Abstract
The repair of DNA double-strand breaks (DSBs) occurs via nonhomologous end-joining (NHEJ) or homologous recombination (HR). These mechanistically distinct pathways are critical for maintenance of genomic integrity and organismal survival. Although inactivation of either pathway leads to embryonic lethality, here we show selective requirements for each DNA DSB repair pathway at different stages of mammalian nervous system development. DNA damage-induced apoptosis resulting from inactivation of HR (Xrcc2 deficiency) only occurred in proliferating neural precursor cells, whereas disruption of NHEJ (DNA ligase IV deficiency) mainly affected differentiating cells at later developmental stages. Therefore, these data suggest that NHEJ is dispensable for a substantial portion of early development because DSB repair during this period utilizes HR. Moreover, DNA damage-induced apoptosis required the ataxia telangiectasia mutated (Atm) kinase after disruption of NHEJ, but not HR, during neurogenesis. However, embryonic lethality arising from disruption of either repair pathway was rescued by loss of p53 and resulted in specific tumor types reflective of the particular DSB repair pathway inactivated. Thus, these data reveal distinct tissue- and cell-type requirements for each DNA DSB repair pathway during neural development and provide insights for understanding the contributions of DNA DSB responses to disease.
Keywords: ataxia telangiectasia mutated, DNA double strand-break, tumorigenesis, neurogenesis, genomic instability
Nonhomologous end-joining (NHEJ) and homologous recombination (HR) are mechanistically distinct DNA repair pathways that ensure the repair of DNA double-strand breaks (DSBs). These pathways differ in their DNA template requirements and the fidelity of the repair process. HR requires a group of RAD51-related proteins that includes XRCC2 to ensure high fidelity DNA repair by using an undamaged homologous DNA template to replace an adjacent damaged one (1, 2). In contrast, NHEJ facilitates direct modification and ligation of the two DNA ends present at the DSB. Efficient NHEJ requires Ku heterodimers (Ku70 and Ku80), DNA-PK catalytic subunit (PKcs), DNA ligase IV (Lig4), and XRCC4 (3, 4). Additionally, other components are also required, amongst which may be Artemis and the recently described XRCC4-binding factor XLF/Cernunnos (5, 6). Whereas NHEJ can participate in repair during all cell cycle phases, HR is functionally important from late synthesis (S) phase to mitosis (7), although there is likely to be cross-talk and cooperation between these two pathways (8–10). Components of each pathway are ubiquitously expressed, and it is assumed that both pathways are competent for and participate in DNA repair, although NHEJ is generally considered the predominant pathway for repairing DSBs in mammalian cells (4). Thus, together, NHEJ and HR provide an effective means of responding to DNA DSBs and act to prevent the accumulation of damaged DNA. However, delineating distinct roles and the functional interrelationship of each pathway during development and homeostasis are unclear. To precisely evaluate the relative contributions of DSB repair pathways during mammalian development, we compared the consequences of germ-line disruption of either HR or NHEJ. Unexpectedly, our data suggest a specific requirement for each pathway at distinct developmental stages during mammalian neurogenesis.
Results
We evaluated the relative contribution of DNA DSB repair pathways during mammalian development. To study each DSB repair pathway, we used mice containing germ-line disruption of either Xrcc2 or Lig4. To disrupt HR, we generated Xrcc2−/− mice by targeted deletion of exon 3 that encompasses ≈75% of the Xrcc2 coding sequence. Xrcc2 is one of a number of mammalian homologues of Saccharomyces cerevisae Rad 51 and plays a critical nonredundant role in HR (11, 12). The Xrcc2−/− mice were embryonically lethal, and mouse embryonic fibroblasts (MEFs) derived from these mice were hypersensitive to DNA damaging agents and underwent spontaneous chromosomal rearrangements in culture (Fig. 6, which is published as supporting information on the PNAS web site). The early embryonic lethality and DNA damage sensitivity in our Xrcc2 mutant is similar to another Xrcc2 mutant (13, 14), and collectively, these data show that Xrcc2 is critical for the cellular response to DNA DSBs and for maintenance of genomic stability.
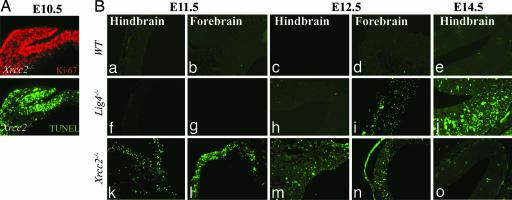
Despite the importance of HR, NHEJ is often considered the predominant pathway for repair of DNA DSBs (4). To further establish the roles for NHEJ and HR during development, we compared Xrcc2−/− mice (defective HR) with Lig4−/− mice (defective NHEJ). We found a striking difference in the spatiotemporal requirements for each DNA repair pathway during development. Inactivation of Xrcc2 affected early embryogenesis and resulted in abundant apoptosis of proliferating cells and resultant lethality around embryonic days 9–10 (E9–E10) (Fig. 1A). Some Xrcc2−/− embryos could be found at later times, and these Xrcc2 knockout embryos also showed widespread apoptosis in proliferating forebrain and hindbrain structures (Fig. 1B k–o). In stark contrast, disruption of NHEJ resulting from Lig4 loss had no obvious consequences until developmental times later than E12, because no apoptosis above control levels (Fig. 1B a–e) was observed in Lig4−/− embryos before E12 (Fig. 1B f–i), suggesting that NHEJ is dispensable during a substantial and critical period of early- to mid-embryonic development.
Fig. 1.
Inactivation of NHEJ or HR affects different embryonic stages during mouse development. Widespread apoptosis can be found in the Xrcc2−/− embryo by E10.5 and is illustrated showing neuroepithelium and adjacent cephalic mesenchyme. (A) Ki67 immunoreactivity (red) marks proliferating cells, and TUNEL staining (green) identifies cells undergoing apoptosis. (B) Although apoptosis is widespread in the proliferative cells of the Xrcc2−/− embryo at E11.5, it is not detectable in WT or Lig4−/− embryos until E12.5. In the differentiating hindbrain region at E14.5, Lig4, but not Xrcc2, loss is associated with apoptosis. (Magnification: ×200.)
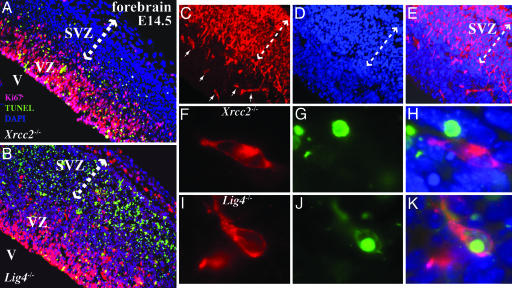
Because the above data indicate the importance of NHEJ from E12, a stage at which regional differentiation of the nervous system is occurring, we determined the respective requirements for NHEJ and HR at later stages of mammalian nervous system development, where there are clear demarcations between proliferating and differentiating tissues. Analysis of Xrcc2−/− and Lig4−/− E14.5 embryos highlighted the difference in the utilization of each DSB pathway during development and confirmed a particular requirement for Xrcc2 during proliferation. TUNEL-positive cells were located almost exclusively in the proliferative ventricular zone (VZ) of the Xrcc2−/− embryos (Fig. 2A). In contrast, apoptosis was largely localized to postmitotic cells of the subventricular zone (SVZ) in Lig4−/− embryos (Fig. 2B). However, apoptosis does occur in the VZ in Lig4−/− mice, albeit substantially less than the SVZ (Fig. 2B). There are two possibilities that could explain the occurrence of apoptotic cells in the VZ in E14 Lig4−/− mice. These cells could be differentiating but have not yet migrated, or these cells are proliferative even though proliferative cells at earlier stages of development are unaffected by Lig4 loss. To distinguish between these possibilities, we further analyzed the cellular identity of the apoptotic cells in the Lig4−/− VZ. We used an early neural differentiation marker, doublecortin (a microtubule-associated protein; see ref. 15), that could distinguish early cell-fate of some neural precursors present in the VZ (Fig. 2 C and E). To identify the nature of the cells undergoing apoptosis, we used double labeling with immunostaining for doublecortin and TUNEL for apoptosis. In the Lig4−/− embryos at E14, we found doublecortin-positive cells that were undergoing apoptosis, suggesting that many of the apoptotic cells in the VZ of the Lig4−/− embryos were differentiating and were not proliferative (Fig. 2 F–H). In contrast, we were unable to identify costaining cells in the VZ of the Xrcc2−/− embryos (Fig. 2 I–K). Thus, these data suggest that the apoptotic cells in the VZ of Lig4−/− embryos were undergoing differentiation.
Fig. 2.
Selective utilization of NHEJ and HR during development. At E14.5, apoptosis is found in the forebrain proliferative VZ of the Xrcc2−/− embryo, whereas in the Lig4−/− embryo, TUNEL-positive cells are predominantly located in the postmitotic SVZ (indicated by arrows). (A and B) Magnification: ×200; V, ventricle. (C and E) Doublecortin identifies scattered differentiating cells in the VZ. (D and E) DAPI staining shows the whole embryonic forebrain. (Magnification: ×200.) (F–K) TUNEL-positive cells did not colocalize with doublecortin-positive cells in Xrcc2−/− embryos (F–H) but did so in Lig4−/− embryos (I–K). (Magnification: ×1,000.)
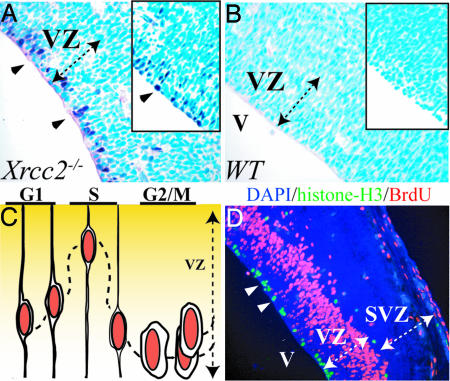
DNA damage in the developing nervous system resulting from dysfunctional NHEJ (Lig4 or Xrcc4 deletion) activates a pathway that is p53 dependent (16, 17). To determine the nature of the DNA damage response in the Xrcc2−/− embryos, we used immunostaining for phosphorylation of p53 ser18 (equivalent to human p53 ser15) to identify whether cells undergo DNA damage-induced p53 activation. We found p53 phosphorylation selectively occurred in the VZ, predominantly in a single row at the ventricular margin in Xrcc2−/− embryos (Fig. 3A), but not in WT littermate controls (Fig. 3B). This location is notable because the nucleus of proliferating cells within the VZ has a defined spatiotemporal relationship with the progression of the cell cycle (18), and, in this position, cells are in either second gap (G2) or mitotic (M) (Fig. 3C). To confirm p53 activation in S phase was the case for Xrcc2−/− embryos, we used immunostaining for phosphorylated histone H3 to identify cells in mitosis and BrdU incorporation to mark cells in S phase. Cells immunopositive for antiphospho-histone H3 were clearly demarcated as a single cell layer in both the WT and Xrcc2−/− ventricular margin and coincided with p53 phosphorylation resulting from Xrcc2 inactivation (Fig. 3D). BrdU also labels a band of S phase cells within the VZ superior to the p53-positive cells (Fig. 3D). The subsequent distribution of apoptotic cells away from the position of the p53 staining cells in the VZ (see Fig. 2A) most likely results from the failed commencement of another cell cycle. Additionally, some antiphospho-histone H3-positive mitotic cells were also present at the VZ/SVZ margin, indicating the occurrence of a laminar population that also coincide with scattered apoptotic cells in this region in Xrcc2−/− embryos (Fig. 2A). Therefore, despite a previous report of Xrcc2 loss leading to death in the SVZ (13, 14), our data now supports a requirement for HR solely in proliferating cells. Thus, activation of p53 in G2/M cells in the Xrcc2−/− embryo reflects DNA damage signaling arising after DNA replication, at a time when HR would normally be available to facilitate DNA repair. Thus, there is a separate requirement for NHEJ and HR at different developmental stages and within different tissues during mouse nervous system development.
Fig. 3.
The DNA damage response in Xrcc2−/− embryos occurs in G2/M cells of the VZ. Xrcc2 deficiency leads to p53 ser18 phosphorylation (A) and p53 stabilization (A Inset) in a layer of cells at the margin (arrowheads) of the ventricle (V) of an E14.5 embryo. (B) No p53 phosphorylation or stabilization (Inset) is found in WT embryo. (C) The cartoon represents the characteristic position of the progenitor cell nucleus within the VZ as the cell cycle progresses. (D) Cells that are immunopositive for phosphorylated histone H3 staining (green signal) indicate cells in mitosis, and BrdU-positive cells (red signal) mark those in S phase (arrowheads). The VZ and the postmitotic SVZ are marked.
To further define the relationship between DNA damage signaling arising from defective NHEJ or HR, we used a genetic approach to determine the involvement of ataxia telangiectasia mutated (Atm), a protein kinase critical for the response to DNA DSBs) (19–22) or p53 in Lig4−/− or Xrcc2−/− embryos. Although Atm has been implicated in HR in in vitro studies (23–25), no data are available in the context of an in vivo setting. Importantly, no relationship between Atm and HR defects has been examined during mammalian development. However, in the case of Lig4−/−, previous work has shown that both Atm and p53 are required for genotoxic stress-induced apoptosis in the Lig4−/− nervous system (17, 26–28). To assess whether a similar signaling scenario exists during neural development after HR inactivation, we generated Xrcc2−/− embryos with associated loss of either Atm or p53, by interbreeding either Xrcc2+/−p53+/− or Xrcc2+/−Atm+/− mice.
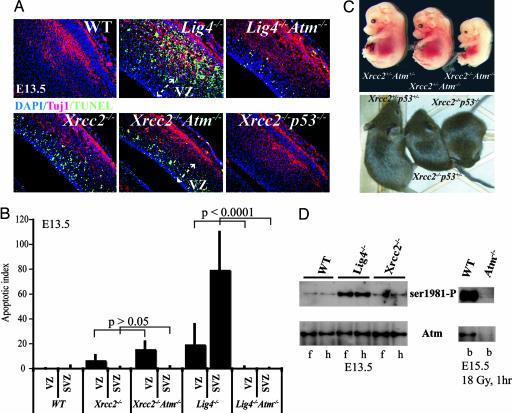
We found that Atm was required for apoptosis after inactivation of NHEJ but not for apoptosis resulting from disruption of HR (Fig. 4). Abundant apoptosis was found throughout the E14.5 Lig4−/− cortex that was dependent on Atm (Fig. 4A) and p53 (data not shown). In contrast to the difference between Lig4−/− and Lig4−/−Atm−/− embryos, Xrcc2−/−Atm−/− embryos showed similar apoptosis to Xrcc2−/− embryos (Fig. 4A). However, loss of p53 prevented apoptosis in Xrcc2−/− embryos (Fig. 4A). WT embryos showed little or no apoptosis, whereas apoptosis was markedly reduced in the Xrcc2−/−p53+/− embryos (data not shown). Apoptosis present in the DNA repair mutants after introduction onto an Atm−/− background was quantified (Fig. 4B). Apoptosis levels in Xrcc2−/− embryos indicates that apoptosis resulting from HR dysfunction is confined to the VZ, and that there is no significant difference between apoptosis in Xrcc2−/− and Xrcc2−/−Atm−/− (P > 0.05), whereas loss of Atm significantly reduced apoptosis after Lig4 loss (P < 0.0001). Thus, although Atm is required for neural apoptosis that occurs in the SVZ of Lig4−/− embryos, it is not required for that resulting from Xrcc2 loss.
Fig. 4.
Atm is not required for DNA damage signal transduction after Xrcc2 loss. (A) Analysis of apoptosis was done by using TUNEL staining in E14.5 using WT, Lig4−/−, Lig4−/−Atm−/−, Xrcc2−/−, Xrcc2−/−Atm−/−, and Xrcc2−/−p53−/− embryos. Tuj1 immunostaining identifies the differentiating neural cell populations. The merged composites are an overlay of Tuj1, TUNEL, and DAPI staining. (B) The amount of apoptosis was quantified in the VZ and the SVZ of the E14.5 nervous system from the indicated mutants. (C) Xrcc2−/− mice can survive on a p53−/− or p53+/− background, whereas Atm loss confers no survival advantage. (D) Phosphorylation (ser1981) of Atm was found in developing brain tissue from Lig4−/− but not Xrcc2−/−; f, forebrain; h, hindbrain isolated from E13.5. Specificity of the Atm antibodies is shown using WT and Atm−/− E15.5 embryonic brain (b) after ionizing radiation exposure.
Consistent with TUNEL analysis, lethality associated with Xrcc2−/− was p53 dependent because Xrcc2−/−p53+/− and Xrcc2−/−p53−/− mice were viable, although in both cases the mice were smaller and less robust than WT littermates (Fig. 4C). Again, in accord with TUNEL analysis, Xrcc2−/−Atm−/− (and Xrcc2−/−Atm+/−) embryos were recovered at the same frequency as Xrcc2−/− embryos, further indicating that loss of Atm provided no survival advantage for Xrcc2−/− deficient embryos. As reported in refs. 26 and 27, Lig4 inactivation is rescued by p53 or Atm deficiency, although Lig4−/−Atm−/− mice do not survive past postnatal day 1 (P1).
We also used a biochemical approach to examine the connection between Atm and DNA damage after HR inactivation by determining Atm autophosphorylation in Xrcc2−/− and Lig4−/− neural tissue. Atm activation requires autophosphorylation of ser1981, and this event is rapidly induced after DNA DSBs (29). Therefore, we isolated developing brain from Xrcc2−/− or Lig4−/− E13.5 embryos and determined Atm activation by using an antibody that recognized phosphorylated ser1981 of Atm (ser1987 in mouse Atm). At E13.5, when there are substantial cellular consequences resulting from Xrcc2 loss, Atm phosphorylation was similar to WT levels. In contrast, at this same developmental stage, loss of Lig4 leads to Atm phosphorylation in developing neural tissues (Fig. 4D). The specificity of the antisera used is shown by an absence of immunoreactivity in Atm−/− developing brain after ionizing radiation exposure compared with robust phosphorylation in irradiated WT tissue (Fig. 4D). Qualitatively similar results were obtained at other developmental times (data not shown). Therefore, although DNA damage arising from dysfunction in HR in Xrcc2−/− embryos presumably includes DNA DSBs, these DNA breaks do not activate Atm in the embryonic nervous system.
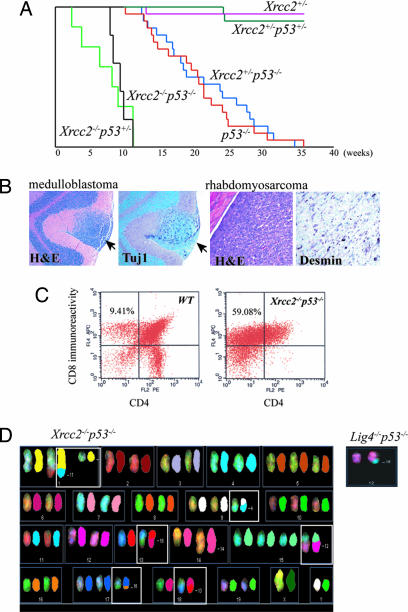
As p53 deficiency rescued the lethality of either Xrcc2−/− or Lig4−/− mice, we monitored these mice as they aged. Compared with the Lig4−/−p53−/− mice, the Xrcc2−/−p53−/− and Lig4−/−p53+/− were substantially less robust and often died perinatally of undetermined causes. However, of the Xrcc2−/−p53−/− mice that survived, all developed multiple tumor types. Tumor onset in Xrcc2−/−p53−/− mice was rapid and with different tumor types present by 10 weeks of age (Fig. 5A). In contrast, Lig4−/−p53−/− mice developed either pro-B cell lymphoma or medulloblastoma or both tumor types by 12 weeks of age (28, 30). Therefore the tumor spectrum in Xrcc2−/−p53−/− mice was broader than that arising from Lig4 loss and included lymphoma, skin tumor, sarcoma (Fig. 5B), and, like Lig4−/−p53−/− mice (28), medulloblastoma. Although Xrcc2−/−p53−/− mice developed lymphomas, these tumors were B220 negative and therefore distinct to the pro-B cell lymphoma (B220+) present in Lig4−/−p53−/− mice (data not shown). CD4/CD8 immunophenotyping of Xrcc2−/−p53−/− lymphomas showed CD8+/CD4− immunoreactivity indicative of a thymoma (Fig. 5C). Spectral karyotyping of primary thymomas from Xrcc2−/−p53−/− showed multiple chromosomal rearrangements (Fig. 5D, boxed). However, unlike the Lig4−/−p53−/− pro-B cell lymphomas that are characterized by an IgH-MYC-C t (12, 15) translocation (Fig. 5D Inset), no recurring chromosomal rearrangements were found between different primary Xrcc2−/−p53−/− tumors. Moreover, flow cytometry analysis of thymocytes and spleenocytes from Xrcc2−/−p53−/− mice by using CD4, CD8, IgM, and B220 cell surface markers showed similar mature B and T cell populations to those of age-matched WT controls, indicating that, in contrast to Lig4 inactivation, the gross development of B and T cells was unaffected by Xrcc2 deficiency (data not shown). Although Lig4+/−p53−/− are prone to sarcomas (31), Lig4−/−p53+/− mice do not develop tumors, at least until their death at ≈6 months (data not shown). In contrast to Lig4−/−, Xrcc2−/−p53+/− mice were extremely cancer prone because thymoma could be found before 12 weeks of age (Fig. 5A). Despite the cancer proneness of Xrcc2−/− mice, there was no evidence for Xrcc2 haploinsufficiency as Xrcc2+/−p53−/− mice developed tumors in a manner indistinguishable from p53−/− mice (Fig. 5A).
Fig. 5.
Inactivation of HR results in multiple tumor types. (A) A Kaplan–Meier curve shows survival of mice with various combinations of Xrcc2 and p53 mutant alleles and also shows Xrcc2+/− is not haploinsufficient for tumors as Xrcc2+/−p53−/− mice developed tumors of the same kind and at the same rate as p53−/− littermates. (B) A medulloblastoma staining immunopositive for the neuronal marker Tuj1 and a sarcoma staining immunopositive for desmin were isolated from a 10-week-old Xrcc2−/−p53−/− mouse. Arrows indicate the medulloblastoma within the cerebellar molecular layer. (C) Lymphomas from Xrcc2−/−p53−/− mice were CD8-immunopositive, indicative of a thymoma. Spectral karyotyping reveals a number of chromosomal rearrangements from a primary Xrcc2−/−p53−/− thymoma. (D) Image shows the spectral color image on the left and the pseudocolor chromosome on the right. (D Inset) Spectral karyotyping (SKY) analysis from a pro-B cell lymphoma from Lig4−/−p53−/− mice showing a typical translocation involving t (12, 15) that affects IgH and c-Myc.
Discussion
Here we have shown that, during mammalian nervous system development, the two major DNA DSB repair pathways, NHEJ and HR, function in a more regional and nonoverlapping manner than previously appreciated. This spatiotemporal distinction also reflects a differential requirement for DNA damage signal transducers and in the tumor spectrum resulting from disruption of either pathway. Our data suggest that HR is the operative DNA DSB repair pathway during early mammalian development and may explain why NHEJ does not appear to be functionally important until around midgestation. Because cellular differentiation will preclude the availability of suitable homologous templates for HR (i.e., sister chromatids), NHEJ becomes a main repair pathway. These data are consistent with lower eukaryotes such as yeast in which the driving cellular process is proliferation, and DNA repair occurs predominantly via HR (1, 4). The usage of HR during this early development would also ensure the greatest chance of error-free repair, thus maintaining embryonic genomic fidelity.
Our finding that inactivation of HR leads to profound defects exclusively in proliferating neural cells clearly contrasts another description of Xrcc2 loss, where it was reported that substantial apoptosis occurred in postmitotic neural cells (14). In reconciling that conclusion with our current data, it seems likely that, in the earlier report, the affected cells were the phospho-H3- or BrdU-positive proliferative cells located at the VZ/SVZ border and not postmitotic neurons (see Fig. 3D). A striking difference between the inactivation of each DSB repair pathway was the absence of obvious defects after Lig4 loss until almost midway through development. Whereas loss of NHEJ leads to a pronounced wave of apoptosis from E12.5 in differentiating and migrating neural cells, we nonetheless also found apoptosis in the VZ where proliferative cells reside. However, many of these TUNEL-positive cells in the VZ colocalized with doublecortin, suggesting the presence of differentiated cells within the VZ area. It is also possible that in Lig4−/− embryos the effects seen in the VZ may be an indirect effect of the substantial apoptosis in the SVZ, or alternatively, they may in some cases be proliferating cells and reflect the fact that NHEJ can be operative during all phases of the cell cycle (7, 9).
The abundant apoptosis in differentiating cells resulting from failure of NHEJ suggests that this pathway is required to address DNA damage present after cell cycle exit, perhaps arising from incomplete repair via HR, and so may therefore act as an additional and final genomic surveillance as cells differentiate and mature. Alternatively, NHEJ may be required for new DNA damage that occurs from cellular metabolism such as free radical damage arising from oxidative stress (32), rather than to address DNA lesions not repaired via HR.
The lack of an observable effect of Atm deficiency in Xrcc2−/− embryos was unexpected, given the reported link to HR (23–25) and the importance of Atm in the DSB response (19–22). This finding implies that Atm is not integral for HR in all settings and may be important for HR in only some tissues, or at later stages of development. In fact the Atm-related protein, ATR, has been implicated in HR and proliferation (33, 34), and may therefore be primarily involved in DSB signaling (e.g., to p53) (35) in the VZ during development.
We also observed a difference in the tumor spectrum resulting from inactivation of each DNA DSB repair pathway. The broader spectrum of tumors present in the Xrcc2−/− animals probably reflects the particular requirement for HR during DNA replication of precursor or stem cells. The one common tumor type, medulloblastoma, that results from loss of either Lig4 or Xrcc2, reflects postnatal development of the cerebellum via generation of granule neurons from an outer proliferative layer (36) where Xrcc2 is functionally important, and a postmitotic differentiating migratory layer where Lig4 is functionally important. Thus, both proliferative precursors and differentiating neurons are present in this organ, and the maintenance of genomic integrity requires the respective contributions of both HR and NHEJ to prevent medulloblastoma.
In summary, we have shown that the two major DNA DSB repair pathways function in a coordinated manner during development, although there is a defined spatiotemporal distinction in the functional importance of each repair pathway during neurogenesis. Therefore, together, these two pathways effectively maintain genomic integrity and suppress genotoxic stress via cooperative, but largely nonoverlapping, roles during mammalian development.
Materials and Methods
Mice.
Xrcc2 was inactivated in the mouse germ line by deletion of exon 3, which contains >75% of the ORF. Full details of the generation of Xrcc2 null mice are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Xrcc2−/−p53−/− and Xrcc2−/−p53+/− mice were obtained by intercrossing of Xrcc2+/−p53+/− animals. Xrcc2−/−Atm−/− mice were obtained by intercrossing of Xrcc2+/−Atm+/− animals. Lig4−/−p53−/− or Lig4−/−Atm−/− mice used in this study have been described (26, 28). The presence of a vaginal plug was designated as E0.5 and the day of birth as P0. All animals were housed in an American Association for the Accreditation of Laboratory Animal Care accredited facility and were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The institutional animal care and use committee at St. Jude Children’s Research Hospital approved all procedures for animal use.
Histological Analysis.
Embryos were fixed in 4% paraformaldehyde, cryoprotected in 25% sucrose/PBS, and cryosectioned (10-μm sagittal sections) by using an HM500 M cryostat (Microm International GmbH, Walldorf, Germany), whereas tissues were fixed in 10% buffered formalin, embedded in paraffin, and sectioned (7-μm sagittal sections) by using an HM325 microtome (Microm International GmbH). These sections were stained with hematoxylin and eosin according to standard procedures. Immunohistological analyses of tissue were performed by using the following antibodies: anti-p53 (CM5; 1:500; NovoCastra); anti-p53 phospho-ser15 (1:100; Cell Signaling Technology, Beverly, MA); antiactive caspase 3 (1:500; BD Biosciences); antiphospho-histone H3 (ser10) (6G3; 1:1,000; Cell Signaling Technology); anti-proliferating cell nuclear antigen (1:100; Calbiochem); anti-Ki67 (1:1,000; NovoCastra); anti-β-tubulin III (Tuj1; 1:1,000; Babco, Richmond, CA); antidesmin (1:500; DAKO); antidoublecortin (1:1,000; Abcam, Inc., Cambridge, MA); and anti-CD4, anti-CD8, and anti-B220 (all were used at 1:50; PharMingen). Antigen retrieval was used for all antibodies, except cell surface markers. Sections were incubated with antibodies overnight after quenching endogenous peroxidase by using 0.6% hydrogen peroxide, and immunoreactivity was visualized with the VIP peroxidase substrate kit (Vector Laboratories) according to the manufacturer’s directions after the tissues were treated with biotinylated secondary antibody and avidin DH-biotinylated horseradish peroxidase-H complex (Vectastain elite kit; Vector Laboratories). Sections were counterstained with 0.1% methyl green (Vector Laboratories), dehydrated, and mounted in DPX (BDH). For fluorescence signals, FITC or indocarbocyanine (Cy3) conjugated secondary antibody (Jackson ImmunoResearch) was used. DAPI (4′,6-diamidino-2-phenylindole) staining was used for counterstaining. Apoptosis was measured by using Apoptag (Chemicon). Quantitative analyses of TUNEL-positive cells in the embryonic forebrain were obtained by using image processing tool kit v5.0 (Reindeer Graphics, Asheville, NC) by using three to five embryos at E13.5 for each genotype. S phase cells were visualized after a 60-min pulse of BrdU (100 μg/g body weight delivered i.p. at E14.5). BrdU incorporation was visualized by using a rat monoclonal anti-BrdU (Abcam, Inc.) after sections were treated with SSC solution for 2 h at 65°C and then 30 min in 2 M HCl at 37°C and rinsed in 0.1 M boric acid (pH 8.5) followed by routine immunohistochemistry. Spectral karyotyping (SKY) was done by using a SKY probe from Applied Spectral Imaging (Vista, CA) with a single cell suspension of primary lymphoma enriched for metaphases after a 4.5-h colcemid incubation. Applied Spectral Imaging protocols were followed for hybridization and detection.
Atm Phosphorylation Analysis.
Atm was immunoprecipitated from microdissected E13.5 embryonic brain regions with Atm antibody D1611 (obtained from Dr. Michael Kastan, St. Jude Children’s Research Hospital, Memphis, TN) by using protein A/G agarose beads. Tissue lysates for immunoprecipitation were prepared in 50 mM Tris (pH 7.5) containing 150 mM NaCl, 50 mM NaF, 1% Tween 20, 0.2% Nonidet P-40, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (Roche), 1 mM DTT, and 1× protease inhibitor mixture (Roche). Beads were rinsed with RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS), split 1:2, run on two separate 3–8% NOVEX (San Diego) Tris-acetate gels, and electroblotted to poly(vinylidene difluoride) (PVDF) membranes. Phosphorylated Atm was visualized by using anti-ser1981 Atm antibody (Rockland Immunochemicals), whereas MAT3 antibody (obtained from Dr. Yosef Shiloh, Tel Aviv University, Tel Aviv) was used to detect total Atm. Two separate sets of embryos were used to determine Atm phosphorylation status.
Flow Cytometry Analysis.
Immunophenotyping of tumor cell suspensions was done as described (26) by using CD4, CD8, B220, and IgM antibodies obtained from PharMingen.
Supplementary Material
Acknowledgments
We thank Suzanne Baker, Gerard Grosveld, and Michael Kastan for discussions and comments on the manuscript. We also thank the Hartwell Center, the Cancer Center Cytogenetics Core, and the Transgenic Core facility at St. Jude Children’s Research Hospital for their support of this work. These studies were supported by National Institutes of Health Grants NS-37956 and CA-21765, Cancer Center Support Grant P30 CA21765, and the American Lebanese and Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
Abbreviations
- Atm
ataxia telangiectasia mutated
- DSB
double-strand break
- En
embryonic day n
- HR
homologous recombination
- NHEJ
nonhomologous end-joining
- Pn
postnatal day n
- SVZ
subventricular zone
- VZ
ventricular zone.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.West S. C. Nat. Rev Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 2.Thompson L. H., Schild D. Mutat. Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 3.Mills K. D., Ferguson D. O., Alt F. W. Immunol. Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 4.Lieber M. R., Ma Y., Pannicke U., Schwarz K. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 5.Ahnesorg P., Smith P., Jackson S. P. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Buck D., Malivert L., de Chasseval R., Barraud A., Fondaneche M. C., Sanal O., Plebani A., Stephan J. L., Hufnagel M., le Deist F., et al. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Rothkamm K., Kruger I., Thompson L. H., Lobrich M. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couedel C., Mills K. D., Barchi M., Shen L., Olshen A., Johnson R. D., Nussenzweig A., Essers J., Kanaar R., Li G. C., et al. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills K. D., Ferguson D. O., Essers J., Eckersdorff M., Kanaar R., Alt F. W. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson C., Jasin M. Mol. Cell. Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R. D., Liu N., Jasin M. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 12.Griffin C. S., Simpson P. J., Wilson C. R., Thacker J. Nat. Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 13.Deans B., Griffin C. S., O’Regan P., Jasin M., Thacker J. Cancer Res. 2003;63:8181–8187. [PubMed] [Google Scholar]
- 14.Deans B., Griffin C. S., Maconochie M., Thacker J. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gleeson J. G., Allen K. M., Fox J. W., Lamperti E. D., Berkovic S., Scheffer I., Cooper E. C., Dobyns W. B., Minnerath S. R., Ross M. E., Walsh C. A. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 16.Abner C. W., McKinnon P. J. DNA Repair (Amst.) 2004;3:1141–1147. doi: 10.1016/j.dnarep.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Frank K. M., Sharpless N. E., Gao Y., Sekiguchi J. M., Ferguson D. O., Zhu C., Manis J. P., Horner J., DePinho R. A., Alt F. W. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 18.Chan W. Y., Lorke D. E., Tiu S. C., Yew D. T. Anat. Rec. 2002;267:261–276. doi: 10.1002/ar.10100. [DOI] [PubMed] [Google Scholar]
- 19.Bakkenist C. J., Kastan M. B. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 20.Lavin M. F., Birrell G., Chen P., Kozlov S., Scott S., Gueven N. Mutat. Res. 2005;569:123–132. doi: 10.1016/j.mrfmmm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 21.McKinnon P. J. EMBO Rep. 2004;5:772–776. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiloh Y. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 23.Bolderson E., Scorah J., Helleday T., Smythe C., Meuth M. Hum. Mol. Genet. 2004;13:2937–2945. doi: 10.1093/hmg/ddh316. [DOI] [PubMed] [Google Scholar]
- 24.Golding S. E., Rosenberg E., Khalil A., McEwen A., Holmes M., Neill S., Povirk L. F., Valerie K. J. Biol. Chem. 2004;279:15402–15410. doi: 10.1074/jbc.M314191200. [DOI] [PubMed] [Google Scholar]
- 25.Morrison C., Sonoda E., Takao N., Shinohara A., Yamamoto K., Takeda S. EMBO J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y., Barnes D. E., Lindahl T., McKinnon P. J. Genes Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiguchi J., Ferguson D. O., Chen H. T., Yang E. M., Earle J., Frank K., Whitlow S., Gu Y., Xu Y., Nussenzweig A., Alt F. W. Proc. Natl. Acad. Sci. USA. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y., McKinnon P. J. Cancer Res. 2002;62:6395–6399. [PubMed] [Google Scholar]
- 29.Bakkenist C. J., Kastan M. B. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 30.Gao Y., Ferguson D. O., Xie W., Manis J. P., Sekiguchi J., Frank K. M., Chaudhuri J., Horner J., DePinho R. A., Alt F. W. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 31.Sharpless N. E., Ferguson D. O., O’Hagan R. C., Castrillon D. H., Lee C., Farazi P. A., Alson S., Fleming J., Morton C. C., Frank K., et al. Mol. Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
- 32.Karanjawala Z. E., Murphy N., Hinton D. R., Hsieh C. L., Lieber M. R. Curr. Biol. 2002;12:397–402. doi: 10.1016/s0960-9822(02)00684-x. [DOI] [PubMed] [Google Scholar]
- 33.Brown E. J., Baltimore D. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Powell S. N., Iliakis G., Wang Y. Cancer Res. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 35.Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., Abraham R. T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldowitz D., Hamre K. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.