Abstract
The molecular mechanism underlying milk fat globule secretion in mammary epithelial cells ostensibly involves the formation of complexes between plasma membrane butyrophilin and cytosolic xanthine oxidoreductase. These complexes bind adipophilin in the phospholipid monolayer of milk secretory granules, the precursors of milk fat globules, enveloping the nascent fat globules in a layer of plasma membrane and pinching them off the cell. However, using freeze-fracture immunocytochemistry, we find these proteins in locations other than those previously inferred. Significantly, butyrophilin in the residual plasma membrane of the fat globule envelope is concentrated in a network of ridges that are tightly apposed to the monolayer derived from the secretory granule, and the ridges coincide with butyrophilin labeling in the globule monolayer. Therefore, we propose that milk fat globule secretion is controlled by interactions between plasma membrane butyrophilin and butyrophilin in the secretory granule phospholipid monolayer rather than binding of butyrophilin–xanthine oxidoreductase complexes to secretory granule adipophilin.
Keywords: freeze-fracture immunocytochemistry, lipid droplet-associated proteins, mammary epithelial cells, milk secretory granules
After parturition, the offspring of all mammals depend on a single foodstuff for initial survival: milk. Although most of the nutritional and appreciable economic value of milk resides in its fat globules, the molecular mechanisms underlying the secretion of milk fat globules have not yet been elucidated (1, 2).
Milk fat globules are secreted by epithelial cells of the mammary glands. They contain mainly triacylglycerols and sterols. The intracellular precursors of milk fat globules, milk secretory granules, arise as milk lipids inundate between the leaflets of the endoplasmic reticulum (ER) membrane, distending the leaflet facing the cytoplasm and causing primary droplets of accumulated lipid to bud off the ER (1, 3–6). Milk secretory granules arise by the fusion of primary lipid droplets and are thus enveloped in a monolayer of phospholipids from the cytoplasmic leaflet of the ER membrane, as are all other lipid droplets (7–15). However, in contrast to all other lipid droplets, milk secretory granules are transported to the cell surface where they are pinched off into the alveolar space entirely surrounded in a layer of plasma membrane (2, 16–18). Accordingly, the envelopes of mature milk fat globules consist of three phospholipid membrane monolayers. On the inside is the cytoplasmic leaflet of the ER membrane arising from the phospholipid monolayer surrounding the milk secretory granule. On the outside is a classic membrane bilayer comprised of the extracellular and cytoplasmic leaflets of the plasma membrane. Variable amounts of cytoplasm are often entrained between the inner monolayer and the outer bilayer. Unfortunately, the historical misnomer “milk fat globule membrane” studiously ignores the complex layered structure of the milk fat globule envelope and its intracellular derivation.
Isolated envelopes of milk fat globules contain typical lipid droplet-associated proteins, such as adipophilin and TIP47, and various other proteins of milk, including butyrophilin and xanthine oxidoreductase (2, 19–24). Adipophilin and TIP47 are lipid droplet-associated proteins that are assumed to reside exclusively in the envelope of lipid droplets and to be involved in droplet formation, but both are also clearly residents of the lipid droplet core (25, 26) and of the plasma membrane (27). Butyrophilin, a type I transmembrane glycoprotein with a cytoplasmic C-terminal tail, is concentrated in the apical plasma membranes of mammary epithelial cells (24, 28–33). Because butyrophilin is expressed only during lactation, it appears to be essential for milk fat globule production (31). Xanthine oxidoreductase, a soluble, homodimeric, cytoplasmic enzyme, is concentrated along the inner surface of the apical plasma membrane, where it is proposed to bind with high affinity to the cytosolic domain of plasma membrane butyrophilin (23, 34–37). Butyrophilin and xanthine oxidoreductase are suggested to link milk secretory granules to the plasma membrane for secretion by interacting with adipophilin at the milk secretory granule surface facilitating the envelopment of the granule with plasma membrane during milk fat droplet formation (2, 22, 38).
These proteins, and probably several others (2), are evidently involved in milk fat globule formation and hence milk production. Their precise locations in milk secretory granules, in the apical plasma membrane of mammary epithelial cells, and notably in mature milk fat globules are uncertain, because the spatial resolution of the techniques hitherto used to detect them has been too low. In the present study, we localized adipophilin, TIP47, butyrophilin, and xanthine oxidoreductase in milk fat globules and mammary epithelial cells using freeze-fracture immunoelectron microscopy. Freeze-fracturing of cells exposes large, unperturbed, planar expanses of biological membranes at high resolution and, combined with immunocytochemistry, permits membrane-bound proteins to be localized precisely. Our findings on the distribution of the proteins yield information for deducing how butyrophilin controls the secretion of milk fat globules.
Results
Antibody Specificity.
In Western blots, the human butyrophilin antibody detected a unique band at 66 kDa representing complete butyrophilin and a band at 62 kDa representing a fragment of butyrophilin lacking the C terminus (24). The antibodies to adipophilin, TIP47, and xanthine oxidoreductase were all confirmed to detect bands at the appropriate molecular masses (52, 47, and 160 kDa, respectively) (see Fig. 4, which is published as supporting information on the PNAS web site).
Fluorescence Microscopy and Electron Microscopy of Ultrathin Cryosections.
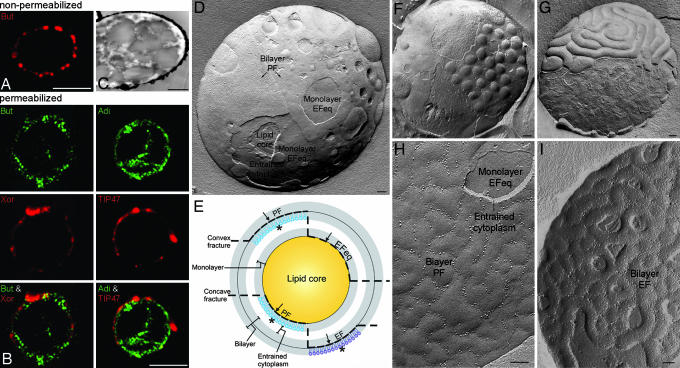
Immunofluorescence microscopy of nonpermeabilized milk fat globules demonstrated labeling of butyrophilin in a punctate ring-like pattern at the surface of the globules (Fig. 1A). A similar pattern of labeling for adipophilin, xanthine oxidoreductase, and TIP47 was observed, but only after permeabilization (Fig. 1B). These results suggest that butyrophilin is exposed at the surface of the globule, the other proteins occurring at a deeper level that becomes accessible only after permeabilization. Electron microscopy of ultrathin cryosections did not permit clear localization of proteins to the monolayer or bilayer, but were informative in revealing the structure of the globule and granule core. With this technique, the core was visualized as an amalgamation of multiple primary lipid droplets, rather than a single uniform structure (Fig. 1C).
Fig. 1.
Proteins and structure of the milk fat globule envelope. (A) Immunofluorescence microscopy reveals butyrophilin (But) on the surface of nonpermeabilized milk fat globules. (B) After permeabilization of milk fat globules, adipophilin (Adi), xanthine oxidoreductase (Xor), and TIP47 are detectable. (C) The lipid core of cryosectioned milk fat globules is comprised of many amalgamated small lipid droplet cores. (D) Overview of a freeze-fractured milk fat globule. The globule has been convexly fractured revealing the P-face (PF) of the envelope bilayer with rough areas populated with intramembrane particles (left arrow) and smooth intramembrane particle-poor regions (right arrow). The fracture has penetrated more deeply in some places exposing the underlying E-face equivalent (EFeq) of the envelope monolayer and the concentrically ordered layers of lipid in the core. Entrained cytoplasm is seen between the bilayer and monolayer. (E) Schematic diagram of fracture planes through the milk fat globule. The envelope is comprised of an inner phospholipid monolayer, derived from the cytoplasmic leaflet of the endoplasmatic reticulum membrane, and an outer bilayer, conferred to the milk secretory granule by the plasma membrane during budding, with variable amounts of entrained cytoplasm between. In convex fractures (upper dashed line), the P-face of the bilayer and the E-face equivalent of the monolayer are exposed. In concave fractures (lower dashed line), the E-face (EF) of the bilayer and the P-face of the monolayer are revealed. Asterisks represent sites at which the proteins are labeled with immunogold, and arrows mark the directions from which immunogold labeling in the replicas is viewed. (F–I) Images illustrating variation in morphology of freeze-fractured milk fat globule envelopes. Convex fractures may expose round bumps in the bilayer P-face (F) or elongated bumps with intervening furrows (G). Sometimes, when the fracture penetrates to the underlying monolayer, the E-face equivalent (EFeq) of the monolayer is exposed in planar view underneath the P-face (PF) of the bilayer (H). Intramembrane particles (proteins) in linear arrays are visible within the furrows, and entrained cytoplasm is seen between the bilayer and monolayer. (I) In concave fractures, the E-face (EF) of the bilayer is displayed. Prominent ridges are visible in the E-face of the bilayer. The ridges in the E-face of the bilayer are complementary to the furrows in the P-face of the bilayer. (Scale bars, 5.0 μm in A and B and 0.1 μm in C–I.)
Interpretation of Freeze-Fractured Milk Fat Globules.
A typical convexly freeze-fractured milk fat globule is shown in Fig. 1D. Fracturing has occurred at different levels within the globule, exposing en face portions of the bilayer, monolayer, and lipid core. These views arise because fracturing tends to split membranes along an interior hydrophobic plane, i.e., between the tails of the phospholipids of the monolayers. An understanding of the topological relationship of these layers and the conventions used in describing freeze-fracture images (39) are essential for interpreting the present results (Fig. 1E). When the globule bilayer is fractured, the fracture preferentially splits the bilayer into its two constituent half-membrane leaflets. One leaflet remains attached to the cytoplasm (designated the P-half), whereas the apposing leaflet remains attached to the extracellular space (E-half). The view of the P-half is referred to as the P-face, and is seen in convexly fractured globules; the view of the E-half is referred to as the E-face, and is seen in concavely fractured globules. Freeze-fracture nomenclature has to be adapted when applied to the underlying monolayer of the globule. Because this structure is apposed to the neutral lipids of the core, the fracture path lies at the interface between the hydrophobic aspects of the monolayer and the core. The monolayer is derived from the cytoplasmic leaflet of the ER, so its hydrophobic aspect revealed in concavely fractured globules is considered to be a P-face view. The complementary aspect seen in convexly fractured globules, which actually represents a view of the outermost aspect of the core, is referred to as the E-face equivalent (25, 27).
Freeze-fracture reveals that the milk fat globule envelope is heterogeneous in appearance. The fracture faces of the bilayer sometimes appear studded with abundant evenly distributed intramembrane particles, replicas of the proteins of the bilayer; adjacent areas may be devoid of particles (Fig. 1D). In other instances, domains showing round bumps (Fig. 1F) and elongated bumps and intervening furrows (Fig. 1G) are apparent. The furrows occur as an interlinking network and are found on the P-face of the bilayer (Fig. 1 G and H); complementary ridges are seen on the E-face (Fig. 1I). Intramembrane particles are generally confined to the ridges and furrows where these are present. The fracture faces of the monolayer consistently exhibit particle-free surfaces (Fig. 1 D and H). The interior of the globule core is also comprised of particle-free lipid layers that are often arranged concentrically like the leaves of an onion (Fig. 1D).
Distribution of Proteins Revealed by Freeze-Fracture Immunocytochemistry.
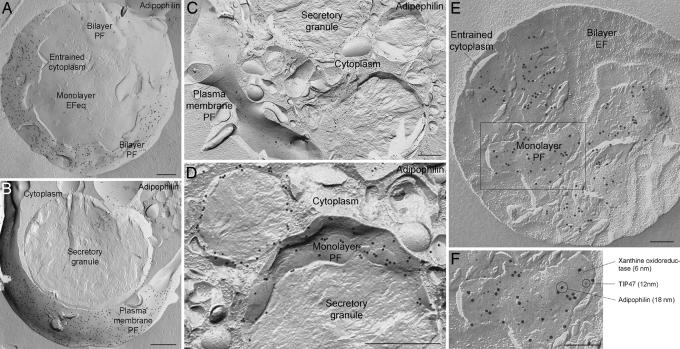
Fig. 2A shows an adipophilin-labeled convexly fractured globule exposing the bilayer P-face and the monolayer E-face equivalent. Abundant gold label for adipophilin is apparent in the globule bilayer. A similar abundance of adipophilin label is seen in large clusters specifically on the portions of the plasma membrane of mammary epithelial cells enveloping milk secretory granules before secretion (Fig. 2B). In both cases, adipophilin label is confined to the P-face; no label is seen on the E-face of the plasma membrane or on the E-face of the bilayer of concavely fractured globules. Apart from the clusters of adipophilin label in the plasma membrane domains apposed to secretory granules, a lower density of label is seen throughout the plasma membrane P-face (Fig. 2C). In concavely fractured secretory granules (Fig. 2D) and mature milk fat globules, fractures that reveal the monolayer also show adipophilin labeling, although at lower levels than those seen in the bilayer and plasma membrane clusters. Adipophilin is similarly detected only on the monolayer P-face, never on the E-face equivalent (see Fig. 5A, which is published as supporting information on the PNAS web site).
Fig. 2.
Distribution of adipophilin, TIP47, and xanthine oxidoreductase in freeze-fractured milk fat globules and in mammary epithelial cells. (A) In the globule abundant gold label for adipophilin is seen on the bilayer P-face (PF); the underlying monolayer E-face equivalent (EFeq) is devoid of label. (B) In a mammary epithelial cell, clusters of gold label for adipophilin occur in plasma membrane areas closely apposed to the secretory granule. (B and C) Noticibly lower concentrations of adipophilin label occur in other regions where the plasma membrane is not directly associated with milk secretory granules. The label is located on the P-face of the plasma membrane, i.e., in an equivalent position to the adipophilin label on the globule bilayer P-face. (D) Concave fractures of granules in epithelial cells reveal adipophilin labeling on the monolayer P-face (PF). (B–D) Gold label at the periphery of cross-fractured granules is attributable to adipophilin in the monolayer. No label is present on the bilayer E-face (EF). (E and F) Triple immunogold labeling of adipophilin (18-nm gold), xanthine oxidoreductase (6-nm gold), and TIP47 (12-nm gold) reveals colocalization of these three proteins in the monolayer P-face (PF). Label for all three proteins is absent in the bilayer E-face (EF). (F) A higher magnification view of the boxed area in E. Note the specificity of labeling and complete lack of background. (Scale bars, 0.5 μm in A–D and 0.2 μm in E and F.)
Unlike adipophilin, labeling for xanthine oxidoreductase and for TIP47 is only observed in the monolayer of the globule, never in the bilayer. However, as with adipophilin, the label on the monolayer is found only on the P-face (see Fig. 5 B and C). Triple immunogold labeling confirmed that adipophilin, xanthine oxidoreductase, and TIP47 are colocalized in the monolayer P-face (Fig. 2 E and F).
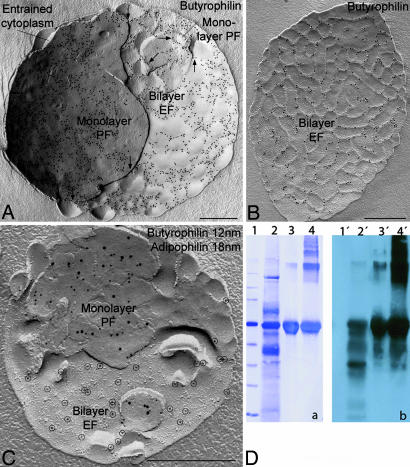
In common with adipophilin, butyrophilin is detected both in the monolayer and the bilayer of milk fat globules (Fig. 3A–C). However, the label is 3.7-fold more abundant in the monolayer than in the bilayer (monolayer, 138.88 ± 0.12 gold particles μm−2; bilayer, 38.24 ± 0.12 gold particles μm−2; P < 0.01). The monolayer P-face is similarly labeled both in globules and secretory granules. In contrast to adipophilin, the bilayer labeling for butyrophilin is confined to the E-face of concavely fractured globules (Fig. 3 A and B); no detectable label is apparent on the P-face. Double labeling for adipophilin and butyrophilin confirms these distinct distribution patterns (Fig. 3 C); the monolayer P-face shows labels both for adipophilin and butyrophilin, whereas the bilayer E-face contains only butyrophilin. The plasma membrane of mammary epithelial cells reveals butyrophilin only on the E-face, i.e., in the corresponding location to that seen in the globule bilayer. The butyrophilin labeling on the globule bilayer is characteristically concentrated along a network of ridges that range from being barely discernible to quite distinct (Fig. 3 A and B). Ridges or furrows are not apparent on the monolayer, but the pattern of butyrophilin labeling on the monolayer is commonly in the form of a similar network. Fractures revealing side-by-side views of the monolayer and the bilayer of the same granule demonstrate that the network of butyrophilin labeling on the bilayer ridges is continuous with the network of labeling on the monolayer (Fig. 3A). This finding indicates that butyrophilin is distributed in mirror image patterns on the monolayer and bilayer. Furthermore, fracture transitions between the monolayer and the bilayer revealed the butyrophilin network to be sites of close contact between the monolayer and bilayer (Fig. 3A).
Fig. 3.
Distribution and analysis of butyrophilin. (A–C) Distribution of butyrophilin in concavely fractured milk fat globules. (A) Intensive labeling of butyrophilin occurs on both the P-face (PF) of the monolayer and the E-face (EF) of the bilayer. Butyrophilin labeling on the bilayer E-face occurs exclusively in a network of more or less prominent ridges, which are especially obvious in B. Note that individual strings of label in the ridges in A are continuous at sites in which the fracture steps between the monolayer and the bilayer (arrows). Thus, the bilayer is strongly inflected toward the monolayer at the ridges; the ridges are sites of apposition between monolayer and bilayer. (C) Double labeling shows that butyrophilin is distributed differently from adipophilin in the globule envelope. Whereas butyrophilin labeling (12-nm gold) occurs on both the P-face of the monolayer and on the E-face of the bilayer (encircled), adipophilin (18-nm gold) is labeled on the P-face of the monolayer, but not on the E-face of the bilayer. The network of ridges is less apparent in this high magnification than in the survey views of A and B. (D) Self-aggregation of butyrophilin from bovine milk fat globule envelopes. Butyrophilin was visualized in gels with Coomassie blue (a) and in blots with anti-bovine butyrophilin antibody (b). Lanes 1 and 1′: protein markers; lanes 2 and 2′, 10 μg of milk fat globule envelopes; lanes 3 and 3′, 10 μg of eluted and precipitated butyrophilin, 1-min treatment at 95°C in SDS-sample buffer; lanes 4 and 4′, 10 μg of eluted and precipitated butyrophilin, 10-min treatment at 95°C in SDS sample buffer. Note that butyrophilin at 65 kDa and aggregates at 200 and 260 kDa. More aggregated butyrophilin is present after longer treatment and heating in SDS-sample buffer. (Scale bars, 0.5 μm.)
Self-Aggregation of Butyrophilin.
Because our freeze-fracture findings raised the possibility of butyrophilin–butyrophilin interactions, we sought biochemical evidence for the existence of such interactions. We found that purified butyrophilin shows a marked tendency to form aggregates in vitro (Fig. 3D). The most prominent aggregates occur at 200 and 260 kDa. Aggregation increases with time and heating.
Discussion
In all mammals, milk fat globules arise when milk secretory granules are enveloped in a layer of the plasma membrane and are shed off mammary epithelial cells (6). Notwithstanding this disarmingly patent mode of origin, the molecular mechanisms underlying milk fat globule secretion are still incompletely resolved (1, 2). At present, milk fat globule formation is thought to be attributable to interactions between specific proteins of the plasma membrane and of the secretory granule surface (22). Specifically, butyrophilin in the plasma membrane of the epithelial cell is believed to bind to cytosolic xanthine oxidoreductase, forming protein complexes that are concentrated in the apical plasma membrane where milk fat globule budding occurs. The butyrophilin–xanthine oxidoreductase complexes bind in turn to adipophilin in the phospholipid monolayer surrounding the secretory granule, drawing the plasma membrane up to and around the secretory granule in zipper-like fashion and provoking the shedding of a mature milk fat globule from the cell.
This hypothesis requires that the proteins involved in the elaboration of milk fat globules be distributed in appropriate positions in the phospholipid monolayer of the milk secretory granule, in the apical plasma membrane of mammary epithelial cells, and in the monolayer and bilayer of the milk fat globule envelope. However, the resolution of techniques previously used to detect these proteins has been insufficient to demonstrate that they are in their presumed locations. The distributions of these proteins have been deduced by comparing the compositions of subcellular fractions, and by immunofluorescence microscopy. However, inferences from biochemical studies on isolated material have to be discounted because pure isolates of milk secretory granule membranes and apical plasma membranes are simply not available (2). Also, separation of the individual components of the globule envelope for study has not been possible. Direct detection of the proteins in milk fat globules has been achieved by immunofluorescence microscopy (34, 40), but at a resolution too low to determine their precise locations in the envelope. In addition, fixatives, detergents, and lipid solvents required for immunofluorescence studies are notorious for altering the size and shape of lipid droplets and the distribution of their associated proteins (41–43). Thus, in which component, the monolayer, the bilayer, the entrained cytoplasm, or even the core, of the milk fat globule the individual proteins reside is uncertain. In contrast, freeze-fracture immunocytochemistry reveals specifically labeled membrane-resident proteins at high resolution and facilitates the unequivocal assignment of the proteins to one or the other membrane leaflet. Thus, in the present context, whether the proteins of the milk fat globule envelope reside in the monolayer or in the bilayer can be determined unambiguously.
We used freeze-fracture immunocytochemistry to localize adipophilin and TIP47, lipid droplet-associated proteins, and butyrophilin and xanthine oxidoreductase, proteins specifically involved in milk fat globule secretion, in envelopes of milk secretory granules and milk fat globules. As predicted by the above hypothesis, we detected adipophilin in monolayers of milk secretory granules and in monolayers of milk fat globule envelopes. We also found butyrophilin in bilayers of milk fat globule envelopes and in plasma membranes of epithelial cells, as expected. Visualization of butyrophilin in nonpermeabilized milk fat globules by immunofluorescence microscopy affirms the presence of butyrophilin in the bilayer of the globule envelope.
Surprisingly, however, we find that adipophilin is present in the inner leaflet (P-face) of the milk fat globule bilayer and clustered in the plasma membrane around milk secretory granules in visually obviously higher concentrations than in the monolayer. Thus, adipophilin in the plasma membrane should compete more successfully for butyrophilin–xanthine oxidoreductase complexes than does adipophilin in the monolayers of milk secretory granules, in effect depressing milk fat globule secretion. For this reason, a key role for adipophilin in milk secretion seems unlikely. On the other hand, secretory granules are unmistakenly physically associated with the regular clusters of adipophilin in the plasma membrane. Because similar clusters of adipophilin and perilipin in the plasma membrane have also been reported near lipid droplets in nonmilk secreting cells (27), the adipophilin clusters in mammary epithelial cells may have the same (unknown) function as in other cells and are therefore not necessarily involved in milk fat globule secretion. In keeping with its reported presence in the phospholipid monolayer of lipid droplets (44–46), TIP47 occurs in the monolayer of milk fat globules. Here as elsewhere, its function is unclear.
We also found that butyrophilin is statistically significantly more abundant in the monolayer than in the bilayer of the milk fat globule envelope, which is unanticipated. Indeed, if milk fat globule secretion depends on the binding of plasma membrane butyrophilin–xanthine oxidoreductase complexes with monolayer adipophilin, then butyrophilin should not be an integral protein of the monolayer at all. Just as unexpected is the presence of xanthine oxidoreductase in the milk fat globule monolayer. Whereas butyrophilin occurs in a well defined network in the monolayer, xanthine oxidoreductase is diffusely distributed. These divergent localizations indicate clearly that butyrophilin is not complexed with xanthine oxidoreductase in the milk fat globule envelope. Similarly, the diffuse distribution of adipophilin in the monolayer does not coincide with the network of butyrophilin. Therefore, butyrophilin binds neither directly nor via butyrophilin–xanthine oxidoreductase complexes to secretory granule adipophilin during milk fat globule secretion. We conclude that binding of plasma membrane butyrophilin–xanthine oxidoreductase complexes to milk secretory granule adipophilin is not responsible for milk fat globule secretion.
Analysis of the distribution of butyrophilin in the milk fat globule envelope provides clues to how butyrophilin may function in milk fat globule secretion. In the bilayer, butyrophilin is localized exclusively in a network of ridges that project to the monolayer. Butyrophilin label in the bilayer is continuous with butyrophilin label in the monolayer. The perfect coincidence of ridges in the bilayer and butyrophilin label in the monolayer indicates close physical apposition between bilayer and monolayer in these regions, suggesting that a network of butyrophilin-containing adhesive sites links the bilayer with the monolayer. Adhesiveness may simply be conferred by butyrophilin–butyrophilin oligomerization, given our finding that isolated butyrophilin molecules exhibit a strong inherent tendency to aggregate. How butyrophilin molecules are organized in the form of a network in the fat globule envelope may be deduced by examining cryosectioned milk fat globules. We found noncoalesced cores of primary lipid droplets inside cryosectioned milk fat globules. Such lipid droplet cores underlying the phospholipid monolayer in secretory granules may impose constraints on the distribution of butyrophilin in the monolayer; butyrophilin, a transmembrane protein with hydrophilic terminals, may be partitioned into the free zones between the hydrophobic lipid droplet cores, hence accounting for the network of butyrophilin in the monolayer. Once established, the network of butyrophilin in the monolayer would be subsequently imprinted onto the bilayer owing to interactions of monolayer butyrophilin and plasma membrane butyrophilin, and the butyrophilin network would be perpetuated in the secreted milk fat globule as we observed. Thus, butyrophilin–butyrophilin binding would be responsible for drawing the plasma membrane around the secretory granule and ultimately for budding of the nascent milk fat globule from the mammary epithelial cell. Accordingly, butyrophilin–butyrophilin interactions would play a decisive role in the control of milk fat globule formation. This mechanism requires that, in accord with our findings, butyrophilin is retained and targeted to two specific locations, the ER and plasma membrane. The 3-fold higher density of label in the ER compared with the plasma membrane suggests that the majority of butyrophilin is retained in the ER, a relatively small proportion of the total pool being adequate for the interaction of the plasma membrane with the monolayer.
In summary, our findings do not support a mechanism of milk fat globule formation involving the binding of plasma membrane butyrophilin–xanthine oxidoreductase complexes to adipophilin in the milk secretory granule. Rather, they suggest that a crucial feature of milk fat globule secretion is the establishment of a network of adhesion sites containing butyrophilin in the secretory granule monolayer and the ensuing binding of monolayer butyrophilin to plasma membrane butyrophilin. Viewed in this way, interactions between milk secretory granule butyrophilin and plasma membrane butyrophilin effectively control milk fat globule secretion in mammals.
Materials and Methods
Antibodies.
Three well characterized antibodies were purchased for these studies, a mouse monoclonal antibody to human adipophilin (AP125; Progen, Heidelberg), a Guinea pig polyclonal antibody to human TIP47 (GP30; Progen), and a rabbit polyclonal antibody to bovine xanthine oxidoreductase with known cross-reactivity to human xanthine oxidoreductase (R1119P; Acris Antibodies, Hiddenhausen, Germany). In addition, we generated two antibodies to butyrophilin (for more details, see Supporting Text, which is published as supporting information on the PNAS web site).
Immunofluorescence Microscopy.
See Supporting Text for more information.
Isolation of Milk Fat Globules and Mammary Epithelial Cells.
Fresh milk was lightly centrifuged and the supernatant containing suspended milk fat globules and a few fortuitously present mammary epithelial cells was collected for immediate use for cryosectioning or freeze-fracturing.
Cryoelectron Microscopy.
Milk supernatants were fixed briefly with equal volumes of 8% paraformaldehyde. After addition of 2.3 M sucrose, the samples were placed on metal pins and rapidly frozen by plunging into liquid nitrogen. Ultrathin cryosections were cut in an UCT ultracryomicrotome (Leitz, Cologne) to a thickness of ≈60 nm (47). They were thawed on 2.3 M sucrose, placed on grids, and stabilized with methyl cellulose containing uranyl acetate essentially as described (48). Preparations were examined in an EM410 electron microscope (Philips) and documented digitally (Ditabis).
Freeze-Fracture Immunocytochemistry.
Milk supernatants were mixed briefly with 30% glycerol (<30 s), snap-frozen in Freon 22 cooled with liquid nitrogen, and freeze-fractured in a BA310 freeze-fracture unit (Balzers) at −105°C under vacuum (2 × 10−6 bar). Replicas of the freshly fractured samples were made immediately by electron beam evaporation of platinum–carbon and carbon at angles of 38° and 90° and to thicknesses of ≈2 and 20 nm, respectively. The replicas were incubated overnight in 5% SDS to remove cellular material except for those molecules adhering directly to the replicas (49, 50). The replicas were washed in distilled water and incubated briefly in 5% BSA before immunolabeling.
Immunolabeling was by incubation with the desired antibody followed by washing and incubation with an appropriate secondary antibody–gold conjugate. Antibody concentrations were chosen empirically to optimize labeling intensity and were usually 5 μg/ml. Double or triple labeling was carried out by using mixtures of the desired antibodies, followed by washing and incubation with mixtures of differently sized appropriate anti-antibody gold conjugates as noted in Figs. 1–3 (see Supporting Text for more information).
Quantitation of Butyrophilin Label in Freeze-Fracture Replicas.
Counts of gold particles were used to estimate the relative concentration of butyrophilin in monolayers and bilayers of milk fat globule envelopes. Significance was evaluated with Student’s t test.
Self-Aggregation of Butyrophilin.
Proteins of isolated bovine milk fat globule envelopes were separated by SDS/PAGE. Bands with butyrophilin (65 kDa) were excised from several gels, eluted and subjected again to electrophoreses after 1 or 10 min of heating in SDS sample buffer. The proteins in the gels were assessed by staining with Coomassie blue or by Western blotting using the antibody to bovine butyrophilin described above.
Supplementary Material
Acknowledgments
We thank Karin Schlattmann, Christina Köppler, Stefanie Winter, and Marianne Opalka for competent and indispensable technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 492.
Abbreviation
- ER
endoplasmic reticulum.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Keenan T. W. J. Mamm. Gland Biol. Neoplasia. 2001;6:365–371. doi: 10.1023/a:1011383826719. [DOI] [PubMed] [Google Scholar]
- 2.Heid H. W., Keenan T. W. Eur. J. Cell Biol. 2005;84:245–258. doi: 10.1016/j.ejcb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Patton S., Keenan T. W. Biochim. Biophys. Acta. 1975;415:273–309. doi: 10.1016/0304-4157(75)90011-8. [DOI] [PubMed] [Google Scholar]
- 4.Zaczek M., Keenan T. W. Protoplasma. 1990;159:179–182. [Google Scholar]
- 5.Keenan T. W., Dylewski D. P., Ghosal D., Keon B. H. Eur. J. Cell Biol. 1992;57:21–29. [PubMed] [Google Scholar]
- 6.Mather J. H., Keenan T. W. J. Mamm. Gland Biol. Neoplasia. 1998;3:259–273. doi: 10.1023/a:1018711410270. [DOI] [PubMed] [Google Scholar]
- 7.Brown D. A. Curr. Biol. 2001;11:R446–R449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 8.Ostermeyer A. G., Paci J. M., Zeng Y., Lublin D., Munro S., Brown D. J. Cell Biol. 2001;151:1071–1078. doi: 10.1083/jcb.152.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Deurs B., Roepstorff K., Hommesgaard A. M., Sandvig K. Trends Cell Biol. 2003;13:92–100. doi: 10.1016/s0962-8924(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 10.Thyberg J. J. Histochem. Cytochem. 2002;50:185–195. doi: 10.1177/002215540205000206. [DOI] [PubMed] [Google Scholar]
- 11.Londos C., Brasaemle D. J., Schultz C. J., Segrest J. P., Kimmel A. R. Semin. Cell Dev. Biol. 1999;10:51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- 12.Robenek M. J., Severs N. J., Schlattmann K., Plenz G., Zimmer K. P., Troyer D., Robenek H. FASEB J. 2004;18:866–868. doi: 10.1096/fj.03-0782fje. [DOI] [PubMed] [Google Scholar]
- 13.Tauchi-Sato K., Ozeki S., Honjou T., Taguchi R., Fujimoto T. J. Biol. Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 14.Peixoto de Menezes A., Pinto da Silva P. J. Cell Biol. 1978;76:767–778. doi: 10.1083/jcb.76.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peixoto de Menezes A., Pinto da Silva P. Lab. Invest. 1979;40:545–553. [PubMed] [Google Scholar]
- 16.Bargmann W., Knoop A. Z. Zellforsch. Mikrosk. Anat. 1959;49:344–388. [PubMed] [Google Scholar]
- 17.Dylewski D. P., Dapper C. H., Valivullak H. M., Deency J. T., Keenan T. W. Eur. J. Cell Biol. 1984;35:99–111. [PubMed] [Google Scholar]
- 18.Murphy D. J. Prog. Lipid Res. 2001;40:325–430. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 19.Mather J. H. J. Dairy Sci. 2000;83:203–247. doi: 10.3168/jds.S0022-0302(00)74870-3. [DOI] [PubMed] [Google Scholar]
- 20.Heid H. W., Schnölzer M., Keenan T. W. Biochem. J. 1996;320:1025–1030. doi: 10.1042/bj3201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heid H. W., Moll R., Schwetlick J., Rackwitz H. R., Keenan T. W. Cell Tissue Res. 1998;294:309–321. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- 22.Murphy D. J., Vance J. Trends Biochem. Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- 23.Harrison R. Drug Metab. Rev. 2004;36:363–375. doi: 10.1081/dmr-120037569. [DOI] [PubMed] [Google Scholar]
- 24.Banghart L. R., Chamberlain C. W., Velande J., Korobko J. V., Ogg S. L., Jack J. L., Vakharia V. N., Mather J. H. J. Biol. Chem. 1998;273:4171–4179. doi: 10.1074/jbc.273.7.4171. [DOI] [PubMed] [Google Scholar]
- 25.Robenek H., Lorkowski S., Schnoor M., Troyer D. J. Biol. Chem. 2005;280:5789–5794. doi: 10.1074/jbc.M407194200. [DOI] [PubMed] [Google Scholar]
- 26.Robenek H., Robenek M. J., Troyer D. J. Lipid Res. 2005;46:1331–1338. doi: 10.1194/jlr.M400323-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Robenek H., Robenek M. J., Buers I., Lorkowski S., Hofnagel O., Troyer D., Severs N. J. J. Biol. Chem. 2005;280:26330–26338. doi: 10.1074/jbc.M413312200. [DOI] [PubMed] [Google Scholar]
- 28.Mather J. H., Jack L. J. W. J. Dairy Sci. 1993;76:3832–3850. doi: 10.3168/jds.s0022-0302(93)77726-7. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen R. C., Andersen M. H., Mabhout P., Berglund L., Petersen T. E., Rasmussen J. T. J. Dairy Sci. 1999;82:2543–2549. doi: 10.3168/jds.S0022-0302(99)75508-6. [DOI] [PubMed] [Google Scholar]
- 30.Ishii T., Aoki N., Noda A., Adachi T., Nakamura R., Matsuda T. Biochim. Biophys. Acta. 1995;1245:285–292. doi: 10.1016/0304-4165(95)00102-6. [DOI] [PubMed] [Google Scholar]
- 31.Ogg S. L., Weldon A. K., Dobbie L., Smith A. J. H., Mather J. H. Proc. Natl. Acad. Sci. USA. 2004;101:10084–10089. doi: 10.1073/pnas.0402930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jack L. J. W., Mather J. H. J. Biol. Chem. 1990;265:14481–14486. [PubMed] [Google Scholar]
- 33.Franke W. W., Heid H. W., Grund C., Winter S., Freudenstein C., Schmid E., Jarasch E. D., Keenan T. W. J. Cell Biol. 1981;89:485–494. doi: 10.1083/jcb.89.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mc Manaman J. L., Palmer C. A., Wright R. M., Neville M. C. J. Physiol. 2002;545:567–579. doi: 10.1113/jphysiol.2002.027185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruder G., Heid H. W., Jarasch E. D., Keenan J. W., Mather J. H. Biochim. Biophys. Acta. 1982;701:357–369. doi: 10.1016/0167-4838(82)90239-4. [DOI] [PubMed] [Google Scholar]
- 36.Jarasch E., Grund C., Bruder G., Heid H. W., Keenan T. W., Franke W. W. Cell. 1981;25:67–82. doi: 10.1016/0092-8674(81)90232-4. [DOI] [PubMed] [Google Scholar]
- 37.Vorbach C., Scriven A., Capecci M. R. Genes Dev. 2002;16:3223–3235. doi: 10.1101/gad.1032702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C. C., Howell K. E., Neville M. C., Yates J. R., McManaman J. L. Electrophoresis. 2000;21:3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 39.Branton D., Bullivant S., Gilula N. W., Karnovsky M. J., Moor H., Mühletahler K., Northcote D., Packer L., Satir B., Satir P., et al. Science. 1975;190:54–56. doi: 10.1126/science.1166299. [DOI] [PubMed] [Google Scholar]
- 40.Mc Manaman J. L., Zabaronick W., Schaack J., Orlicky D. J. J. Lipid Res. 2003;44:668–673. doi: 10.1194/jlr.C200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Ohsaki Y., Maeda T., Fujimoto T. Histochem. Cell Biol. 2005;124:445–452. doi: 10.1007/s00418-005-0061-5. [DOI] [PubMed] [Google Scholar]
- 42.DiDonato D., Brasaemle D. L. J. Histochem. Cytochem. 2003;51:773–780. doi: 10.1177/002215540305100608. [DOI] [PubMed] [Google Scholar]
- 43.Fukumoto S., Fujimoto T. Histochem. Cell Biol. 2002;118:423–428. doi: 10.1007/s00418-002-0462-7. [DOI] [PubMed] [Google Scholar]
- 44.Hickenbottom S. J., Kimmel A. R., Londos C., Hurly J. H. Structure (London) 2004;12:1199–1207. doi: 10.1016/j.str.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Than N. G., Sumegi B., Bellyei S., Berki T., Szekeres G., Janaky T., Szigetti A., Bohn H., Than G. M. Eur. J. Biochem. 2003;270:1176–1188. doi: 10.1046/j.1432-1033.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- 46.Wolins N. E., Rubin B., Brasaemle D. L. J. Biol. Chem. 2001;276:5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 47.Tokuyasu K. T. Histochem. J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- 48.Robenek M. J., Schlattmann K., Zimmer K. P., Plenz G., Troyer D., Robenek H. FASEB J. 2003;17:1940–1942. doi: 10.1096/fj.03-0008fje. [DOI] [PubMed] [Google Scholar]
- 49.Fujimoto K. J. Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 50.Fujimoto K. Histochem. Cell Biol. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.