Abstract
cDNA microarray technology allows the “profiling” of gene expression patterns for virtually any cellular material. In this study, we applied cDNA microarray technology to profile changes in gene expression associated with human prostate tumorigenesis. RNA prepared from normal and malignant prostate tissue was examined for the expression levels of 588 human genes. Four different methods for data normalization were utilized. Of these, normalization to ACTB expression proved to be the most rigorous technique with the least probability of producing spurious results. After normalization to ACTB expression, 15 of 588 (2.6%) genes examined by array analysis were differentially expressed by a factor of 2x or more in malignant compared to normal prostate tissues. The expression patterns for 8 of 15 genes have been reported previously in prostate tissues (TGFβ3, TGFBR3, IGFII, IGFBP2, VEGF, FGF7, ERBB3, MYC), but those of seven genes are reported here for the first time (MLH1, CYP1B1, RFC4, EPHB3, MGST1, BTEB2, MLP). These genes describe at least four metabolic and signaling pathways likely disrupted in human prostate tumorigenesis. Reverse transcriptase polymerase chain reaction (RT-PCR) and Northern blot analyses quantitated with reference to ACTB expression levels verified the trends in gene expression levels observed by array analysis for 14/15 and 8/8 genes, respectively. However, RT-PCR and Northern blot analyses accurately verified the “fold” differences in expression levels for only 6/15 (40%) and 7/8 (88%) of genes examined, respectively, demonstrating the need to better validate quantitative differences in gene expression revealed by array-based techniques.
Keywords: microarray, prostate, expression, RNA, cancer
Introduction
The imminent complete elucidation of the human transcriptome has necessitated the development of various technologies designed to examine gene expression in tissues, xenografts and cell lines. One of these technologies involves the construction of microarrays whereby cDNAs, ESTs or oligonucleotides are deposited onto nylon membranes or glass slides, then hybridized with radiolabeled or fluorescently labeled reverse-transcribed RNA to reveal the identities and relative expression levels of specific transcripts [1]. This technology permits the “profiling” of the gene expression pattern, or transcriptome, of virtually any source of cellular material. Transcriptome “profiling” of the changes in gene expression that occur throughout malignant progression would greatly elucidate the complex biology involved in human tumorigenesis. In this study, we applied array technology to profile the expression patterns of 588 genes in normal and malignant prostate tissues derived from the same surgical specimens. These experiments identified specific genes differentially expressed in malignant, compared to normal, prostate tissues, and implicated the involvement of at least four metabolic and signaling pathways in prostate tumorigenesis. We also examined the robustness of the array-derived data using different means to normalize gene expression levels across the array, and the validity of the array-derived data using reverse transcriptase polymerase chain reaction (RT-PCR) and Northern blot analysis to verify differential gene expression levels.
Materials and Methods
Tissue Acquisition and Characterization
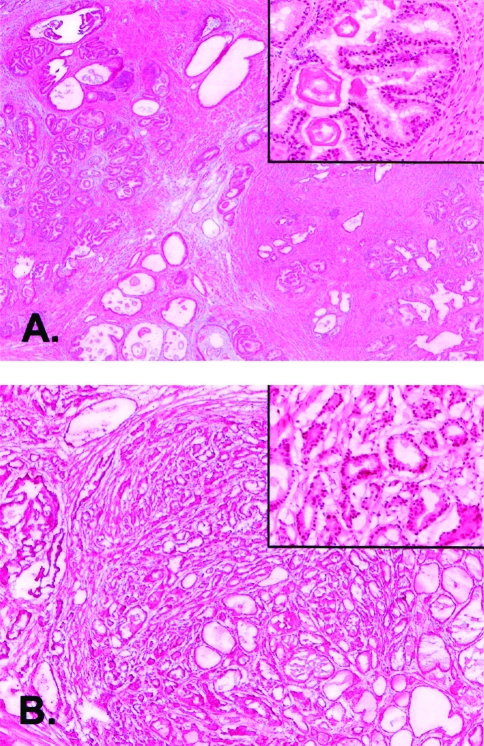
Prostate tissue was obtained after radical prostatectomy from a patient diagnosed with prostate cancer. After an initial pathologic evaluation of radical prostatectomy tissue, presumed malignant and normal tissues were snap-frozen in liquid nitrogen and stored at -70°C. One section each from normal and malignant tissues was examined after staining with hematoxylin/eosin. The tumor specimen was comprised of >80% malignant cells and the benign specimen comprised of an approximately equal admixture of normal epithelial and stromal cells (Figure 1). The specimen was acquired with full Institutional Review Board approval. The pathology of the prostate tumor was established as combined Gleason Score 6 (3+3), stage T2a, with focal involvement of the surgical margin.
Figure 1.
Histology of prostate tissue specimens. Photomicrographs of hematoxylin/eosin stained sections of normal benign (A) and malignant (B) human prostate tissues from the same radical prostatectomy specimens are shown at x50 and at x400 (inset) magnifications.
Array Analysis
RNA was purified from minced frozen tissue using Trizol reagent (Life Technologies, Inc., Rockville, MD). Total RNA was briefly treated with DNAse, then 4 µg each of total RNA from normal and malignant tissue was used to synthesize cDNA probes according to manufacturer's protocols (Clontech Atlas cDNA Expression Arrays User Manual PT3140-1). The Clontech Broad Coverage Human nylon arrays (#7740-1) were prehybridized in 15 ml Express Hyb and 1.5 mg denatured salmon testis DNA for 30 minutes at 68°C. The probes were denatured and added to the prehybridization mixtures. The filters were hybridized 16 hours at 68°C, then washed as directed by the manufacturer. Arrays were visualized on a Molecular Dynamics STORM phosphorimager and interpreted using the Clontech Atlas Image v1.0 software. The data were analyzed without normalization, with global normalization, and with normalization to ACTB or GAPDH expression.
RT-PCR Analysis
RT-PCR was carried out as described in the Atlas Array Custom Primers User Manual (Clontech, Palo Alto, CA). cDNA was prepared by reverse transcription using an oligo(dT) primer. PCR cycling conditions were as follows: one cycle of 94°C for 60 seconds; followed by 28 or 32 cycles (primer-dependent) of 94°C for 30 seconds, 68°C for 30 seconds, 72°C for 60 seconds, then 72°C for 5 minutes. PCR was carried out using primers specific for each gene. ACTB amplifications were carried as described for 22 cycles to avoid saturation of the reaction product. An aliquot of each reaction product was subjected to electrophoresis on 2% agarose gels and visualized after staining with ethidium bromide.
Northern Blot Analysis
Ten micrograms each of total RNA was electrophoresed through 1.2% agarose formaldehyde gels and transferred onto nylon membranes by capillary blotting overnight. Each blot was probed with radiolabeled PCR products amplified for the gene of interest. Blots were hybridized and washed as described in the Clontech ExpressHyb user manual.
Quantitation of Transcript Expression by RT-PCR and Northern Blot Analysis
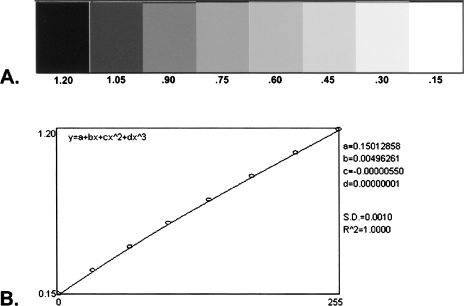
In order to properly quantitate separate RT-PCR and Northern blot experiments, the optical density of all reaction products on RT-PCR gels and hybridization signals on Northern blot autoradiographs were evaluated with reference to the same gray scale step tablet (see Figure 2). The step tablet was generated using a scale of 0.15 to 1.20. The value “0.15” was used rather than “0” to represent zero optical density in order to avoid the generation of irrational numbers for signal intensity ratios with “0” in the denominator (e.g., where no PCR reaction product or Northern blot hybridization signal was evident). RT-PCR and Northern blot autoradiographs were evaluated using Scion Image, the PC version of NIH Image, from Scion Corporation (http://www.scioncorp.com/), Frederick, MD. Use of the step tablet permitted the generation of a standard curve against which signal intensities of all RT-PCR and Northern blot analyses were measured (Figure 2). For RT-PCR gels, the signal intensities of all PCR products were obtained and the ratio values for normal/tumor pairs were determined for each transcript. The ratio values were averaged when two or more RT-PCR assays were accomplished to evaluate the same transcript.
Figure 2.
Step tablet and standard curve. (A) Eight-step gray scale step tablet and (B) standard curve generated from the step tablet shown in (A) used as a reference standard for the signal intensities of gene transcripts measured by RT-PCR and Northern blot analysis. The standard curve line equation and constant values are also shown in (B).
ACTB expression was assayed in triplicate by RT-PCR for 22 cycles. The ratio of ACTB expression in malignant compared to normal tissues was calculated as 1.270, 0.820 and 0.860, for an average value of 0.983. This ratio value was very close to 1.0, the value expected for equivalent expression of ACTB in normal and malignant tissues. The similarity in quantitative values for the expression levels of the 15 genes of interest, and the approximation of ACTB values to 1.0 between replicate experiments, suggested that normalization of the average expression values for each gene to ACTB expression was appropriate. Therefore, all average gene expression ratio values were multiplied by 1.017 (the inverse of 0.983) to normalize them against ACTB and to adjust the ACTB ratio itself to 1.0 (see Table 1). This normalization allowed the direct comparison of array-derived and RT-PCR derived data.
Table 1.
Gene Expression in Malignant Relative to Normal Prostate Tissues by Array, RT-PCR and Northern Blot Analysis.
| Gene Symbol | Primer Set* | Expression Pattern (by Array) | Comparison (RT-PCR, Array, Northern) | Clontech Array, T/N Normalized to ACTB¶ | Northern Blot T/N# | Northern Blot T/N for ACTB | Northern Blot T/Nx1/ACTB (Normalized to ACTB)** | Gene Description | |||||
| RT-PCR | |||||||||||||
| Ratio, T/N† | Ratio, T/N† | Ratio, T/N† | Average T/N‡ | Normalized to ACTB (x1.017)§ | |||||||||
| TGFB3 | 4 | ↓ in tumor | consistent | -2.745 | -2.422 | -2.583 | -2.627 | -4.801 | -4.240 | +0.890 | -4.759 | Transforming growth factor-beta 3 | |
| VEGF | 21 | ↓ in tumor | consistent | -6.939 | -6.939 | -6.937 | -2.618 | Vascular endothelial growth factor | |||||
| IGF2 | 20 | ↓ in tumor | consistent | -1.277 | -1.429 | -1.353 | -1.376 | -2.364 | -14.06 | +0.951 | -14.784 | Insulin-like growth factor II | |
| FGF7 | 23 | ↓ in tumor | consistent | -2.1079 | -2.108 | -2.144 | -2.028 | FGF7 | |||||
| MLH1 | 14 | ↓ in tumor | array>rt pcr | +1.593 | +1.376 | +1.485 | +1.510 | -2.133 | MutL (Escherichia coli) homolog 1 | ||||
| MLP | 8 | ↑ in tumor | consistent | +1.290 | +1.271 | +1.299 | +1.287 | +1.309 | +4.082 | +3.460 | +1.225 | +2.825 | MacMarcks |
| MYC | 1 | ↑ in tumor | consistent | +1.406 | +1.764 | +1.585 | +1.612 | +3.509 | +1.577 | +0.855 | +1.845 | c-Myc oncogene | |
| TGFBR3 | 2 | ↑ in tumor | consistent | +1.028 | +1.078 | +1.053 | +1.071 | +3.378 | Transforming growth factor beta receptor III | ||||
| BTEB2 | 15 | ↑ in tumor | consistent | +13.333 | +4.950 | +9.142 | +9.297 | +3.049 | +3.145 | +0.922 | +3.413 | Basic transcription element-binding protein 2 | |
| MGST1 | 11 | ↑ in tumor | consistent | +1.795 | +1.508 | +1.652 | +1.680 | +2.801 | +4.256 | +0.807 | +5.274 | Glutathiones S-transferase 12 | |
| IGFBP2 | 3 | ↑ in tumor | consistent | +1.250 | +1.580 | +1.415 | +1.439 | +2.747 | +2.238 | +0.857 | +2.610 | Insulin-like growth factor binding protein 2 | |
| EPHB3 | 6 | ↑ in tumor | consistent | +1.698 | +3.257 | +2.478 | +2.520 | +2.331 | Tyrosine kinase receptor | ||||
| RFC4 | 13 | ↑ in tumor | consistent | +6.849 | +6.849 | +6.965 | +2.294 | Replication factor C 37-kDa subunit | |||||
| ERBB3 | 16 | ↑ in tumor | consistent | +2.283 | +2.427 | +2.355 | +2.400 | +2.193 | +1.326 | +0.932 | +1.422 | Epidermal growth factor receptor; HER3 | |
| CYP1B1 | 7 | ↑ in tumor | consistent | +1.553 | +3.650 | +2.601 | +2.646 | +2.174 | Dioxin-inducible cytochrome P450 | ||||
| ACTB | 1.270 | 0.820 | 0.860 | 0.983 | 1.000 | Beta actin | |||||||
Identifiers for forward and reverse oligonucleotides used in RT-PCR experiments (consistent with those in Figure 3).
The ratio of the optical densities of RT-PCR products from malignant (T) and normal (N) tissues.
The average of all RT-PCR ratios for a particular gene.
The inverse (1.017) of the average (0.983) of the optical densities of RT-PCR products from malignant (T) and normal (N) tissues for the ACTB gene. All average RT-PCR values were multiplied by 1.017 in order to set T/N to 1.0 for ACTB, and to normalize all other average RT-PCR ratios to ACTB expression.
The ratio of expression levels of malignant (T) /normal (N) tissues for each gene after normalization to ACTB expression by array analysis.
The ratio of the optical densities of transcripts identified using Northern blot analysis for malignant (T) /normal (N) tissues for each gene.
The ratios in the preceding column (#) after multiplication by 1 /ACTB for the purpose of normalization to ACTB expression for each blot.
For Northern blot analysis, the hybridization signal intensities of each transcript were determined for normal and malignant tissues. The ratio of tumor/normal expression values for ACTB for the same blots was determined and ranged from 0.816 to 1.239. Therefore, these values were multiplied by their inverses to set the ACTB ratios to 1.0. The corresponding gene-specific ratio was also multiplied by the inverse ACTB expression ratio (1/ACTB) to normalize the data to ACTB expression (Table 1).
Results
RNA Profiling of Normal and Malignant Prostate Tissues by cDNA Array Analysis
Phosphorimages of array membranes hybridized with radiolabeled RNA prepared from normal or malignant tissues from a single radical prostatectomy specimen were analyzed using the AtlasImage version 1.0 software. We chose to perform the analysis using four different methodologies. The first method omitted any type of normalization function from the analysis so that the raw signal from both arrays was used to calculate signal intensity ratios. The second method utilized global normalization such that the average values of all the genes on the array were used to calculate a normalization coefficient, which was then applied across the array. The third and fourth methods utilized the expression levels of the abundantly and constitutively expressed GAPDH or ACTB genes, respectively, to calculate normalization coefficients, which were then applied across the arrays. These methodologies allowed the user to define specific values as criteria for differential expression. In all four cases, “downregulation” of gene expression in malignant compared to normal prostate tissues was defined when the ratio of tumor/normal gene expression was ≤0.5, for a -2.0x level of expression. “Upregulation” of gene expression in malignant compared to normal prostate tissues was defined when the ratio of tumor/normal gene expression was ≥2.0. The results of all four analyses are shown in Table 2.
Table 2.
Expression in Tumor Relative to Normal Human Prostate Epithelium.
| Gene Code | Not Normalized | Globally Normalized | Normalized to GAPDH | Normalized to ACTB | Gene Code | Not Normalized | Gene Code | Not Normalized |
| TGFβ3 | -8.000 | -5.144 | -6.000 | -4.801 | HSP27 | -2.753 | IGFBP3 | -2.149 |
| VEGF | -4.450 | -2.843 | -3.248 | -2.618 | CD44 | -2.689 | CLU | -2.137 |
| IGF2 | -4.000 | -2.600 | -2.972 | -2.364 | EPHA3 | -2.612 | IL6ST | -2.125 |
| MLH1 | -3.623 | -2.313 | -2.648 | -2.133 | BMP1 | -2.588 | NFIX | -2.114 |
| FGF7 | -3.442 | -2.209 | -2.509 | -2.028 | GSTT1 | -2.587 | SLC14A1 | -2.086 |
| RNH | -3.131 | -2.000 | -2.322 | ADRB2 | -2.550 | SCYA1 | -2.071 | |
| GSTP1 | -2.013 | -2.301 | BNIP3 | -2.500 | ITGB1 | -2.071 | ||
| DDIT3 | -3.050 | -2.260 | GSR | -2.478 | CDC25B | -2.071 | ||
| PRKM9 | -2.857 | -2.105 | GSTM1 | -2.418 | BST1 | -2.068 | ||
| IGF1R | -2.703 | -2.095 | ITGA6 | -2.394 | ILK | -2.067 | ||
| CASP7 | -2.840 | -2.088 | FGFR1 | -2.370 | CSBP1 | -2.053 | ||
| HSPB1 | -2.004 | CDKN1C | -2.333 | MNAT1 | -2.043 | |||
| PTPRG | -2.7370 | -2.000 | PRKDC | -2.307 | NFRKB | -2.032 | ||
| ERCC5 | -2.304 | HMR | -2.030 | |||||
| JUND | +2.000 | PDGFRA | -2.294 | PRKM6 | -2.029 | |||
| CYP1B1 | +2.174 | TNFRSF1A | -2.273 | PTN | -2.000 | |||
| ERBB3 | +2.012 | +2.193 | DDR1 | -2.235 | ||||
| RFC4 | +2.119 | +2.294 | CASP2 | -2.222 | ||||
| EPHB3 | +2.128 | +2.347 | CTNNB1 | -2.218 | ||||
| IGFBP2 | +2.531 | +2.212 | +2.747 | NR2F1 | -2.206 | |||
| MGST1 | -3.153 | +2.584 | +2.257 | +2.801 | DAP3 | -2.188 | ||
| BTEB2 | +2.793 | +2.457 | +3.049 | S100A9 | -2.182 | |||
| TGFBR3 | +3,098 | +2.174 | +3.378 | AR | -2.180 | |||
| MYC | +2.062 | +3.226 | +2.833 | +3.509 | CCND2 | -2.172 | ||
| MLP | +2.398 | +3.759 | +3.320 | +4.082 | POR | -2.160 | ||
Analysis Without Normalization
The omission of any normalization methodology produced the largest list of differentially expressed genes, with 52 genes downregulated, and only two genes upregulated, in malignant compared to normal prostate tissues. Therefore, a total of 54/588 genes, or 9.2% of all transcripts examined, demonstrated differential expression at a two-fold or higher level in malignant compared to normal human prostate tissues. The gene transcripts downregulated in malignant compared to normal tissues included TGFβ3 (-8x), VEGF (-4.5x), IGF2 (-4x), MLH1 (-3.6x) and FGF7 (-3.4x), whereas the two upregulated transcripts were MYC (+2.0) and MLP (+2.4x) (where “x” designates “fold difference”).
Analysis With Global Normalization
Global normalization across all genes on the array revealed seven downregulated and 10 upregulated transcripts in malignant compared to normal tissues, for a total of 17/588, or 2.9%, of all transcripts examined. The five genes most dramatically downregulated in expression were identical to those previously revealed without prior normalization. The two genes identified as upregulated without prior normalization, MYC and MLP, were also identified as upregulated using global normalization. Other genes also dramatically upregulated in malignant compared to normal tissues included TGFBR3 (+3.0x), BTEB2 (+2.8x) and MGST1 (+2.6x) (Table 2).
Analysis with Normalization to GAPDH
Normalization of all genes to expression of the constitutively and abundantly expressed GAPDH gene revealed 13 genes that were downregulated, and six that were upregulated, in malignant compared to normal tissues, for a total of 19/588, or 3.2%, of all transcripts examined. Again, the same five genes found to be downregulated without prior normalization or with prior global normalization were found to be downregulated by factors of -6.0x (TGF/β3) to -2.5x (FGF7). In addition, eight other genes were identified as downregulated, including five previously identified without prior normalization (DDIT3, PRKM9, IGF1R, CASP7 and RNH), one previously identified using global normalization (GSTP1) and two genes not classified as downregulated by prior analysis (HSPB1 and PTPRG). Normalization to GAPDH also identified six genes upregulated in malignant tissues by factors of +2.2x to +3.3x, all of which were previously identified using no or global normalization analyses (Table 2).
Analysis with Normalization to ACTB
Normalization of all genes to expression of the constitutively and abundantly expressed ACTB gene revealed five genes that were downregulated, and 10 that were upregulated, in malignant compared to normal tissues, for a total of 15/588, or 2.6%, of all transcripts examined. The downregulated genes identified using this analysis were limited to five genes previously identified by other analyses. All 10 genes found to be upregulated in expression in malignant compared to normal tissues were previously identified by other analyses, with the exception of CYP1B1 (+2.2x) (Table 2).
Comparison of Normalization Techniques
Based upon the number of transcripts identified as differentially expressed, the four techniques used to normalize the array data could be ordered by the number of genes identified as differentially expressed as follows: ACTB (15 genes) >Global (17 genes) >GAPDH (19 genes) >None (54 genes). By these criteria, normalization to ACTB proved to be the most rigorous normalization technique with the least probability of producing spurious results. Therefore, all subsequent verification analyses were performed so that transcript expression levels were normalized to ACTB expression (Table 2).
Although the four array normalization methodologies differed in the number of genes defined as down- or upregulated in expression in malignant compared to normal prostatic tissues, all identified the same five downregulated genes (TGFβ3, VEGF, IGF2, MLH1 and FGF7) and the same two upregulated genes (MYC and MLP). This finding suggested that at least these seven genes were clearly differentially expressed in normal and malignant prostatic tissues. To test this hypothesis, two other techniques — RT-PCR and Northern Blot analysis — were performed to determine whether the array results could be verified using independent techniques to measure gene expression.
Verification of Array Results by RT-PCR
RT-PCR was performed to examine the expression levels of all 15 genes identified as differentially expressed by array analysis after normalization to ACTB. All 15 transcripts were reverse-transcribed from RNA template purified from the same malignant and normal prostatic tissues utilized in the array analyses, then amplified with sequence-specific primers (Figure 3). These reactions were repeated for 12 of the 15 transcripts and a third time for the MLP gene transcript. ACTB was also assayed in each amplification set to provide a means for normalization.
Figure 3.
RT-PCR validation of array-derived data. RT-PCR products for transcripts of the 15 genes identified as differentially expressed by array analysis (1–23) and the ACTB gene amplified from the same normal and malignant tissues examined by array analysis are shown after electrophoresis through 2% agarose and visualization with ethidium bromide staining. The transcripts are identified as follows: (1) MYC; (2) TFGBR3; (3) IGFBP2; (4) TGF/33; (6) EPHB3; (7) CYP1B1; (8) MLP; (11) MGST1; (13) RFC4; (14) MLH1; (15) BTEB2; (16) ERBB3; (20) IGF2; (21) VEGF; (23) FGF7. Mw indicates the 100-bp ladder molecular weight marker.
After quantitative assessment of reaction product yields, the ratio of transcript levels in normal and malignant prostate tissues was calculated and the averages were taken. As seen in Table 1, the ratio of transcript expression levels for malignant/normal tissues differed by less than 2x after repeated measures for 10/12 genes, with replicate measures differing by <10% for 8/12 genes. However, 2/12 genes, BTEB2 and CYP1B1, differed in expression levels between replicate measures by >2x. For BTEB2, the RT-PCR assays clearly demonstrated dramatic upregulation in malignant compared to normal tissues, by 13x in one reaction set and by 5x in the other. In the case of CYP1B1, both measures again demonstrated upregulation in malignant compared to normal tissues, by 1.6x in one reaction set and by 3.7x-fold in the other.
As may be seen in Table 1, the trends toward up- or downregulation in gene expression levels observed by array analysis after normalization to ACTB were consistent with those observed by RT-PCR after normalization to ACTB for 14/15 genes. These results suggested that the array-derived data were largely reliable and verifiable by other means. However, the actual quantitative values associated with these trends differed by >2x between array-derived and RT-PCR derived data for 8/13 genes, disallowing attempts to correlate “fold differences” in transcript expression levels obtained using the two methodologies. It should be noted that differences of less than two-fold were observed for the TGFβ3, IGF2, FGF7, EPHB3, ERBB3 and CYP1B1 genes by array and RT-PCR analysis. This group included three genes — TGFβ3, IGF2 and FGF7 — consistently identified as downregulated in malignant compared to normal prostate tissue by all four array normalization methodologies, reinforcing the identity of these genes as differentially expressed. The one exception to the observed consistency in expression “trend” between array-derived and RT-PCR data was MLH1, which was downregulated in tumor by array analysis (-2.1x) but upregulated by RT-PCR analysis ( + 1.5x), though the fold differences in both cases were near 2x, very close to the criteria used to define differential expression.
Verification of Array Results by Northern Blot Analysis
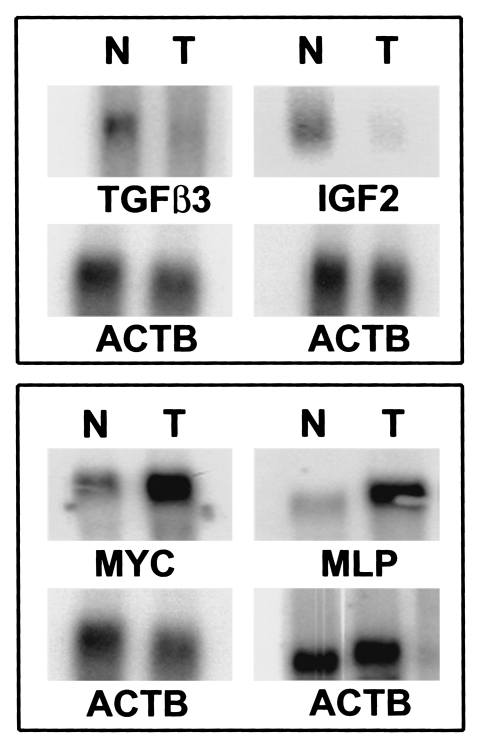
The RT-PCR experiments largely corroborated the trends in gene expression observed by array analysis for malignant and normal prostate tissues. However, these experiments had not corroborated the actual fold differences in differential gene expression. Therefore, another quantitative technique for the measurement of transcript expression, Northern blot analysis, was utilized to determine whether the actual fold differences in differential gene expression could be verified. It was possible to examine the expression levels of 8/15 genes with the amount of RNA still available from the prostate tissues, including two genes downregulated (TGFβ3, IGF2) and six genes upregulated (MLP, MYC, BTEB2, MGST1, IGFBP2 and ERBB3) in malignant compared to normal prostate tissues. These analyses were carried out on separate Northern blots, and each blot was subsequently hybridized to a probe for ACTB to allow normalization to the same “control” gene used for the array and RT-PCR experiments (Figure 4). As may be seen in Table 1 from the normalized data, identical expression “trends” were maintained for all eight genes examined, and gene expression levels differed by <2x, between the array and Northern blot data for the TGFβ3, MLP, MYC, BTEB2, MGST1, IGFBP2 and ERBB3 genes. Therefore, quantitation of array-derived fold differences in gene expression levels was verified for 88% (7/8) of genes examined by Northern blot analysis compared to 40% (6/15) of genes examined by RT-PCR. However, downregulation of IGF2 expression in malignant compared to normal prostate tissues was quantitated at -2.4x by array and at -14x by Northern blot analysis. This suggests that although the “trend” in gene expression may be defined reliably by array or Northern data, quantitation of the actual fold difference in differential expression may be problematic in some cases.
Figure 4.
Northern blot validation of array-derived data. Ten micrograms each of total RNA purified from the same normal and malignant tissues examined by array analysis was electrophoresed through 1.2% agarose formaldehyde gels, blotted and hybridized as described in the text with probes for the TGFβ3, IGF2, MYC, MLP and ACTS genes, as shown.
Discussion
Verification of Array-Derived Data
The normalization of array, RT-PCR and Northern blot data to the same internal standard, ACTB gene expression, permitted a direct comparison of the abilities of all three methods to quantitate gene expression. Using this systematic approach, we were able to demonstrate that array-derived data are verifiable using other means to detect and quantify transcript levels. Specifically, the “trends” toward up- or downregulation in gene expression in malignant compared to normal prostate tissues observed from the array date for the expression of specific genes were also observed using either RT-PCR (14/15 genes) or Northern blot (8/8 genes) analyses. However, the quantitation of transcript levels observed by array analysis was replicated most accurately by Northern blot analysis, and differed by less than two-fold for 7/8 genes examined. This can be compared to conventional RT-PCR analysis, where transcript levels differed by less than two-fold from those observed by array analysis for only 6/15 genes examined. It is possible that conventional RT-PCR methodologies, even those using normalization to an internal standard such as the experiments reported here, lack the degree of sensitivity and reproducibility required to exactly quantify transcript levels. If so, then other RT-PCR methodologies, including real-time PCR, may provide a more accurate means to measure transcript levels, hence produce results more consistent with those obtained using array technology.
Specific Genes Involved in Prostate Tumorigenesis Identified by Array Analysis
After normalization to ACTB expression, 15 of 588 genes examined by array analysis were differentially expressed by a factor of 2x or more in malignant compared to normal prostate tissues. Of the 15 genes differentially expressed in malignant compared to normal prostate tissues, eight have been reported previously (TGFβ3, TGFBR3, IGFII, IGFBP2, VEGF, FGF7, ERBB3, MYC), and seven are reported here for the first time (MLH1, CYP1B1, RFC4, EPHB3, MGST1, BTEB2, MLP).
TGFβ3 pathway The ACTB-normalized array data demonstrated downregulation of TGFβ3 gene expression by -4.8x, and upregulation of TGFβ receptor III gene expression by +3.4x, in malignant compared to normal prostate tissues. These results are consistent with those of other studies reporting that TGFβ3 mRNA is downregulated in prostate carcinoma compared to normal prostate. Moreover, picomolar quantities of TGFβ3 inhibit the proliferation of cultured stromal and epithelial prostate cells; conversely, proliferating cells fail to express TGFβ3 [2,3]. Functionally, TGFβ3 normally mediates growth inhibition. Therefore, downregulation of TGFβ3 expression is permissive for growth and is associated with the malignant phenotype [4]. In addition, upregulation of the TGFβ receptor III may comprise a cellular response to reduced TGF/33 levels, though other studies have failed to examine this possibility in prostatic or other neoplasms.
IGF pathway Insulin-like growth factor 2 (IGF II) was downregulated in malignant compared to normal prostate tissues by -2.4 x, whereas insulin-like growth factor binding protein 2 (IGFBP2) was upregulated by +2.7x from the ACTB-normalized array data. Upregulation of IGFBP2 in malignant prostate tissues has been reported previously [5]. However, examination of IGFII expression in prostatic tissues has proven more problematic. IGFII is secreted by stromal cells and is mitogenic for both stromal and epithelial cells in the prostate [6,7]. Therefore, the apparent downregulation of IGFII transcript in malignant compared to normal prostatic tissues may actually be a function of the percent of stromal cells present in each tissue type. As seen in the hematoxylin/eosin stained tissue sections shown in Figure 1, the tumor tissue was almost entirely epithelial in cell type, whereas the normal benign tissue was approximately 50% epithelial and 50% stromal cells. Therefore, it is possible that the larger stromal compartment in the normal tissue is responsible for the apparent upregulation of IGFII transcript in normal tissue.
Mismatch repair pathways and DNA damage repair pathways Both the RFC4 and MLH1 genes are involved in mismatch repair in eukaryotic cells. The expression profiles of these genes in malignant and normal prostatic tissues have not been previously reported. RFC4 encodes the p70 subunit of the five-subunit replication factor C multimeric protein complex. Together, the five RFC subunits are required to “load” PCNA (proliferating cell nuclear antigen) onto DNA, thus recruit DNA polymerases to the site of DNA replication [8]. The ACTB-normalized array data showed that the level of RFC4 gene transcript was upregulated by +2.3x in malignant compared to normal prostatic tissues, suggesting that RFC4 functions may be enhanced in prostate tumors. The MLH1 gene, which also functions in mismatch repair, encodes a protein involved in the G2-M cell cycle checkpoint [9]. Mutations of the gene have been associated with the microsatellite instability phenotype in several types of cancer, and transcription of the gene is downregulated via promoter methylation in some tumors [10,11]. Though not previously described for prostate cancer, we found that MLH1 transcription was downregulated -2.3x in malignant compared to normal prostatic tissues, suggesting that MLH1 activity may be reduced in prostate tumors. Interestingly, both the RFC4 and MLH1 gene products are part of the putative BASC, or BRCA1-Associated Surveillance Complex, a group of proteins that physically associate with the BRCA1 protein. Other proteins within this complex include MSH2, MSH6; ATM, BLM; and the RAD50-MRE11-NBS1 protein complex, and function in the recognition of damaged DNA or abnormal DNA structures, or in DNA replication-associated repair [12]. These data suggest that the dysfunction of mismatch repair pathways contributes to tumorigenesis in the prostate.
The transcript of the MLP gene (MacMARCKS, or MARCKS Related Protein) was upregulated +4x in malignant compared to normal prostate tissues by array data. Upregulation of the MLP gene transcript has not been described previously in human prostate cancer. The MLP protein is associated with the cell membrane and actin cytoskeleton. It is a substrate for protein kinase C and also binds calmodulin in a manner that is regulated by phosphorylation of PKC. Due to these activities, MLP is thought to mediate cross talk between calmodulin and PKC signal transduction pathways. However, Poly(ADP)-ribose binding to MARCKS and MacMARCKS inhibits the ability of these proteins to bind to PKC or calmodulin, thus inhibiting membrane association and actin filament formation. This suggests that MARCKS proteins (and actin) could be targets of the Poly(ADP-ribose) DNA damage signal pathway, and that these proteins, along with other Poly(ADP)-binding proteins, such as p21, p53, and Ku70/86, may participate in a “DNA Break Signal Mechanism [13].” In this mechanism, poly(ADP-ribose) polymerase (PARP) signals DNA breaks and produces PARP-bound polymers, e.g., poly(ADP-ribose), at the site of a DNA break. These polymers then recruit proteins that have polymer recognition/binding sites to the DNA breakage site, which allows these proteins to participate in DNA strand repair activities. PARP then degrades the polymers, which releases the polymer-bound proteins [14]. It is possible that increased levels of MLP in malignant tissues may act to bind up and essentially sequester all or most of the Poly(ADP-ribose) polymers as they are synthesized by PARP. If so, MLP would render Poly(ADP-ribose) unavailable for binding to other DNA Break Signal Mechanism proteins, thus interfering with DNA repair activities and contributing to expression of the transformed phenotype.
Oxidative stress response pathway(s) The transcripts of two genes involved in oxidative stress response pathways, MGST1 and CYP1B1, were upregulated in malignant prostate tissue compared to normal tissues. The promoter region of the MGST1 upstream of the dominant first exon responds to oxidative stress as a detoxification mechanism [15]. MGST1 may also be involved in cytoprotection; as a close homologue, MST1-L1 is involved in redox regulation and is upregulated in response to p53 expression [16]. CYP1B1 is a dioxin-inducible cytochrome P450 enzyme that catalyzes oxidative metabolism of many compounds, including xenobiotic procarcinogens [17]. It is not expressed by most normal tissues and is upregulated in many cancers, including those of the bladder, brain, breast, colon, esophagus lung, ovary, skin, stomach, lymph node, testis and uterus [18]. This is the first report of MGST1 and CYP1B1 upregulation in malignant prostate tissue, suggesting that upregulation of oxidative stress response pathways occurs during prostate tumorigenesis.
Growth factors, growth factor receptors and transcription factors Two growth factors downregulated in malignant tissues by array analysis, VEGF (-2.6x) and FGF7 (-2.0x), have been studied previously in prostate cancer. Several studies have reported overexpression of VEGF in prostate cancer cells [19,20]. However, another recent study found that normal prostate cells secrete VEGF and other angiogenic cytokines, suggesting that the ability of prostate cells to stimulate angiogenesis is an intrinsic trait, rather than one that must be acquired during tumorigenesis [21]. The apparent down regulation of VEGF in malignant compared to normal prostate tissue was observed in both the array and RT-PCR experiments, suggesting that malignant cells were not expressing VEGF at higher levels than normal cells. FGF7 was also apparently downregulated in malignant compared to normal prostate tissue. FGF7 is expressed by stromal cells in the prostate and is mitogenic for epithelial cells [22,23]. Therefore, the apparent downregulation of FGF7 expression in malignant prostate tissue observed by array and RT-PCR analysis may actually reflect differences in cell type composition, as the normal prostate tissue used in these experiments was composed of both epithelial and stromal cells, whereas the malignant tissue was predominantly epithelial. It is likely that expression of FGF7 by the stromal cell compartment of the normal prostate tissue is responsible for the higher levels of FGF7 observed in normal compared to malignant prostate tissues.
Other growth factors and growth factor receptors upregulated in malignant compared to normal prostate tissues by array analysis included MYC, ERBB3 and EPHB3. Upregulation of MYC gene transcription has been associated with immortalization and malignant transformation in prostate and other cancers [24,25]. Array, RT-PCR and Northern blot analysis all demonstrated upregulation of MYC transcript in malignant compared to normal prostate tissues, consistent with previous reports. Array analysis also showed that ERBB3 and EPHB3 transcripts were upregulated in malignant compared to normal prostate tissues. Both ERBB3 and EPHB3 encode growth factor receptors with tyrosine kinase activities. Whereas ERBB3 encodes both secreted and transmembrane forms due to alternative splicing, EPHB3 occurs only as a transmembrane molecule [26,27]. ERBB3 is expressed constitutively in LNCaP prostate cancer cells and is phosphorylated in response to IL-6 stimulation [28]. However, the current study is the first report to examine ERBB3 expression in normal and malignant prostate tissues. The EPHB3 gene encodes a tyrosine protein kinase receptor, EPH-3, that is part of a large 14-member Eph family. Ephrin receptors are expressed prominently in the developing adult nervous system, suggesting roles in neural development or function. Their ligands, “ephrins,” all have close sequence homology and are also membrane-associated [29]. Ephrin receptor and ligand transcripts have been observed in small cell lung cancer cell lines and tumors, suggesting that these molecules are involved in autocrine loops during development and tumorigenesis [30]. This is the first report of ephrin receptor overexpression in malignant prostate tissues. However, it should be noted that overexpression of other ephrin receptors (EPHA1, EPHA3) and ligands (EPLG3, EPLG4) on the array was not observed and that the functional significance of EPHB3 upregulation in prostate tumors is not known.
Expression of the basic transcription element-binding protein 2, BTEB2, was upregulated 3.1 x by array, 3.3x by Northern blot and 6.9x by RT-PCR analysis in malignant compared to normal prostate tissues. BTEB2 encodes a zinc-finger protein that is normally expressed in vascular smooth muscle cells, often is response to injury. The promoter of the BTEB2 gene is activated by bFGF and EGF1; thus, BTEB2 expression is regulated by the MAP kinase pathway [31]. With MYC and EGR1, BTEB2 expression is upregulated in keratinocyte cells exposed to factor Vila, suggesting that BTEB2 may play a role in the epithelial proliferative aspect of wound healing [32]. Functional studies of BTEB2 expression in prostate cells would be required to ascertain whether BTEB2 upregulation is merely reflective of, or directly involved in, the proliferative state of malignant prostate epithelium.
Functional Significance of Differential Gene Expression in Malignant Prostate Tissues
Few other studies have attempted to profile prostate tissue expression patterns using array technology. Bubendorf et al. [5] recently reported the results of a study profiling the RNA expression patterns of a hormone-dependent (CWR22) and derivative-independent (CWR22R) xenograft of a human prostate tumor. Using a cut-off of two-fold or higher differences in expression levels to define transcriptional up- or downregulation, they report differential expression of 3.3% of genes examined, comparable to the 2.6% reported here. Gene expression levels were not normalized to those of housekeeping genes on the arrays (ACTB or GAPDH), but were determined using a methodology similar to that of global normalization. Few genes defined as differentially expressed in Bubendorf et al. were also defined as differentially expressed in the current study. This is likely due to the difference in tissues profiled in the two studies since the CWR22 xenograft was derived from a high grade, metastatic tumor whereas the malignant prostate tissue profiled in the current study was moderately differentiated and localized to the prostate. In another study, Vaarala et al. [33] utilized array analysis to profile the transcript expression patterns of the androgen-responsive LNCaP cultured malignant prostate cell line and an androgen-insensitive derivative. The data were not normalized, and comparison of these data with those of Bubendorf et al. or the current study reveals few similarities. This, again, may be due to the inability to directly compare the RNA expression profiles of a diverse set of tissues and cell lines. Elek et al. examined the RNA expression profiles of one normal and three malignant human prostate tissues using the same cDNA microarray used in the current study. They observed that 19/588, or 3.2% of genes examined, were differentially expressed between normal and malignant prostate, similar to the 2.6% observed in the current study. However, the data were analyzed visually without background subtraction or normalization, making comparison with the current study difficult [34]. These types of difficulties encountered when attempting to compare array-derived datasets should be eliminated as data analysis methods utilized become more widespread and standardized.
Conclusion
The results reported here demonstrate that cDNA microarray methodology provides a means to identify specific genes and functional cellular pathways likely disrupted in human prostate tumorigenesis. They also suggest that the accurate profiling of differential gene expression using cDNA microarrays requires further validation through other means, such as RT-PCR or Northern blot analysis. Moreover, the normalization of cDNA microarray-derived gene expression levels to one or more constitutively expressed genes (e.g., ACTB or GAPDH) provides a more rigorous methodology for the evaluation of differential gene expression by array analysis, and also provides standards by which array-derived data may be validated by other means. The consistent use of appropriate data normalization strategies should enable both the validation and comparison of multiple cDNA microarray-based studies and an accurate depiction of gene expression changes during prostate tumorigenesis.
Abbreviations
- RT-PCR
reverse transcriptase polymerase chain reaction
Footnotes
This work was supported by the NIH/NCI 1P50 CA69568 SPORE in Prostate Cancer (JAM., M.A.R.).
References
- 1.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JF. Expression profiling using cDNA microarrays. Nat Gen. 1999;21:10–14. doi: 10.1038/4434. supplement. [DOI] [PubMed] [Google Scholar]
- 2.Story MT, Hopp KA, Molter M. Expression of transforming growth factor beta 1 (TGF beta1), -beta 2, and -beta 3 by cultured human prostate cells. J Cell Phys. 1996;169:97–107. doi: 10.1002/(SICI)1097-4652(199610)169:1<97::AID-JCP10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 3.Djonov V, Ball RK, Graf S, Mottaz AE, Arnold AM, Flanders K, Studer UE, Merz VW. Transforming growth factor-beta 3 is expressed in nondividing basal epithelial cells in normal human prostate and benign prostatic hyperplasia, and is no longer detectable in prostate carcinoma. Prostate. 1997;3:103–109. doi: 10.1002/(sici)1097-0045(19970501)31:2<103::aid-pros5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 4.Lucia MS, Sporn MB, Roberts AB, Stewart LV, Danielpour D. The role of transforming growth factor-beta1, -beta2, and -beta3 in androgen-responsive growth of NRP-152 rat prostatic epithelial cells. J Cell Phys. 1998;175:184–192. doi: 10.1002/(SICI)1097-4652(199805)175:2<184::AID-JCP8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 5.Bubendorf L, Kolmer M, Kononen J, Koivisto P, Mousses S, Chen Y, Mahlamaki E, Schraml P, Moch H, Willi N, Elkahloun AG, Pretlow TG, Gasser TC, Mihatsch MJ, Sauter G, Kallioniemi OP. Hormone therapy failure in human prostate cancer: analysis by complementary DNA and tissue microarrays. J Natl Cancer Inst. 1999;91:1758–1764. doi: 10.1093/jnci/91.20.1758. [DOI] [PubMed] [Google Scholar]
- 6.Grant ES, Ross MB, Ballard S, Naylor A, Habib FK. The insulin-like growth factor type I receptor stimulates growth and suppresses apoptosis in prostatic stromal cells. J Clin Endocrinol Metab. 1998;83:3252–3257. doi: 10.1210/jcem.83.9.5119. [DOI] [PubMed] [Google Scholar]
- 7.Boudon C, Rodier G, Lechevallier E, Mottet N, Barenton B, Sultan C. Secretion of insulin-like growth factors and their binding proteins by human normal and hyperplastic prostatic cells in primary culture. J Clin Endocrinol Metab. 1996;81:612–617. doi: 10.1210/jcem.81.2.8636277. [DOI] [PubMed] [Google Scholar]
- 8.Mossi R, Hubscher U. Clamping down on clamps and clamp loaders — the eukaryotic replication factor C. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- 9.Davis TW, Wilson Van-Patten C, Meyers M, Kunugi KA, Cuthill S, Reznikoff C, Garces C, Boland CR, Kinsella TJ, Fishel R, Boothman DA. Defective expression of the DNA mismatch repair protein, MLH1, alters G2-M cell cycle checkpoint arrest following ionizing radiation. Cancer Res. 1998;58:767–768. [PubMed] [Google Scholar]
- 10.Fleisher AS, Esteller M, Harpaz N, Leytin A, Rashid A, Xu Y, Liang J, Stine OC, Yin J, Zou TT, Abraham JM, Kong D, Wilson KT, James SP, Herman JG, Meltzer SJ. Microsatellite instability in inflammatory bowel disease-associated neoplastic lesions is associated with hypermethylation and diminished expression of the DNA mismatch repair gene, hMLH1. Cancer Res. 2000;60:4864–4868. [PubMed] [Google Scholar]
- 11.Esteller KM, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14(8):927–939. [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz AA, Pleschke JM, Kleczkowska HE, Althaus FR, Vergeres G. Poly(ADP-ribose) modulates the properties of MARCKS proteins. Biochemistry. 1998;37(26):9520–9527. doi: 10.1021/bi973063b. [DOI] [PubMed] [Google Scholar]
- 14.Althaus FR, Kleczkowska HE, Malanga M, Muntener CR, Pleschke JM, Ebner M, Auer B. Poly ADP ribosylation: a DNA break signal mechanism. Mol Cell Biochem. 1999;193(1–2):5–11. [PubMed] [Google Scholar]
- 15.Kelner MJ, Bagnell RD, Montoya MA, Estes LA, Forsberg L, Morgenstern R. Structural organization of the microsomal glutathione S-transferase gene (MGST1) on chromosome 12p13,1-13,2. Identification of the correct promoter region and demonstration of transcriptional regulation in response to oxidative stress. J Biol Chem. 2000;275:13000–13006. doi: 10.1074/jbc.275.17.13000. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsson PJ, Morgenstern R, Mancini J, Ford-Hutchinson A, Persson B. Membrane-associated proteins in eicosanoid and glutathione metabolism (MAPEG), A widespread protein superfamily. Am J Respir Crit Care Med. 2000;161:S20–S24. doi: 10.1164/ajrccm.161.supplement_1.ltta-5. [DOI] [PubMed] [Google Scholar]
- 17.Kaminsky LS, Spivack SD. Cytochromes P450 and cancer. Mol Aspects Med. 1999;20:70–84. [PubMed] [Google Scholar]
- 18.Murray GI, Taylor MC, McFadyen MC, McKay JA, Greenlee WF, Burke MD, Melvin WT. Tumor-specific expression of cytochrome P450 CYP1B1. Cancer Res. 1997;57:3026–3031. [PubMed] [Google Scholar]
- 19.Chen HJ, Treweeke AT, Ke YQ, West DC, Toh CH. Angiogenically active vascular endothelial growth factor is overexpressed in malignant human and rat prostate carcinoma cells. Br J Cancer. 2000;82:1694–1701. doi: 10.1054/bjoc.2000.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harper ME, Glynne-Jones E, Goddard L, Thurston VJ, Griffiths K. Vascular endothelial growth factor (VEGF) expression in prostatic tumours and its relationship to neuroendocrine cells. Br J Cancer. 1996;74:910–916. doi: 10.1038/bjc.1996.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell CL, Savarese DM, Quesenberry PJ, Savarese TM. Expression of multiple angiogenic cytokines in cultured normal human prostate epithelial cells: predominance of vascular endothelial growth factor. Int J Cancer. 1999;80:868–874. doi: 10.1002/(sici)1097-0215(19990315)80:6<868::aid-ijc12>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Story M, Hopp K, Molter M, Meier D. Characteristics of FGF receptors expressed by stromal and epithelial cells cultured from normal and hyperplastic prostates. Growth Factors. 1994;10:269–280. doi: 10.3109/08977199409010993. [DOI] [PubMed] [Google Scholar]
- 23.Giri D, Ropiquet F, Ittmann M. Alterations in expression of basic fibroblast growth factor (FGF) 2 and its receptor FGFR-1 in human prostate cancer. Clin Cancer Res. 1999;5:1063–1071. [PubMed] [Google Scholar]
- 24.Fuhrmann G, Rosenberger G, Grusch M, Klein N, Hofmann J, Krupitza G. The MYC dualism in growth and death. Mutat Res. 1999;437:205–217. doi: 10.1016/s1383-5742(99)00084-8. [DOI] [PubMed] [Google Scholar]
- 25.Latil A, Vidaud D, Valéri A, Fournier G, Vidaud M, Lidereau R, Cussenot O, Biàche I. htert expression correlates with MYC overexpression in human prostate cancer. Int J Cancer. 2000;89:172–176. [PubMed] [Google Scholar]
- 26.Katoh M, Yazaki Y, Sugimura T, Terada M. c-erbB3 gene encodes secreted as well as transmembrane receptor tyrosine kinase. BBRC. 1993;192:1189–1197. doi: 10.1006/bbrc.1993.1542. [DOI] [PubMed] [Google Scholar]
- 27.Bombe B, VandenBos T, Cerretti DP, Park LS, Holtrich U, Rubsamen-Waigmann H, Strebhardt K. Cell-cell adhesion mediated by binding of membrane-anchored ligand LERK-2 to the EPH-related receptor human embryonal kinase 2 promotes tyrosine kinase activity. J Biol Chem. 1996;271:24747–24752. doi: 10.1074/jbc.271.40.24747. [DOI] [PubMed] [Google Scholar]
- 28.Qiu Y, Ravi L, Kung HJ. Requirement of ErbB2 for signalling by interleukin-6 in prostate carcinoma cells. Nature. 1998;393:83–85. doi: 10.1038/30012. [DOI] [PubMed] [Google Scholar]
- 29.Bergemann AD, Zhang L, Chiang MK, Brambilla R, Klein R, Flanagan JG. Ephrin-B3, a ligand for the receptor EphB3, expressed at the midline of the developing neural tube. Oncogene. 1998;16:471–480. doi: 10.1038/sj.onc.1201557. [DOI] [PubMed] [Google Scholar]
- 30.Tang XX, Brodeur GM, Campling BG, Ikegaki N. Coexpression of transcripts encoding EPHB receptor protein tyrosine kinases and their ephrin-B ligands in human small cell lung carcinoma. Clin Cancer Res. 1999;5:455–460. [PubMed] [Google Scholar]
- 31.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ Res. 1999;85:787–795. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 32.Camerer E, Gjernes E, Wiiger Mn, Pringle S, Prydz H. Binding of factor VIIa to tissue factor on keratinocytes induces gene expression. J Biol Chem. 2000;275:6580–6585. doi: 10.1074/jbc.275.9.6580. [DOI] [PubMed] [Google Scholar]
- 33.Vaarala MH, Porvari K, Kyllonen A, Vihko P. Differentially expressed genes in two LNCaP prostate cancer cell lines reflecting changes during prostate cancer progression. Lab Invest. 2000;80:1259–1268. doi: 10.1038/labinvest.3780134. [DOI] [PubMed] [Google Scholar]
- 34.Elek J, Park KH, Narayan R. Microarray-based expression profiling in prostate tumors. In Vivo. 2000;14:173–182. [PubMed] [Google Scholar]