Abstract
Many cellular and viral processes depend on site-specific proteolysis. Here, a genetic system for the identification of such proteases and inhibitors is described. The system utilizes the temperature- sensitive Saccharomyces cerevisiae CDC25-2 mutant strain and exploits the strict requirement of membrane localization of a constitutively active Ras mutant for the complementation of the yeast growth defect at the non-permissive temperature. Expres sion of a fusion protein in which a substrate peptide of the TEV protease separates a myristoylation signal from a constitutively active human Ras protein confers temperature insensitivity. Co-expression of the protease results in release of the Ras mutant from the membrane and growth arrest at the non-permissive temperature. This non-transcriptional assay represents a new approach to the in vivo analysis of site-specific proteases and may be a valuable alternative to existing methods. It has significant potential for the selection of inhibitors of cytoplasmic and membrane-associated proteases of biotechnical and clinical relevance.
INTRODUCTION
Site-specific cleavage of proteins plays a crucial role in cellular processes such as signal transduction (1–3), apoptosis (4–6) and development (7–10). Furthermore, virion formation in retroviruses and most eukaryotic positive strand RNA viruses requires cleavage of precursor polypeptides (11,12).
In the yeast Saccharomyces cerevisiae, growth depends on the production of cyclic adenosine monophosphate (cAMP) by adenylyl cyclase, which is stimulated by activated Ras proteins. The guanyl nucleotide exchange factor (GEF) CDC25, the homologue of human ‘son-of-sevenless’, activates Ras proteins by stimulating the release of GDP and the uptake of GTP. The S.cerevisiae strain CDC25-2 harbours a mutation in the CDC25 gene, rendering its product temperature sensitive, with nearly wild-type activity at 25°C and almost completely diminished activity at 36°C (13). Growth at 36°C can be restored by expression of a membrane-associated, constitutively active human Ras mutant. Membrane association is strictly required for this complementation and can be achieved by the endogenous C-terminal CAAX box for farnesoylation or by a heterologous N-terminal signal for myristoylation.
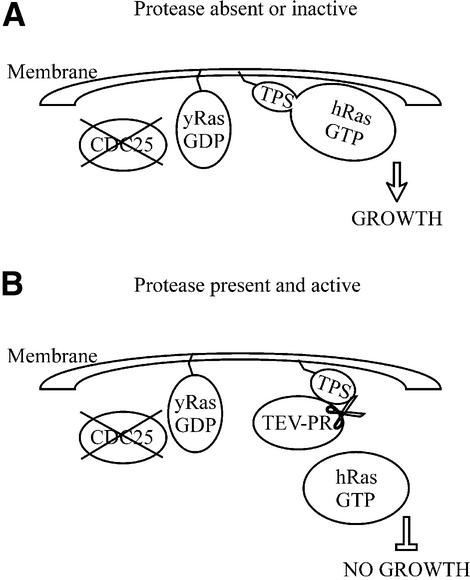
We took advantage of this effect to devise an in vivo system for the detection of site-specific proteases and their inhibitors as illustrated in Figure 1. Expression of a fusion protein, in which the substrate peptide of a protease separates a signal for myristoylation from a constitutively active Ras mutant (lacking its farnesoylation signal) [hRas(Q61L)ΔF], leads to growth at the non-permissive temperature of 36°C if no protease is co-expressed or if a co-expressed protease is inhibited. Presence of an active protease leads to release of the Ras mutant from the membrane and the inability to grow at 36°C.
Figure 1.
Schematic representation of the assay. CDC25-2 cells are growth arrested at 36°C due to a mutation in the Ras GEF CDC25. The yeast Ras function can be complemented by a membrane-localized constitutively active human Ras mutant. Membrane localization is strictly required for this complementation. A substrate (TPS) for the TEV protease (TEV-PR) separates a myristoylation signal from the mutant Ras protein (hRas,GTP). (A) In the absence of active protease, the substrate is not cleaved and the mutant Ras protein allows growth at 36°C. (B) In the presence of active protease, the mutant Ras is released from the membrane, leading to growth arrest at 36°C.
We used the nuclear inclusion protease of the tobacco etch virus (TEV), a member of the potato virus Y group (potyviridae) (14), as a model system. It is highly specific and recognizes the seven amino acid consensus sequence E-X-X-Y-X-Q↓G/S (15). The recent finding that the P1′ specificity (G or S in the consensus sequence) is not as stringent as anticipated further increases the usefulness of the TEV protease. This allows the removal of N-terminal tags without leaving a novel amino acid at the N-terminus, which potentially alters a given protein’s characteristics (16).
Biochemical methods, including resonance energy transfer (17), release of radioactive products (18) and spectrophotometry of chromogenic peptide substrates (19), have been used for the study of site-specific proteases. Since these assays are laborious and inconvenient for cloning and selection procedures, several in vivo protease assays have been developed. These include methods using phages or bacterial cells (20–23) and yeast-based methods (24–30). Basically, available yeast assays fall into two classes: assays in which a transcription factor is inactivated by a proteolytic event and assays in which a transcription factor is released from the membrane by site-specific proteolysis. Both classes rely on protease- mediated alteration of the expression of a reporter gene, e.g. β-galactosidase. Most of these assays utilize the yeast transcription factor Gal4 and corresponding reporter cassettes to detect proteolysis. Thus, yeast strains need to be used in which the endogenous Gal4 system has been deleted. Furthermore, assays which exploit endogenous signal transduction cascades may have the potential to be more sensitive than those based on reporter gene expression (22).
We sought to develop a yeast-based assay for site-specific proteases that uses interference with an endogenous signal transduction pathway rather than changes in reporter gene expression. Our approach adds to available techniques for studying site-specific proteases and may represent a useful alternative in situations when existing methods are not applicable. Furthermore, it offers the possibility to confirm results obtained with existing approaches in a second, independent in vivo assay.
MATERIALS AND METHODS
Construction of plasmids
The myristoylated substrate–Ras constructs were cloned into pYES2 (Invitrogen). The myristoylation signal is derived from v-Src (31). The TEV protease substrate sequences were generated by oligonucleotide synthesis. M-TPSo-Ras was constructed using the coding oligonucleotide 5′-TGG CCA ACC ACA GAA AAC CTC TAC TTT CAG TCC GGA ACC GTT GAC GCC GGC GGC CGC CCC-3′ annealed to a complementary oligonucleotide. The annealed oligonucleotides contain a 5′ BalI site, a 3′ NotI site and a 3′ SmaI half-site and encode an optimal recognition sequence for the TEV protease (TTENLYFQ↓SGTVDA) (32). They were ligated into the SmaI site downstream of the myristoylation signal of pYES-M-Ras. M-TPSs-Ras was constructed accordingly, encoding a suboptimal recognition sequence (coding oligonucleotide 5′-TGG CCA ACC ACA GAA AAC CTC TAC TTT CAG CGC GGA ACC GTT GAC GCC GGC GGC CGC CCC-3′; peptide TTENLYFQ↓RGTVDA) (32). M-TPSoi-Ras and M-TPSsi-Ras contain the optimal and suboptimal substrate sequence, respectively, in the inverse orientation, giving rise to ‘random’ peptide sequences, which are unrelated to the protease substrate. These peptides are not recognized by the TEV protease and served as uncleavable negative controls. pYES-M-Ras encodes amino acids 1–185 of the human H-Ras mutant Q61L fused to the myristoylation signal of v-Src. All substrate constructs are expressed from the Gal1 promoter and confer complementation of a uracil-negative phenotype.
The TEV-PR coding sequence was subcloned into BamHI and XhoI sites of the low copy number shuttle vector p414Gal1 [TEV-PR(l.c.)] and the high copy number shuttle vector p424Gal1 [TEV-PR(h.c.)] conferring complementation of a tryptophan-negative phenotype (33). The expression of both the substrate constructs and the TEV-PR constructs is induced by galactose and repressed by glucose.
Yeast growth and manipulation
CDC25-2 (MATα, ura3, lys2, leu2, trp1, hisΔ200, ade2-101, cdc25-2) were transformed using a standard lithium acetate procedure (34) and plated on synthetic dropout (SD) medium containing 2% glucose, 0.5% ammonium sulfate, 0.015% adenine sulfate, 0.17% yeast nitrogen base, 2.5% agar and complete supplement mixture lacking tryptophan and uracil and incubated at 25°C. Three independent clones were grown to late log phase at 25°C in SD+Gluc–TU liquid medium (identical to SD+Gluc–TU plates without agar). Cells were transferred to plates or liquid medium containing either glucose or galactose (SD+Gal–TU; 3% galactose, 2% raffinose and 2% glycerol instead of glucose) and incubated at 25 or 36°C for 24 h. Growth in liquid medium was quantified by measuring the optical density at 600 nm.
RESULTS AND DISCUSSION
We developed an in vivo assay for site-specific proteases based on the Ras/cAMP signal transduction pathway in yeast. Fusion protein substrates for the TEV protease were created, which will localize at the plasma membrane and complement the growth defect of CDC25-2 at the non-permissive temperature of 36°C. Co-expression of the active protease should lead to release of the substrate from the membrane and the restoration of the mutant phenotype.
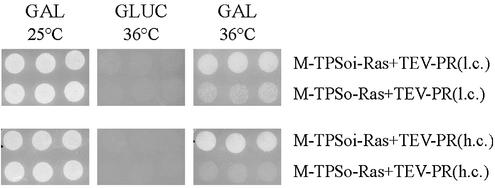
At 36°C under conditions allowing the expression of the foreign proteins, no growth was observed, when the protease was expressed from the high copy plasmid together with the myristoylated Ras substrate containing the optimal recognition sequence, whereas robust growth was observed in conjunction with the uncleavable substrate construct. The same holds true for the samples transformed with the uncleavable substrate construct together with the low copy TEV protease expression plasmid. Co-expression of the latter with the cleavable substrate construct led to greatly reduced growth (Fig. 2, GAL 36°C). To show that the observed effect is due to the expressed foreign proteins, cells were incubated at 36°C under non-inducing conditions on plates containing glucose. No growth was observed, when the expression of the protease and the substrate protein was repressed by glucose (Fig. 2, GLUC 36°C). The growth observed at 36°C under inducing conditions, with the uncleavable substrate protein, suggests that even high level expression of the protease is tolerated by the CDC25-2 cells. This is further underlined by the finding that no differences in growth are observed when incubating the cells at the permissive temperature of 25°C on galactose plates (Fig. 2, GAL 25°C). These results show that the system is suitable for the detection of the activity of a site-specific protease.
Figure 2.
Differential growth of CDC25-2 at 25 and 36°C. Three independent clones of CDC25-2 transformed with the indicated plasmids were spotted on plates containing 2% glucose (GLUC) or 3% galactose, 2% raffinose and 2% glycerol (GAL), respectively, and incubated at the indicated temperatures for 24 h. TPSoi, inverted optimal TEV protease substrate; TPSo, optimal TEV protease substrate; TEV-PR(l.c.), TEV-PR(h.c.), TEV protease expressed from a plasmid with low copy number (CEN/ARS origin) and with high copy number (2µ origin).
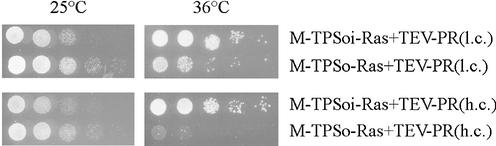
Since growth of cells at 36°C, expressing the protease from a low copy plasmid, was greatly diminished, but not completely impaired, a dilution series of one of the three clones analyzed in Figure 2 was performed in order to show that the growth rate is dependent on the expression level of the protease. Samples were diluted to an OD600 of 6.0 and these, together with four 10-fold dilutions, were spotted on two galactose-containing plates and incubated at either 25 or 36°C (Fig. 3). The growth difference at 36°C between samples expressing the protease from the low copy plasmid, compared to expression from the high copy plasmid, was evident. This was not due to a general growth reduction under these conditions, since the samples containing the uncleavable substrate showed no differences in growth (Fig. 3, 36°C). Surprisingly, at 25°C, a small reverse effect was observed. Samples containing an uncleavable substrate construct displayed reduced growth compared to the ones in which the Ras protein is released from the membrane (Fig. 3, 25°C). This may be due to competition of the foreign Ras protein with the endogenous, activated Ras proteins. For example, the foreign Ras protein might be efficient in binding certain components of the Ras/cAMP signal transduction pathway, but less efficient in activating them. This might make them unavailable for activation by the endogenous Ras protein, resulting in reduced intracellular cAMP levels. On the other hand, high level expression of a constitutively active Ras protein might lead to an overproduction of cAMP which could impair yeast growth. This effect was also observed in other experimental settings, when a constitutively active Ras protein is localized at the membrane of CDC25-2 and cells were incubated at 25°C (personal observations). Though speculative, it may be possible, by tuning expression levels of the protease and the Ras substrate, to generate a system in which expression of an active protease leads to growth at 25°C and incubation of the same yeast cells at 36°C also leads to growth, but only if the protease is inactive. This would allow for screening for proteases as well as for their inhibitors using the same yeast at different temperatures.
Figure 3.
Differential growth of CDC25-2 at 25 and 36°C. CDC25-2 transformed with the indicated plasmids was grown to late log phase in liquid medium for 2 days at 25°C and diluted to an OD600 of 6.0. These and four 10-fold dilutions were spotted on plates and incubated at the indicated temperatures for 24 h. TPSoi, inverted optimal TEV protease substrate; TPSo, optimal TEV protease substrate. TEV protease is expressed from plasmids with low copy number [CEN/ARS origin; TEV-PR(l.c.)] and with high copy number [2µ origin; TEV-PR(h.c.)].
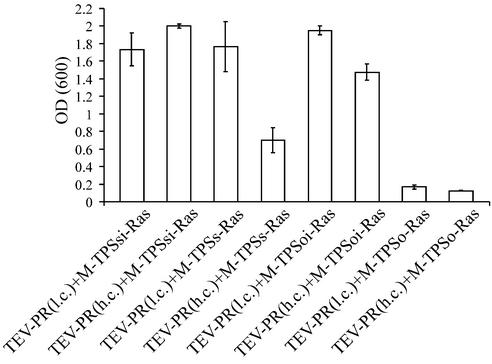
In order to quantify the observed effects, and to show that different cleavage efficiencies can be discriminated by the assay, liquid cultures of cells transformed with different substrate and protease constructs were diluted to an OD600 of 0.1 and growth was determined by measuring OD600 after incubation at 36°C for 24 h. Figure 4 shows that the assay discriminates between optimal and suboptimal substrate sequences. Comparison of samples expressing the protease from the high copy plasmid together with the suboptimal substrate with those expressing the protease together with the optimal substrate clearly reveals that growth is dependent on the nature of the substrate sequence. These results are in good agreement with the results obtained by Dougherty et al. (32), who performed mutational analyses of the cleavage efficiency of the TEV protease with substrates derived from the 54 kDa Mr nuclear inclusion protein/30 kDa Mr capsid protein junction.
Figure 4.
Quantification of growth at 36°C. CDC25-2 transformed with the indicated plasmids was diluted to an OD600 of 0.1 and incubated at 36°C. After 24 h incubation, OD600 was determined. Values represent means of three independent clones.
Differences in growth with the suboptimal substrate construct were only observed at high expression levels of the protease. Although in most applications of this assay, an optimal substrate sequence will be used, there might be situations in which substrate sequences with different cleavage efficiencies are to be analysed. In this respect, the question arises as to whether the assay is applicable to other, less site-specific proteases or proteases that cleave endogenous substrates. High levels of expression of such proteases might cause intolerable toxicity, as has been observed for members of the caspase family of proteases (28). However, it has been shown that TEV protease rapidly cleaves itself to generate a truncated protein with greatly diminished activity (35). This inactivation increases with the concentration of the protease (36). In addition, the TEV protease has a temperature optimum of 30°C and its activity is reduced at higher temperatures. Thus, it is likely that there is no need for high level expression when analysing proteases which do not display auto- inactivation or which are optimally active at 36°C. Furthermore, there is a certain threshold for the amount of activated membrane-associated Ras protein to effect complementation of the CDC25-2 phenotype (personal observations). The expression level used in this study is far beyond this threshold. Therefore, reduction of the expression level of the substrate–Ras fusion protein will further increase the sensitivity of the assay without compromising its discriminatory parameters. Potential toxicity caused by expression of proteases in the cytoplasm of a living cell is an inherent problem which the presented system shares with other in vivo assays for proteases. Future studies are required to determine if this technique is applicable to a wide range of less selective proteases.
These data show that an in vivo protease assay can be designed by coupling proteolytic activity to an essential signal transduction pathway of the yeast S.cerevisiae.
Several in vivo protease assays have been described. Some of them utilize bacterial or yeast cells (20–30). For cDNA library screening approaches, protease assays based on phages or bacterial cells may be advantageous due to high transformation efficiencies. However, many post-translational modifications are not carried out by these organisms. Furthermore, it has been noted that site-specific proteases are highly regulated and their activity often requires interaction with additional cellular components (37). In this respect, yeast is a highly suitable organism as it combines the characteristics of a eukaryotic cell with an ease of manipulation comparable to that of bacteria.
Since in this assay, proteolysis takes place at the plasma membrane, it may be specially useful for the analysis of membrane-associated proteases or proteases that process membrane-bound substrates. Many such proteases play critical roles in normal (38–40) and pathological cellular conditions (41,42). These proteases often strictly require membrane association for activity or cleave their substrates only within or close to the membrane (43,44). This makes them highly refractory to biochemical analyses that include protein purification steps. Furthermore, most of the in vivo assays described to date require the expression of soluble proteins (20–26). Available yeast-based assays suitable for the analysis of membrane-associated site-specific proteases all function by releasing a transcription factor from the membrane, which then translocates to the nucleus and stimulates transcription of a reporter gene (27–30). These assays produce false positive results when introducing proteins that bind to the regulatory regions of the reporter gene and activate transcription. Although readily backchecked, this is a limitation that is not encountered with the presented approach. The main source of false negatives in our system may be the introduction of heterologous Ras proteins, which can be eliminated by co-expression of a GTPase-activating protein (45).
When dealing with proteins that are potentially toxic when expressed in yeast, the ability to tightly regulate their expression is highly desirable. Most of the available yeast assays for site-specific proteases are based on Gal4-mediated transcriptional activation in a yeast strain devoid of a functional endogenous Gal system. In these strains, Gal promoters for the inducible expression of foreign proteins, which are extremely tightly regulated, cannot be used. This problem can be circumvented by using transcription factors other than Gal4 (e.g. LexA/B42) in a Gal(+) strain or by choosing other regulatable promoters (e.g. copper- or methionine-responsive promoters) in a Gal(–) strain (30). However, these systems are by far less widespread and less well characterized compared to Gal-based systems. These limitations originate from the use of transcription-based readouts reporting protease activity. Potentially, assay systems based on endogenous signal transduction pathways may display increased sensitivity as compared to reporter gene assays (22). Since the non-transcriptional assay presented in this study exploits interference with an endogenous signal transduction pathway, it may be applicable to a wider range of yeast assay scenarios. This new assay approach will not replace current assay techniques, but may provide a useful additional route for looking at protease function in yeast.
The system presented here overcomes some of the constraints of previously described assays and may contribute to the cloning and characterization of biotechnically or clinically relevant site-specific proteases and their inhibitors.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank A. Aronheim for the gift of the CDC25-2 strain and the original RasQ61LΔF construct and M. Hager and A. Zimmermann for helpful discussions. This work was supported by a grant from the State of Baden-Württemberg, Germany.
REFERENCES
- 1.Lee J.J., Ekker,S.C., von Kessler,D.P., Porter,J.A., Sun,B.I. and Beachy,P.A. (1994) Autoproteolysis in hedgehog protein biogenesis. Science, 266, 1528–1537. [DOI] [PubMed] [Google Scholar]
- 2.Coughlin S.R. (1994) Protease-activated receptors start a family. Proc. Natl Acad. Sci. USA, 91, 9200–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollenberg M.D. (1996) Protease-mediated signalling: new paradigms for cell regulation and drug development. Trends Pharmacol. Sci., 17, 3–6. [DOI] [PubMed] [Google Scholar]
- 4.Porter A.G., Ng,P. and Janicke,R.U. (1997) Death substrates come alive. BioEssays, 19, 501–507. [DOI] [PubMed] [Google Scholar]
- 5.Lazebnik Y.A., Kaufmann,S.H., Desnoyers,S., Poirier,G.G. and Earnshaw,W.C. (1994) Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature, 371, 346–347. [DOI] [PubMed] [Google Scholar]
- 6.Boldin M.P., Goncharov,T.M., Goltsev,Y.V. and Wallach,D. (1996) Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell, 85, 803–815. [DOI] [PubMed] [Google Scholar]
- 7.Stein D. (1995) Pattern formation: the link between ovary and embryo. Curr. Biol., 5, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 8.Song Z., McCall,K. and Steller,H. (1997) DCP-1, a Drosophila cell death protease essential for development. Science, 275, 536–540. [DOI] [PubMed] [Google Scholar]
- 9.Smith C.L., Giordano,H., Schwartz,M. and DeLotto,R. (1995) Spatial regulation of Drosophila snake protease activity in the generation of dorsal-ventral polarity. Development, 121, 4127–4135. [DOI] [PubMed] [Google Scholar]
- 10.Murugasu-Oei B., Rodrigues,V., Yang,X. and Chia,W. (1995) Masquerade: a novel secreted serine protease-like molecule is required for somatic muscle attachment in the Drosophila embryo. Genes Dev., 9, 139–154. [DOI] [PubMed] [Google Scholar]
- 11.Weiss R.A., Teich,N.M., Varmus,H.E. and Coffin,J.M. (eds) (1982) Molecular Biology of Tumor Viruses: RNA Tumor Viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Kräusslich H.G. and Wimmer,E. (1988) Viral proteinases. Annu. Rev. Biochem., 57, 701–754. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean A., Higler,F. and Tatchell,K. (1990) Comparison of thermosensitive alleles of the CDC25 gene involved in the cAMP metabolism of Saccharomyces cerevisiae. Genetics, 124, 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcifull D.E. and Hiebert,E. (1982) Tobacco etch virus. In Gibbs,A., Harrison,B. and Murant,T. (eds), Description of Plant Viruses (258). Commonwealth Mycological Institute and Association of Applied Biologists (CMI/AAB), Kew, Surrey, UK, p. 258.
- 15.Dougherty W.G., Cary,S.M. and Parks,T.D. (1989) Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology, 171, 356–364. [DOI] [PubMed] [Google Scholar]
- 16.Kapust R.B., Tözsér,J., Copeland,T.D. and Waugh,D.S. (2002) The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun., 294, 949–955. [DOI] [PubMed] [Google Scholar]
- 17.Matayoshi E.D., Wang,G.T., Krafft,G.A. and Erickson,J. (1990) Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science, 247, 954–958. [DOI] [PubMed] [Google Scholar]
- 18.Billich A., Hammerschmid,F. and Winkler,G. (1990) Purification, assay and kinetic features of HIV-1 proteinase. Biol. Chem. Hoppe Seyler, 371, 265–272. [PubMed] [Google Scholar]
- 19.Richards A.D., Phylip,L.H., Farmerie,W.G., Scarborough,P.E., Alvarez,A., Dunn,B.M., Hirel,P.H., Konvalinka,J., Strop,P., Pavlickova,L., Kostka,V. and Kay,J. (1990) Sensitive, soluble chromogenic substrates for HIV-1 proteinase. J. Biol. Chem., 265, 7733–7736. [PubMed] [Google Scholar]
- 20.Sices H.J. and Kristie,T.M. (1998) A genetic screen for the isolation and characterization of site-specific proteases. Proc. Natl Acad. Sci. USA, 95, 2828–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kupiec J.J., Hazebrouck,S., Leste,L.T. and Sonigo,P. (1996) Conversion of thymidylate synthase into an HIV protease substrate. J. Biol. Chem., 271, 18465–18470. [DOI] [PubMed] [Google Scholar]
- 22.Dautin N., Karimova,G., Ullmann,A. and Ladant,D. (2000) Sensitive genetic screen for protease activity based on a cyclic AMP signaling cascade in Escherichia coli. J. Bacteriol., 182, 7060–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stebbins J., Deckman,I.C., Richardson,S.B. and Debouck,C. (1996) A heterologous substrate assay for the HIV-1 protease engineered in Escherichia coli. Anal. Biochem., 242, 90–94. [DOI] [PubMed] [Google Scholar]
- 24.Smith T.A. and Kohorn,B.D. (1991) Direct selection for sequences encoding proteases of known specificity. Proc. Natl Acad. Sci. USA, 88, 5159–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das Mahapatra B., DiDomenico,B., Dwyer,S., Ma,J., Sadowski,I. and Schwartz,J. (1993) A genetic system for studying the activity of a proteolytic enzyme. Proc. Natl Acad. Sci. USA, 89, 4159–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray M.G., Hung,W., Sadowski,I. and Das Mahapatra,B. (1993) Inactivation of a yeast transactivator by the fused HIV-1 proteinase: a simple assay for inhibitors of the viral enzyme activity. Gene, 134, 123–128. [DOI] [PubMed] [Google Scholar]
- 27.Steiner H., Pesold,B. and Haass,C. (1999) An in vivo assay for the identification of target proteases which cleave membrane-associated substrates. FEBS Lett., 463, 245–249. [DOI] [PubMed] [Google Scholar]
- 28.Hawkins C.J., Wang,S.L. and Hay,B.A. (1999) A cloning method to identify caspases and their regulators in yeast: identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc. Natl Acad. Sci. USA, 96, 2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S.Y., Park,K.W., Lee,Y.J., Back,S.H., Goo,J.H., Park,O.K., Jang,S.K. and Park,W.J. (2000) In vivo determination of substrate specificity of hepatitis C virus protease: genetic assay for site-specific proteolysis. Anal. Biochem., 284, 42–48. [DOI] [PubMed] [Google Scholar]
- 30.Kang H., Kim,S.Y. and Park,W.J. (2001) An improved strategy for a genetic assay for site-specific proteolysis. Mol. Cells, 11, 263–266. [PubMed] [Google Scholar]
- 31.Aronheim A., Zandi,E., Hennemann,H., Elledge,S.J. and Karin,M. (1997) Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol., 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dougherty W.G., Carrington,J.C., Cary,S.M. and Parks,T.D. (1988) Biochemical and mutational analysis of a plant virus polyprotein cleavage site. EMBO J., 7, 1281–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mumberg D., Müller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose M., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Parks T.D., Howard,E.D., Wolpert,T.J., Arp,D.J. and Dougherty,W.G. (1995) Expression and purification of a recombinant tobacco etch virus NIa proteinase: biochemical analyses of the full-length and a naturally occurring truncated proteinase form. Virology, 210, 194–201. [DOI] [PubMed] [Google Scholar]
- 36.Kapust R.B., Tözsér,J., Fox,J.D., Anderson,D.E., Cherry,S., Copeland,T.D. and Waugh,D.S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng., 14, 993–1000. [DOI] [PubMed] [Google Scholar]
- 37.Dougherty W.G. and Semler,B.L. (1993) Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev., 57, 781–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alnemri E.S., Livingston,D.J., Nicholson,D.W., Salvesen,G., Thornberry,N.A., Wong,W.W. and Yuan,J. (1996) Human ICE/CED-3 protease nomenclature. Cell, 87, 171. [DOI] [PubMed] [Google Scholar]
- 39.Schroeter E.H., Kisslinger,J.A. and Kopan,R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–385. [DOI] [PubMed] [Google Scholar]
- 40.Struhl G. and Adachi,A. (1998) Nuclear access and action of notch in vivo. Cell, 93, 649–660. [DOI] [PubMed] [Google Scholar]
- 41.Haass C. (1997) Presenilins: genes for life and death. Neuron, 18, 687–690. [DOI] [PubMed] [Google Scholar]
- 42.Vito P., Lacana,E. and D’Adamio,L. (1996) Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science, 271, 521–525. [DOI] [PubMed] [Google Scholar]
- 43.Citron M., Teplow,D. and Selkoe,D.J. (1995) Generation of amyloid beta protein from its precursor is sequence specific. Neuron, 14, 661–670. [DOI] [PubMed] [Google Scholar]
- 44.Tischer E. and Cordell,B. (1996) Beta-amyloid precursor protein. Location of transmembrane domain and specificity of gamma-secretase cleavage. J. Biol. Chem., 271, 21914–21919. [DOI] [PubMed] [Google Scholar]
- 45.Aronheim A. (1997) Improved efficiency Sos recruitment system: expression of the mammalian GAP reduces isolation of Ras GTPase false positives. Nucleic Acids Res., 25, 3373–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]