myr. Notably, this agrees with extensive palynological evidence (17) that suggests cold upland habitats first appeared 2–4 Myr ago, after the final uplift of the northern Andes 3–5 Myr ago (15). Age estimates for the Andean radiation are relatively insensitive to older calibration dates for the Lupinus stem node. For example, using the maximum age of 21.16 Myr (25) in place of the mean of 16.01 Myr for the stem node only increases the estimated age of the Andean radiation by 0.46 Myr to 1.93 ± 0.35 Myr.
The average per-lineage species diversification rate for the Andean Lupinus radiation is 2.50–3.72 species per Myr. Using the older age estimate of 1.93 Myr for the Andean node yields a more conservative estimate of 1.93–2.78 species per Myr. However, even this minimum estimate exceeds reported species diversification rates for plants either on islands (10, 23) or continents (4, 6, 11, 26), providing the most spectacular example of explosive species diversification in plants documented to date. Low sequence divergence, lack of resolution and support, and/or sparse taxon sampling in phylogenetic studies of other high-elevation Andean plant groups (20, 21, 27, 28) mean that the monophyly of other putative Andean plant radiations has not been rigorously demonstrated. Despite these difficulties, two recent studies estimate approximate diversification rates of 0.8–1.34 species per Myr for Andean Valeriana (20) and 1.71–3.2 species per Myr for Andean Gentianella (21). These high diversification rates, as well as high endemic Andean species diversity in other high-elevation plant genera, such as Draba, Espeletia, Huperzia, Hypericum, and Lysipomia, suggest that lupines are just one of a set of spectacular, but as yet poorly documented, plant radiations that followed the final uplift of the northern Andes, and that a high proportion of the 3,400 plant species recorded in the northern Andean páramos (14) have evolved in the very recent evolutionary past.
It is notable that there are no novel morphological or physiological traits obviously associated with the Andean Lupinus clade that might have precipitated rapid diversification. Although the existence of such a key innovation cannot be ruled out pending more detailed anatomical and physiological analyses, this finding suggests that lupine diversification was driven by ecological opportunities similar to those on islands created by the emergence of largely unoccupied habitats after Andean uplift and subsequent Pleistocene glaciation. This idea is reinforced by the observation that the great diversity of life forms displayed by Andean lupines (Fig. 3, a–j) mirrors that seen in many plant radiations on islands (10, 23). Accelerated morphological divergence of this sort is often associated with island radiations and has been attributed to opportunities provided by depauperate environments where competitors are fewer (1). We identify four factors in the Andes that may explain the exceptional Lupinus diversification rate compared with other island radiations. First, the continental scale of the Andean Lupinus radiation, spanning >4,000 km from Venezuela to Argentina (Figs. 1 and 3, k), far exceeds typical island radiations, which generally occupy much more confined geographical areas. This scale creates extensive opportunities for geographical isolation, which is thought to be one of the main factors underlying the higher species diversification rates found on larger islands (29). Second, repeated fragmentation of high-altitude habitats due to altitudinal shifts in Andean vegetation zones prompted by Quaternary climate fluctuations are likely to have contributed to geographical isolation and diversification in the Andes (16, 17, 22, 30). Third, the extremely dissected topography in the high Andes and steep tropical montane environmental gradients associated with altitude (31) further enhance opportunities for geographical isolation that is evident from the prevalence of narrowly restricted endemics among the Andean Lupinus species today. Finally, resource heterogeneity in terms of habitats and environmental conditions is very high in the mid- to high-elevation Andes.
For plants, the idea that island-type diversifications are faster than mainland radiations has been hotly debated without conclusion (4, 6, 10, 11, 23, 26). The exceptional rates of diversification found for high-elevation Andean plants suggest that island-like diversification driven by ecological opportunity may indeed outpace mainland-type radiations. This result adds to the growing weight of evidence from diverse groups, including fish (2, 7, 8), plants (4, 6), and birds (5), that opportunity, in the form of extrinsic circumstances and events rather than evolutionary innovation, plays a dominant role in driving peak episodes of species diversification.
The age of the Andean Lupinus clade and our estimate of diversification rate are comparable to those previously reported for cichlid fish in east African lakes, which show the highest diversification rate estimates across the tree of life (2, 7). This result demonstrates that the high fish diversification rates are not unique and that radiations measured over similarly short time periods can match these rates. Clearly, much more densely sampled and robust phylogenies are needed to accurately measure peak episodes of diversification embedded within older radiations, as, for example, in ref. 32. The overall Lupinus phylogeny (Fig. 3) is highly resolved and robustly supported because of the fact that the LEGCYC1A locus is among the most rapidly evolving DNA sequence regions used to resolve plant species relationships. However, our gene trees show little resolution among the Andean species despite the diversity of life forms (Fig. 3, a–j), environmental adaptations, and broad geographic span encompassed by the radiation (Figs. 1 and 3, k). This lack of resolution suggests that the branching order for such rapidly evolving plant radiations will be difficult to resolve using conventional DNA sequence data (33), just as it has been for east African cichlid fish (34). As we unravel and document the history of plant diversification in the high Andes we can expect similarly heated debate about the monophyly and exact timing of radiations to that surrounding the cichlid fish radiations (2, 7, 8, 34). However, it is clear that the high-elevation Andean flora, comprising several thousand plant species, provides a system that rivals the most charismatic documented radiations, including cichlid fish, in terms of opportunities for understanding explosive species diversification.
Methods
Andean Species Diversity.
Altitudinal patterns of extant Andean species diversity for 31 plant genera with putative north temperate (16–18) or north Andean origins are summarized in Fig. 2. Data on numbers of Andean species were assembled from widely scattered sources, including taxonomic monographs, species checklists, and, in some cases, studies that identify monophyletic groups. Representative Andean altitudinal ranges are derived from the Checklist of Ecuador (35) in the form of frequency distributions of numbers of species present in 500-m elevation bands. The mid-point of the elevation band with maximum species diversity is plotted for each genus. The genera included are Alnus, Ambrosia, Bartsia, Berberis, Boehmeria, Buddleja, Calamagrostis, Cornus, Diplostephium, Draba, Geranium, Gentianella, Halenia, Huperzia, Hypericum, Lachemilla, Lupinus, Lysipomia, Myrica, Niphogeton, Prunus, Quercus, Rhamnus, Ribes, Salix, Sambucus, Senecio, Vaccinium, Valeriana, and Viburnum.
Species Delimitation.
Our estimate of 85 Andean species of Lupinus is based on comprehensive reexamination of species boundaries as part of taxonomic work that aims to bring consistency of species delimitation across the genus. This work has involved 8 months’ fieldwork in the Andes, examination of 8,000 herbarium sheets of Andean material, and observation of greenhouse-grown material. With 480 Lupinus species names based on Andean plants and recent estimates of as many as 170 species in the Andes (18), our estimate represents a substantial rationalization of the chaos surrounding the taxonomy of Andean lupines. Furthermore, our estimate is conservative in recognizing variation at infraspecific rank within widespread polymorphic species. Andean species not included in the analysis were assigned to the two New World clades based on morphology and chromosome number, which differ consistently between these groups (our unpublished data).
Sequence Data and Phylogenetic Analysis.
One hundred forty-eight accessions representing 98 species covering previously recognized Lupinus clades (24), the geographic range, and eight outgroup species representing five genera from tribe Genisteae were sampled. To test the monophyly of the Andean species, 53 accessions representing 36 species were sampled spanning the range of life forms (Fig. 3, a–j), geography, and altitudes. (Fully annotated parsimony and Bayesian trees, locality and voucher details, and GenBank accession nos. are available in supporting information, which is published on the PNAS web site.) The 5.8S subunit and flanking ITS (ITS1+ITS2) of nuclear ribosomal DNA and one copy (LEGCYCIA) of the rapidly evolving regulatory gene CYCLOIDEA were sequenced by using standard protocols. Two copies of CYCLOIDEA-like LEGCYCI genes have been characterized and sequenced in the Genistoid legumes including Lupinus (36, 37). These sequences were used to design complementary locus-specific primers located between the conserved TCP and R domains (38), which can be used in combination with general LEGCYCI primers to amplify the entire LEGCYCIA ORF of ≈1,000 bp in two fragments. An additional pair of locus-specific primers (available upon request) was designed to amplify LEGCYCIA for taxa in the lowland eastern New World clade (Fig. 3, clade A), which have a deletion at the original locus-specific primer site. The LEGCYCIA paralogue was chosen because it is more variable than LEGCYCIB (37, 38) and, unlike LEGCYCIB, does not show allelic length variation. ITS and LEGCYCIA sequences from Ree et al. (37) were included in the analysis. New sequences have been submitted to the GenBank database (see supporting information). Sequences were aligned by using clustalx (39) and manually adjusted. Indels were coded by using the simple gap-coding method in seqstate (40) and included in the parsimony analysis. Parsimony analysis of the combined ITS/LEGCYCIA data set was conducted with nona (41) by using 1,000 random addition sequences, tree bisection, and reconnection, holding 100 trees per replication and attempting to swap to completion (see supporting information). A reduced data set of 89 sequences representing one outgroup and one sequence per species was compiled for Bayesian analysis (42) using the GTR+G and GTR+I models (43) for the ITS and LEGCYCIA data partitions, respectively. This analysis involved 3,000,000 Metropolis-coupled Markov Chain Monte Carlo permutations of tree parameters and four chains heated to 0.01. The consensus Bayesian tree was virtually congruent with the parsimony tree but slightly more resolved (see supporting information). Clade stability tests involved Bayesian posterior probabilities and parsimony bootstrap resampling.
Evolutionary Rates Analysis.
The relative rate test implemented in mr bayes rejected a molecular clock for the consensus Bayesian tree (likelihood ratio = 3313.72, df = 142, P = 0.00). The program r8s (44) was used to assess variation in substitution rates and incorporate this into the estimation of ages of lineages. Branching order and branch lengths from 100 Bayesian trees sampled every 10,000 generations after stationarity were analyzed to obtain means and standard deviations of ages of clades. Cross-validation and penalized likelihood analysis yielded an average smoothing parameter of 13, which was used in final dating analyses. Relative ages estimated with r8s were converted to absolute ages by fixing the age of the Lupinus stem node using mean (16.01 Myr) and maximum (21.16 Myr) dates for the most recent common ancestor of Spartium and Lupinus from the legume-wide analysis of matK sequences calibrated using 13 well studied fossils (25).
Species Diversification Rate.
Species diversification rates, assuming an equal rate of random speciation Yule model, were calculated as SR = (ln n1 − ln n0)/t, where n1 is the number of extant species, n0 is the initial species diversity, here taken as 1, and t is time in Myr. Upper and lower standard deviations of age estimates were used in calculations of speciation rate.
Supplementary Material
Acknowledgments
We thank Helene Citerne for primers; Rosemary Wise for artwork; Simon Ho and Matt Lavin for advice; Matt Lavin for age estimates based on but not presented in ref. 25; Donovan Bailey, Stephan Beck, Tim Budden, Ruth Clark, Aniceto Daza, Alfonso Delgado, Chris Fagg, Rob Forrest, Martin Gardner, Greg Kenicer, Bente Klitgaard, Gwil Lewis, Helga Ochoterena, David Neill, Arely Palabral, John Pannell, Terry Pennington, Toby Pennington, Carolyn Proenca, Carlos Reynel, Mario Sousa, Salvador Talavera, John Wood, Marty Wojciechowski, and especially Chris Drummond (University of Idaho, Moscow), Richard Spellenberg (New Mexico State University, Las Cruces), and Silvia Miotto and Maria-Teresa Schifino-Wittman (Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil) for assistance with fieldwork and provision of plant samples; the authorities in Ecuador, Peru, Bolivia, and Brazil for permission to collect material; the B, CAS, CGE, E, F, FHO, G, GH, ISC, K, LPB, M, MO, MOL, MSB, NY, QCNE, UB, UC, US, USM, and USZ herbaria (Index Herbariorum acronyms) for loan of material; and Julie Hawkins, Jane Langdale, John Pannell, Mike Sanderson, and an anonymous reviewer for comments. R.E. created Figs. 1 and 3. This work was supported by the Royal Society, Biotechnology and Biological Sciences Research Council, the Genetics Society, and the Stanley Smith Horticultural Trust.
Abbreviations
- Myr
million years
- ITS
internal transcribed spacer.
Footnotes
References
- 1.Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford Univ. Press; 2000. [Google Scholar]
- 2.Kocher T. D. Nat. Rev. Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- 3.Gravilets S., Vose A. Proc. Natl. Acad. Sci. USA. 2005;102:18040–18045. doi: 10.1073/pnas.0506330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kay M. K., Reeves P. A., Olmstead R. G., Schemske D. W. Am. J. Bot. 2005;92:1899–1910. doi: 10.3732/ajb.92.11.1899. [DOI] [PubMed] [Google Scholar]
- 5.Ricklefs R. E. Proc. Biol. Sci; 2003. pp. 2285–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson J. E., Pennington R. T., Pennington T. D., Hollingsworth P. M. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- 7.McCune A. R. In: Molecular Evolution and Adaptive Radiation. Givinish T. J., Sytsma K. J., editors. Cambridge, U.K.: Cambridge Univ. Press; 1997. pp. 585–610. [Google Scholar]
- 8.Verheyen E., Salzburger W., Snoeks J., Meyer A. Science. 2003;300:325–329. doi: 10.1126/science.1080699. [DOI] [PubMed] [Google Scholar]
- 9.Nee S., Mooers A. O., Harvey P. H. Proc. Natl. Acad. Sci. USA. 1994;89:8322–8326. doi: 10.1073/pnas.89.17.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin B. G., Sanderson M. J. Proc. Natl. Acad. Sci. USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klak C., Reeves G., Hedderson T. Nature. 2003;427:63–65. doi: 10.1038/nature02243. [DOI] [PubMed] [Google Scholar]
- 12.Smith J. M. B., Cleef A. M. J. Biogeogr. 1988;15:631–645. [Google Scholar]
- 13.Myers N., Mittermeier R. A., Mittermeier C. G., Da Fonseca G. A. B., Kent J. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 14.Luteyn J. L. Mem. N.Y. Bot. Gard. 1999;84:1–278. [Google Scholar]
- 15.Gregory-Wodzicki K. M. Geol. Soc. Am. Bull. 2000;112:1091–1105. [Google Scholar]
- 16.Simpson B. B. Paleobiology. 1975;1:273–294. [Google Scholar]
- 17.Van der Hammen T., Cleef A. M. In: High Altitude Tropical Biogeography. Vuilleumier F., Monasterio M., editors. New York: Oxford Univ. Press; 1986. pp. 153–201. [Google Scholar]
- 18.Burnham R. J., Graham A. Ann. Mo. Bot. Gard. 1999;86:546–589. [Google Scholar]
- 19.Monasterio M., Sarmiento L. Trends Ecol. Evol. 1991;6:387–391. doi: 10.1016/0169-5347(91)90159-U. [DOI] [PubMed] [Google Scholar]
- 20.Bell C. D., Donoghue M. J. Organisms Divers. Evol. 2005;5:147–159. [Google Scholar]
- 21.Von Hagen K. B., Kadereit J. W. Organisms Divers. Evol. 2001;1:61–79. [Google Scholar]
- 22.Vuilleumier F. Am. Nat. 1970;104:373–388. [Google Scholar]
- 23.Böhle U.-T., Hilger H. H., Martin W. F. Proc. Natl. Acad. Sci. USA. 1996;93:11740–11745. doi: 10.1073/pnas.93.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ainouche K., Bayer R. Am. J. Bot. 1999;86:590–607. [PubMed] [Google Scholar]
- 25.Lavin M. T., Herendeen P. S., Wojciechowski M. F. Syst. Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 26.Richardson J. E., Weitz F. M., Fay M. F., Cronk Q. C. B., Linder H. P., Reeves G., Chase M. W. Nature. 2001;412:181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- 27.Koch M., Al-Shehbaz I. A. Ann. Mo. Bot. Gard. 2002;89:88–109. [Google Scholar]
- 28.Wojciechowski M. F., Sanderson M. J., Hu J.-H. Syst. Bot. 1999;24:409–437. [Google Scholar]
- 29.Losos J., Schluter D. Nature. 2000;408:847–850. doi: 10.1038/35048558. [DOI] [PubMed] [Google Scholar]
- 30.Hooghiemstra H., Van der Hammen T. Philos. Trans. R. Soc. London B Biol. Sci. 2004;359:173–181. doi: 10.1098/rstb.2003.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janzen D. H. Am. Nat. 1967;101:233–249. [Google Scholar]
- 32.Kozak K. H., Weisrock D. W., Larson A. Proc. R. Soc. London Biol. Sci; 2005. pp. 539–546. [Google Scholar]
- 33.Hughes C. E., Eastwood R. J., Bailey C. D. Philos. Trans. R. Soc. London B Biol. Sci. 2006;361:211–225. doi: 10.1098/rstb.2005.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kocher T. D. Nature. 2003;423:489–491. doi: 10.1038/423489a. [DOI] [PubMed] [Google Scholar]
- 35.Jørgensen P. M., León-Yánes S., editors. Catalogue of the Vascular Plants of Ecuador. St. Louis: Missouri Botanical Garden Press; 1999. [Google Scholar]
- 36.Citerne H., Luo D., Pennington R. T., Coen E., Cronk Q. C. B. Plant Physiol. 2003;131:1042–1053. doi: 10.1104/pp.102.016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ree R. H., Citerne H. L., Lavin M. T., Cronk Q. C. B. Mol. Biol. Evol. 2004;21:321–331. doi: 10.1093/molbev/msh022. [DOI] [PubMed] [Google Scholar]
- 38.Citerne H. L. Edinburgh J. Bot. 2006;62:119–126. [Google Scholar]
- 39.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller K. Appl. Bioinformatics. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- 41.Goloboff P. nona: A Tree Searching Program. Argentina: Fundación e Instituto Miguel Lillo Tucumán; 2000. [Google Scholar]
- 42.Huelsenbeck J. P., Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 43.Nylander J. A. A. mrmodel test. Uppsala: Uppsala Univ.; 2004. Version 2. [Google Scholar]
- 44.Sanderson M. J. r8s Users Manual. Davis: Univ. of California; 2004. Version 1.7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.