Abstract
Caenorhabditis elegans has a single lamin gene, designated lmn-1 (previously termed CeLam-1). Antibodies raised against the lmn-1 product (Ce-lamin) detected a 64-kDa nuclear envelope protein. Ce-lamin was detected in the nuclear periphery of all cells except sperm and was found in the nuclear interior in embryonic cells and in a fraction of adult cells. Reductions in the amount of Ce-lamin protein produce embryonic lethality. Although the majority of affected embryos survive to produce several hundred nuclei, defects can be detected as early as the first nuclear divisions. Abnormalities include rapid changes in nuclear morphology during interphase, loss of chromosomes, unequal separation of chromosomes into daughter nuclei, abnormal condensation of chromatin, an increase in DNA content, and abnormal distribution of nuclear pore complexes (NPCs). Under conditions of incomplete RNA interference, a fraction of embryos escaped embryonic arrest and continue to develop through larval life. These animals exhibit additional phenotypes including sterility and defective segregation of chromosomes in germ cells. Our observations show that lmn-1 is an essential gene in C. elegans, and that the nuclear lamins are involved in chromatin organization, cell cycle progression, chromosome segregation, and correct spacing of NPCs.
INTRODUCTION
The nuclear lamina is a filamentous meshwork that is present between the inner nuclear membrane and the peripheral chromatin. The inner nuclear membrane and the nuclear lamina are involved in organizing nuclear structure and regulating nuclear events. These include the organization of the higher order structure of chromatin and regulation of nuclear assembly and disassembly. The nuclear lamina is a primary target for caspases in apoptosis (reviewed in Goldberg et al., 1999b). Lamins are the major proteins of the nuclear lamina. They are classified as type-V intermediate filaments and are composed of an α-helical rod domain flanked by a short amino (head) and a long carboxy (tail) domains. The rod domain of lamins is 52-nm long and contains four α-helices, each composed of heptad repeats. Coiled-coil interactions and head-to-tail associations between lamin monomers form 10- to 200-nm thick lamin filaments (reviewed in Stuurman et al., 1998)
In vivo, lamin filaments are closely associated with the chromatin fibers (Belmont et al., 1993). In vitro, lamins can bind interphase chromatin (Hoger et al., 1991; Yuan et al., 1991; Taniura et al., 1995; Ulitzur et al., 1997; Goldberg et al., 1999a), mitotic chromosomes (Glass and Gerace, 1990; Glass et al., 1993), or specific DNA sequences (Shoeman and Traub, 1990; Luderus et al., 1992; Luderus et al., 1994; Baricheva et al., 1996; Zhao et al., 1996). The binding site of vertebrate lamins to chromatin is localized to specific sequences in the tail domain and can be displaced with the core histones H2A and H2B (Taniura et al., 1995; Goldberg et al., 1999a).
The composition of the nuclear lamina varies in different cell types and is under developmental regulation in both vertebrate and Drosophila cells (Stuurman et al., 1998). Metazoan cells contain between one to seven lamin proteins. Mammalian lamin A, C, AΔ10, and C2 proteins result from alternative splicing of the lamin A gene. Separate genes encode lamin B1 and B2. Lamin B3 is a germ cell-specific splicing variant of lamin B2. Mutations in the human lamin A gene cause the autosomal-dominant form of Emery-Dreifuss muscular dystrophy (EDMD) (Bonne et al., 1999) and Dunningan-type familial partial lipodystrophy (Cao and Hegele, 2000). Homozygous mice with a knockout in the lamin A gene appear normal at birth but retarded in their growth and die at 4 to 8 wk. These lamin A-deficient mice exhibit a cardiac and skeletal myopathy, with symptoms similar to human EDMD patients, and lack fat cells (Sullivan et al., 1999).
Two lamin genes are known in Drosophila, coding for lamins Dm0 and C (Gruenbaum et al., 1988; Bossie and Sanders, 1993). Lamin Dm0 is an essential gene encoding a type-B lamin that is required for nuclear organization. Flies homozygous for mutations in the lamin Dm0 gene have aberrant nuclear structure and die after 9 to 14 h of development, as the maternal pool of lamin Dm0 is diluted (Harel et al., 1998). Hypomorphic mutation in the lamin Dm0 gene (<20% of lamin expression) causes reduced viability, defective nuclear envelopes, and accumulation of annulate lamellae (Lenz-Bohme et al., 1997).
A search of the nearly complete genome of C. elegans reveals a single lamin gene, termed lmn-1. The lmn-1 gene is located on chromosome I, is 2.7-kb long, and is composed of 6 exons and 5 short introns (Riemer et al., 1993). The lmn-1 gene encodes a putative 64-kDa type-B lamin protein, termed Ce-lamin.
C. elegans has several features for analysis of the biological roles of the nuclear lamina: (A) C. elegans has only one lamin gene, and (B) C. elegans gene expression can be readily manipulated during early embryogenesis. In this study we have characterized Ce-lamin in wild-type C. elegans and investigated the effects of partial or complete loss of Ce-lamin protein on cell and nuclear organization.
MATERIALS AND METHODS
Antibodies
We produced a polypeptide that included coil 2 in the rod domain plus the tail domain of Ce-lamin (amino acid residues D-217 to F-550), by expressing a 1,087-bp EcoRI-BglII cDNA fragment in Escherichia coli JM109, using pRSET (Invitrogen, Leek, The Netherlands). The resulting His-tagged fusion protein was isolated by affinity purification on a nickel-NTA column (Qiagen, Hilden, Germany) in buffer containing 8 M urea and further purified by preparative gel electrophoresis. Rabbits were immunized with the purified Ce-lamin fusion peptide. The Ce-lamin-antibody was affinity purified by incubating the serum with the purified Ce-lamin peptide bound to CN-bromide-activated Sepharose (Pharmacia). Bound antibodies were eluted using a glycine buffer (pH 2.5), and immediately adjusted to neutral pH with NaOH. MAb414, which recognizes a subset of nucleoporins, was purchased from Babco (Richmond, California). T-9026, which recognizes alpha tubulin, was purchased from Sigma (St. Louis, MO). Cy3-conjugated goat-antimouse and goat-antirabbit antibodies were purchased from Jackson laboratories (West Grove, PA).
Immunostaining
Immunostaining was performed essentially as described (Miller and Shakes, 1995). Mixed stages or adults C. elegans were placed on polylysine-treated slides, and 45-mm coverslips were placed above the nematodes. The slides were placed either in liquid N2 or on dry ice, and the coverslips were immediately removed. The nematodes were fixed for 4 min at − 20°C in methanol and then incubated for 30 min at room temperature in PBST (phosphate buffer saline containing 0.1% Tween 20) with 3.7% formaldehyde. Animals were then washed once in PBST, incubated for 10 min at room temperature in PBST containing either 10% low-fat milk or 5% nonfat dry milk, washed once with PBST, and incubated overnight at 4°C with the primary antibody diluted in PBST (1:400 for Ce-lamin and 1:1000 for MAb414). Excess antibody was removed by washes in PBST: once for 1 min, once for 10 min, and twice for 30 min each. The nematodes were then incubated for 2 h at 22°C with the Cy3-conjugated goat-antirabbit (for anti-Ce-lamin) or Cy3-conjugated goat-antimouse (for MAb414), diluted in PBST. Double-label immunostaining for mAb414 and Ce-lamin was performed as follows: Animals were first stained with antibodies to Ce-lamin, followed by FITC-conjugated secondary antibody, and then washed in PBST (once for 1 min, once for 10 min, and twice for 30 min each). The animals were then incubated for 2 h at 22°C with mAb414, rewashed as above, and incubated for 2 h with Cy3-conjugated secondary antibodies. For both double- and single-label immunostaining, excess secondary antibody was then removed by washes in PBST: once for 1 min, once for 10 min, and twice for 30 min each. C. elegans were then incubated for 10 min in PBS containing 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI), washed once with PBS, and mounted in glycerol containing 2% n-propyl gallate. C. elegans were viewed either with an Olympus IX70 microscope equipped with epifluorescence or a Bio-Rad MRC-1024 confocal scanhead coupled to a Zeiss Axiovert 135 M inverted microscope with a 40x/NA = 1.3 oil immersion objective. Excitation light was provided by a 100-mW air-cooled argon ion laser run in the multi-line mode. Both 488-nm and 514-nm excitation were used, as described below. The emission filter in the Cy3 detection channel was a D580/32 interference filter (32-nm bandpass centered on 580 nm). In the GFP channel, a D522/35 interference filter (522-nm center wavelength, 35-nm bandwidth) was used with 488-nm excitation, and a D540/30 interference filter (540-nm center wavelength, 30-nm bandwidth) was used with 514-nm excitation. The confocal iris diameter was 2.5 to 3 mm, with the larger opening used for weaker signals. Vertical resolution was ∼ 1 μm. If necessary, 2 to 4 images were averaged in order to reduce noise. Images of 512 × 512 pixels were acquired, using a hardware zoom of 1.0 (0.308 μm/pixel) or 1.8 (0.175 μm/pixel).
Quantification of Fluorescence Intensities
Fluorescence levels were quantified using a DAGE 300ET-RC CCD camera, with a series of neutral density filters used to confirm linearity of measurements in the range of the assay. Six embryos each were measured for wild-type and lmn-1(RNAi).
Generating lmn-1-GFP Transgenic Lines
Three different lmn-1-GFP constructs were prepared (Figure 1A). The first construct, termed pDRNL1, contained a GFP-Ce-lamin fusion with GFP fused to the N-terminus of lmn-1. To prepare the first construct, a 6.5-kb HindII–SalI genomic fragment positioned between − 7.95 kb and − 1.45 kb upstream to the lmn-1 coding region, was cloned into the pPD95.77 vector. Precise structures of GFP vectors ppD95.77 and pPD102.33 are described on the Fire lab web site, www.ciwenb.edu. A 1.45-kb fragment, just 5′ to the lmn-1 coding region, was amplified by PCR with a novel NcoI cloning site introduced at its 3′ end, and inserted 3′ to the 6.5-kb genomic fragment. The 0.75-kb KpnI–SmaI fragment of the GFP gene was PCR amplified from pEGFP (Clontech) and inserted into the NcoI and SmaI sites. A 2.2-kb KpnI–SmaI fragment of the lmn-1 genomic region, containing the complete lamin coding region, was PCR amplified and inserted 3′ in frame to the GFP gene.
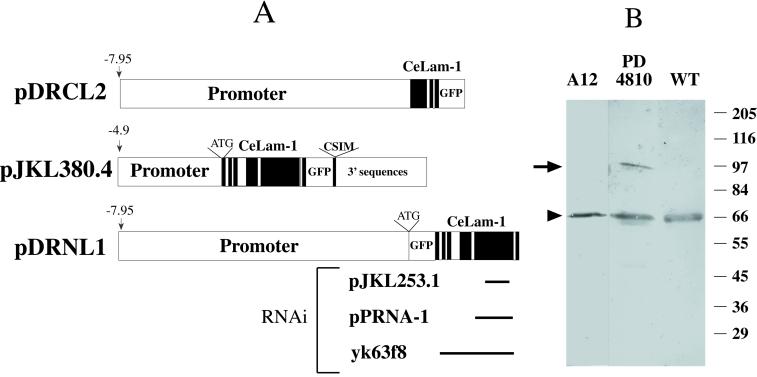
Figure 1.
Ce-lamin encodes a 64-kDa protein. (A) Different lmn-1:gfp constructs used to generate Ce-lamin-GFP lines and clones used to produce dsRNA for the RNAi experiments. For lmn-1:gfp constructs, exons are presented as black boxes. The positions of promoter, GFP, and 3′ sequences are marked. The pDRCL2 construct lacks exons 4 to 6 of lmn-1. The pJKL380.4 construct was used to generate the integrated line PD4810. The pDRNL1 construct was used to generate the unintegrated line A12. The ends of the different constructs used for the RNAi experiments correspond to the positions in the lmn-1 insert in the pDRNL1 construct. (B) Western blot analysis of mixed stages: wild-type (WT), PD4810, and A12 animals probed with Ce-lamin antibodies. The position of Ce-lamin in wild-type nematodes is marked with an arrowhead. A band corresponding in size to the Ce-lamin-GFP fusion protein, which is present in the PD4810 line (marked with an arrow), is below the level of detection in the A12 line. The positions of the size markers in kilodaltons are shown on the right.
The second construct is a fusion of GFP to the C-terminus of Ce-lamin. To prepare this construct, termed pJKL380.4, long-range PCR (Expand long template PCR system, Boehringer Mannheim) was used to amplify from wild-type genomic DNA a 9.6-kb lamin genomic fragment, containing 4.7 kb of 5′ sequence, the entire coding sequence plus introns, and 2.8 kb of 3′ sequence. This 9.6-kb fragment was engineered such that unique NotI and SmaI sites were present at its ends and used for cloning into the pBluescript II SK+ vector. The resulting plasmid, in which the BamHI site in the vector backbone was destroyed, contained a single BamHI site 12 amino acids before the stop codon of lamin. This site was used to insert GFP (from pPD102.33) in frame into the lmn-1 gene.
A third construct, termed pDRCL2, contained 7.95 kb of 5′ promoter sequence of lmn-1, the first three exons, and two introns of lmn-1 fused in frame to the GFP gene.
To make transgenic lines, all three constructs were first linearized. pJKL380.4 construct was coinjected with linearized pMH86 marker plasmid into the temperature-sensitive dpy-20 (e1282) animals using the complex array injection method (Kelly et al., 1997). pDRNL1 and pDRCL2 constructs were each coinjected with the pRF4 plasmid containing the dominant rol(6) marker (Mello et al., 1991) into wild-type (N2) animals with no carrier DNA. Multiple transgenic lines were obtained for each construct, with GFP localized as expected in the nuclear envelopes in lines expressing pJKL380.4 and pDRNL1, and in the cytoplasm in lines expressing pDRCL2. One of the pDRNL1-derived transgenic lines, A12, was kept as an unintegrated line and used for further characterization.
For most of the transgenic lines, the earliest stage at which GFP expression could be detected was in embryos containing around 60 cells. Several of the pJKL380.4-derived transgenic lines initially showed GFP expression in the germ line also, including germ cells in the syncytial gonad, oocytes, and early embryos. The pJKL380.4 transgene from one of these lines was then integrated using the standard gamma-irradiation method (Mello and Fire, 1995). Three independent integrated lines were generated, and all were out crossed three times and mapped to the X chromosome. One such line, PD4810 (ccIs4810), was used for further studies. The germ line expression of lmn-1-GFP was lost after several generations even in the integrated lines. To circumvent this problem, the lmn-1-GFP transgene from PD4810 was crossed into the mut-7 (pk204) mutator background. mut-7 has been shown to desilence transgenes in the germ line to some extent (Tabara et al., 1999).
RNAi Injections
To assess the function of lamin in C. elegans development, RNAi was employed to deplete both the maternal and zygotic product of the lamin gene. Three different constructs were used for making double-stranded RNA in vitro (Figure 1A). yk63f8, a gift of Dr. Yuji Kohara (Japan), contained a 2.13-kb, nearly full-length, lmn-1 cDNA; pJKL253.1 contained a 659-bp PCR fragment from exon 5 (the largest exon) of the lmn-1 gene; pPRNA-1 contained a 995-bp BglII-EcoRI fragment of the lmn-1 gene (amino acids 217–550). Both pPRNA-1 and pJKL253.1 contained a pBluescript II SK+ vector backbone. Double-stranded RNA was made in vitro following the protocol described by Fire et al., (1998). The dsRNA was injected at concentrations between 0.1 to 1.0 μg/μl into wild-type (N2) hermaphrodites. The injected animals were transferred to new plates every 6 to 12 h, and their progeny were scored for abnormalities during development.
Four-dimensional Time-lapse Microscopy
One-cell embryos from wild-type or lmn-1 dsRNA injected hermaphrodites were mounted on agarose pads, and 4-D time-lapse recording was performed as described (Fire, 1994), with a complete series of 28 images every 1.2 microns in the Z axis taken at 30-s intervals. Complete image series were analyzed from three lmn-1(RNAi) embryos and several wild-type embryos. All nuclei from five additional embryos were analyzed by direct observation in real time with a similar phenotype. Confirming data was also obtained from a number of additional embryos from which shorter series of images were collected.
RESULTS
lmn-1 Encodes a Single Type-B Lamin Protein
The C. elegans genome contains an open reading frame encoding a putative 64-kDa protein with homology to lamins. This protein has been termed Ce-lamin (Riemer et al., 1993). Analysis of the near-completed genome sequence data now available for C. elegans suggests that this is the only lamin homologue in this organism. Portions of the rod and tail domains of Ce-lamin (amino acids 217 to 550) were expressed in bacteria, purified to homogeneity by gel electrophoresis, and used to raise polyclonal antibodies in rabbits. Western blot analysis, using affinity-purified Ce-lamin antibodies, showed a single band of 64 kDa in SDS-PAGE of complete C. elegans extract from a mixed stage nematode population (Figure 1B).
Immunostaining of C. elegans at different developmental stages with the Ce-lamin antibodies showed that Ce-lamin is predominantly localized to the nuclear periphery (Figure 2). Additional confocal microscopy analysis revealed subregions of the nuclear envelope with a higher intensity staining (our unpublished results). Such subregions of intense staining are characteristic of lamin staining in other metazoans (Paddy et al., 1990) (Figure 2). The localization of Ce-lamin to the nuclear periphery, its pattern in the nuclear periphery, and its sequence homology to other known type-B lamins, all suggest that lmn-1 is the unique type-B lamin ortholog in C. elegans.
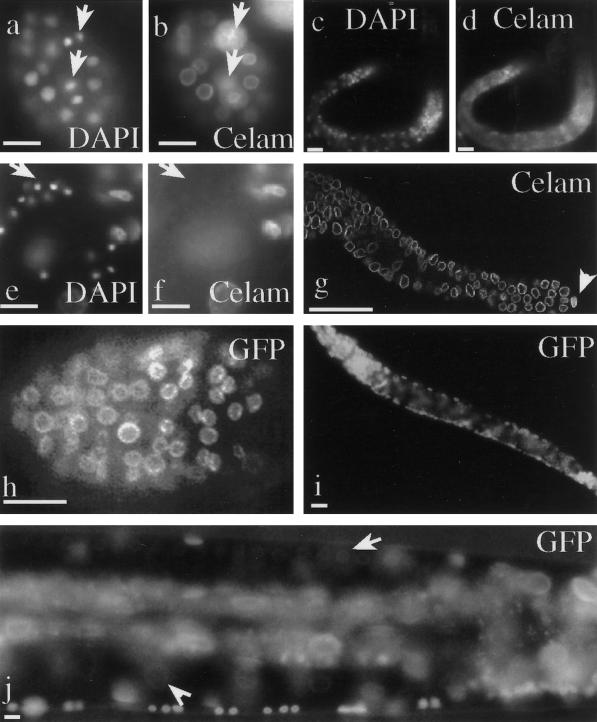
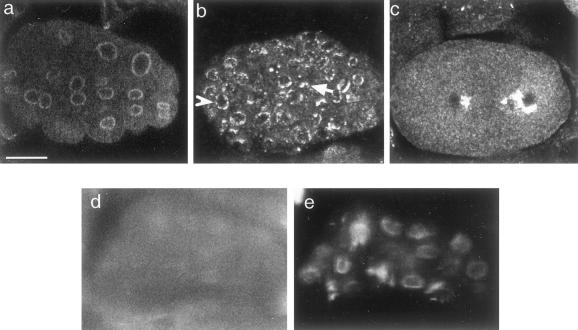
Figure 2.
Ce-lamin protein distribution. Wild-type C. elegans (N2) were stained with affinity purified polyclonal antibodies to Ce-lamin (primary), and Cy3-conjugated antirabbit antibodies (secondary) (b,d,f,g). DNA was stained with DAPI (a,c,e). Embryo (a,b). Arrows in (a) are pointing toward nuclei in telophase where some Ce-lamin is dispersed in the cytoplasm (arrows in b). L1 larvae (c,d). The arrow in (e) points to a cluster of sperm cells in an adult hermaphrodite's gonad. Ce-lamin is detected only in the surrounding cells. Ce-lamin is expressed in all other cells in the gonad (g). Note the intense lamin staining in the distal tip cell (arrowhead in g). Expression of Ce-lamin-GFP can be detected in the nuclear envelope of all somatic cells in PD4810 embryo (h), larva (i), and adult (j). Arrow in (j) points toward germ cell nuclei in the gonad; Arrowhead points toward an oocyte nucleus. Bar in each panel represents 10 μm.
Ce-lamin Is Present in the Nuclear Envelope of Essentially All Cells during C. elegans Development
Antibodies to Ce-lamin stained the nuclear envelope at all developmental stages and in essentially every region where the Ce-lamin antibodies penetrated, including embryos, all larval stages, and adults (Figure 2a-d,g). Nuclear envelope staining of Ce-lamin was also detected in all cells in the gonad (Figure 2g), with one exception: antibody staining was not detected in cells undergoing spermiogenesis that had condensed chromatin (Figure 2e-f).
Transgenic lines expressing the GFP fused to Ce-lamin (Ce-lamin-GFP) were prepared in order to study the spatial and developmental distribution of Ce-lamin in living cells. With the exception of mature sperm, Ce-lamin-GFP fusion protein was localized to the nuclear periphery of all cells throughout the development of the nematode (Figure 2h-i). The Ce-lamin-GFP expression in these lines reproduced the antibody staining pattern of the native protein, including the presence of subregions in the nuclear envelope with higher fluorescence intensity (Figure 3, bottom panel).
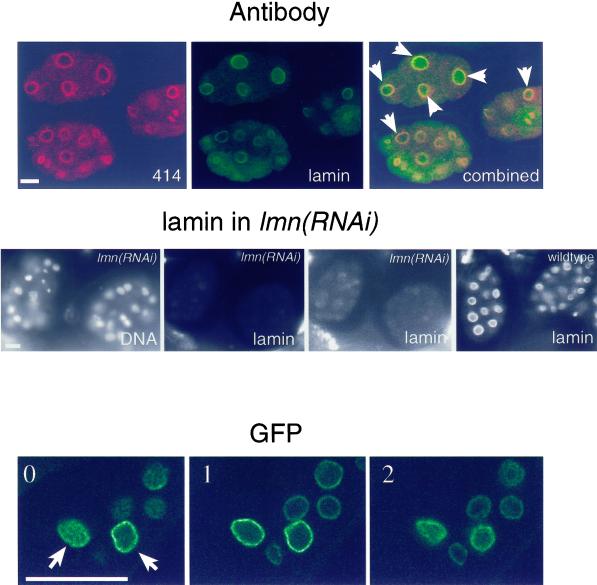
Figure 3.
Ce-lamin is present in the nuclear interior. Top panel: embryos were double stained with monoclonal antibody MAb414 (red) and affinity-purified Ce-lamin antibodies (green), and viewed with a confocal microscope. Overlap between Ce-lamin and nucleoporins appears in yellow. Most embryonic nuclei contained internal Ce-lamin, while MAb414 staining was confined to the nuclear envelope and the cytoplasm. Examples of nuclei with internal Ce-lamin staining are marked with arrows. Middle panel: DNA staining, left image; lamin staining, all other images. Both lmn-1(RNAi) images show the same confocal image of Ce-lamin staining, of the corresponding DNA-stained embryos. Intensity levels in the right lmn-1(RNAi) image were elevated, in order to detect residual lamin staining (as compared with the normal intensity levels in the left lmn-1(RNAi) image). Ce-lamin staining was eliminated in lmn-1(RNAi) embryos from both the nuclear envelope and the nuclear interior. The right image shows lamin staining in the control wild-type embryos. Bottom panel: three consecutive confocal sections, (one micron apart, 0 to 2), of adult tissue in the A12 line, which expresses low levels of Ce-lamin-GFP (see Figure 1B). Examples of nuclei with internal Ce-lamin-GFP staining are marked with arrows. Bar in each panel represents 10 μm and applies to all images in that panel.
Ce-lamin Is also Present in the Nuclear Interior
Intranuclear presence of Ce-lamin was also observed in most embryonic nuclei and in a fraction of nuclei in adult tissues, including the distal tip cell in the gonad (Figure 2g). In order to verify that the internal lamin staining was specific, embryonic and adult tissues were stained with both Ce-lamin antibodies and monoclonal antibody mAb414 (Davis and Blobel, 1987), which recognizes the nuclear pores in many eukaryotes, including C. elegans (Pitt et al., 2000). As expected, both MAb414 and Ce-lamin brightly stained the nuclear envelope. However, only Ce-lamin showed intranuclear location (top panels in Figure 3), while MAb414 gave a higher cytoplasmic background, as described (Pitt et al., 2000). The internal staining disappeared as well as the nuclear envelope staining in lmn-1(RNAi) embryos (Figure 3).
To further verify the presence of Ce-lamin in the nuclear interior, the spatial distribution of the GFP-Ce-lamin fusion protein was analyzed by confocal microscopy in vivo in line A12, which expresses the lowest levels of Ce-lamin-GFP fusion (Figure 1B). Consistent with the antibody staining, a subset of cells showed GFP-Ce-lamin protein in the nuclear interior, in addition to the nuclear envelope (bottom panels in Figure 3). The intranuclear staining was not uniform and was always weaker than that seen on the nuclear envelope.
lmn-1 Is an Essential Gene
To understand the function of Ce-lamin in vivo, RNA interference experiments (RNAi) aimed at inhibiting lmn-1 expression, were performed by injecting lmn-1 dsRNA into the gonads of adult hermaphrodites. Equivalent results were obtained with three different dsRNA constructs (Figure 1A), injected at several different concentrations (0.1 to 1.0 μg/μl).
RNAi effects are most potent in a time window of 18 to 36 h after injection. All embryos laid during this time window arrested during development, indicating lamin is required for embryonic viability. We use the nomenclature the lmn-1 (RNAi) to describe the F1 progeny of hermaphrodites injected with dsRNA of lmn-1.
To monitor the extent of residual Ce-lamin expression in the lmn-1 (RNAi) embryos, embryos were stained at different stages with lamin antibodies. There was some variability between lmn-1 (RNAi) embryos and between regions in the same embryo (our unpublished results). However, quantitative fluorescence analysis carried out as described in Materials and Methods revealed a decrease of > 60-fold in intensity of Ce-lamin staining in the lmn-1 (RNAi) embryos (Figure 4a,b).
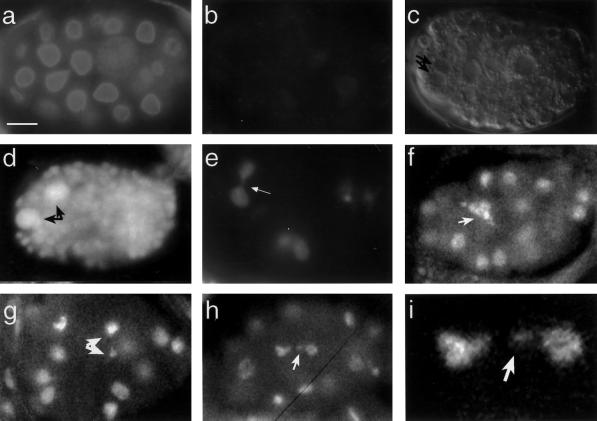
Figure 4.
Arrested embryos injected with lmn-1 dsRNA. Ce-lamin indirect immunofluorescence staining of wild-type (a) and lmn-1 (RNAi) embryos (b). The lmn-1 (RNAi) embryos showed significantly reduced levels of Ce-lamin staining as compared with wild-type embryos. Fluorescence quantification of this difference revealed an effect > 60-fold (c). Nomarski image of a lmn-1 (RNAi) embryo (c). Note the uneven size of the nuclei and a cell with two nuclei (arrows in c). DAPI staining of lmn-1 (RNAi) embryos (d-i). At the time of arrest, abnormalities included nuclei with higher DNA content (black arrows in panel d and regions with fewer nuclei (our unpublished results). Abnormalities in nuclear morphology were already obvious in embryos undergoing the first nuclear divisions (e-i), although most embryos did develop further. These abnormalities included chromosome bridges between two nuclei (arrow in e), abnormal chromosome morphology (arrow in f), an unequal distribution of chromatin between daughter nuclei (arrows in g), and a chromatin or micronuclei left outside the daughter nuclei after nuclear division (arrows in i show the affected nuclei in h are magnified in j). The bar in panel (a) represents 10 μm and also applies to panels (b-h) and 2.5 μm in panel (i).
The intensity of the residual Ce-lamin staining in lmn-1(RNAi) embryos decreased as the embryos continued to develop; Ce-lamin was not detectable upon developmental arrest. A dramatic reduction in Ce-lamin staining in lmn-1(RNAi) embryo was also detected in the nuclear interior (Figure 3, middle panels), proving further evidence for the presence of Ce-lamin inside the nucleus.
Although some lmn-1(RNAi) embryos arrested with < 100 nuclei, DIC microscopy revealed that most lmn-1(RNAi) embryos were able to form several hundred nuclei before arrest (Figure 4c,d). All lmn-1(RNAi) embryos were abnormal and contained nuclei that varied in size (Figure 4d). Further incubation of the lmn-1 (RNAi) embryos caused them to degrade and to form regions in the embryos that were devoid of nuclei (our unpublished results). The reduced lmn-1 activity and embryonic arrest in the lmn-1 (RNAi) embryos indicated that lamins are an essential component of the nuclear envelope.
lmn-1 Is Required for Nuclear Organization and Cell Cycle Progression
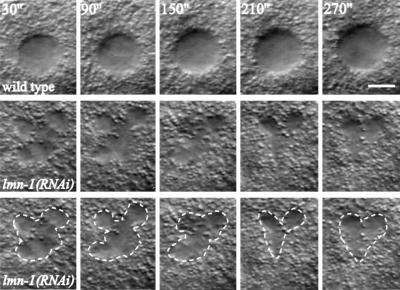
The phenotype of lmn-1(RNAi) embryos was examined by 4-D time-lapse microscopy on live embryos (Fire, 1994). The earliest observed defect was as early as the two-cell stage. While nuclei in the wild-type embryo had well defined and stable boundaries, the nuclei in lmn-1(RNAi) embryos appeared plastic, with rapid changes over time (see Figure 5 for an example of the AB nucleus in a two-cell embryo). Despite the gross defects in nuclear morphology, nuclear divisions still occurred in lmn-1(RNAi) embryos, with these embryos eventually producing several hundred cells.
Figure 5.
Nuclear morphology in lmn-1(RNAi) embryos. Five consecutive images of the AB cell nucleus in a two-cell embryo taken at 1-min intervals using 4-D time-lapse recording (Fire, 1994). The central focal plane for each nucleus is shown. The top panels are from a wild-type embryo, and the bottom two sets are from a lmn-1(RNAi) embryo, with the bottom set showing the outline of the nucleus in dotted line. Bar represents 5 μm.
The nuclear morphology of the lmn-1 (RNAi) embryos was further examined by staining with the DNA-specific dye DAPI. Whereas control embryos of mock-injected hermaphrodites or wild-type embryos had no apparent nuclear abnormalities, a fraction of nuclei in > 95% of the lmn-1 (RNAi) embryos appeared abnormal, with defects as early as the first few nuclear divisions (Figure 4e-i). These phenotypes included inability to complete the cell cycle and distribution of unequal amounts of chromatin into the daughter nuclei. The most common phenotypes in the progression of the cell cycle were bridges of chromatin between nuclei (arrow in Figure 4e) and chromatin that expanded abnormally (Figure 4f). Problems in the distribution of chromatin between daughter nuclei included large differences in the amounts of chromatin in the daughter nuclei (arrows in Figure 4g) and chromosomes that were outside the daughter nuclei (arrows in Figure 4h,i). These abnormalities were not due to gross defects in microtubule organization, since the pattern of staining of the lmn-1 (RNAi) embryos with tubulin antibodies was similar to that of wild-type embryos (our unpublished results).
Arrested embryos showed regions where the DNA was absent (our unpublished results), or regions with a large increase in the amount of DNA (black arrows in Figure 4d show ‘giant’ nuclei that are 8 times the size of wild-type gut nuclei in a comma stage embryo). In summary, the RNAi experiments revealed a broad requirement role for lmn-1 in cell cycle progression, and chromatin organization.
An Intact Nuclear Lamina Is Required for the Correct Spacing of NPCs
Reduction in levels of lamin Dm0 activity in Drosophila and elimination of lamin A gene in mouse have both shown to yield abnormalities in the pattern of NPCs (Lenz-Bohme et al., 1997; Harel et al., 1998; Sullivan et al., 1999). We therefore compared the spatial organization of the NPCs in lmn-1 (RNAi) embryos and normal C. elegans embryos by staining them with monoclonal antibody MAb414, which recognizes the FG repeats present in many nucleoporins (Davis and Blobel, 1987). While wild-type or mock-injected embryos showed a typical patchy nuclear rim staining, characteristic of nuclear pores in most species examined (Figure 6a), the NPCs in many (but not all) nuclei of the lmn-1 (RNAi) embryos showed an abnormal clustering pattern to one side of the nucleus (arrows in Figure 6b and both nuclei in Figure 6c; a “normal” distribution of NPCs is shown by an arrowhead in Figure 6b). Staining of the lmn-1(RNAi) embryos with both Ce-lamin antibodies and monoclonal antibody MAb414 revealed that NPCs clustering does not correlate with residual Ce-lamin; indeed, most NPC clusters were observed in nuclei where residual Ce-lamin could not be observed (Figure 6d,e). These results confirm the importance of lmn-1 in C. elegans for proper spacing of nuclear pore complexes.
Figure 6.
Clustering of NPCs in lmn-1 (RNAi) embryos. Embryos were stained with monoclonal antibody MAb414 (a-c) and viewed with a confocal microscope. Wild-type embryo (a); lmn-1 (RNAi) embryos, (b-c). The single-head arrows in panel (b) point toward regions where NPCs are clustered on one side of the nucleus. The embryo in panel (c) has two nuclei, both of which have clustered NPCs, never seen in wild-type embryos. Most embryos also contain nuclei with normal spacing of NPCs (arrowhead). The lmn-1(RNAi) embryos were also stained with both lamin antibodies (d) and monoclonal antibody MAb414 (e). Bar in panel (a) represents 10 μm and applies to all panels.
Analysis of lmn-1(RNAi) Effects on Germ Line Development
We were able to examine germline development after conditions of Ce-lamin insufficiency by studying a few rare F1 animals that escaped the lethal effects of lmn-1 (RNAi). These animals resulted from eggs laid outside the most potent window of RNAi activity (Figure 7). Significantly, most of these F1 adults were sterile or semisterile. Examination of the gonads of the sterile animals by DIC microscopy and DAPI staining revealed dramatic reductions in the number of germ cells (Figure 7a-b). In a fraction of such animals, small numbers of abnormal oocyte-like cells were observed (some with multiple nuclei and large vacuoles; Figure 7c,g); nuclei with condensed chromatin (potentially spermatocytes or spermatocyte-like cells) were also observed (Figure 7h). Some of the F2 embryos from semisterile animals developed into fertile adults. Among these progeny, there was a high incidence of males (31 males out of 253 progeny or 12.25%, compared with 0.1% in wild-type animals; Hodgkin et al., 1979). These males were fertile. Their genotype was XO, since mating with dpy-11 unc-60 hermaphrodites produced wild-type progeny with 1:1 ratio of males and hermaphrodites.
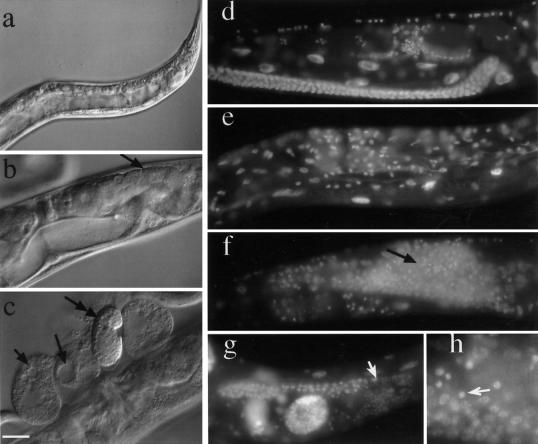
Figure 7.
Sterile and semisterile animals from the RNAi experiments. Nomarski images of progeny that escaped lmn-1(RNAi) lethality. These adults developed into either sterile (a) or semisterile animals (b,c). The adults had very few germ cells (arrow in b) and accumulated yolk. These embryos were laid in time windows 6 to 18 h, or > 36 h after injection, when RNAi effects are generally incomplete. These animals frequently produced abnormal oocytes with vacuoles (arrows in c). Occasionally, some of these animals gave rise to normal-looking embryos, which developed normally (double-headed arrow) (c). DAPI staining of wild-type (d), sterile (e) or semisterile animals (f-h). Some animals had few or no germ cells (e), whereas others had germ cells with both condensed nuclei and normal looking nuclei, starting from the most distal region of the germ line (arrows in f,g). The region shown in (h) is a higher magnification of the distal arm of the gonad shown in f. Arrow points to a condensed nucleus. Bar in panel c represents 20 μm for panels b-g, 40 μm for panel a, and 8 μm for panel h.
DISCUSSION
Ce-lamin Is a Component of the Nuclear Envelope in Essentially all Cells during C. elegans Development
In both vertebrates and Drosophila, the type-A lamins have a cell- and tissue-specific pattern of expression (reviewed in Stuurman et al., 1998), while type-B lamins are present in essentially all cells. The single C. elegans lamin protein, Ce-lamin, is similar to type-B lamins in both Drosophila and vertebrates (Goldberg et al., 1999b). With the exception of mature sperm cells, Ce-lamin is detected in the nuclear envelope of essentially all cells during all developmental stages of C. elegans. C. elegans is a multicellular animal, which undergoes complicated differentiation processes to give rise to differentiated tissues and a nervous system. We therefore propose that Ce-lamin perform activities that in other animals are performed by both type-B and type-A lamins, and these activities maybe regulated by posttranslational modifications.
The only cells that do not contain detectable Ce-lamin are sperm cells with condensed chromatin. However, we cannot rule out the possibility that technical issues prevent expression of lamin-GFP and lamin antibody staining in sperm cells. Drosophila sperm cells have also been reported to lack a type-B lamin (lamin Dm0) and its associated protein, otefin (Ashery Padan et al., 1997). In contrast, vertebrate sperm cells contain both type-A and type-B lamins, which are probably involved in nuclear organization during meiosis (Furukawa and Hotta, 1993; Furukawa et al., 1994; Alsheimer et al., 1999).
Ce-lamin Is also Detected in Nuclear Interior
In addition to the nuclear periphery, Ce-lamin was also detected in the nuclear interior. Internal Ce-lamin staining was not an immunofluorescence artifact, since (a) it was observed both in fixed animals stained with Ce-lamin antibodies and in GFP patterns from live animals that express low amounts of a GFP-Ce-lamin fusion protein, and (b) it was eliminated in lmn-1(RNAi) embryos. Most embryonic cells contained intranuclear lamin, while adult cells with internal nuclear lamin staining were found next to cells in which Ce-lamin could be detected only at the nuclear periphery. Internal type-A lamins have been reported in HeLa cells (Hozak et al., 1995) and in mammalian cells expressing lamin A-GFP fusion proteins (Broers et al., 1999). Type-A and type-B lamins are also seen as intranuclear foci in mammalian cultured cells (Bridger et al., 1993; Moir et al., 1994). These foci are thought to be intermediate stages of lamin processing, before their incorporation into the peripheral lamina (Goldman et al., 1992). Lamins also colocalize at sites of DNA replication, where they are required for the elongation stage of DNA replication (Spann et al., 1997). In addition, during apoptosis, lamins are distributed relatively uniformly in the interior of the nucleus (Rao et al., 1996b). Thus, the roles of interior lamins will be interesting topic for a future study.
lmn-1 Is an Essential Gene
The RNAi experiments revealed that lmn-1 is an essential gene. The lethality caused by the lmn-1 dsRNA was specific since (A) embryos laid by mock-injected adults developed normally; (B) similar phenotypes were seen with three different RNAi constructs and different concentrations of lmn-1 dsRNA ranging between 0.1 to 1.0 μg/μl; and (C) the observed phenotypes were associated with a dramatic reduction in levels of Ce-lamin. The Ce-lamin staining became undetectable as the lmn-1 (RNAi) embryos continued to develop. In contrast to Ce-lamin levels, the level of nucleoporins was apparently unchanged after lmn-1 RNAi. It was not surprising that loss of Ce-lamin is lethal, since mutations in the Drosophila lamin Dm0 gene (Lenz-Bohme et al., 1997; Harel et al., 1998) and in mouse lamin A gene (Sullivan et al., 1999) also cause nuclear defects and lethality. The results of this study extend the results in Drosophila and mice in several ways. Unlike Drosophila or mice, the genome of the C. elegans contains only one lamin gene; the maternal contribution of this gene can be largely eliminated in a gene-specific manner by RNA-mediated interference (Fire et al., 1998). This allows the only example to date in which one can exam the effects of reducing all lamin on cell growth and development. In addition, due to large maternally contributed pools of Drosophila lamin Dm0 protein and RNA, the phenotypes did not appear until relatively late in embryogenesis, when lamin C is also being expressed. Likewise, the delayed phenotypes of mice lacking lamin A (4 to 8 wk after birth) were probably due to complementation by the lamin B1 and B2 genes.
We attribute the weak residual Ce-lamin signal in the lmn-1 (RNAi) embryos to the maternal contribution of stable proteins that cannot be eliminated completely by dsRNA. There is a large maternal pool of Ce-lamin in wild-type oocytes (our unpublished results), and lamins are very stable proteins (Harel et al., 1998). We consider it likely that the residual Ce-lamin probably allows nuclei to continue dividing in lmn-1 (RNAi) embryos until there is not enough lamin activity to support normal, or even abnormal, nuclear divisions. Consistent with this, residual Ce-lamin could not be detected in lmn-1 (RNAi) embryos with > 100 to 200 nuclei (our unpublished results).
Lamins Are Required for Proper Spacing of NPCs
The nuclear lamina anchors the NPCs (reviewed in Stoffler et al., 1999). We found that loss of Ce-lamin caused NPCs to cluster. These results suggest that the association of the nuclear lamina with the NPCs is required for their correct spacing. The role of the nuclear lamina in spacing NPCs is probably conserved in evolution since similar phenotypes are seen in lines with reduced Drosophila lamin Dm0 and in mice lacking the lamin A gene (Lenz-Bohme et al., 1997; Harel et al., 1998; Sullivan et al., 1999).
A Role for Lamins in Nuclear Morphology
A remarkable phenotype in the lmn-1 (RNAi) embryos is the rapid change in nuclear morphology in vivo. This change clearly demonstrates the role of nuclear lamina in determining the shape of the nucleus. However, the abnormality in nuclear shape did not interfere with the ability of most nuclei to undergo mitosis. In addition, the timing of nuclear divisions appeared similar in the lmn-1 (RNAi) and wild-type embryos (our unpublished results). We consider it likely that the instability of nuclear morphology in the lmn-1 (RNAi) embryos is a primary defect eventually leading to defects in cell cycle progression and chromatin organization (see below).
Lamins May Be Involved in Chromosome Segregation
It was surprising to find that lamin activity may be required for the proper segregation of chromosomes. In the lmn-1 (RNAi) embryos, there were many examples of unequal segregation of chromatin to daughter nuclei, and of chromosomes that must have been lost from one of the daughter nuclei during the cell cycle. The difference in the amount of chromatin in daughter nuclei may be also the consequence of incomplete DNA replication, since lamins have been proposed to play an essential role in the elongation step of DNA replication (Moir et al., 2000). This difference can be also due to either nuclear fragmentation or fusion of chromatin during interphase. However, the high incidence of male offspring from the F1 animals that survived the initial RNAi indicates that Ce-lamin activity is required, either directly or indirectly, for proper segregation of chromosomes. Although Ce-lamin is present in a spindle envelope structure until early anaphase (Lee et al., 2000), its role in chromosome segregation is probably not through a change in microtubule distribution, since the spatial organization of microtubules in lmn-1 (RNAi) embryos is similar to wild-type.
Lamins Are Required for Proper Cell Cycle Progression
A high fraction of the lmn-1 (RNAi) embryos contained some nuclei that were unable to complete the cell cycle. A requirement for lamins and lamin-associated proteins to complete the cell cycle in mammalian cells was suggested from a previous study, in which lamin antibodies injected into mammalian cells caused chromatin arrest in a telophase-like configuration (Benavente and Krohne, 1986). The roles of nuclear lamins in nuclear assembly are still a matter of debate (reviewed in Gant and Wilson, 1997). Lamins were suggested to play a role in assembling the nuclear envelope in Drosophila (Ulitzur et al., 1997), Xenopus (Dabauvalle et al., 1991) and mammalian cells (Burke and Gerace, 1986). The results presented here demonstrate that lamins, either directly or by affecting the distribution of their associated proteins, are required for proper cell cycle progression, which may reflect a defect in nuclear assembly in vivo.
Lamins and Chromatin Organization
Many nuclei in the arrested lmn-1 (RNAi) embryo and in gonads of the lmn-1 (RNAi) F1 adults had abnormally condensed chromatin. We propose that chromatin might condense in these cells due to insufficient attachment to the nuclear envelope. Given that many nuclear membrane proteins bind directly to a chromatin partner, our results suggest that (A) attachments between chromatin and nonlamin components of the nuclear membrane are insufficient to keep the chromatin decondensed; (B) the membrane proteins may not be fully functional in the absence of lamins; or (C) loss of the lamina may trigger the activity of apoptotic factors that cause chromatin condensation, which normally only occurs after caspase-induced lamin degradation. Indeed, in C. elegans the nuclear envelope is a primary docking site for CED-4 (Chen et al., 2000), and in mammalian cells lamin degradation is required for the dissociation of chromatin from the nuclear membranes during apoptosis (Rao et al., 1996a). Further experiments will be aimed at examining apoptotic patterns in C. elegans lacking normal Ce-lamin levels.
ACKNOWLEDGMENTS
This study was funded in part by grants from the USA-Israel Binational Science Foundation (BSF to Y.G.), the Israel Science Foundation (BRF to Y.G.), and the German-Israel Foundation (GIF #1–573-036.13 to Y.G. and K.W). Support to J.L. and A.F. was provided by the NIH grants F32HD08331 and GM37706.
We thank Margalit Eshel and Jamie Fleenor for technical help, Aryeh M Weiss and Naomi Melamed-Book for helping with the confocal microscope, Bill Kelly for C. elegans strains, and Mary Osborn and Kenneth Lee for critical reading of the manuscript. We are also in great debt to Kathy Wilson for her wonderful comments and inspiring discussions.
Abbreviations used:
- NPCs
nuclear pore complexes
- DAPI
4′,6′-diamidino-2-phenylindole
REFERENCES
- Alsheimer M, von Glasenapp E, Hock R, Benavente R. Architecture of the nuclear periphery of rat pachytene spermatocytes: distribution of nuclear envelope proteins in relation to synaptonemal complex attachment sites. Mol Biol Cell. 1999;10:1235–1245. doi: 10.1091/mbc.10.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery Padan R, Ulitzur N, Arbel A, Goldberg M, Weiss AM, Maus N, Fisher PA, Gruenbaum Y. Localization and posttranslational modifications of otefin, a protein required for vesicle attachment to chromatin, during Drosophila melanogaster development. Mol Cell Biol. 1997;17:4114–4123. doi: 10.1128/mcb.17.7.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baricheva EA, Berrios M, Bogachev SS, Borisevich IV, Lapik ER, Sharakhov IV, Stuurman N, Fisher PA. DNA from Drosophila melanogaster beta-heterochromatin binds specifically to nuclear lamins in vitro and the nuclear envelope in situ. Gene. 1996;171:171–176. doi: 10.1016/0378-1119(96)00002-9. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente R, Krohne G. Involvement of nuclear lamins in postmitotic reorganization of chromatin as demonstrated by microinjection of lamin antibodies. J Cell Biol. 1986;103:1847–1854. doi: 10.1083/jcb.103.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery- Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Bossie CA, Sanders MM. A cDNA from Drosophila melanogaster encodes a lamin C-like intermediate filament protein. J Cell Sci. 1993;104:1263–1272. doi: 10.1242/jcs.104.4.1263. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, O'Farrell M, Hutchison CJ. Internal lamin structures within G1 nuclei of human dermal fibroblasts. J Cell Sci. 1993;104:297–306. doi: 10.1242/jcs.104.2.297. [DOI] [PubMed] [Google Scholar]
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, Ramaekers FC. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Burke B, Gerace L. A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz HR. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Dabauvalle MC, Loos K, Merkert H, Scheer U. Spontaneous assembly of pore complex-containing membranes (“annulate lamellae”) in Xenopus egg extract in the absence of chromatin. J Cell Biol. 1991;112:1073–1082. doi: 10.1083/jcb.112.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci USA. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. A four-dimensional digital image archiving system for cell lineage tracing and retrospective embryology. Comput Appl Biosci. 1994;10:443–447. doi: 10.1093/bioinformatics/10.4.443. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Hotta Y. cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J. 1993;12:97–106. doi: 10.1002/j.1460-2075.1993.tb05635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Inagaki H, Hotta Y. Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp Cell Res. 1994;212:426–430. doi: 10.1006/excr.1994.1164. [DOI] [PubMed] [Google Scholar]
- Gant TM, Wilson KL. Nuclear assembly. Annu Rev Cell Dev Biol. 1997;13:669–695. doi: 10.1146/annurev.cellbio.13.1.669. [DOI] [PubMed] [Google Scholar]
- Glass CA, Glass JR, Taniura H, Hasel KW, Blevitt JM, Gerace L. The alpha-helical rod domain of human lamins A and C contains a chromatin binding site. EMBO J. 1993;12:4413–4424. doi: 10.1002/j.1460-2075.1993.tb06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JR, Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, Gruenbaum Y. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc Natl Acad Sci USA. 1999a;96:2852–2857. doi: 10.1073/pnas.96.6.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Harel A, Gruenbaum Y. The nuclear lamina: molecular organization and interaction with chromatin. Crit Rev Eukaryot Gene Expr. 1999b;9:285–293. doi: 10.1615/critreveukargeneexpr.v9.i3-4.130. [DOI] [PubMed] [Google Scholar]
- Goldman AE, Moir RD, Montag LM, Stewart M, Goldman RD. Pathway of incorporation of microinjected lamin A into the nuclear envelope. J Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Landesman Y, Drees B, Bare JW, Saumweber H, Paddy MR, Sedat JW, Smith DE, Benton BM, Fisher PA. Drosophila nuclear lamin precursor Dm0 is translated from either of two developmentally regulated mRNA species apparently encoded by a single gene. J Cell Biol. 1988;106:585–596. doi: 10.1083/jcb.106.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Goldberg M, Ulitzur N, Gruenbaum Y. Structural organization and biological roles of the nuclear lamina. In: Boulikas T, editor. Textbook of Gene Therapy and Molecular Biology: From Basic Mechanism to Clinical Applications“. Palo Alto, CA: Gene Therapy Press; 1998. , 1, 529–542. [Google Scholar]
- Hodgkin J, Horvitz RH, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoger TH, Krohne G, Kleinschmidt JA. Interaction of Xenopus lamins A and LII with chromatin in vitro mediated by a sequence element in the carboxyterminal domain. Exp Cell Res. 1991;197:280–289. doi: 10.1016/0014-4827(91)90434-v. [DOI] [PubMed] [Google Scholar]
- Hozak P, Sasseville AM, Raymond Y, Cook PR. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J Cell Sci. 1995;108:635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins Ce-Emerin, Ce-Man, Ce-lamin and nucleoporins reveal developmentally-regulated timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. Insertional mutation of the Drosophila nuclear lamin dm(0) gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderus ME, de Graaf A, Mattia E, den BJ, Grande MA, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992;70:949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- Luderus ME, den Blaauwen J, de Smit O, Compton DA, van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol Cell Biol. 1994;14:6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Shakes DC. Immunofluorescence microscopy. Vol. 48. In: Epstein FH, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. New York: Academic Press; 1995. [Google Scholar]
- Moir RD, Montag LM, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. The dynamics of the nuclear lamins during the cell cycle- relationship between structure and function. J Struct Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- Paddy MR, Belmont AS, Saumweber H, Agard DA, Sedat JW. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62:89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996a;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996b;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer D, Dodemont H, Weber K. A nuclear lamin of the nematode Caenorhabditis elegans with unusual structural features: cDNA cloning and gene organization. Eur J Cell Biol. 1993;62:214–223. [PubMed] [Google Scholar]
- Shoeman RL, Traub P. The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. J Biol Chem. 1990;265:9055–9061. [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalente-Alcalde D, Bhatt H, Anver M, Naryan B, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitzur N, Harel A, Goldberg M, Feinstein N, Gruenbaum Y. Nuclear membrane vesicle targeting to chromatin in a Drosophila embryo cell-free system. Mol Biol Cell. 1997;8:1439–1448. doi: 10.1091/mbc.8.8.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Simos G, Blobel G, Georgatos SD. Binding of lamin A to polynucleosomes. J Biol Chem. 1991;266:9211–9215. [PubMed] [Google Scholar]
- Zhao K, Harel A, Stuurman N, Guedalia D, Gruenbaum Y. Binding of matrix attachment regions to nuclear lamin is mediated by the rod domain and depends on the lamin polymerization state. FEBS Lett. 1996;380:161–164. doi: 10.1016/0014-5793(96)00034-8. [DOI] [PubMed] [Google Scholar]