Abstract
Gene expression studies using microarrays have great potential to generate new insights into human disease pathogenesis, but data quality remains a major obstacle. In particular, there does not exist a method to determine prior to hybridization whether an array will yield high quality data, given good study design and target preparation. We have solved this problem through development of a three-color cDNA microarray platform where printed probes are fluorescein labeled, but are spectrally compatible with Cy3 and Cy5 dye-labeled targets when using confocal laser scanners possessing narrow bandwidths. This approach enables prehybridization evaluation of array/spot morphology, DNA deposition and retention and background levels. By using these measurements and the intra-slide coefficient of variation for fluorescence intensity we show that slides in the same batch are not equivalent and measurable prehybridization parameters can be predictive of hybridization performance as determined by replicate consistency. When hybridizing target derived from two cell lines to high and low quality replicate pairs (n = 50 pairs), a direct and significant relationship between prehybridization signal-to-background noise and post-hybridization reproducibility (R2 = 0.80, P < 0.001) was observed. We therefore conclude that slide selection based upon prehybridization quality scores will greatly benefit the ability to generate reliable gene expression data.
INTRODUCTION
The cDNA microarray platform has great potential to generate new insights into human disease (1–7). The use of cDNA microarrays begins with construction of the array where, typically, hundreds to thousands of cDNA probes are amplified by PCR, purified and printed onto coated glass slides (typically poly-l-lysine). Slides are fixed, blocked and are finally hybridized with Cy3- and Cy5-labeled cDNA targets derived from the two biological samples being compared for differential gene expression. After hybridization, the array is analyzed with a fluorescence scanner and the relative amounts of a mRNA species in the original two samples is defined as a ratio between the two fluorophores at the homologous array element using specially designed software (1,2,8–10).
This useful technology, however, possesses recognized data quality/reproducibility issues that can limit its application to complex biological systems (11,12). High experimental variability can arise through laboratory technical problems as well as normal biological variation (13). Yue et al. (14), using Saccharomyces cerevisiae probes and complementary in vitro transcripts, demonstrated that the amount of DNA bound to the glass slide is dependent, in part, on the concentration of the DNA printed and that the amount retained by the slide is critical for good quality differential expression data. The range of detected values of known transcript ratios was compressed when elements were printed at concentrations <100 ng/µl in water. Printing at more dilute printing concentrations exacerbated ratio compression to the point where input transcript ratios of 30:1 or 1:30 were detected as output ratios close to 1:1, illustrating that limiting bound probe results in an underestimation or failure to detect differential gene expression (14). The concentration of DNA printed, the printing buffer selected and the glass coating will influence the amount of DNA retained by the slide after processing. Commonly used printing solutions include 3× SSC (saline sodium citrate), 50% dimethyl sulfoxide (DMSO) and water (8,14). Diehl et al. (15) found that the addition of the PCR additive betaine, which is known to normalize base pair stability differences, increases solution viscosity and reduces evaporation rates and also greatly enhances probe binding to poly-l-lysine-coated slides (15–17). Furthermore, probe saturation of the glass slide was obtained at a lower printing concentration of 250 ng/µl when betaine was present versus >500 ng/µl in printing solutions without betaine, which can greatly increase the number of potential slides produced from a single library amplification (15).
Controlling array fabrication variables is difficult since the array is typically invisible until after it has been hybridized. Therefore, we have generated probe arrays directly labeled with fluorescein as a means of visualizing element/array morphology and quantifying DNA deposition/retention on the slide prior to hybridization. Direct labeling of probes separates slide coating, printing and processing from hybridization and has facilitated evaluation and optimization of methods. In this report, we make the important observation that slides coated, printed and processed together are not necessarily equal and that prehybridization imaging is predictive of hybridization performance. Therefore, prehybridization slide evaluation and selection can improve data reproducibility and quality, since slides that do not meet minimum standards can be avoided.
MATERIALS AND METHODS
The Research Genetics (Huntsville, AL) sequence-verified human library, consisting of 41 472 clones, was used as a source of probe DNA. The library was reformatted from 96 to 384 format and subsequently manipulated using 0.5 and 5 µl volume 96 and 384 slot pin replicator tools (VP Scientific, San Diego, CA). Clone inserts were directly amplified in 384-well format from 0.5 µl bacterial culture using 0.26 µM each vector primer [array F, 5′-fluorescein-CTGCAAG GCGAT-(fluorescein)TAAGTTGGGTAAC-3′; array R, 5′-fluorescein-GTGAGCGGAT-(fluorescein)AACAATTTCAC ACAGGAAACAGC-3′] (Integrated DNA Technologies, Coralville, IA) in a 20 µl reaction consisting of 10 mM Tris–HCl, pH 8.3, 3.0 mM MgCl2, 50 mM KCl, 0.2 mM each dNTP (Amersham, Piscataway, NJ), 1 M betaine and 0.25 U Taq polymerase (Roche, Indianapolis, IN). Reactions were incubated at 95°C for 5 min and 35 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 1 min, and terminated with a 7 min hold at 72°C. PCR products were routinely analyzed for quality by 1% agarose gel electrophoresis analysis. Products were purified by size exclusion filtration using Multiscreen 384 PCR filter plates (Millipore, Bedford, MA) to remove unincorporated primer and PCR reaction components. Forty wells of each 384-well probe plate were quantified by the PicoGreen assay (Molecular Probes, Eugene, OR) according to the manufacturer’s instructions, dried down and reconstituted at 125 ng/µl in 3% DMSO/1.5 M betaine.
Microarrays possessing a density of 10 000 probes/slide were printed onto poly-l-lysine slides using a GeneMachines Omni Grid printer (San Carlos, CA) with eight Telechem International SMP3 pins (Sunnyvale, CA). Slides were post-processed using the previously described aqueous (8) or non-aqueous (15) protocols. Slide coating, isolation of mRNA, labeling and hybridization were performed as described previously (http://cmgm.stanford.edu/pbrown/mguide/index.html). After hybridization, arrays were scanned with a ScanArray 5000 (GSI Lumonics, Billerica, MA) and image files were obtained. Array image files were analyzed with the Matarray software (10).
RESULTS AND DISCUSSION
A number of approaches have been described to address the problem of determining DNA deposition/retention and array element morphology prior to experimental use of slides. It is possible to stain the fixed slide prior to hybridization with a DNA-binding fluorescent dye, such as SYBR Green II or SYTO 61 (14,18). However, investigational use of the slide after quality control analysis requires destaining, and potential changes in slide performance after destaining must be considered. The use of ‘universal’ targets which will hybridize to every element of a microarray have also been reported (14). While these hybridization-based techniques provide information as to the amount of DNA present within each element of the array, they require sacrificing a slide from a batch of printed slides for quality control analysis and do not completely assure the investigator that the arrays actually used for experimentation are equivalent to those evaluated during quality control. Our initial objective was to co-hybridize a fluorescein-labeled vector-specific oligonucleotide with Cy3- and Cy5-labeled targets to every element such that array quality control was incorporated into each experiment. This approach was aborted due to the fact that optimal hybridization conditions (Tm) for the oligonucleotide differed considerably from those for labeled cDNA targets, which on co-hybridization resulted in low fluorescein signals (data not shown).
To circumvent this problem, we have developed a means of directly visualizing printed arrays by generating probes with fluorescein-labeled primers (excitation 488 nm, emission 508 nm), which are spectrally compatible with the Cy5 and Cy3 dyes typically used for target labeling (Cy3 excitation 543 nm, emission 570 nm; Cy5 excitation 633 nm, emission 670 nm) when using the GSI Luminonics ScanArray 5000 confocal laser scanner. The narrow 10 nm bandwidth of this instrument allows for excitation of Cy3 at 543 nm without co-excitation of fluorescein, which would contaminate the Cy3 emission with its broad emission tail (http://www.probes.com/handbook). Our approach, which separates slide coating, printing and processing from hybridization, provides a method for (i) probe amplification control, (ii) direct examination of array/element morphology, (iii) determination of post-processed probe retention and (iv) a means of bound probe quantitative quality control for improved differential gene expression analysis.
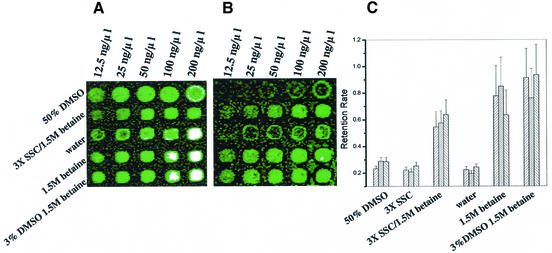
An advantage of this approach is the existence of a direct relationship between detected relative fluorescence units (RFUs) and the amount of DNA probe present on the slide, once unincorporated primer has been removed from the amplified probe, making DNA retention studies possible. Human probes for glyceraldehyde 3-phosphate deydrogenase-1 (GAPDH), β-actin and glutamate receptor-2 (HBGR2) (IMAGE Consortium 50117, 34357 and 43622, respectively) were serially diluted and printed in 50% DMSO, 3× SSC, water, 1.5 M betaine, 1.5 M betaine/3× SSC (15) and 1.5 M betaine/3.1% DMSO. Arrays were evaluated for spot morphology (size/shape) and DNA retention was measured by scanning arrays immediately after printing and again after post-processing. Only 30% of probe is retained by poly-l-lysine-coated glass slides after post-processing when the commonly used printing solutions water, 50% DMSO or 3× SSC are used (Fig. 1A and B). Probes printed with 50% DMSO resulted in 151.1 ± 5.9 µm diameter array elements compared to 120.6 ± 5.4 µm diameter elements for those printed in water or 3× SSC (with or without 1.5 M betaine), therefore, DMSO was titrated in an effort to control spot size. The use of 3% DMSO/1.5 M betaine resulted in the highest average probe retention on the slide (>70%), more than twice what is observed with commonly used printing solutions, as well as optimal average spot size (<130 µm) (Fig. 1C). Preparation of DNA probe is the most time consuming and expensive component of high-density array construction and making efficient use of prepared probe through high retention is an important ongoing issue.
Figure 1.
Evaluation of spotting solutions for post-blocking probe retention. A dilution series (indicated) of three different probes (from top to bottom: GAPDH, β-actin, β-actin, HBGR2, GAPDH) was printed in six different printing solutions (indicated). (A) Fluorescein image immediately after printing. (B) Fluorescein image [same array as (A)] after aqueous post-processing. (C) Plotted are the percent retention values determined from the 100 ng/µl dilution element for each of the three genes for the six printing solutions (n = 35 elements, distributed over 35 slides). Bar graphs ordered: GAPDH; β-actin, HBGR2. 3% DMSO/1.5 M betaine was superior.
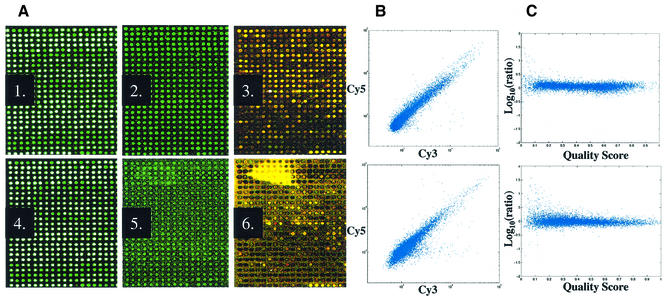
The critical post-arraying blocking process, where unreacted primary amines are converted to carboxylic moities, is typically performed with succinic anhydride in aqueous borate buffered 1-methyl-2-pyrrolidinone (1,2,8,19). Generation of fluorescein-labeled arrays enabled direct hybridization-free comparison of this traditional blocking process to blocking with succinic anhydride in the non-polar, non-aqueous solvent 1,2-dichloroethane (15). Processing with the non-aqueous method resulted in arrays with very low background fluorescein signal levels compared to the aqueous blocking method [Fig. 2A(2) versus 2A(5)]. The array in Figure 1B was aqueously post-processed and illustrates our observation that inter-element background levels increase as a function of printed DNA concentration with this method. The prehybridization image quality was predictive of slide performance in homotypic hybridizations employing UACC903 RNA where arrays processed with the non-aqueous method generated images with higher overall signal intensity and fewer outliers [Fig. 2A(3) versus 2A(6) and 2B]. Image quality was assessed with Matarray software, which employs a spatial and intensity-dependent algorithm for spot detection and signal segmentation. Matarray also generates a composite quality score (qcom) that is defined for each spot on the array according to size, signal-to-noise value [signal/(signal + noise)], background uniformity and saturation status (10). Variation in Cy5/Cy3 intensity ratio values correlated with the fluorescein qcom score and revealed an overall lower spot quality with the aqueous method that impacts data quality (Fig. 2C). Using simultaneously produced 10 000 probe arrays, mean fluorescein signal-to-noise quality score [signal/(signal + noise)] per element of 0.93 ± 0.04 (n = 15) were observed with the non-aqueous method versus 0.71 ± 0.02 (n = 15) with the aqueous method. Probe fluorescein signal measurements of 6- to 9-fold over noise were observed on arrays processed with the non-aqueous blocking method and values slightly less for those arrays aqueously processed; these values are sufficient for credible measurement of bound probe. These observations are consistent with the notion that aqueous blocking methods result in partial redissolving and redeposition of printed DNA, generating higher background.
Figure 2.
Processed array fluorescein image is reflective of hybridized array performance. The experiment employs human 10K probe cDNA arrays. (A) A1–A3, non-aqueous blocking; A4–A6, aqueous blocking. Array image immediately after printing (A1, A4), post-processing (A2, A5) and homotypic hybridization with Cy5 and Cy3 direct labeled UACC903 RNA (A3, A6). (B) Scatter plots of homotypic hybridizations on arrays processed with non-aqueous (top) and aqueous (bottom) methods. (C) The variability in Cy3/Cy5 intensity ratio measurement (y-axis) is correlated with fluorescein qcom (x-axis); non-aqueous (top) and aqueous (bottom) methods. Images were collected using the same laser and PMT settings and are illustrated under the same parameters using GenePix Pro Software. [Note: loss of DNA after processing step (A1 versus A2; A4 versus A5); white elements in panels A1 and A4 are saturated.]
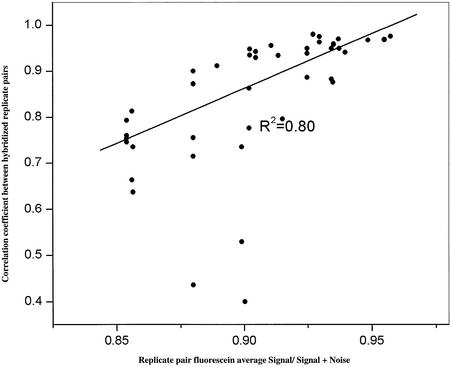
Slides that are coated, printed and processed together do not necessarily result in equivalent arrays. One hundred slides each possessing a 10 000 human probe array were simultaneously printed, non-aqueously processed and evaluated. The average fluorescein signal per slide varied between processed slides from 4500 to 20 000 RFU (10 770 ± 4202); while overall slide signal-to-noise values ranged from 0.85 to 0.95 (mean 0.92 ± 0.03). Competitive hybridizations between UACC903 and Jurkat cDNA on arrays, selected from three independent printings of the same probe set, with high average DNA/element and low background values were compared to those performed on arrays with low DNA/element and/or high background values. When comparing hybridization results between replicate pairs of differing quality (n = 50 pairs), a direct and significant relationship (R2 = 0.80, P < 0.001) was observed between prehybridization fluorescein image quality and replicate consistency, illustrating that microarray data quality can be improved through prehybridization slide selection based upon quality analysis. This is illustrated in Figure 3, where the average fluorescein signal-to-noise value [signal/(signal + noise)] for each replicate pair is plotted against the Pearson correlation coeffient of the Cy3/Cy5 ratio data. The observation of a relationship between pre- and post-hybridized image/data quality is completely consistent with our previous report in that prehybridized arrays possessing low signal-to-noise scores give rise to hybridized arrays with low signal-to-noise scores and hybridization data from such arrays do not correlate well with each other (10). Selection of quality arrays does not necessarily guarantee a high replicate Cy5/Cy3 ratio correlation, since RNA samples, target labeling, hybridization, washing, laboratory technique and image collection are sources of variation, as indicated by the three outliers observed in Figure 3. It must be emphasized that the 100 hybridizations represented in Figure 3 were performed by multiple laboratory personnel utilizing multiple labeling reactions of the same RNA.
Figure 3.
Fluorescein signal-to-noise score (x-axis) of 50 replicate pairs (100 slides) is predictive of correlation coefficient of Cy3/Cy5 ratio data between hybridized replicate arrays (y-axis). All hybridizations are between Jurkat and UACC903 cDNA.
Fluorescein derivatives have been the most commonly used label for biological molecules. In addition to its relatively high absorption properties, excellent fluorescence quantum yield and good water solubility, fluorescein has an excitation maximum (494 nm) that closely matches the 488 nm spectral line of the argon ion laser, making it a useful fluorophore for confocal laser scanning microscopy applications. Our selection of fluorescein as the third dye was first driven by the fact that it is compatible with Cy3 and Cy5 when using the ScanArray 5000 and, secondly, by the fact that this fluorophore is relatively inexpensive and readily available as a 5′ end-label on oligonucleotide primers. Unfortunately, not many confocal laser scanners possess the performance specifications to support the use of a three-color system as we describe here using fluorescein. This situation is likely to change as both the fluorescent labels and instrumentation continue to improve, allowing more flexibility in dye and instrument selection in three-color applications. Nonetheless, the strategy as described in this report performs well; we are confident fluorescein labeling of the probes does not interfere with the subsequent detection of Cy3 and Cy5 hybrids, since: (i) scanning of slides prior to hybridization shows no signal for either Cy3 or Cy5; (ii) Cy3/Cy5 scatter plots pass through the origin with no evidence of the detected Cy3 or Cy5 signal being negatively influenced by a quenching effect nor positively influenced by carryover signal. Furthermore, all of our arrays (including those shown in Fig. 2) possess a series of fluorescein-labeled Arabidopsis thaliana probes to be used as positive (in combination with homologous in vitro transcript) and negative controls. These probes generate no signal under Cy3 or Cy5 scanning conditions either before or after hybridization in the absence of labeled in vitro transcript.
Direct measurement of the bound probe available for hybridization has other important advantages. Electrophoretic analysis of probe amplification efficiency can be greatly reduced since failed PCRs can be identified and recorded through analysis of fluorscein signal intensity. Precious clinical target material can be conserved through reduction of replicates necessary since poor slides can be avoided. Quality-based prehybridization selection results in a higher probability of successful experiments and reduced overall cost. Assuming a compatible scanner does not need to be purchased, this approach can be implemented for the added cost of labeled PCR primers (<$200 per library amplification), purification (which is a routine step in many probe preparation protocols) and the labor costs associated with third dye image collection and analysis. Currently under investigation is the ability to filter data and to normalize intra-slide and inter-slide variability using the third dye since we have observed that Cy3/Cy5 ratio correlation coefficients between replicate arrays can be improved by dropping elements with the lowest fluorescein signal. We are currently working to identify, in general, how much bound probe is necessary to obtain highly reproducible results across high density arrays, however, for key experiments we are selecting arrays with signal-to-noise ratios >0.90, average element fluorescein intensity >3000 and coefficient of variation of element fluorescein intensity <10%. In conclusion, this direct visualization strategy, which is applicable to both cDNA- and oligonucleotide-based arrays, provides a means of quantitative quality control for improved gene expression analysis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jill Waukau, Kami Montgomery, Kate Frederick, Obrad Kokanovich and Shehnaz Khan for their excellent technical efforts on this project. We also thank Drs Howard Jacob and Yasuhiro Kita for numerous insightful discussions. This work has been supported by funding awarded to M.H. from the Medical College of Wisconsin Research Affairs Committee and by a special fund from the Children’s Hospital Foundation, Children’s Hospital of Wisconsin.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R. and Brown,P. (1995) Quantitative monitoring of gene expression patterns with complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.Schena M., Shalon,D., Heller,R., Chai,A., Brown,P.O. and Davis,R.W. (1996) Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garber M.E., Troyanskaya,O.G., Schluens,K., Petersen,S., Thaesler,Z., Pacyna-Gengelbach,M., van de Rijn,M., Rosen,G.D., Perou,C.M., Whyte,R.I. et al. (2001) Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl Acad. Sci. USA, 98, 13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedenfalk I., Duggan,D., Chen,Y., Radmacher,M., Bittner,M., Simon,R., Meltzer,P., Gusterson,B., Esteller,M., Kallioniemi,O.P. et al. (2001) Gene-expression profiles in hereditary breast cancer. N. Engl. J. Med., 344, 539–548. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T., Perou,C.M., Tibshirani,R., Aas,T., Geisler,S., Johnsen,H., Hastie,T., Eisen,M.B., van de Rijn,M., Jeffrey,S.S. et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclass with clinical implications. Proc. Natl Acad. Sci. USA, 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegde P., Qi,R., Gaspard,R., Abernathy,K., Dharap,S., Earle-Hughes,J., Gay,C., Nwokekeh,N.U., Chen,T., Saeed,A.I. et al. (2001) Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray. Cancer Res., 61, 7792–7797. [PubMed] [Google Scholar]
- 7.Dhanasekaran S.M., Barrette,T.R., Ghosh,D., Shah,R., Varambally,S., Kurachi,K., Pienta,J., Rubin,M.A. and Chinnaiyan,A.M. (2001) Delineation of prognostic biomarkers in prostate cancer. Nature, 412, 822–826. [DOI] [PubMed] [Google Scholar]
- 8.Eisen M. and Brown,P. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 9.Hegde P., Qi,R., Abernathy,K., Gay,C., Dharap,S., Gaspard,R., Hughes,J.E., Snesrud,E., Lee,N. and Quackenbush J. (2000) A concise guide to cDNA microarray analysis. Biotechniques, 29, 548–556. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Ghosh,S. and Guo,S.-W. (2001) Quantitative quality control in microarray image processing and data acquisition. Nucleic Acids Res., 29, e75–e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr M. and Churchill,G. (2001) Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc. Natl Acad. Sci. USA, 98, 8961–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M., Kuo,F., Whitmore,G. and Sklar,J. (2000) Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl Acad. Sci. USA, 97, 9834–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard C.C., Hsu,L., Delrow,J. and Nelson,P.S. (2001) Project normal: defining normal variance in mouse gene expression. Proc. Natl Acad. Sci. USA, 98, 13266–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue H., Eastman,P.S., Wang,B.B., Minor,J., Doctolero,M.H., Nuttall,R.L., Stack,R., Becker,J.W, Montgomery,J.R., Vainer,M. et al. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res., 29, e41–e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl F., Grahlmann,S., Beier,M. and Hoheisel,J. (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res., 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henke W., Herdel,K., Jung,K., Schnorr,D. and Loening,S. (1997) Betaine improves the PCR amplification of GC-rich sequences. Nucleic Acids Res., 25, 3957–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rees W., Yager,T., Korte,J. and Von Hippel,P. (1993) Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry, 32, 137–144. [DOI] [PubMed] [Google Scholar]
- 18.Battaglia C., Salani,G., Consolandi,C., Bernardi,L.R. and De Bellis,G. (2000) Analysis of DNA microarrays by non-destructive fluorescent staining using SYBR green II. Biotechniques, 29, 78–81. [DOI] [PubMed] [Google Scholar]
- 19.Dolan P.L., Wu,Y., Ista,L.K., Metzenberg,R.L., Nelson,M.A. and Lopez,G.P. (2001) Robust and efficient synthetic method for forming DNA microarrays. Nucleic Acids Res., 29, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]