Abstract
The gene encoding human spectrin Src homology domain binding protein 1, or Hssh3bp1, which is a marker of macropinocytic vesicles and a potential regulator of macropinocytosis, co-localizes to a YAC containing chromosome 10p sequences at loci D10S89 and D10S111 that are frequently deleted in prostate tumors. Expression of Hssh3bp1 was evaluated at the protein level in 17 paired normal and malignant prostate tumor samples using the monoclonal antibody 2G8 to Hssh3bp1. These experiments demonstrated that 4/6 tumors (67%) with 10p deletion failed to express Hssh3bp1 protein compared to 5/11 (46%) tumors with intact 10p. Thus, loss of Hssh3bp1 expression is concordant with allelic loss of adjacent 10p sequences in human prostate tumors. In addition, two prostate tumor cell lines contain an exon skipping mutation in the Hssh3bp1 gene that leads to the abnormal splicing of the mRNA and loss of a portion of Abl tyrosine kinase SH3 domain binding site in the protein. These data are consistent with a role for Hssh3bp1 as a candidate tumor suppressor gene inactivated during prostate tumorigenesis.
Keywords: spectrin, Src homology domain, tumorigenesis, tyrosine kinase, prostate cancer
Introduction
The deletion of specific chromosomal regions has been reported in human prostatic tumors. For chromosome 10, both the 10p and 10q arms have been reported as frequently deleted. Deletions on 10q often involve the 10q23–24 region, including sequences mapped to the candidate prostate tumor suppressor gene, PTEN [1] or MMAC1 [2] (reviewed in Refs. [3,4]). Loss of heterozygosity (LOH) on the short arm of chromosome 10, 10p, has also been observed in prostate tumors. Several studies performed using polymorphic markers indicated high rates of LOH specifically in the 10p11.2 region [5–8]. The rate of LOH varies among the studies, dependent upon the marker used and the stages of the cancers examined. Genetic alterations on 10p are often present in conjunction with the changes on 10q [6,9–11]. We previously performed an extensive deletion mapping of 10p in human prostate tumors at 13 highly polymorphic loci. In that study, 57% of 35 tumors examined demonstrated loss of 10p sequences. The highest concentration of allelic losses on 10p spanned a 4- to 7-cM region and included loci D10S211, D10S89, and D10S111, which defined a minimal common region of deletion on 10p in human prostate tumors. Moreover, this study suggested that one or more deletion domains may map to 10p, as some tumors were deleted exclusively at D10S211 or D10S89–D10S111 [6]. These studies were confirmed by Ittmann [7] who observed LOH of 3.2% in localized (Stage B) and LOH of 27% in advanced prostate cancer (Stages C and D) using the marker D10S111. Taken together, these allelotyping studies suggest that one or more tumor suppressor genes map to 10p in human prostate tumors. Functional studies supporting this hypothesis were provided by studies in prostate cell lines supplemented with portions of chromosome 10p. The introduction of subchromosomal fragments encompassing 10pter-q11 into the PC3 prostate adenocarcinoma cell line reduced tumorigenicity following injection of hybrid clones in nude mice [12]. Similar results were obtained by another group [13] using the PPC-1 cell line, a subline of PC3 [14], in which decreased colony formation in soft agar was observed following introduction of 10p sequences.
Thus, both allelotyping and functional studies suggest that one or more tumor suppressor genes critical for prostate tumorigenesis map to 10p inclusive of the D10S89 and D10S111 loci. We now show that the minimal common region of deletion on 10p in human prostate tumors contains a gene encoding a candidate human spectrin SH3 domain binding protein 1, Hssh3bp1 [15]. Hssh3bp1 binds to SH3 domains of spectrin and Abl tyrosine kinase [15], associates with macropinocytic vesicles in cultured cells, and is a potential regulator of macropinocytosis [16]. E3b1, a protein identified independently by another group as Eps8 binding protein [17] and which is identical to isoform 2 of Hssh3bp1 [15], was recently implicated in transmission of signals from Ras to Rac [18]. Hssh3bp1 maps near loci D10S89 and D10S111 within the 10p minimal region of deletion observed in prostate cancer, and all three sequences localize to a single YAC, 961C7. Moreover, Hssh3bp1 protein expression is downregulated in prostate tumors deleted for D10S89 or D10S111. Two prostate tumor cell lines contain a mutation in Hssh3bp1 gene leading to expression of the aberrant form of Hssh3bp1. These data are consistent with a role for Hssh3bp1 as a candidate tumor suppressor gene inactivated during prostate tumorigenesis.
Materials and Methods
DNA Analysis
Colonies from each of 11 CEPH YACs (965D10, 746D9, 815C7, 747H10, 857C9, 934E11, 796F8, 899E10, 875B4, 746G7, and 961C7), comprising a complete contig of the 10p minimal region of deletion, were picked and incubated in 10 µl of lyticase solution (1.2 M sorbitol, 10 mM sodium phosphate, pH 7.4 [1:4 v/v monobasic: dibasic from 1 M stocks]) and 2.5 mg/ml lyticase (Sigma, St. Louis, MO) at 37°C for 5 minutes. Five microliters of each digestion mixture was used in subsequent polymerase chain reaction (PCR) comprising 200 µM each dGTP, dATP, dTTP, and dCTP; 1x PCR buffer (50 mM KCI, 10 mM Tris-HCI, pH 8.3, 2.5 mM MgCl2); 1 µM each forward and reverse primers, and 0.6 U Taq Polymerase (Life Technologies, Rockville, MD) using an annealing temperature of 55°C. The linkage order of these markers has been reported as: pter-D10S211 - WI4906 - 10S553 - D10S1789 - D10S550 - WI4133 - D10S582 - D10S1673-D10S586-D10S1749-D10S1747-D10S572-D10S89-D10S111-centromere. Primer sequences and linkage information were obtained from databases maintained by the Human Genome Data Base E-mail: (http://gdbww-w.gdb.org/), Center for Genome Research at the Whitehead Institute for Biomedical Research E-mail: (http://www-genome.wi.mit.edu/), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) as accessed through the Internet. The Hssh3bp1 forward primer used was PROM 35′, 5′-CTGCAGAGACCCATGATTGTGCC-3′, and the reverse primer used was PROM 53′, 5′-CAAGTTGAGTACGAATACTCCGTAC-3′. Reaction products were electrophoresed on 2.5% agarose and visualized after ethidium bromide staining.
The Hssh3bp1 exon 6 sequences were amplified from genomic DNA isolated from prostate cell lines using forward primer Ex615′, 5′-CAAAGGGAGACTCACATATTTTTGG-3′, and the reverse primer Ex613′, 5′-TCCATAGGAGTTTGTCGCCAGTCAG-3′ and sequenced. The primer sequences were derived from Contig NT 008730 (Gen-Bank) containing the entire Hssh3bp1 gene (see also AceView of the gene at NCBI web site at the address indicated above).
Analysis of Hssh3bp1 Expression
Frozen paired normal and tumor prostatic tissues previously characterized for 10p dosage status were utilized [6]. Monoclonal antibody (mAb) 2G8 was raised to the recombinant C-terminal portion of Hssh3bp1, plasmid C5 [15] at the Institute for Basic Research in Developmental Disabilities Antibody Facility using standard techniques. Immunohistochemistry was performed in 5-µ sections, which were cut from paired frozen normal and malignant tissues from radical prostatectomy specimens, fixed for 10 minutes in ice-cold acetone, then air-dried briefly at 4°C. The slides were stained with a 1:2000 dilution of mAb 2G8 using a Ventana 320 ES Automatic Immunohistochemistry/IPOX Staining Station according to manufacturer's protocols. The antibody staining was evaluated by a pathologist (M.A.R.), and the degree of staining was assessed as 0 (absent), 1 (weak), 2 (moderate), or 3 (strong).
Protein Truncation Test (PTT)
Prostate cell lines LNCAP.FGC-10 (CRL-10995), LNCaP.FGC (CRL-1740), and PC3 (CRL 1435) were obtained from ATCC and were grown according to ATCC instructions. RNA from cultured cells was prepared using Tri-Reagent (Molecular Research Center, Cincinnati, OH). RT-PCR was performed using Hssh3bp1-specific primers T7-M (5′-GATTAATACGACTCACTATAGGGACGCGAGAGGAAGCGATGCAGAG-3′) (5′ primer) and P3 (5′-CTTGAATTCAAGCAAATCAGTGAAGGAAAGGAC-3′) (3′ primer). In vitro translation of gel-purified PCR products (200 ng) was performed using T7-h1 primer and T7 TNT System (Promega, Madison, WI). SDS-PAGE protein electrophoresis and Western blotting were performed as described [19]. Polyclonal antibody Ab-2 to Hssh3bp1 [16] was used in the analysis.
Results
Hssh3bp1 Maps to the 10p Minimal Common Region of Deletion in Prostate Tumors
Each of 11 CEPH YACs was amplified for 14 loci mapping within the 10p minimal common region of deletion previously described by our laboratory [6]. These experiments ordered the YACs into a contig spanning this region (Table 1). The Hssh3bp1 gene localized exclusively to YAC 961C7, which also contains sequences specific for markers D10S89 and D10S111. Because D10S89 also localizes to a more telomeric YAC, 875B4, the likely sequence order is: 10pter-D10S89-Hssh3bp1/D10S111-10cen, where “/” indicates that the actual orientation is unclear (Table 1). The relatively small size of YAC 961C7, which is 1.67 Mb, suggested the possibility that the Hssh3bp1 gene may be co-deleted in tumors deleted for D10S89 or D10S111.
Table 1.
YAC Contig of 10p Prostate Cancer Minimal Deletion Region.
| YAC | Chromosome 10p Loci | ||||||||||||||
| Designation | D10S211 | WI-4906 | D10S553 | D10S1789 | D10S550 | WI-4133 | D10S582 | D10S1673 | D10S586 | D10S1749 | D10S1747 | D10S572 | D10S89 | Hssh3bp1 | D10S111 |
| 965-D-10 | + | ||||||||||||||
| 746-D-9 | + | ||||||||||||||
| 815-C-7 | + | + | |||||||||||||
| 747-H-10 | + | + | |||||||||||||
| 857-C-9 | + | + | + | + | + | + | |||||||||
| 934-E-11 | + | + | + | + | + | + | + | + | |||||||
| 796-F-8 | + | + | + | + | + | ||||||||||
| 899-E-10 | + | + | + | ||||||||||||
| 875-B-4 | + | + | |||||||||||||
| 746-G-7 | + | ||||||||||||||
| 961-C-7 | + | + | + | ||||||||||||
YAC clones are listed on the left and chromosome 10p loci are listed on the top. YAC 961-C-7 contains D10S89, Hssh3bp1, and D10S111. The analysis was done by PCR using specific primers.
Hssh3bp1 Expression is Downregulated in Prostate Tumors Deleted for Adjacent 10p Sequences
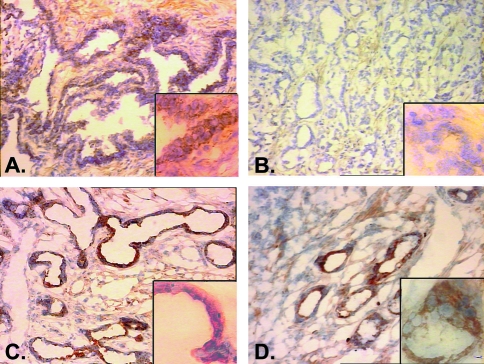
Immunohistochemical analysis of prostate tissues using a mAb to Hssh3bp1 was performed to determine whether Hssh3bp1 protein expression correlated with D10S89 or D10S111 dosage in prostate tumors. Seventeen paired normal and malignant prostate specimens previously characterized for dosage at D10S89 and D10S211 were utilized for these studies [6]. Of the 17 tumor tissues, six were characterized by deletions at D10S89 or D10S111 (Table 2). The remaining 11 tumors retained normal diploid dosage at D10S89, D10S111, or both loci (Table 2). Immunohistochemical staining of epithelial cytoplasm was graded into four groups: absent (0), weak (1), moderate (2), or strong (3). Moderate or strong expression of Hsshb3p1 was detected in 82% (14/17) of normal tissues examined (Table 2). In contrast, moderate or strong expression of Hssh3bp1 was detected in only 41% (7/17) of malignant tissues examined. Moreover, 4/6 (67%) tumors deleted for 10p sequences at D10S89 or D10S111, within the minimal common region of deletion, failed to express Hssh3bp1 protein compared to 5/11 (46%) tumors that retained normal diploid dosage at these loci. An example of Hssh3bp1 staining in normal and malignant is shown in Figure 1.
Table 2.
Expression of Hssh3bp1 in Prostate Tumors.
| Case Number | 10p Deletion* | Antibody Staining† | Correlation with 10p Status | Tumor Grade‡ | Tumor Stage§ | |
| Normal | Tumor | |||||
| 404 | yes | 3 | 3 | no | 3+3 | T2b |
| 334 | yes | 2 | 0 | yes | 3+3 | T2b |
| 340 | yes | 2 | 0, 2¶ | yes | 3+4 | T2b |
| 398 | yes | 1–2 | 0 | yes | 3+3 | T2 |
| 408 | yes | 3 | 0 | yes | 3+4 | T2 |
| 394 | yes | 2 | 3 | no | 3+3 | T1b |
| 344 | no | 1 | 1 | yes | 3+3 | T2 |
| 402 | no | 3 | 3 | yes | 4+3 | T3N1 |
| 244 | no | 0 | 2 | no | 3+3 | T3 |
| 390 | no | 3 | 3 | yes | 3+3 | T3 |
| 260 | no | 2–3 | 1 | no | 3+4 | T2b |
| 392 | no | 1 | 0 | no | 3+3 | T2 |
| 400 | no | 3 | 3 | yes | 3+3 | T3 |
| 386 | no | 3 | 0 | no | 3+3 | T3N1 |
| 380 | no | 2 | 1 | no | 3+4 | T3 |
| 320 | no | 2 | 2 | yes | 3+4 | T2 |
| 268 | no | 3 | 1, 3¶ | no | 3+3 | T2b |
10p deletion was characterized as described [6]. “no” indicates that the tumors were not deleted at 10p or were uninformative at one or more loci.
Mab 2G8 was used in all cases. (0) Absent; (1) weak; (2) moderate; (3) strong.
According to Gleason score.
According to TNM system.
Two areas with different pathology were identified in these tumors.
Figure 1.
Staining of normal (A, C) and malignant (B, D) prostate tissues from cases 398 (A, B) 400 (C, D) with mAb 2G8 antibody to Hsshb3p1. Staining is intense in both normal tissues shown (A, C) and in tumor tissue from case 400 (D), which was not deleted for 10p loci. Note the complete absence of staining in the malignant tissue from case 398 (B), which also demonstrated deletion of sequences adjacent to the Hsshb3p 1 locus on 10p. The large panels in (A), (B), (C), and (D) are shown at x100 magnification; insets are at x400 magnification.
Taken together, these experimental results show that the loss of Hsshb3p1 protein expression was clearly associated with the deletion of adjacent loci on 10p deletion in human prostate tumors.
Prostate Tumor Cell Lines Contain Mutations in Hssh3bp1
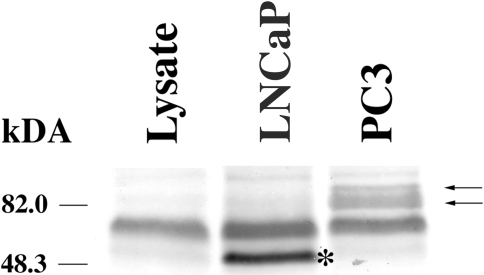
We performed PTT in two prostate tumor cell lines, LNCaP (CRL-10995) and PC3 (CRL-1435). PTT test using an antibody to the C-terminus of Hssh3bp1 indicated the presence of a truncated polypeptide in LNCaP cell line in comparison to two polypeptides in the PC3 cell line (Figure 2). We determined by DNA sequencing that the LNCaP cell line, but not the PC3 cell line, contains deletion of nucleotides 660–800 (total of 141 nucleotides) of the Hssh3bp1 cDNA [15]. This results in the in-frame deletion of amino residues 194 through 240 of Hssh3bp1 and is consistent with the observation of a smaller translation of product in the PTT. Identical deletion of Hssh3bp1 sequences was observed in another tumor cell line, CRL-1740 (not shown). Sequencing of Hssh3bp1 gene from these cell lines revealed the presence of a heterozygous point mutation in exon 6 of the gene located near the 3′ splice junction of the preceding intron: the sequence TAG↓ACGG is now TAG↓ACAG, where italic indicates intronic sequence, an arrow splice site, and underline/bold mutated residue. The mutation causes codon 194 change from R (CGG) to Q (CAG). This missense mutation apparently led to exon 6 skipping in the splicing of Hssh3bp1 mRNA, which may be explained by at least two independent mechanisms: a missense-induced exon skipping or by a conformational change in Hssh3bp1 mRNA near the 3′ splice site (see Discussion section). PCR analysis of Hssh3bp1 cDNA in PC3 cells using isoform-specific primers (not shown) revealed expression of isoforms 2 and 3 of Hssh3bp1, which is consistent with the observation of two closely spaced polypeptides (Figure 2). Apparent migration of the polypeptide from LnCAP cells corresponds to migration of isoform 5 of Hssh3bp1 (not shown) lacking amino acid sequence encoded by exon 6.
Figure 2.
Protein truncation test of Hssh3bp1 in prostate tumor cell lines. (B) In vitro translation of Hssh3bp1 cDNA using rabbit reticulocyte lysate. Reaction mixtures following in vitro translation of PCR products were separated on 7% Tricine SDS poly aerylamide gel; the gel was blotted with the polyclonal antibody Ab-2 to Hssh3bp1. Lane 1, reaction mixture without addition of exogenous DNA; lane 2, reaction mixture containing Hssh3bp1 cDNA from LNCaP; lane 3, reaction mixture containing Hssh3bp1 cDNA from PC3. Apparently the rabbit reticulocyte lysate contains an Ab-2 antibody reactive band (lane 1). Arrows indicate the full-length Hsshb3p1 polypeptides representing isoforms 2 and 3 of Hsshb3p1 [15] as confirmed by PCR analysis (not shown). Asterisk indicates a truncated polypeptide.
Discussion
Previous allelotyping and functional studies suggested that one or more tumor suppressor genes important for prostate tumorigenesis mapped to the 10p chromosomal region. In the current study, we mapped the Hssh3bp1 gene adjacent to loci D10S89 and D10S111 within the 10p minimal region of deletion previously defined by our laboratory for prostate tumors [6]. Moreover, expression of the Hssh3bp1 protein was reduced in the majority of prostate tumors deleted for either D10S89 or D10S111. These studies suggest that the observed reduction of Hssh3bp1 protein expression may be due to the allelic inactivation of the gene through the deletion of one copy and mutation of the remaining copy. This mechanism is consistent with that of the “two-hit” model originally proposed for the “prototype” tumor suppressor gene, retinoblastoma [20]. Therefore, Hssh3bp1 may comprise a candidate tumor suppressor gene important for prostate tumorigenesis.
The co-localization of Hsshb3p1 sequences with D10S89 and D10S111 within the 10p minimal common region of deletion in prostate cancer led us to the hypothesis that expression of Hssh3bp1 may be lost in prostate tumors. The results of immunohistochemical studies presented here support this hypothesis and show that Hssh3bp1 protein expression is absent or reduced in the majority (5/6, 83%) of prostate tumors examined with deletions of D10S89 or D10S111, but is expressed in the majority of normal tissues and prostate tumors intact at these loci.
Two mechanisms that may account for the observed reduced expression of Hssh3bp1 protein in conjunction with the deletion of adjacent 10p loci include deletion and/or mutation of Hssh3bp1 sequences. Other mechanisms may also be involved, however. For example, five tumors (cases 344, 260, 392, 386, and 380) failed to express Hssh3bp1 protein, though they did not exhibit deletion of D10S89 or D10S111. In these cases, it is possible that Hssh3bp1 expression was downregulated through other means, e.g., small interstitial deletions, mutations involving both Hssh3bp1 alleles, transcriptional downregulation, or reduced protein stability. Significantly, these proposed events are apparently specific to prostate tumors, as the vast majority of normal prostate epithelial specimens (14/17, 82%) exhibited moderate or strong Hssh3bp1 protein expression. It should also be noted, as an exception, that one tumor (case 404), deleted for one allele at both D10S89 and D10S111, expressed high levels of Hssh3bp1 protein. It is possible that deletion affected only one allele of Hssh3bp1 in this tumor, allowing normal expression of the remaining allele.
Two prostate tumor LnCAP cell lines, CRL-10995 and CRL-1740, contain a missense mutation in exon 6 of Hssh3bp1. These cell lines are derivative of each other, and this is probably why we observe the same mutation in the gene. This mutation leads to an apparent exon 6 skipping resulting in expression of aberrant form of Hssh3bp1 mRNA and protein. Although the exon 6 mutation is observed in one allele of Hsshb3p1 gene, only expression of the truncated polypeptide is observed in these cells, suggesting downregulation of the normal allele by mechanisms mentioned in the previous paragraph. Exon skipping was demonstrated to be due to nonsense or missense mutations located in exonic splice enhancers present in both constitutively and alternatively spliced exons in genes such as BRCA1 and others [21]. The identified mutation may also affect RNA secondary structure and conformation required for appropriate splicing of exon 6 since it is located at +3 position from the 3′ splice junction of the preceding intron. Whether the mutation is part of the functional splicing enhancer and/or changes RNA conformation remains to be determined, but the presence of exon 6 sequences is invariably observed in Hssh3bp1 cDNA obtained from various tissues including primary prostate cells (not shown), PC3 cells (this report), human brain [15], as well as several cultured cell lines (not shown). Alternative splicing of Hssh3bp1 in brain leads to five isoforms of the mRNA coding region with additional possibility of alternative splicing of the 3′ untranslated region [15]. Expression of specific Hssh3bp1 isoforms in different cells may be functionally significant and is different in PC3 than in LnCAP cell lines.
Lack of exon 6 sequences that encode amino residues 194 through 240 of Hssh3bp1 results in the loss of a portion of Abl tyrosine kinase SH3 domain binding site (amino residues 144–260 of Hssh3bp1 [15]) in Hssh3bp1 in LnCAP cell lines. Such deletion may affect subcellular localization of Abl tyrosine kinase [15] and alter its kinase activity, resulting in cellular transformation. This hypothesis is consistent with the fact that mutations of the Abl SH3 domain lead to the increased transformation ability of Abl [22,23]. Consistent with the proposed tumor suppressor function, Hsshb3p1 may be a negative regulator of the function of Abl tyrosine kinase, an oncogene.
Previous studies in our laboratory identified Hsshb3p1 as a marker of macropinocytic vesicles [16]. In addition, overexpression of Hssh3bp1 decreased endocytosis of a fluorescent dye, suggesting a potential negative regulatory role of Hssh3bp1 in macropinocytosis. Macropinocytosis is upregulated in tumor cell lines by stimulation with growth factors [24,25], and PI-3 kinase is a positive regulator of the process [26,27]. LY294002, a specific inhibitor of PI-3 kinase [28], blocks endocytosis of fluorescent dyes into Hssh3bp1 macropinosomes and dramatically affects their morphology [16]. This suggests that Hssh3bp1 is involved in a transduction pathway involving PI-3 kinase. PI-3 kinase function is, in turn, opposed by PTEN/MMAC [29,30], a tumor suppressor gene implicated in prostate tumorigenesis (reviewed in Ref. [31]). Thus, Hssh3bp1 and PTEN may be located in the same signal transduction pathway affected in prostate cancer. Thus, down-regulation of Hssh3bp1 may be a downstream event of abberant regulation of PTEN or PI-3 kinase. Our studies show that downregulation of Hssh3bp1 protein expression may also occur independently through a deletion and, possibly, mutation mechanism. Future studies will examine this pathway further through the identification of molecular events involving Hssh3bp1, PI-3 kinase, and PTEN.
Acknowledgements
We thank Elizabeth J. Luna (University of Massachusetts, Worcester, MA) for helpful discussions and suggestions. We would like to thank K. S. Kim and Richard Kascsak (NYS IBR, Staten Island, NY) for help in the development of the mAb 2G8 to Hssh3bp1.
Abbreviations
- CEPH
Centre d′Etude Polymorphisme Humain
- YAC
yeast artificial chromosome
- LOH
loss of heterozygosity
- mAb
monoclonal antibody
- PCR
polymerase chain reaction
- PI-3 kinase
phosphoinositide-3 kinase
- PTT
protein truncation test
Footnotes
This work was supported by the National Institute of Neurological Disorders and Stroke grant R29 NS32874 (L.K.) and, in part, by funds provided by the New York State Office of Mental Retardation and Developmental Disabilities.
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner S, Giovanella B, Ittmann M, Tycko B, Hibshoosh H, Wigler M, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;274:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck P, Pershouse M, Jasser S, Yung A, Lin H, Ligon A, Langford L, Baumgard M, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng D, Tavtigian S. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Ittmann MM. Chromosome 10 alterations in prostate adenocarcinoma (Review) Oncol Rep. 1998;5:1329–1335. doi: 10.3892/or.5.6.1329. [DOI] [PubMed] [Google Scholar]
- 4.Gray IC, Phillipa SMA, Lee SJ, Neoptolemos JP, Weissenbach J, Spurr NK. Loss of the chromosomal region 10q23–25 in prostate cancer. Cancer Res. 1995;55:4800–4803. [PubMed] [Google Scholar]
- 5.Komiya A, Suzuki H, Ueda T, Emi M, Ito H, Shimazaki J. Allelic losses at loci on chromosome 10 are associated with metastasis and progression of human prostate cancer. Genes, Chromosomes Cancer. 1996;17:245–253. doi: 10.1002/(SICI)1098-2264(199612)17:4<245::AID-GCC6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Trybus T, Burgess A, Wojno K, Glover T, Macoska J. Distinct areas of allelic loss on chromosomal regions 10p and 10q in human prostate cancer. Cancer Res. 1996;56:2263–2267. [PubMed] [Google Scholar]
- 7.Ittmann M. Allelic loss on chromosome 10 in prostate adenocarcinomas. Cancer Res. 1996;56:2143–2147. [PubMed] [Google Scholar]
- 8.Ittmann M. Loss of heterozygosity on chromosomes 10 and 17 in clinically localized prostate carcinoma. Prostate. 1996;28:275–281. doi: 10.1002/(SICI)1097-0045(199605)28:5<275::AID-PROS1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Phillips S, Morton D, Lee S, Wallace D, Neoptolemus J. Loss of heterozygosity of the retinoblastoma and adenomatous polyposis gene loci and in chromosomes 10p, 10q and 16q in human prostate cancer. Br J Urol. 1994;73:390–395. doi: 10.1111/j.1464-410x.1994.tb07602.x. [DOI] [PubMed] [Google Scholar]
- 10.Bergerheim U, Kunimi K, Collins V, Ekman P. Deletion mapping of chromosomes 8, 10 and 16 in human prostatic carcinoma. Genes, Chromosomes Cancer. 1991;3:215–220. doi: 10.1002/gcc.2870030308. [DOI] [PubMed] [Google Scholar]
- 11.Carter BS, Ewing CM, Ward WS, Treiger BF, Aalders TW, Schalken JA, Epstein JI, Isaacs WB. Allelic loss of chromosomes 16q and 10q in human prostate cancer. Proc Natl Acad Sci USA. 1990;87:8751–8755. doi: 10.1073/pnas.87.22.8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez Y, Lovell M, Marin M, Wong P, Wolf-Ledbetter M, McDonnell T, Killary A. Tumor suppression and apoptosis of human prostate carcinoma mediated by a genetic locus within human chromosome 10pter;q11. Proc Natl Acad Sci USA. 1996;93:2551–2556. doi: 10.1073/pnas.93.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshinori M, Albertsen H, Brothman A, Leach R, White R. Suppression of the malignant phenotype of human prostate cancer cell line PPC-1 by introduction of normal fragments of human chromosome 10. Cancer Res. 1996;56:2157–2160. [PubMed] [Google Scholar]
- 14.Chen R. Chromosome identity of human prostate cancer cell lines, PC-3 and PPC-1. Cytogenet Cell Genet. 1993;62:183–184. doi: 10.1159/000133468. [DOI] [PubMed] [Google Scholar]
- 15.Ziemnicka-Kotula D, Xu J, Gu H, Potempska A, Kim KS, Jenkins EC, Trenkner E, Kotula L. Identification of a candidate spectrin SH3 binding protein suggests a general mechanism of association of tyrosine kinases with the spectrin-based membrane skeleton. J Biol Chem. 1998;273:13681–13692. doi: 10.1074/jbc.273.22.13681. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Ziemnicka D, Merz G, Kotula L. Human spectrin Src homology 3 domain binding protein 1 regulates macropinocytosis in NIH 3T3 cells. J Cell Sci. 2000;113:3805–3814. doi: 10.1242/jcs.113.21.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biesova Z, Piccoli C, Wong WT. Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene. 1997;14:233–241. doi: 10.1038/sj.onc.1200822. [DOI] [PubMed] [Google Scholar]
- 18.Scita G, Nordstrom J, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegard M, Betsholtz C, Di Fiore PP. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 1999;401:290–293. doi: 10.1038/45822. [DOI] [PubMed] [Google Scholar]
- 19.Kotula L, DeSilva T, Speicher D, Curtis PJ. Functional characterization of recombinant human erythrocyte a spectrin polypeptides containing the tetramer binding site. J Biol Chem. 1993;268:14788–14793. [PubMed] [Google Scholar]
- 20.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 22.Franz WM, Berger P, Wang JY. Deletion of an N-terminal regulatory domain of the c-abl tyrosine kinase activates its oncogenic potential. EMBO J. 1989;8:137–147. doi: 10.1002/j.1460-2075.1989.tb03358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson P, Baltimore D. N-terminal mutations activate the leukemogenic potential of the myristoylated form of c-abl. EMBO J. 1989;8:449–456. doi: 10.1002/j.1460-2075.1989.tb03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haigler HT, McKanna JA, Cohen S. Rapid stimulation of pinocytosis in human carcinoma cells A-431 by epidermal growth factor. J Cell Biol. 1979;83:82–90. doi: 10.1083/jcb.83.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West MA, Bretscher MS, Watts C. Distinct endocytic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J Cell Biol. 1989;109:2731–2739. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Racoosin EL, Swanson JA. M-CSF-induced macro-pinocytosis increases solute endocytosis but not receptor-mediated endocytosis in mouse macrophages. J Cell Sci. 1992;102:867–880. doi: 10.1242/jcs.102.4.867. [DOI] [PubMed] [Google Scholar]
- 27.Araki N, Johnson MT, Swanson JA. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol. 1996;135:1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-AH-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 29.Haas-Kogan D, Shaley N, Wong M, Mills G, Yount G, Stokoe D. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 30.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 31.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4250. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]