Abstract
Herein, we report the identification of a unique HIV-1 integrase (IN) inhibitor-binding site using photoaffinity labeling and mass spectrometric analysis. We chemically incorporated a photo-activatable benzophenone moiety into a series of coumarin-containing IN inhibitors. A representative of this series was covalently photo-crosslinked with the IN core domain and subjected to HPLC purification. Fractions were subsequently analyzed by using MALDI-MS and electrospray ionization (ESI)-MS to identify photo-crosslinked products. In this fashion, a single binding site for an inhibitor located within the tryptic peptide 128AACWWAGIK136 was identified. Site-directed mutagenesis followed by in vitro inhibition assays resulted in the identification of two specific amino acid residues, C130 and W132, in which substitutions resulted in a marked resistance to the IN inhibitors. Docking studies suggested a specific disruption in functional oligomeric IN complex formation. The combined approach of photo-affinity labeling/MS analysis with site-directed mutagenesis/molecular modeling is a powerful approach for elucidating inhibitor-binding sites of proteins at the atomic level. This approach is especially important for the study of proteins that are not amenable to traditional x-ray crystallography and NMR techniques. This type of structural information can help illuminate processes of inhibitor resistance and thereby facilitate the design of more potent second-generation inhibitors.
Keywords: drug design, mass spectrometry, photoaffinity labeling
HIV-1 integrase (IN) mediates the insertion of viral DNA into the host genome. This process occurs through two separate events, both catalyzed by IN. In the 3′-processing reaction, IN cleaves a dinucleotide adjacent to a conserved CA on each terminus of the reverse-transcribed viral DNA. This cleavage results in two 3′ hydroxyl groups that are used for a subsequent nucleophilic attack. IN then inserts this DNA product into the host genome in the second reaction, termed strand transfer (1, 2). IN reactions can be carried out in vitro by using purified protein, a DNA substrate with ends mimicking the U3 or U5 viral DNA termini, and Mg2+ or Mn2+ as a cofactor (3).
Structural information detailing the association between IN and inhibitors under development is of enormous therapeutic importance. Knowledge of key amino acid residues involved in the binding of potential drugs, and therefore which residues are likely to mutate under therapeutic pressure, would inevitably help researchers stay one step ahead of drug-resistant viral strains. A co-crystal structure of one of our inhibitors was previously solved with the ASV-IN (4, 5). This complex was subsequently used as a surrogate structure to discover IN inhibitors through high-throughput docking studies (6). Thus far, only two examples of co-crystal structures of HIV-1 IN core in complex with inhibitors have been reported (7, 8). Despite our own repeated attempts, solving co-crystal structures of IN with our potent inhibitors has failed. This paucity of structural information necessitates a need to seek alternative technologies to obtain reliable information at the atomic level.
The use of affinity-labeled inhibitors to covalently modify the site of interaction and subsequent analysis of the protein have been very effective in providing useful information about inhibitor binding for a multitude of therapeutic target proteins. Application of this approach to IN has elucidated a small number of inhibitor-binding sites to atomic resolution. Two separate studies have used this approach to map different nucleotide inhibitor-binding sites of IN. The first used an AZT analog for photo-affinity labeling and proteolytic mapping to identify amino acid residues K156, K159, and K160 as the key residues involved in nucleotide binding (9). Mutational analysis confirmed their involvement by decreasing photo-crosslinking selectively. The second study identified residue K244 as a possible nucleotide analog-binding site (10). Recently, K173 was identified to selectively interact with acetylated chicoric acid (11).
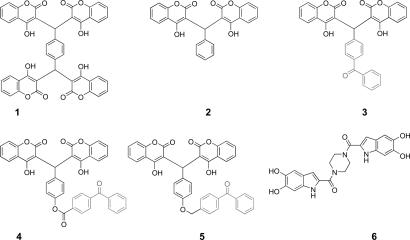
Coumarins represent a potent class of IN inhibitors that do not contain a catechol moiety. Previous catechol-containing IN inhibitors were highly cytotoxic, rendering therapeutic development impractical (12). Certain hydroxycoumarin analogues within this class also exhibit antiviral activity (13). Compound 1 inhibited IN at an IC50 = 1.5 μM and inhibited disintegration activity of the catalytic core domain (IN50–212) at a concentration of ≥15 μM. This result provides evidence that compound 1 binds the catalytic core domain, and binding this region alone is responsible for inhibition activity. Compound 1 also exhibits antiviral activity (13). Subsequent structure-activity relationship (SAR) studies were performed to define the minimal chemical features of compound 1 required for inhibitory activity (14). In the simplest case, the SAR deconstruction resulted in the generation of compound 2. Compound 2, with the removal of two coumarin units, exhibited a reduction in potency, with IC50 values of 43.4 ± 23.7 μM and 38.8 ± 25.9 μM for 3′-processing and strand transfer, respectively (14). Compound 2 represents the unmodified parent compound for the benzophenone-linked IN inhibitors 3–5. Chemical structures of compounds 1 and 2, along with the inhibitors used in this study, are depicted in Fig. 1.
Fig. 1.
Structures of earlier antiviral hydroxycoumarin compound 1 and the unmodified compound 2, with the benzophenone-linked coumarins (compounds 3–5) and positive control compound 6.
Here, we report the binding site characterization of a coumarin-containing IN inhibitor (compound 3; Fig. 1). Compound 3 contains a photo-activatable benzophenone moiety, which was then used to crosslink the inhibitor to its binding site. Subsequent trypsin digest and MS analysis identified the peptide fragment located in a region of IN never before implicated in inhibitor binding. Four residues within the region of protein were chosen for site-directed mutagenesis, by using both conservative and nonconservative substitutions. Residues C130 and W132, showed a marked resistance to the inhibitors when substituted. On the basis of molecular modeling studies, we conclude that the coumarins exhibit their inhibitory effects by causing a steric obstruction at the dimer interface of IN. This obstruction possibly leads to an arrest in the formation of an enzymatically functional multimeric complex.
Results
Compound 3 Binds to the IN Core Domain at a Stoichiometric Ratio of 1:1.
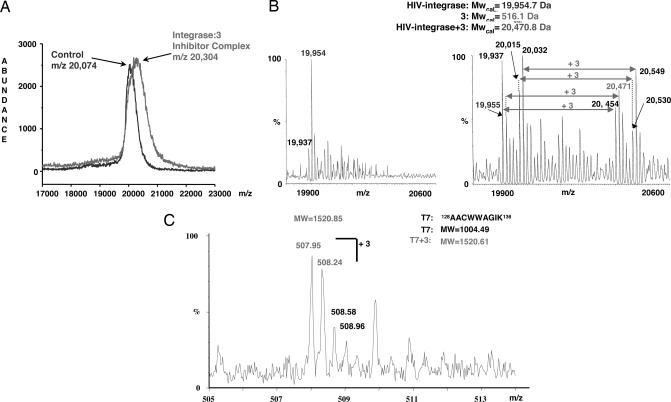
Chemical structures of IN inhibitors 3–5, conjugated to benzophenone moieties, are depicted in Fig. 1. Benzophenone is an ideal photo-affinity probe for selective protein labeling. It is activated at wavelengths between 350 and 360 nm, a range that avoids damage to the target protein. Once activated, it alkylates the protein backbone (for review, see ref. 15). IC50 values for the unmodified compound 2 (14) are in agreement with values obtained for benzophenone-linked analogues 3–5 using WT IN, indicating that the benzophenone moiety does not affect compound binding. Additionally, the benzophenone moiety alone did not show any inhibitory activity at concentrations up to 1 mM (data not shown). We used compound 3 as a representative inhibitor for crosslinking experiments. After crosslinking, the sample was analyzed by using both MALDI-MS and electrospray ionization (ESI)-MS. Fig. 2A shows a representative MALDI-MS spectrum of the IN core:compound 3 complex. A comparison of IN core domain alone with the IN core:compound 3 complex is shown in Fig. 2B. The ESI-MS technique revealed that the molecular weight shift corresponds to a single molecule of compound 3. This result indicates that the stoichiometric ratio of IN core domain to inhibitor is 1:1 and provides direct evidence for a single binding site.
Fig. 2.
Compound 3 binds to IN at a 1:1 ratio. (A) MALDI-TOF of an HPLC fraction containing the IN core complexed to compound 3 along with the IN core domain alone (control). (B) Two ESI-MS spectra showing the IN core domain alone, and complexed with compound 3. (C) ESI-MS spectrum showing the tryptic peptide T7 bound to one molecule of compound 3.
Compound 3 Interacts with a Region of IN Never Before Implicated in Inhibitor Binding.
Tryptic digestion of the complex was performed to narrow down which peptide region of the IN core domain interacts with the inhibitor. Resulting peptides exhibiting a difference in UV absorbance compared with control were purified, and the fragment (T7) 128AACWWAGIK136 was identified as the site of photo-crosslinking. The predicted molecular weight of T7 is 1,004.49, which, crosslinked to compound 3, gives a molecular weight of 1,520.61. Fig. 2C depicts the ESI-MS spectrum of the IN tryptic peptide T7 crosslinked to compound 3. This region provides a unique drug-binding site that can be exploited in the design of new IN inhibitors.
Catalytic Activities of IN Mutants.
Four different residues were chosen for site-directed mutagenesis: C130, W131, W132, and K136. We also selected a residue (Q148) outside the implicated peptide to be included in the inhibition assays. Multiple substitutions at position W132 were constructed to obtain a mutant with both 3′-processing and strand transfer catalytic activities. This effort resulted in the production of three nonconservative and one conservative substitution. Mutants were synthesized in the context of an IN double mutant (F185K/C280S) that exhibits increased solubility and that has been previously shown to be as catalytically active as the WT enzyme in vitro (16). All compounds showed similar inhibition profiles in the WT IN (data not shown) as well as the soluble double mutant. Fig. 6, which is published as supporting information on the PNAS web site, shows a representative gel depicting the enzymatic activities of the mutant IN proteins, including the soluble double mutant, versus the WT IN. Table 1 lists the relative activities of each mutant. With the exception of Q148A, all of the 3′-processing activities of the mutant IN proteins were fairly robust. On the other hand, strand transfer activity of the mutants varied significantly. The nonconservative substitutions made at position W132 abolished strand transfer activity of these IN mutants. Experiments using other metal cofactors (Ca2+ and Zn2+) in the reaction buffer failed to restore activity (data not shown). Additionally, both C130S and Q148A exhibited diminished strand transfer capabilities (10–40%).
Table 1.
Relative activities of integrase proteins
| IN protein | 3′-Processing* | Strand transfer* |
|---|---|---|
| SolMut† | +++ | +++ |
| C130A | +++ | +++ |
| C130S | +++ | + |
| W131G | ++ | ++ |
| W132A | +++ | − |
| W132G | +++ | − |
| W132R | +++ | − |
| W132Y | +++ | ++ |
| K136R | +++ | ++ |
| Q148A | + | + |
*Activity designation relative to WT: −, 0–10%; +, 10–40%; ++, 40–80%; +++, 80–100%.
†F185K/C280S soluble double mutant.
Substitutions at IN Residues C130 and W132 Confer Resistance to Inhibitors 3–5.
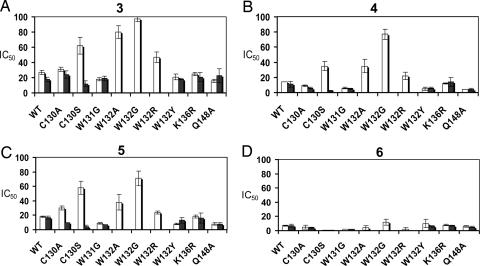
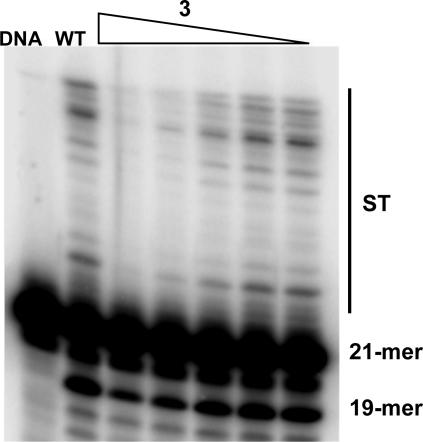
Inhibition of IN-mediated catalytic activities by all compounds is shown in Fig. 3, with IC50 values given in Table 2. We used the IN inhibitor 6 (17) as the positive control (Fig. 1). Compound 6 is structurally dissimilar to the coumarin set and therefore is predicted to have a different binding site. A comparison of the mutant IN inhibition profiles with WT indicates two residues, C130 and W132, that are critical for the inhibitory effects of the coumarins. In the case of the C130 substitutions, resistance was observed in the serine substitution mutant, but not the alanine mutant. C130A displayed a similar inhibition profile to that of WT. C130S showed an increased resistance specific for 3′-processing only. C130S showed an ≈2-fold increase in 3′-processing IC50 values for compounds 3 and 4 (P < 0.001 for each, respectively) and an ≈3-fold increase in the IC50 value for compound 5 (P < 0.001). Nonconservative substitutions at position W132 generated a pronounced decrease in inhibition of 3′-processing activity by compounds 3–5. The most radical substitution, from tryptophan to glycine, created the most resistant IN protein to these inhibitors. For 3′-processing activity, the W132G mutant exhibited IC50 values that increased nearly 4-fold for compound 3 (P < 0.001), 5-fold for compound 4 (P < 0.001), and >3-fold for compound 5 (P < 0.001). The resistance was decreased to a certain degree for W132A, with IC50 values increasing by 3-fold for compound 3 (P < 0.001), close to 2-fold for compound 4 (P < 0.001), and an IC50 value one and a half times that of WT for compound 5 (P = 0.016). Resistance decreased even more for the W132R mutant, with an IC50 value for compound 3 (P = 0.004) being one and a half times that of WT, and with values for compound 4 (P = 0.439) and compound 5 (P = 0.949) being quite similar to WT. None of the above W132 mutants retained their ability to catalyze strand transfer, and therefore the effects of each substitution on drug potency concerning this activity could not be assessed. The last W132 mutant, which contained a conservative tryptophan to tyrosine substitution, retained strand transfer activity. All compounds exhibited inhibition profiles comparable with that of WT when tested with W132Y. After statistical analysis, two of the nonconservative W132 substitutions (G and A) and C130S showed significant mean IC50 value deviations for 3′-processing activity when tested with compounds 3–5. W132R showed significant deviations when tested with compound 3 only. None of the mutants showed any significant mean IC50 value deviations for 3′-processing inhibition by the positive control, compound 6. Concerning strand transfer inhibition, C130A (compound 6), C130S (compounds 4–6), and W131G (compounds 5 and 6) each showed significant mean IC50 value deviations from WT. All significant mean strand transfer IC50 values were lower than WT. This finding indicates a possible increase in inhibitor susceptibility of these mutants, or it may be linked to a reduction in basal level strand transfer activity, concerning C130S and W131G. Resistance to the coumarins compounds 3–5 in C130S and the nonconservative W132 mutants was observed only for 3′-processing activity, not strand transfer. The absence of strand transfer resistance may be linked to the successive nature (strand transfer activity depends on prior 3′-processing activity) of the in vitro IN assay. To directly address the inhibitory effect of the coumarins on the strand transfer activity, we tested compound 3 against WT, the soluble double mutant, C130S, and W132G (representative of nonconservative W132 mutants) using a 3′-processed oligonucleotide substrate. Compound 3 strand transfer IC50 values for each IN protein were in general agreement with results obtained with the unprocessed substrate (Fig. 7, which is published as supporting information on the PNAS web site). Calculated IC50 values of 17, 18, and 3.2 μM were obtained for WT, the soluble double mutant, and C130S, respectively. This result demonstrates that the biased resistance observed for these mutants is not an artifact of the IN in vitro assay. The mutant W132G still did not retain strand transfer activity.
Fig. 3.
Resistance to coumarins observed in nonconservative W132 mutants and C130S. Comparison of the IC50 values for IN inhibitory compounds 3 (A), 4 (B), 5 (C), and 6 (D) in different mutants. 3′-processing values are indicated by open bars, whereas strand transfer values are indicated by filled bars.
Table 2.
Integrase proteins with IC50 values (μM) for inhibitory compounds and corresponding P values* after comparison with WT
| Compound | WT | C130A | C130S | W131G | W132A | W132G | W132R | W132Y | K136R | Q148A |
|---|---|---|---|---|---|---|---|---|---|---|
| 3′ Processing | ||||||||||
| 3 | 27 ± 3 | 31 ± 3 | 62 ± 11 | 18 ± 2 | 80 ± 8 | 97 ± 3 | 47 ± 7 | 21 ± 4 | 25 ± 2 | 16 ± 2 |
| (0.993)† | (<0.001) | (0.446) | (<0.001) | (<0.001) | (0.004) | (0.882) | (1.0) | (0.286) | ||
| 4 | 14 ± 0 | 9 ± 1 | 35 ± 6 | 6 ± 1 | 35 ± 9 | 77 ± 7 | 22 ± 5 | 5 ± 2 | 12 ± 1 | 4 ± <1 |
| (0.872) | (<0.001) | (0.46) | (<0.001) | (<0.001) | (0.439) | (0.352) | (1.0) | (0.187) | ||
| 5 | 18 ± 1 | 30 ± 3 | 58 ± 9 | 9 ± 1 | 38 ± 11 | 71 ± 10 | 24 ± 2 | 8 ± 1 | 18 ± 2 | 8 ± 2 |
| (0.322) | (<0.001) | (0.614) | (0.016) | (<0.001) | (0.949) | (0.558) | (1.0) | (0.502) | ||
| 6 | 7 ± 1 | 5 ± 3 | 1 ± <1 | 2 ± <1 | 4 ± 3 | 12 ± 4 | 2 ± 2 | 10 ± 6 | 8 ± 1 | 6 ± 1 |
| (0.974) | (0.219) | (0.278) | (0.847) | (0.479) | (0.445) | (0.983) | (1.0) | (0.999) | ||
| Strand transfer | ||||||||||
| 3 | 18 ± 3 | 24 ± 5 | 12 ± 4 | 19 ± 3 | NA‡ | NA | NA | 18 ± 1 | 21 ± 6 | 23 ± 9 |
| (0.752) | (0.761) | (1.0) | (1.0) | (0.966) | (0.87) | |||||
| 4 | 11 ± 4 | 6 ± 1 | 2 ± 1 | 5 ± 1 | NA | NA | NA | 6 ± 1 | 14 ± 6 | 4 ± 2 |
| (0.456) | (0.034) | (0.204) | (0.448) | (0.935) | (0.180) | |||||
| 5 | 17 ± 2 | 10 ± <1 | 4 ± 2 | 7 ± <1 | NA | NA | NA | 13 ± 4 | 16 ± 7 | 8 ± 2 |
| (0.185) | (0.005) | (0.043) | (0.815) | (1.0) | (0.061) | |||||
| 6 | 7 ± 2 | 4 ± 1 | <1 ± <1 | 2 ± <1 | NA | NA | NA | 6 ± 1 | 7 ± 1 | 5 ± 1 |
| (0.017) | (<0.001) | (<0.001) | (0.539) | (0.992) | (0.139) | |||||
*P values generated by using a Tukey multiple comparison test, after a statistical test of variance (P = 0.05). Mean IC50 values of three independent IN mutant experiments were compared with that of the WT IC50 mean value (in triplicate) for each inhibitory compound.
†P values are in parentheses.
‡NA, not applicable. Strand transfer activity for nonconservative substitutions at position W132 was abolished. Therefore, IC50 values could not be obtained and statistical analyses could not be conducted.
Docking Studies Show That Inhibitor 3 Binds Close to the Dimeric Interface of the IN Core Domain.
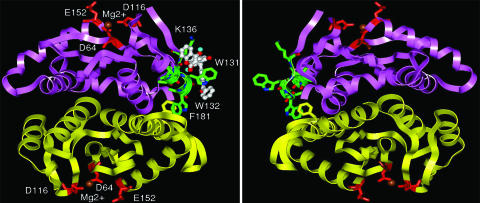
To distinguish key binding interactions between the IN core domain, particularly with respect to the peptide 128AACWWAGIK136 region and compound 3, we docked the compound onto the IN core domain dimer (PDB 1BL3) (18) carrying the WT sequence and two W132 mutations. Compound 3 binds to the peptide 128AACWWAGIK136 region on the surface of the IN core domain in WT as well as the mutants W132G and W132Y. The benzophenone moiety of compound 3 occupies an area close to the backbone of amino acid residues A133 and G134. This finding supports the formation of a covalent linkage between the backbone of the peptide 128AACWWAGIK136 region and the benzophenone moiety of 3 upon photo activation. The bound conformation of 3 on the surface of the dimeric IN core domain is shown in Fig. 4. A hypothetical tetrameric arrangement for catalytically functional IN core domain formed from two dimeric core domains is also depicted in Fig. 4. Compound 3 binds to the peptide 128AACWWAGIK136 and most likely disrupts the formation of a catalytically functional tetrameric IN. From the analysis of IN dimeric core domain crystal structure, it is observed that a part of the peptide 128AACWWAGIK136 contributes to the interfacial surface of the IN dimer. In particular, the amino acid residue W132 is buried in the interfacial surface and establishes a stacking interaction with residue F181 from the other IN monomer (the shortest interplanar atom–atom distance is 3.65 Å). The predicted bound conformations of compound 3 on the core domain surface of WT and IN mutants W132G and W132Y are shown in Fig. 8 a–c, which is published as supporting information on the PNAS web site. Compound 3 adopted a similar bound conformation on these three core domain structures, suggesting that the coumarins still bind the resistant IN mutants but are unable to inhibit 3′-processing activity. To gain further insight, we examined the activity of compound 3 against preassembled WT IN–DNA complexes in the presence of Ca2+. Ca2+ facilitates IN–DNA complex formation without proceeding to enzymatic cleavage of the DNA (19, 20). The inhibitory potency of compound 3 was drastically reduced against preassembled IN–DNA complexes, ≈4-fold increase to an IC50 = 100 μM for 3′-processing only. The strand transfer IC50 value of compound 3 decreased ≈2-fold to 6.3 μM (Fig. 5). This result supports our hypothesis that the coumarin compounds exhibit their inhibitory mechanism of action through disruption of IN multimer formation.
Fig. 4.
Coumarins bind to a region that may disrupt formation of active IN multimers. Shown are symmetric dimers with a tetrameric arrangement for a catalytically functional IN core domain. Compound 3 is shown in white ball and stick, the side chains of the 128AACWWAGIK136 residues are shown as green stick, the magenta and yellow ribbon shows two IN monomers, whereas red sticks and brown spheres represent the conserved DDE motif and Mg2+ ion, respectively.
Fig. 5.
Coumarins are less potent against Ca2+-induced preassembled IN-DNA complexes. Shown are the inhibitory effects of compound 3 on preassembled WT IN complex. Concentration of compound 3 decreases successively by a fraction of two-thirds from 100 μM to 1.2 μM.
Antiretroviral Activity.
Compounds 3–5 were found to inhibit HIV-1(IIIB) replication in the presence of 0.2 μg/ml S-1360 at EC50 values of 5.96, 6.47, and 3.46 μg/ml for compounds 3, 4, and 5, respectively, in MT-4 cells. Cytotoxicity was observed at 5- to 7-fold higher concentrations [50% cytotoxic concentration (CC50) values: 28.7, 30.9, and 24.3 μg/ml, respectively, in mock-infected MT-4 cells].
Discussion
We identified the peptide region 128AACWWAGIK136 of IN as the binding site for a set of coumarins. This region is located on an α-helical segment adjacent to the IN dimer interface. An analogous α-helix contributes to the dimerization of the RuvC resolvase protein, a holliday junction-specific endonuclease involved in homologous DNA recombination (21). Coumarins interact specifically with the peptide backbone of this region, potentially disrupting the formation of active multimeric complexes of IN. The C130S and nonconservative W132 substitutions create structural perturbations that disrupt multimer formation and therefore the binding interactions between these inhibitors and the peptide region 128AACWWAGIK136 in the context of a multimeric IN structure. Structural perturbations caused by C130S have been documented and proposed to disrupt other processes, but not inhibitor binding (22–24). Substitutions made at amino acid residue W131 (W131D and W131E) have been shown to increase the solubility of IN fragments, enhancing their ability to crystallize, without effecting protein structure (25, 26). The mutant W131G exhibited no resistance to the coumarins. After analysis of both of the two domain x-ray crystal structures of IN (26, 27), a critical π–π stacking interaction between W132 and F181 was observed between two IN monomers. To our knowledge, no previous reports implicate F181 and W132 as important for any IN processes. Making conservative and nonconservative amino acid substitutions at position W132, thereby either retaining or abolishing the π–π stacking interaction between the two residues, produced very interesting effects on IN phenotype. The conservative substitution, tryptophan to tyrosine, where an aromatic ring system is retained, was the only W132 mutant that preserved strand transfer activity. It seems that having an aromatic ring for interaction with F181 on a complementary IN monomer is crucial for the strand transfer reaction. Additionally, only the W132Y mutant displayed an inhibition profile similar to that of WT, suggesting that the π–π stacking interaction at the dimer interface promotes IN multimer formation and optimal coumarin inhibition. The nonconservative substitutions generated at position W132 eliminate the π–π stacking interaction with F181 between monomers, possibly creating structural defects that may arrest the formation of fully functional IN multimers. This may be the reason why the strand transfer activity is abolished, even though the ability to conduct 3′-processing is preserved, with, however, more resilience to coumarin inhibition. Indeed current models and experimental evidence on retroviral IN composition indicate that a dimeric species may be sufficient for 3′-processing, but a tetrameric arrangement, stabilized through target DNA binding at the dimer–dimer interface, is necessary for the integration process and more relevant to the in vivo nucleo–protein complex (28, 29). Results on avian sarcoma virus IN also suggest that tetramer formation is an obligatory step that occurs during strand transfer catalysis (28). Our results using compound 3 on “preassembled” IN–DNA complexes also support the above sequential model for HIV-1 IN multimer formation. Ca2+-induced IN-DNA “preassembly” may constitute the formation of only an IN dimer bound to the DNA substrate. The addition of compound 3 afterward would be less effective on 3′-processing, but still effective at inhibiting multimer formation, and therefore strand transfer, once catalysis is initiated (Mn2+ or Mg2+ addition) and multimerization is stimulated. Our results also indicate the π–π stacking interaction at the dimer interface is an additional requirement for multimerization (tetramerization) and the integration process. We hypothesize that an aromatic ring system at position 132 is required for formation of an active IN multimer. The requirement of the π–π stacking interaction at the dimer interface for strand transfer catalytic activity, but not 3′-processing, is an interesting observation. K136 is an exposed surface residue. An arginine substitution at this position created no effect on inhibitor binding. Q148 is located outside the coumarin-binding peptide region (128–136). Q148A mutant IN proteins did not show any severe resistance to these molecules.
In conclusion, we successfully identified an IN inhibitor-binding site through photo-affinity labeling/MS analysis and validated the site through site-directed mutagenesis and in vitro assays. Concerning IN mutants with increased resistance, IC50 values of the coumarins increased from 2-fold up to 5-fold. The observed coumarin resistance at the protein level is consistent with other studies on IN inhibitor resistance. A prior study examined HIV-1 viral resistance to L-708,906 and S-1360, which are derivatives from the diketo acid class of IN inhibitors. Although a 10-fold viral resistance to these IN inhibitors was the observed phenotype, the corresponding triple mutant (T66I/L74M/S230R) protein exhibited only 2- to 3-fold resistance in vitro to the same compounds (30). The coumarins interacted directly with the peptide backbone in the region 128AACWWAGIK136. This peptide is located at the dimer interface of IN. We also located a π–π stacking interaction at the dimer interface between W132 of this region and F181 on a separate monomer that is critical for IN multimer structure and function. Upon binding to the peptide backbone, the inhibitors are hypothesized to disrupt the formation of active IN multimers. Substitutions at amino acid positions that structurally alter IN multimer formation also exhibited resistance to the inhibitors in in vitro inhibition assays using full-length IN.
Several molecules bearing the coumarin structural unit have been reported to show activity against a variety of therapeutic targets. Compounds 3–5 are among a class of coumarins that show a range of inhibitory activities against IN (14). The extent of IN inhibition, however, is directly correlated to definite structural variations in each analogue. This observed structure-activity relationship (SAR) supports that compounds 3–5 selectively disrupt IN catalytic activity and are specific for this enzyme (14). Early hydroxycoumarins like the tetrameric compound 1 have shown antiviral activity at 11.5 μM (13). The compounds used in this study inhibit HIV-1(IIIB) replication in the presence of 0.2 μg/ml S-1360 at EC50 values of 5.96, 6.47, and 3.46 μg/ml for compounds 3, 4, and 5, respectively, in MT-4 cells. Compound 3 does display excellent activity in the presence of Mg2+ (Fig. 9, which is published as supporting information on the PNAS web site), although the presence of polyethylene glycol (PEG) in the reaction buffer does seem to affect activity. Compound 3 IC50 values in the absence of PEG are 9 ± 1 μM and 4 ± 1 μM for 3′-processing and strand transfer, respectively. These values are significantly lower than those obtained using Mn2+ as a cofactor. With the inclusion of PEG, compound 3 IC50 values for 3′-processing and stand transfer were 36 μM and 6.8 μM, respectively. This compound is less effective against the 3′-processing activity of IN in the presence of Mg2+ and PEG.
Members of the diketo acid class of compounds were the first important developmental candidates suspected of binding the IN active site. The V(D)J recombinase protein RAG1/2, which is important for T and B cell development, has mechanistic similarities to HIV-1 IN. Certain IN diketo inhibitors suspected to bind the active site have the potential to interfere with RAG1/2 cellular function, posing serious toxicity issues. Indeed, there has been a report showing a member of the diketo acid class of IN inhibitors that was able to inhibit RAG1/2 in vitro, although at a much higher concentration than what is required for IN inhibition (31). With this potential diketo acid nonspecificity in mind, the field has slightly shifted focus on the more IN-exclusive naphthyridine carboxamide class of inhibitors. Potential IN inhibitors designed to target the binding site discovered here should exhibit effective multimer disruption, affording a very different mechanism of action than active site binding. Additionally, a cocrystal structure of the binding domains of IN and lens-epithelium-derived growth factor (LEDGF) (PDB 2B4J) (32), an important IN cellular cofactor thought to facilitate chromatin tethering, has recently been reported in the literature. The hotspots of protein–protein interaction contained several critical contacts directly corresponding to our identified inhibitor-binding site, including IN residues A128, A129, W131, and W132. This independent result indicates that molecules specifically designed to target the allosteric binding site presented here may also disrupt LEDGF interaction in vivo. These converging results highlight a legitimate allosteric binding site that will enable high-resolution, structure-based drug design strategies to develop small molecules with further selectivity and potency. The designed molecules have the capability to exhibit two simultaneous inhibitory mechanisms, disruption of HIV-1 IN-LEDGF cofactor interaction and IN multimer formation.
Materials and Methods
Chemistry.
The benzophenone moiety was chemically attached as described (33).
Photoaffinity Labeling.
The catalytic core domain of IN was used for photo-crosslinking based on previous results demonstrating that hydroxycoumarins inhibit disintegration activity of the IN catalytic core domain (13). To form the protein–ligand complex, IN was incubated with a 25 molar excess of compound 3 at a final protein concentration of 1.3 × 10−5 M in a buffer containing 2.5 mM MnCl2, 50 mM 2-mercaptoethanol, 0.2% CHAPS, and 33% DMSO for 30 min at room temperature. The protein–ligand complex was radiated for 30 min with UV (360 nm) at a 3-cm distance at 4°C under the conditions described (34, 35).
For further details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Zahrah Zawahir, Tino Sanchez, Kennth B. Tomer, An Nijs, Barbara Van Remoortel, and Elisabeth O. Hochleitner for superb technical assistance. This work was financially supported by funds from a University of Southern California (USC) Zumberge Award, the Universitywide AIDS Research Program, a GlaxoSmithKline Drug Discovery and Development Award, the Gustavus and Louise Pfeiffer Research Foundation (N.N.), and, in part, by the USC Training Program in Genetic, Molecular, and Cellular Biology (L.Q.A.-M.).
Abbreviations
- IN
integrase
- ESI
electrospray ionization.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brown P. O. Integration. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 2.Asante-Appiah E., Skalka A. M. Adv. Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 3.Bushman F. D., Craigie R. Proc. Natl. Acad. Sci. USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubkowski J., Yang F., Alexandratos J., Wlodawer A., Zhao H., Burke T. R., Jr., Neamati N., Pommier Y., Merkel G., Skalka A. M. Proc. Natl. Acad. Sci. USA. 1998;95:4831–4836. doi: 10.1073/pnas.95.9.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicklaus M. C., Neamati N., Hong H., Mazumder A., Sunder S., Chen J., Milne G. W., Pommier Y. J. Med. Chem. 1997;40:920–929. doi: 10.1021/jm960596u. [DOI] [PubMed] [Google Scholar]
- 6.Chen I. J., Neamati N., Nicklaus M. C., Orr A., Anderson L., Barchi J. J., Jr., Kelley J. A., Pommier Y., MacKerell A. D., Jr. Bioorg. Med. Chem. 2000;8:2385–2398. doi: 10.1016/s0968-0896(00)00180-2. [DOI] [PubMed] [Google Scholar]
- 7.Goldgur Y., Craigie R., Cohen G. H., Fujiwara T., Yoshinaga T., Fujishita T., Sugimoto H., Endo T., Murai H., Davies D. R. Proc. Natl. Acad. Sci. USA. 1999;96:13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molteni V., Greenwald J., Rhodes D., Hwang Y., Kwiatkowski W., Bushman F. D., Siegel J. S., Choe S. Acta Crystallogr. D. Biol. Crystallogr. 2001;57:536–544. doi: 10.1107/s0907444901001652. [DOI] [PubMed] [Google Scholar]
- 9.Drake R. R., Neamati N., Hong H., Pilon A. A., Sunthankar P., Hume S. D., Milne G. W., Pommier Y. Proc. Natl. Acad. Sci. USA. 1998;95:4170–4175. doi: 10.1073/pnas.95.8.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams K. L., Zhang Y., Shkriabai N., Karki R. G., Nicklaus M. C., Kotrikadze N., Hess S., Le Grice S. F., Craigie R., Pathak V. K., Kvaratskhelia M. J. Biol. Chem. 2005;280:7949–7955. doi: 10.1074/jbc.M413579200. [DOI] [PubMed] [Google Scholar]
- 11.Shkriabai N., Patil S. S., Hess S., Budihas S. R., Craigie R., Burke T. R., Jr., Le Grice S. F., Kvaratskhelia M. Proc. Natl. Acad. Sci. USA. 2004;101:6894–6899. doi: 10.1073/pnas.0400873101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanwell C., Ye B., Yuspa S. H., Burke T. R., Jr. Biochem. Pharmacol. 1996;52:475–480. doi: 10.1016/0006-2952(96)00250-x. [DOI] [PubMed] [Google Scholar]
- 13.Mazumder A., Wang S., Neamati N., Nicklaus M., Sunder S., Chen J., Milne G. W., Rice W. G., Burke T. R., Jr., Pommier Y. J. Med. Chem. 1996;39:2472–2481. doi: 10.1021/jm960074e. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H., Neamati N., Hong H., Mazumder A., Wang S., Sunder S., Milne G. W., Pommier Y., Burke T. R., Jr. J. Med. Chem. 1997;40:242–249. doi: 10.1021/jm960450v. [DOI] [PubMed] [Google Scholar]
- 15.Dorman G., Prestwich G. D. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins T. M., Engelman A., Ghirlando R., Craigie R. J. Biol. Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 17.Sechi M., Angotzi G., Dallocchio R., Dessi A., Carta F., Sannia L., Mariani A., Fiori S., Sanchez T., Movsessian L., et al. Antiviral Chem. Chemother. 2004;15:67–81. doi: 10.1177/095632020401500203. [DOI] [PubMed] [Google Scholar]
- 18.Maignan S., Guilloteau J. P., Zhou-Liu Q., Clement-Mella C., Mikol V. J. Mol. Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 19.Ellison V., Brown P. O. Proc. Natl. Acad. Sci. USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazuda D. J., Felock P. J., Hastings J. C., Pramanik B., Wolfe A. L. J. Virol. 1997;71:7005–7011. doi: 10.1128/jvi.71.9.7005-7011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ariyoshi M., Vassylyev D. G., Iwasaki H., Nakamura H., Shinagawa H., Morikawa K. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 22.Lu R., Limon A., Devroe E., Silver P. A., Cherepanov P., Engelman A. J. Virol. 2004;78:12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit C., Schwartz O., Mammano F. J. Virol. 1999;73:5079–5088. doi: 10.1128/jvi.73.6.5079-5088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu K., Dobard C., Chow S. A. J. Virol. 2004;78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldgur Y., Dyda F., Hickman A. B., Jenkins T. M., Craigie R., Davies D. R. Proc. Natl. Acad. Sci. USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J. C., Krucinski J., Miercke L. J., Finer-Moore J. S., Tang A. H., Leavitt A. D., Stroud R. M. Proc. Natl. Acad. Sci. USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J. Y., Ling H., Yang W., Craigie R. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao K. K., Wang H., Miller J. K., Erie D. A., Skalka A. M., Wong I. J. Biol. Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 29.Karki R. G., Tang Y., Burke T. R., Jr., Nicklaus M. C. J. Comput. Aided Mol. Des. 2004;18:739–760. doi: 10.1007/s10822-005-0365-5. [DOI] [PubMed] [Google Scholar]
- 30.Fikkert V., Van Maele B., Vercammen J., Hantson A., Van Remoortel B., Michiels M., Gurnari C., Pannecouque C., De Maeyer M., Engelborghs Y., et al. J. Virol. 2003;77:11459–11470. doi: 10.1128/JVI.77.21.11459-11470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melek M., Jones J. M., O’Dea M. H., Pais G., Burke T. R., Jr., Pommier Y., Neamati N., Gellert M. Proc. Natl. Acad. Sci. USA. 2002;99:134–137. doi: 10.1073/pnas.012610699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherepanov P., Ambrosio A. L., Rahman S., Ellenberger T., Engelman A. Proc. Natl. Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H., Neamati N., Pommier Y., Burke T. R., Jr. Heterocycles. 1997;45:2277–2282. [Google Scholar]
- 34.Wine R. N., Dial J. M., Tomer K. B., Borchers C. H. Anal. Chem. 2002;74:1939–1945. doi: 10.1021/ac011041w. [DOI] [PubMed] [Google Scholar]
- 35.Borchers C., Boer R., Klemm K., Figala V., Denzinger T., Ulrich W. R., Haas S., Ise W., Gekeler V., Przybylski M. Mol. Pharmacol. 2002;61:1366–1376. doi: 10.1124/mol.61.6.1366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.