Abstract
Chitinase-like proteins have long been proposed to play roles in normal plant growth and development, but no mutations in chitinase-like genes have been obtained previously to support this hypothesis. In this study, we have shown that the gene responsible for the elp1 mutation in Arabidopsis encodes a chitinase-like protein (AtCTL1). Mutation of this chitinase-like gene caused ectopic deposition of lignin and aberrant shapes of cells with incomplete cell walls in the pith of inflorescence stems. The AtCTL1 gene was expressed in all organs during normal plant growth and development, but it was not induced by wounding, salicylic acid, pectin fragments, or ethylene. Consistent with its ubiquitous expression pattern, mutation of the AtCTL1 gene affected many aspects of plant growth and development, including exaggerated hook curvature, reduced length and increased diameter of hypocotyls in dark-grown seedlings, and reduced root length and increased number of root hairs in light-grown seedlings. The mutant phenotypes could be rescued partially by ethylene inhibitors, and ethylene production in the mutant was significantly greater than in the wild type. Together, these results suggest that AtCTL1, a chitinase-like gene, is essential for normal plant growth and development in Arabidopsis.
INTRODUCTION
Plant chitinases are a group of enzymes that presumably hydrolyze chitin, a biopolymer of GlcNAc in a β-1,4 linkage. Only a few of them have been proven to possess chitinase activities. Most of the reported chitinases have been identified based on their sequence similarity to known chitinases. Plant chitinases are grouped into six different classes based on sequence similarity to tobacco chitinases (Meins et al., 1994). The two common classes of chitinases are class I and class II, which differ in the presence (class I) or absence (class II) of a conserved N-terminal cysteine-rich lectin domain. All classes of chitinases possess some conserved amino acid residues in the catalytic domains (Levorson and Chlan, 1997).
Because the chitinase substrate chitin is the main component of many fungal walls and expression of many chitinase genes is induced by pathogens, chitinases have long been proposed to play roles in defense. It has been shown that overexpression of some chitinases alone or together with other antifungal proteins leads to enhanced protection against pathogen attack (Broglie et al., 1991; Zhu et al., 1994; Jach et al., 1995). The enhanced protection induced by chitinases probably results from the direct inhibition of fungal growth or from induction of the plant defense response by GlcNAc oligomers generated by chitinases. In addition to the fungus-induced expression, chitinases have been shown to be induced by a variety of biotic and abiotic cues, such as wounding, ethylene, salicylic acid, pectin fragments, and heavy metals (Collinge et al., 1993; Graham and Sticklen, 1994). Although the exact functions of chitinases in these responses are not known, some of the responses induced by factors such as ethylene and salicylic acid might be part of the mechanisms involved in protection against pathogen attack.
Much attention has been paid recently to those chitinases that are expressed constitutively in plant organs not challenged by pathogens or stress. Chitinases from different plant species have been shown to be expressed in organs such as flowers, leaves, seed, and roots (Samac and Shah, 1991; Graham and Sticklen, 1994; Wemmer et al., 1994; Van Hengel et al., 1998; Ancillo et al., 1999; Domon et al., 2000; Regalado et al., 2000; Takakura et al., 2000; Passarinho et al., 2001). Some of these chitinases also exhibit tissue-specific expression patterns (Ancillo et al., 1999; Takakura et al., 2000). It is suggested that some of these constitutively expressed chitinases likely are involved in defense, which might make it difficult for pathogens to initialize colonization (Graham and Sticklen, 1994). Chitinases that are expressed in a tissue-specific manner and are not induced by pathogens or stress may have roles in normal plant growth and development. Direct evidence for a role of chitinases in normal plant development came from the study of the temperature-sensitive carrot mutant cell line ts11 (De Jong et al., 1992). At the restrictive temperature, carrot ts11 cells failed to undergo normal somatic embryogenesis because of a transient reduction of secretion of a chitinase into cell walls (De Jong et al., 1992, 1995). Interestingly, GlcNAc-containing lipooligosaccharides or nodulation factors were found to be able to rescue the embryogenesis-defective phenotype of carrot ts11 cells (De Jong et al., 1993). On the basis of these findings, it has been proposed that the carrot chitinase might be involved in the production of a nodulation factor–like signal molecule that is essential for normal embryo development (De Jong et al., 1993; Van Hengel et al., 1998).
Although the studies from the carrot mutant cell line indicate that chitinases are involved in normal plant development in addition to their roles in defense response, many questions remain to be answered. The foremost question concerns the endogenous substrates of chitinases in plants, because plant cells apparently do not produce chitin (Graham and Sticklen, 1994). In addition, little is known about the functions of those chitinases that are expressed constitutively in different tissues or at different developmental stages. To date, no mutations in chitinase-like genes have been identified in plants. The carrot mutant cell line ts11 is defective in the secretion of a chitinase, but the enzymatic activity of the chitinase is normal (De Jong et al., 1995). It is conceivable that genetic knockout of chitinase-like genes could establish whether any of the constitutively expressed chitinase-like genes is essential for normal plant growth and development.
Previously, we isolated and characterized an Arabidopsis elp1 (ectopic deposition of lignin in pith) mutant with alteration of lignin deposition patterns (Zhong et al., 2000). In elp1 mutant stems, lignin is deposited ectopically in patches of parenchyma cells in the pith that are not lignified in the wild type. The ectopic deposition of lignin in the pith of the elp1 mutant was identified by histological staining of lignin, and it correlated with increased klason lignin content and ectopic expression of caffeoyl CoA O-methyltransferase, an enzyme involved in monolignol biosynthesis (Zhong et al., 2000). In this article, we show that the ELP1 gene encodes a chitinase-like gene (AtCTL1). In addition to ectopic lignin deposition, we find that some pith cells in the elp1 mutant have aberrant shapes of cells with incomplete cell walls. We further demonstrate that mutation of the AtCTL1 gene results in growth alterations in both dark- and light-grown seedlings that are caused partially by overproduction of ethylene in the mutant. Our results provide direct evidence that a chitinase-like protein plays a role in normal plant growth and development in Arabidopsis.
RESULTS
Positional Cloning of the ELP1 Gene
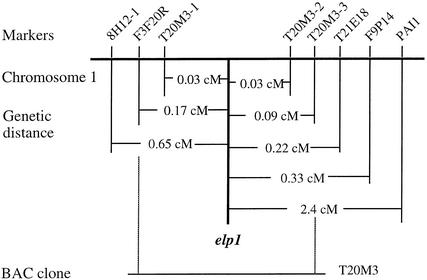
To investigate the molecular mechanisms underlying the ectopic deposition of lignin in the elp1 mutant (Zhong et al., 2000), we proceeded to isolate the ELP1 gene using a map-based cloning strategy. The elp1 mutant was generated with ethyl methanesulfonate mutagenesis, and the elp1 locus was mapped previously to a region between markers 8H12-1 and PAI1 on chromosome 1 (Zhong et al., 2000). To further map the elp1 locus for positional cloning, we collected 2200 F2 mapping plants from a cross of the elp1 mutant and the wild-type ecotype Landsberg plant. New cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993) (Figure 1) were developed by polymerase chain reaction (PCR) amplification of genomic DNA from ecotypes Columbia and Landsberg and by digestion of the amplified DNA with various restriction enzymes. With these new CAPS markers, the elp1 locus was narrowed progressively to a 26-kb region between CAPS markers T20M3-1 and T20M3-2 located in the bacterial artificial chromosome (BAC) clone T20M3 (Figure 1).
Figure 1.
Fine Mapping of the elp1 Locus.
CAPS markers were used to locate the elp1 locus to a 26-kb region between T20M3-1 and T20M3-2 in the BAC clone T20M3. The genetic distance was calculated based on the percentage of recombinant chromatids in a mapping population of 2200 plants. cM, centimorgan.
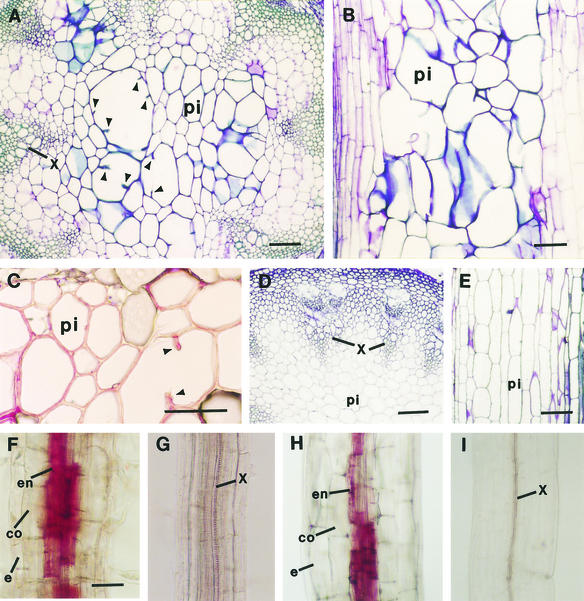
According to the sequence annotation information of BAC clone T20M3, six putative genes reside in the 26-kb region between markers T20M3-1 and T20M3-2. After sequencing all six putative genes, we found a G-to-A mutation within a gene encoding a putative basic endochitinase gene. To determine whether the point mutation in the putative chitinase gene corresponded to the elp1 locus, the wild-type putative chitinase gene was ligated into the binary vector pBI101 and the construct was introduced into the elp1 mutant by Agrobacterium transformation. Transgenic plants containing the wild-type putative chitinase gene rescued the elp1 phenotype completely, that is, it eliminated the ectopic deposition of lignin in pith (Figures 2A to 2E). It also was found that the primary roots (Figure 2F) and hypocotyls (Figure 2H) of the elp1 mutant had ectopic deposition of lignin in some endodermal cells, and this too was rescued by the wild-type putative chitinase gene (Figures 2G and 2I). These results demonstrated unequivocally that the G-to-A point mutation in the putative chitinase gene was directly responsible for the elp1 phenotypes. Thus, the ELP1 gene was renamed as a chitinase-like gene (AtCTL1).
Figure 2.
Complementation of the elp1 Mutant Phenotypes by the AtCTL1 Gene.
Sections were stained with toluidine blue ([A], [B], [D], and [E]) or phloroglucinol HCl ([C] and [F] to [I]). Lignified cell walls were stained blue by toluidine blue and red by phloroglucinol HCl.
(A) Cross-section of an elp1 stem showing aberrant shapes and enlargement of some lignified pith cells. Note that some large pith cells had partial cell walls protruding inward (arrowheads).
(B) Longitudinal section of an elp1 stem showing the disorganized arrangement of lignified pith cells.
(C) Cross-section of an elp1 stem showing lignin staining in pith cell walls. Note that one large pith cell had partial cell walls protruding inward (arrowheads).
(D) and (E) Cross-section (D) and longitudinal section (E) of the stems of elp1 plants transformed with the AtCTL1 gene showing lack of lignin staining in pith cells and the regular shape and arrangement of pith cells.
(F) Root of a light-grown elp1 seedling showing lignin staining in endodermal cells.
(G) Root of a light-grown elp1 plant transformed with the AtCTL1 gene showing lack of lignin staining in endodermal cells.
(H) Hypocotyl of a dark-grown elp1 seedling showing lignin staining in endodermal cells.
(I) Hypocotyl of a dark-grown elp1 seedling transformed with the AtCTL1 gene showing lack of lignin staining in endodermal cells.
co, cortex; e, epidermis; en, endodermis; pi, pith; x, xylem. Bars in (A) to (E) = 112 μm; bar in (F) = 56 μm for (F) to (I).
Sequence Analysis of the AtCTL1 Gene
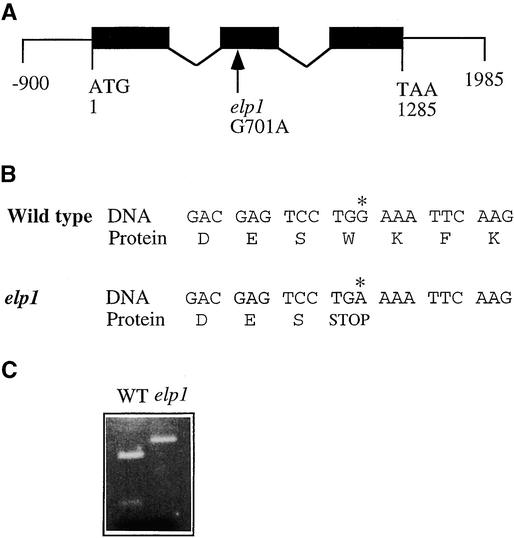
To analyze the exon and intron organization of the AtCTL1 gene, we isolated and sequenced its cDNA from an Arabidopsis stem cDNA library (Zhong and Ye, 1999). Comparison of the AtCTL1 gene and cDNA sequences revealed that the AtCTL1 gene is 1285 bp long from the start codon to the stop codon and contains three exons and two introns (Figure 3A). The three exons are located between nucleotide residues 1 to 388, 607 to 766, and 868 to 1285. The longest open reading frame in the AtCTL1 cDNA encodes a polypeptide of 321 amino acids with a predicted molecular mass of 35,556 D and a predicted pI of 7.49.
Figure 3.
Structure of the AtCTL1 Gene and Nature of the elp1 Mutation.
(A) Exon and intron organization of the AtCTL1 gene. The AtCTL1 gene is 1285 bp long from the start codon (nucleotide 1) to the stop codon (nucleotide 1285). A single nucleotide mutation was found at nucleotide 701 in elp1. Black boxes indicate exons, and lines between exons indicate introns.
(B) Effect of the single nucleotide mutation in elp1 on the translation of the predicted protein. Shown are residues of nucleotides and amino acids around the mutation site. The G-to-A transition (asterisk) changes the wild-type codon that encodes a tryptophan residue to a stop codon.
(C) Elimination of a ScrF1 site in the mutant elp1 cDNA. The single nucleotide mutation in elp1 happens to occur at a ScrF1 restriction endonuclease cleavage site. This is revealed readily by digesting reverse transcriptase–mediated PCR-amplified cDNA fragments with ScrF1, which shows that the ScrF1 site is missing in the elp1 cDNA compared with the wild-type (WT) AtCTL1 cDNA.
The elp1 mutation changed the G at nucleotide 701 in the second exon to an A (Figure 3A). This G-to-A mutation changed the codon TGG to TGA, which created a premature stop codon (Figure 3B). This resulted in a truncated protein with only 180 amino acid residues. The G-to-A point mutation created a polymorphic sequence between the wild type and elp1, which was revealed by ScrF1 digestion of PCR-amplified genomic DNA (Figure 3C).
Comparison of the deduced amino acid sequence of AtCTL1 with sequences in GenBank showed that AtCTL1 exhibits significant similarity to chitinases. The highest sequence identity occurs in the catalytic region of chitinases, with ∼40% sequence identity and ∼60% sequence similarity between AtCTL1 and chitinases from Arabidopsis, potato, tomato, rice, and tobacco (Figure 4A). It was found that the Arabidopsis genome contains another AtCTL1-like protein, tentatively named AtCTL2, which shows 70% identity to AtCTL1 in amino acid sequence. The known consensus amino acid residues for chitinases (Levorson and Chlan, 1997) also are conserved in AtCTL1. AtCTL1 does not have a lectin domain because the conserved cysteine-rich residues are not present (Figure 4A). This finding suggests that AtCTL1 is more closely related to class II chitinases, because class II chitinases lack a conserved N-terminal cysteine-rich lectin domain (Meins et al., 1994). We expressed AtCTL1 fusion protein in Escherichia coli and attempted to assay its possible activity using chitin as a substrate. However, we failed to detect any appreciable activity, probably because of an inability of AtCTL1 to degrade chitin or improper folding and modification of the fusion protein. Hydropathy analysis showed the presence of a signal secretion peptide at the N terminus of AtCTL1 (Figure 4B), indicating that AtCTL1 enters the secretory pathway. Analysis of the AtCTL1 amino acid sequence with the PSORT program (http://psort.nibb.ac.jp) did not reveal any vacuole- or other organelle-targeting sequences, suggesting that AtCTL1 likely is secreted into cell walls.
Figure 4.
Analysis of the Deduced Amino Acid Sequences of AtCTL1.
(A) Amino acid sequence alignment of the catalytic regions of AtCTL1 and a few other chitinase-like proteins from Arabidopsis (AtCTL2 and BCHI), potato (PotCHI), tomato (TomCHI), rice (RiCHI), and tobacco (TobCHI). Identical amino acid residues are indicated by colons. Dashed lines are gaps introduced to maximize the identity. The putative secretion signal peptide sequence in AtCTL1 is underlined. Amino acid residues known to be conserved among chitinases are indicated by asterisks.
(B) Hydropathy plot of AtCTL1 showing a secretion signal peptide sequence at the N terminus.
Expression Pattern of the AtCTL1 Gene
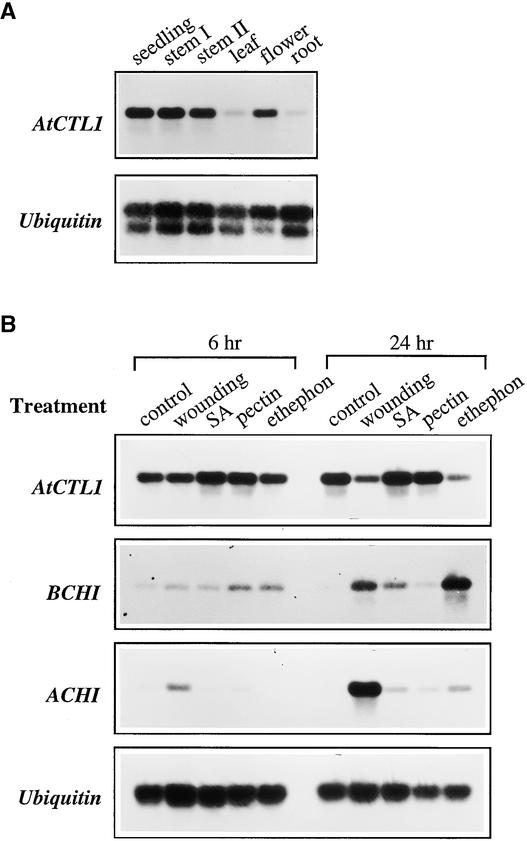
Plant chitinase genes have been shown to be either inducible by pathogens and stress or regulated developmentally (Graham and Sticklen, 1994). We analyzed the expression patterns of the AtCTL1 gene during normal plant development and under different stress conditions. Under normal growth conditions, the AtCTL1 gene was highly expressed in seedlings, stems, and flowers (Figure 5A). It also was detected in mature leaves and mature roots at a much lower level. The expression of AtCTL1 was not induced by various treatments, including wounding, salicylic acid, pectin fragments, and the ethylene-generating agent 2-chloroethylphosphonic acid (ethephon) (Figure 5B). The AtCTL1 mRNA level was decreased significantly 24 hr after wounding or treatment with ethylene. In contrast, the expression of two other basic and acidic chitinase genes (Samac et al., 1990) was altered significantly by these treatments (Figure 5B). Both the basic and acidic chitinase genes had little expression under normal growth conditions. The expression of the basic chitinase gene was found to be induced dramatically by wounding, salicylic acid, and ethephon, and the acidic chitinase gene was induced mainly by wounding (Figure 5B). These results indicated that the expression of AtCTL1 was not stress inducible; instead, it was regulated during normal plant growth and development.
Figure 5.
Expression of the AtCTL1 Gene.
Total RNA was isolated from different plant organs or stress-treated seedlings and used for reverse transcriptase–mediated PCR analysis. The ubiquitin gene was used as an internal control for PCR.
(A) Ubiquitous expression of the AtCTL1 gene in different organs. The seedlings were 3 weeks old. Mature leaves, mature roots, and flowers were from 8-week-old plants. Stem I and stem II were from 4- and 8-week-old plants, respectively.
(B) Expression of the AtCTL1, basic chitinase (BCHI), and acidic chitinase (ACHI) genes in response to wounding, salicylic acid (SA), pectin fragments, and ethephon. Three-week-old seedlings were treated with water (control) or other conditions for 6 and 24 hr before being used for expression analysis. The BCHI and ACHI genes were highly induced by various treatments, whereas the gene expression of AtCTL1 was not altered.
Effects of the elp1 Mutation on Pith Cell Shapes
We focused previously on the ectopic lignin deposition pattern in the elp1 mutant because the elp1 mutant was isolated based on its dramatic lignin staining in pith cells (Zhong et al., 2000). Close examination showed that the elp1 mutation also led to a dramatic change in the shape and organization of some pith cells in the inflorescence stems (Figure 2). Many lignified pith cells had irregular shapes and increased diameters (Figures 2A and 2B) compared with the wild type (Figures 2D and 2E). In addition, some of the giant cells in the pith appeared to be formed by two to three cells with incomplete cell walls, because they had partial cell walls protruding inward (Figures 2A and 2C). This finding suggested that the elp1 mutation might affect the normal formation of cell walls in some pith cells. The shapes of lignified endodermal cells in primary roots (Figure 2F) and hypocotyls (Figure 2H) of the elp1 mutant did not show any dramatic alteration compared with the wild type (Figures 2G and 2I).
Effects of the elp1 Mutation on Seedling Development
The observation that the AtCTL1 gene was expressed in all organs (Figure 5A) prompted us to examine further whether the elp1 mutation caused any other alterations in plant growth and development in addition to the ectopic deposition of lignin and aberrant cell shapes. Profound phenotypic changes were observed in both dark- and light-grown seedlings (Figure 6). In dark-grown seedlings, the elp1 mutation resulted in shortened length and increased diameter in both hypocotyls and roots and an exaggerated hook curvature (Figure 6B) compared with the wild type (Figure 6A). Quantitative measurement showed that the length of elp1 hypocotyls was reduced to 39% of that of the wild type, and the hypocotyl diameter was increased to 148% of that of the wild type (Table 1). In light-grown seedlings, the elp1 mutation also dramatically altered the growth of primary roots (Figure 6D) compared with the wild type (Figure 6C). The length of elp1 roots was only 58% of that of the wild type, and its diameter was increased to 131% of that of the wild type (Table 1). The length of inflorescence stems and their corresponding internodes of the elp1 mutant also were reduced dramatically compared with the wild type (Figure 6K, Table 1). It was apparent that the reduced length and increased diameter of etiolated hypocotyls and light-grown roots of elp1 were correlated with the reduced length and increased width of epidermal and cortical cells in these organs (Figures 6F and 6J, Table 1) compared with the wild type (Figures 6E and 6I). These results clearly indicated that the elp1 mutation caused a dramatic alteration in normal seedling development.
Figure 6.
Morphology of the Wild Type and the elp1 Mutant.
(A) and (B) Etiolated seedlings showing decreased length and increased width of hypocotyls of elp1 (B) compared with the wild type (A).
(C) and (D) Light-grown seedlings showing a reduction in root length of elp1 (D) compared with the wild type (C).
(E) and (F) Visualization of etiolated hypocotyl cells showing a decrease in length of epidermal cells in elp1 (F) compared with the wild type (E).
(G) and (H) Close-up of roots of light-grown seedlings showing a dramatic increase in root hair number and length in elp1 (H) compared with the wild type (G).
(I) and (J) Visualization of root cells showing a decrease in length of epidermal cells in elp1 (J) compared with the wild type (I).
(K) Five-week-old plants showing a reduction in length of inflorescence stems of elp1 (right) compared with the wild type (left).
Bars in (A) and (B) = 1.4 mm; bars in (C) and (D) = 1.0 mm; bar in (E) = 80 μm for (E), (F), (I), and (J); bars in (G) and (H) = 0.47 mm.
Table 1.
Measurement of Length and Diameter of Roots, Hypocotyls, and Inflorescence Stems of the Wild Type and the elp1 Mutant
| Organs | Wild Type | elp1 |
|---|---|---|
| Light-grown seedlinga | ||
| Root length (mm) | 10.6 ± 0.8 | 6.2 ± 0.6 |
| Root diameter (μm) | 130 ± 10 | 170 ± 20 |
| Root hair number (mm) | 13.5 ± 2.0 | 45.2 ± 2.4 |
| Root hair length (μm) | 80 ± 30 | 460 ± 60 |
| Cortical cells in roots (μm) | ||
| Length | 230 ± 30 | 130 ± 40 |
| Width | 18 ± 3 | 23 ± 4 |
| Etiolated seedlingb | ||
| Hypocotyl length (mm) | 13.3 ± 1.6 | 5.2 ± 0.9 |
| Hypocotyl diameter (μm) | 230 ± 50 | 340 ± 80 |
| Cortical cells in hypocotyls (μm) | ||
| Length | 331 ± 53 | 171 ± 29 |
| Width | 23 ± 4 | 54 ± 5 |
| Main inflorescence stemc | ||
| Height (cm) | 8.3 ± 0.8 | 3.1 ± 0.4 |
| Internode length (cm) | ||
| First internode | 1.5 ± 0.3 | 1.2 ± 0.2 |
| Second internode | 3.7 ± 0.6 | 1.0 ± 0.3 |
Data are means ±se from 15 plants.
Light-grown seedlings used for measurement were five days old.
Etiolated seedlings were grown for four days in the dark.
Five-week-old plants grown on agar were used for measurement.
Although the elp1 mutation inhibited root elongation, it greatly enhanced root hair formation (Figure 6H). When wild-type seedlings were grown on Murashige and Skoog (1962) medium without sucrose, a few short root hairs were produced along the surface of the root (Figure 6G). However, the elp1 roots produced three times more root hairs than the wild type, and the length of the elp1 root hairs was nearly six times longer than that of the wild type (Table 1), indicating that the elp1 mutation affected root hair formation and elongation.
To confirm that the phenotypes exhibited by the elp1 mutant were caused directly by mutation of the AtCTL1 gene, we examined seedlings of the elp1 plants transformed with the wild-type AtCTL1 gene. Both dark- and light-grown seedlings showed wild-type morphology (data not shown), indicating that the wild-type AtCTL1 gene completely rescued the abnormal seedling development of the elp1 mutant. Therefore, the abnormal growth of the elp1 mutant seedlings was caused by mutation of the AtCTL1 gene.
Partial Rescue of the elp1 Phenotypes by Ethylene Inhibitors
It appeared that the phenotypes exhibited by etiolated elp1 seedlings resembled the triple response of Arabidopsis seedlings treated with ethylene. In addition, the reduced root elongation and enhanced root hair formation in light-grown seedlings also were similar to responses caused by ethylene (Johnson and Ecker, 1998). Therefore, we attempted to determine whether the elp1 phenotypes could be rescued by the ethylene inhibitors 1-aminoethoxyvinyl glycine (AVG) and Ag+. AVG is known to inhibit the activity of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, thereby reducing the production of ethylene. Ag+ is thought to be a competitive inhibitor of ethylene (Johnson and Ecker, 1998). We also used ACC, a precursor of ethylene synthesis, to examine the responses of both wild-type and elp1 mutant seedlings. Different concentrations of AVG, Ag+, and ACC were used to test the optimal concentrations for rescue of the elp1 phenotypes (data not shown). Representative data from the treatment of plants with the optimal concentration of each drug are shown (Figures 7 to 9).
Figure 7.
Effects of ACC, Ag+, and AVG on the Morphology of elp1 Seedlings.
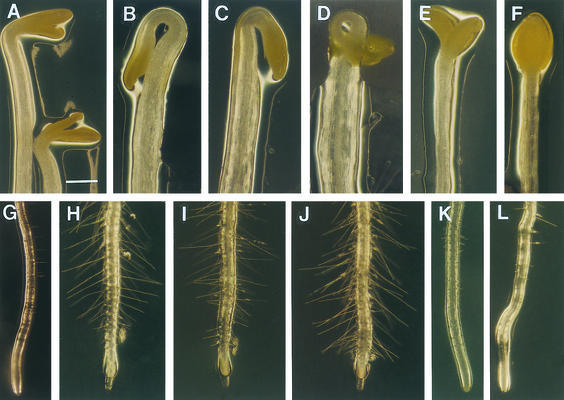
Four-day-old etiolated seedlings ([A] to [F]) and 5-day-old light-grown seedlings ([G] to [L]) in the absence or presence of ACC (0.1 μM for light-grown seedlings and 5 μM for dark-grown seedlings), Ag+ (1 μM), or AVG (0.5 μM) were used for observation of their morphology.
(A) Etiolated hypocotyls of the wild type.
(B) Etiolated hypocotyls of the wild type grown in the presence of ACC.
(C) Etiolated hypocotyls of elp1 showing increased hook curvature and hypocotyls width compared with the wild type (A).
(D) Etiolated hypocotyls of elp1 grown in the presence of ACC showing further increased hook curvature and hypocotyl width.
(E) and (F) Etiolated hypocotyls of elp1 grown in the presence of Ag+ (E) and AVG (F) showing decreased hook curvature and hypocotyl width.
(G) Roots of light-grown seedlings of the wild type.
(H) Roots of the wild-type seedlings grown in the presence of ACC showing increased number of root hairs.
(I) Roots of light-grown elp1 seedlings showing increased number of root hairs compared with the wild type (G).
(J) Roots of elp1 seedlings grown in the presence of ACC showing increased number of root hairs.
(K) and (L) Roots of the elp1 mutant seedlings grown in the presence of Ag+ (K) and AVG (L) showing a dramatic decrease in root hair number and length.
Bar in (A) = 0.47 mm for (A) to (L).
Figure 9.
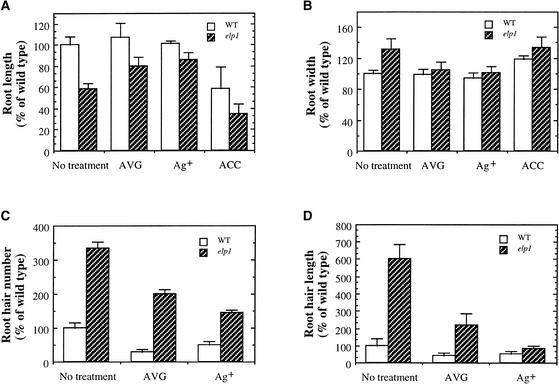
Measurement of Roots and Root Hairs of Light-Grown Wild-Type (WT) and elp1 Mutant Seedlings Treated with AVG, Ag+, or ACC.
Seed were germinated and grown in the light for 5 days in the presence of AVG (0.5 μM), Ag+ (1 μM), or ACC (0.1 μM) before measurement. Parameters of roots or root hairs of the wild type without drug treatment were taken as 100, and those of the wild type with drug treatment and all elp1 samples were expressed as percentages of the wild type without drug treatment. Data are means ±se from 20 samples.
(A) and (B) Root length (A) and width (B) of the wild type and elp1.
(C) and (D) Root hair number (C) and length (D) of the wild type and elp1.
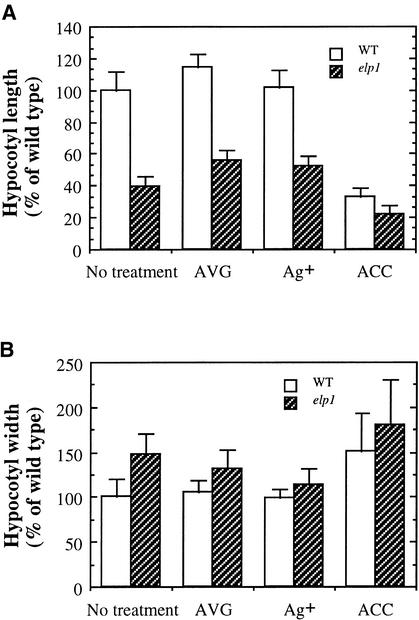
In dark-grown hypocotyls, it was shown that both AVG and Ag+ completely restored the hook curvature of the elp1 mutant (Figures 7C, 7E, and 7F) to the same level as in the wild type (Figure 7A). Treatment of the dark-grown elp1 seedlings with AVG and Ag+ also caused an increase in the length of elp1 hypocotyls from 39% (untreated) to ∼55% (P < 0.001) of that of the wild type (Figure 8A) and a decrease in the diameter from 148% (untreated) to ∼125% (P = 0.03) of that of the wild type (Figure 8B). Treatment of the etiolated wild-type seedlings with 5 μM ACC caused a phenotype (Figures 7B and 8) similar to that of the elp1 mutant without ACC treatment (Figures 7C and 8). Treatment of the elp1 mutant with the same amount of ACC further exaggerated the hook curvature, further reduced the hypocotyl length, and increased the hypocotyl diameter (Figures 7D and 8). A higher concentration of ACC (50 μM) caused more severe effects on the hypocotyl length and width of the elp1 mutant than of the wild type, although no difference was seen in terms of hook curvature (data not shown).
Figure 8.
Measurement of the Length and Width of Etiolated Hypocotyls of the Wild Type (WT) and the elp1 Mutant Treated with AVG, Ag+, or ACC.
Seed were germinated and grown in the dark for 4 days in the presence of AVG (0.5 μM), Ag+ (1 μM), or ACC (5 μM) before measurement. The length or width of the wild-type hypocotyls without drug treatment were taken as 100, and those of the wild type with drug treatment and all elp1 samples were expressed as percentages of the wild type without drug treatment. Data are means ±se from 20 samples.
(A) Hypocotyl length of the wild type and elp1.
(B) Hypocotyl width of the wild type and elp1.
Similar effects of AVG and Ag+ on root growth and root hair formation were observed in light-grown elp1 plants (Figures 7 and 9). In the presence of AVG and Ag+, the length of the elp1 roots increased from 58% (untreated) to ∼85% (P < 0.001) of that of the wild type (Figure 9A), whereas the diameter was restored to nearly the same as that of the wild type (Figure 9B). Treatment of the elp1 mutant with AVG and Ag+ also reduced the number of root hairs from 350% (untreated) to ∼175% (P < 0.001) of that of the wild type (Figures 7I, 7K, 7L, and 9C) and reduced the length of root hairs from 600% (untreated) to ∼150% (P < 0.001) of that of the wild type (Figures 7I, 7K, 7L, and 9D). Application of ACC appeared to cause more severe effects on root growth in the elp1 mutant (Figures 7I and 7J) than in the wild type (Figures 7G and 7H). The fact that AVG and Ag+ partially rescued the elp1 mutant phenotypes suggested that the elp1 mutation might cause overproduction of ethylene.
Induction of Ethylene Overproduction and Basic Chitinase Expression in the elp1 Mutant
To ascertain whether the elp1 mutation caused increased production of ethylene, we measured the amount of ethylene released by both dark- and light-grown plants. Quantitative measurement showed that dark- and light-grown elp1 mutant plants produced 10 and 2 times more ethylene, respectively, than the wild type (Table 2), which was consistent with the observation from ethylene inhibitor experiments. This finding suggested that the elp1 mutation resulted in overproduction of ethylene, thereby causing some phenotypes resembling those of wild-type plants treated with exogenous ethylene.
Table 2.
Quantitative Measurement of Ethylene Released by Dark- and Light-Grown Plants of the Wild Type and the elp1 Mutant
| Plants | Wild Type | elp1 |
|---|---|---|
| Etiolated seedlingsa | 3.5 ± 0.5 | 35.3 ± 3.0 |
| Light-grown plantsb | 1.7 ± 0.4 | 3.7 ± 0.3 |
Data are means ±se (μL/g fresh weight) from five samples.
Etiolated seedlings were grown for four days in the dark.
Light-grown plants used for ethylene measurement were three weeks old.
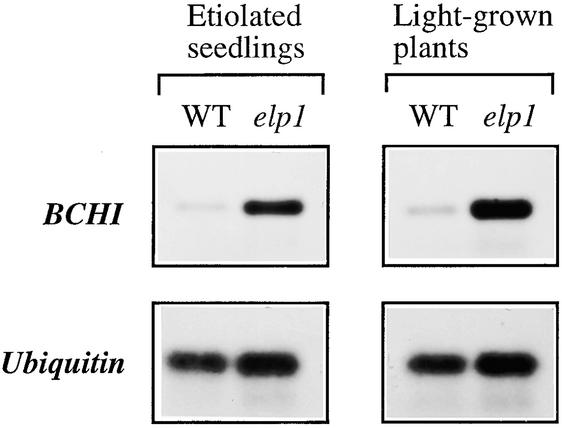
We further used a basic chitinase as a marker to determine whether its gene expression was upregulated in the elp1 mutant. The Arabidopsis basic chitinase is known to be induced by ethylene (Samac et al., 1990) and often has been adopted as a positive marker to confirm the effects of ethylene overproduction on endogenous gene expression (Kieber et al., 1993). As expected, the expression level of the basic chitinase gene was increased dramatically in both etiolated and light-grown plants of the elp1 mutant compared with the wild type (Figure 10). The induction level of the basic chitinase gene expression in the elp1 mutant was comparable with that in the wild type treated with the ethylene-releasing agent ethephon (Figure 5B). This finding further supported the observation that the elp1 mutation caused increased production of ethylene.
Figure 10.
Expression of the Basic Chitinase Gene in the Wild Type (WT) and the elp1 Mutant.
Four-day-old etiolated seedlings or 3-week-old light-grown plants were used for total RNA isolation and subsequent reverse transcriptase–mediated PCR analysis of the basic chitinase (BCHI) gene. The ubiquitin gene was used as an internal control for PCR. Although the expression of the BCHI gene was barely detectable in the wild type, it was highly induced in the elp1 mutant.
DISCUSSION
Many chitinase-like genes that exhibit tissue- and development-specific expression patterns have been identified in plants. It has been hypothesized that some of these chitinases are involved in the regulation of normal plant growth and development other than defense against pathogens (Collinge et al., 1993; Graham and Sticklen, 1994). Proof of the roles of different chitinases in normal plant development awaits the identification of chitinase mutants. Our demonstration that mutation of a chitinase-like gene alters normal plant growth and development is an important step in this direction. Mutation of the AtCTL1 gene caused a number of conspicuous phenotypes, including ectopic lignin deposition, aberrant shapes of cells with incomplete cell walls, reduced length and increased width of hypocotyls and roots, increased number and length of root hairs, and increased production of ethylene. In addition, the AtCTL1 gene was shown to be not induced by stress, indicating that AtCTL1 is a development-associated rather than a pathogenesis-related chitinase-like protein.
Chitinase-Like Proteins and Their Possible Substrates
AtCTL1 shows significant amino sequence similarity to a group of chitinases, so it is designated a chitinase-like protein (Graham and Sticklen, 1994). However, AtCTL1 appears not to belong to the pathogenesis-related chitinases, which use chitin as their substrates, because AtCTL1 is not induced by any stress stimuli such as wounding, salicylic acid, pectin fragments, and ethylene. Considering the facts that AtCTL1 is essential for normal plant growth and plants do not have chitin, it is unlikely that AtCTL1 uses chitin as its substrate. The next obvious question concerns the possible endogenous substrates for chitinase-like proteins such as AtCTL1 that are involved in normal plant growth and development. Although no endogenous substrates have been established for any developmentally regulated plant chitinases, early work suggests that plants may have molecules that chitinases might use as substrates.
One of the likely candidates is a group of glycoproteins called arabinogalactan proteins, which are thought to be important for plant growth and development (Cassab, 1998). Chitinase-like proteins isolated from embryogenic cell lines of pine (Domon et al., 2000) and carrot (Passarinho et al., 2001) have been shown to be able to cleave arabinogalactan proteins, suggesting that arabinogalactan proteins could be endogenous substrates for these chitinase-like proteins. Some glycoproteins from rice have been shown to contain GlcNAc dimers attached to asparagines (Hoson and Wada, 1986). Chitinase-like proteins also might act on these GlcNAc-containing glycoproteins. Although the functions of these glycoproteins are not known, it is possible that chitinase-like proteins such as AtCTL1 could act on these glycoproteins to modify their activities or to generate oligosaccharide signaling molecules important for normal plant growth.
Because GlcNAc-containing lipooligosaccharides can rescue the embryogenesis-defective phenotype of the carrot mutant cell line ts11, it has been proposed that plants may use nodulation factor–like molecules to regulate plant growth and development (De Jong et al., 1993). Although no direct evidence is available to support the existence of such molecules in plants, several studies suggest that plants might produce chitooligosaccharide-like molecules and use them as signaling molecules. Early work searching for putative substrates of plant chitinases led to the finding of the existence of abundant GlcNAc residues mainly in secondary cell walls of various plant species (Benhamou and Asselin, 1989). It also has been shown that Lathyrus plants contain lipophilic compounds that migrate similarly to rhizobial nodulation factors on reverse phase thin layer chromatography and that, when treated with a chitinase, are digested to form products similar to those of nodulation factors (Spaink et al., 1993). Further evidence came from transgenic studies showing that overexpression of the rhizobial NodB gene, which encodes a chitooligosaccharide deacetylase (John et al., 1993), resulted in altered plant morphology, indicating that chitooligosaccharide-like substrates might exist in plants (Schmidt et al., 1993). If plants do have chitooligosaccharide-like molecules, chitinase-like proteins such as AtCTL1 could be involved in modification of the activities of these molecules. Such a role of plant chitinases in the regulation of activities of lipochitooligosaccharides has been suggested for normal nodule development (Goormachtig et al., 1998).
It is worth noting that although it is not conclusive whether plants produce chitooligosaccharides, N-acetylchitooligosaccharides have been shown to be produced by zebrafish and frog and have been suggested to be patterning agents in zebrafish embryogenesis (Semino et al., 1996; Semino and Allende, 2000). It was found that removal of chitooligosaccharides by the introduction of an exogenous chitinase caused developmental defects in zebrafish embryos. Moreover, inhibition of the endogenous extracellular chitinase activity resulted in developmental defects in embryos, suggesting that both the synthesis and the degradation of chitooligosaccharides are essential for the normal development of zebrafish embryos (Semino and Allende, 2000). Genes that encode proteins with sequence similarity to the rhizobial NodC protein have been isolated from Xenopus and zebrafish and have been shown to be able to catalyze the synthesis of chitooligosaccharides (Semino and Robbins, 1995).
Mutation of the AtCTL1 Gene Causes Ectopic Lignin Deposition and Aberrant Cell Shapes
It is surprising that the ELP1 gene responsible for the ectopic deposition of lignin encodes a chitinase-like protein (AtCTL1). It seems unlikely that AtCTL1 is involved directly in the regulation of lignin biosynthesis. Instead, mutation of the AtCTL1 gene likely affects other cellular processes, which in turn cause ectopic lignin formation. One possibility is that ethylene overproduction in the mutant might induce ectopic lignification. A correlation has been observed between ethylene overproduction and lignification in response to stress (Prasad and Cline, 1987; Walter, 1992). However, ectopic deposition of lignin in the elp1 mutant appears not to be correlated closely with increased ethylene production, because ethylene inhibitors did not affect the ectopic deposition of lignin and treatment of the wild type with ACC did not induce ectopic lignification (data not shown). Another possibility is that aberrant cell expansion in the elp1 mutant might induce ectopic lignification. Ectopic deposition of lignin in several other mutants has been shown to be correlated with altered cell expansion (Cano-Delgado et al., 2000). In the elp1 mutant, lignified pith cells had irregular shapes and much larger sizes compared with the wild type. However, ectopic lignification in roots and hypocotyls of elp1 does not occur in cortical and epidermal cells, which are reduced dramatically in length, suggesting that ectopic deposition of lignin is not always associated with altered cell expansion. It will be interesting to use other ethylene overproduction mutants to examine whether ectopic lignin deposition and irregular cell shapes are associated with ethylene overproduction.
Mutation of the AtCTL1 Gene Leads to Increased Production of Ethylene
It is extremely interesting that ethylene production is enhanced in the elp1 mutant. This is in sharp contrast to pathogenesis-related chitinases, whose activities may generate signal molecules that induce ethylene production and lignification (Graham and Sticklen, 1994). According to the nature of the AtCTL1 protein, it is unlikely that AtCTL1 is involved directly in the biosynthesis of ethylene. Instead, it is possible that products or substrates of AtCTL1 might be involved in regulating ethylene biosynthesis, and mutation of the AtCTL1 gene might alter this regulation, thus leading to increased ethylene production. Previous studies have shown that GlcNAc-containing lipooligosaccharides can induce ethylene overproduction in Vicia sativa, which causes thick and short root phenotypes, including reduced length and increased diameter of roots and an increased number of root hairs, indicating that these molecules are able to regulate ethylene biosynthesis (Zaat et al., 1989; Van Spronsen et al., 1995). If GlcNAc-containing oligosaccharides were possible endogenous substrates of AtCTL1, mutation of the AtCTL1 gene could result in an accumulation of GlcNAc-containing oligosaccharides that could induce ethylene overproduction. Identification of the endogenous substrates of AtCTL1 will be essential for further understanding of the molecular mechanisms regulating ethylene production by AtCTL1.
elp1 Mutant Phenotypes Are Caused by Both Ethylene-Dependent and Ethylene-Independent Processes
The elp1 mutation causes a number of phenotypes, including exaggerated hook curvature and radial swelling of dark-grown hypocotyls, enhanced root hair formation, reduced elongation of roots and hypocotyls, and ectopic deposition of lignin. Our findings that the elp1 mutation resulted in increased ethylene production and that ethylene inhibitors partially rescued the elp1 phenotypes indicate that ethylene overproduction in the mutant caused some of the elp1 phenotypes. It appears that the increased root hair formation was caused by the increased production of ethylene in the mutant, because treatment with AVG and Ag+ significantly reduced the number and length of root hairs in the elp1 mutant, which is consistent with the known effects of ethylene on root hair formation (Tanimoto et al., 1995). The rescue of the hook curvature of dark-grown mutant hypocotyls by ethylene inhibitors also indicates that ethylene likely is the factor causing this phenotype.
However, ethylene appears to be partially responsible for the reduction in the length of roots and hypocotyls in the elp1 mutant, because ethylene inhibitors only partially rescued these phenotypes. This finding indicates that the elp1 mutation might affect some other pathways that also are important for normal cell elongation. Based on the observations that mutation of the AtCTL1 gene causes ectopic deposition of lignin, increased production of ethylene, and reduced elongation of some plant organs, we propose that the substrates or products of the AtCTL1-mediated reaction are essential for normal plant growth and development. Further understanding of the molecular mechanisms responsible for these phenotypes awaits identification of the endogenous substrates for AtCTL1.
METHODS
Map-Based Cloning
For fine mapping of the elp1 locus, 2200 F2 mapping Arabidopsis thaliana plants segregated from a cross of the elp1 mutant and wild-type ecotype Landsberg erecta were used to delimit the boundaries of the locus using cleaved amplified polymorphic sequence markers (Konieczny and Ausubel, 1993). For complementation analysis, the wild-type AtCTL1 gene was amplified by polymerase chain reaction (PCR), confirmed by sequencing, and then ligated into the binary vector pBI101 (Clontech, Palo Alto, CA). The construct was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The AtCTL1 gene was introduced into the elp1 mutant plants using the vacuum infiltration–mediated transformation procedure (Bechtold and Bouchez, 1994; Bent et al., 1994). Transgenic plants were selected on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 50 mg/L kanamycin and then examined by lignin staining in stem sections.
The AtCTL1 full-length cDNA was isolated from an Arabidopsis cDNA library (Zhong and Ye, 1999) and sequenced using a dye-based cycle sequencing kit (Applied Biosystems, Foster City, CA).
Histology
Plant tissues were fixed in 2% glutaraldehyde in 50 mM phosphate buffer at 4°C overnight. Tissues were dehydrated in a gradient of ethanol, cleared in Histochoice clearing agent (Amresco, Solon, OH), and then embedded in paraffin (Polysciences, Warrington, PA). Thin sections were cut with a microtome and stained with toluidine blue or phloroglucinol HCl.
For visualization of epidermal cells, fresh roots and hypocotyls were stained with 0.1 μg/mL FM1-43 (Molecular Probes, Eugene, OR) for 10 min, and cells in maturation regions of roots and hypocotyls were imaged with a confocal microscope.
Reverse Transcriptase–Mediated PCR
Total RNA was isolated from tissues using an RNA isolation kit (Qiagen, Valencia, CA). RNA was first treated with DNase and then used for first-strand cDNA synthesis. Gene-specific primers were used to amplify the cDNA of interest. PCR products were separated on an agarose gel and transferred to a nylon membrane. The membrane was hybridized with a digoxigenin-labeled gene-specific probe, and the hybridized signals were detected with a chemiluminescence detection kit (Roche Molecular Biochemicals, Indianapolis, IN).
Seedling Growth and Treatment
Seed were surface-sterilized and grown on MS medium without sucrose either in total darkness or under a 16-hr-light/8-hr-dark cycle at 24°C in a plant growth chamber. For observation of roots and hypocotyls, plates were placed vertically. The lengths of roots and hypocotyls were measured with a digital caliper. The widths of roots and hypocotyls and the lengths of root hairs were measured with a dissection microscope with a micrometer fitted onto an eyepiece. For observation of inflorescence stems, plants were grown in MS medium in magenta jars.
For drug treatment, various concentrations of 1-aminoethoxyvinyl glycine, Ag+, and 1-aminocyclopropane-1-carboxylic acid were added to MS medium after sterilization. Seedlings were grown on the medium containing drugs until used.
The quantitative differences of length and width of hypocotyls and roots between the untreated and treated seedlings were analyzed statistically using the Simple Interactive Statistical Analysis t test program (http://home.clara.net/sisa/t-test.htm) and shown to be significant (P < 0.05).
Stress Treatment
Three-week-old seedlings grown in soil were sprayed with water as a control, 1 mM salicylic acid, 100 μg/mL pectin fragments, or 0.1% (w/v) 2-chloroethylphosphonic acid. Pectin fragments were prepared by digesting citrus fruit pectin (Sigma, St. Louis, MO) with pectinase according to Nühse et al. (2000). For wounding treatment, leaves were cut into small pieces with a razor blade and leaf pieces were kept in water until used.
Measurement of Ethylene
For measurement of ethylene released from etiolated seedlings, sterilized seed were germinated on MS medium in a 20-mL vial stopped with a rubber cap. For measurement of ethylene released from light-grown plants, plants were grown on MS medium in a 20-mL vial and the vial was sealed to air tightness with a rubber cap 1 day before measurement. Ethylene was determined by gas chromatography. One milliliter of air from each container was taken to detect the presence of ethylene. After measurement, the fresh weight of seedlings was determined and the ethylene production was quantitated based on the fresh weight of plants.
Accession Numbers
The GenBank accession numbers for the sequences described in the text and in Figure 4 are AC009999 (BAC clone T20M3), AF422178 and AF422179 (AtCTL1), BAA94976 (AtCTL2), B45511 (BCHI), X67693 (PotCHI), U30465 (TomCHI), X56787 (RiCHI), and X64518 (TobCHI).
Acknowledgments
We thank W. Herbert Morrison for his suggestion on ethylene measurement, John Shields for his help with confocal microscopy, Alan Darvill for stimulating discussions and helpful suggestions, and the reviewers and editor for their constructive comments and suggestions.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010278.
References
- Ancillo, G., Witte, B., Schmelzer, E., and Kombrink, E. (1999). A distinct member of the basic (class I) chitinase gene family in potato is specifically expressed in epidermal cells. Plant Mol. Biol. 39, 1137–1151. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Benhamou, N., and Asselin, A. (1989). Attempted localization of a substrate for chitinases in plant cells reveals abundant N-acetyl-d-glucosamine residues in secondary walls. Biol. Cell 67, 341–350. [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Broglie, K., Chet, I., Hollyday, M., Cressman, R., Biddle, P., Knowlton, S., Mauvais, C.J., and Broglie, R. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Cano-Delgado, A.I., Metzlaff, K., and Bevan, M.W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127, 3395–3405. [DOI] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 281–309. [DOI] [PubMed] [Google Scholar]
- Collinge, D.B., Kragh, K.M., Mikkelsen, J.D., Nielsen, K.K., Rasmussen, U., and Vad, K. (1993). Plant chitinases. Plant J. 3, 31–40. [DOI] [PubMed] [Google Scholar]
- De Jong, A.J., Cordewener, J., Lo Schiavo, F., Terzi, M., Vandekerckhove, J., Van Kammen, A., and De Vries, S.C. (1992). A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4, 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, A.J., Heidstra, R., Spaink, H.P., Hartog, M.V., Meijer, E.A., Hendriks, T., Lo Schiavo, F., Terzi, M., Bisseling, T., Van Kammen, A., and De Vries, S.C. (1993). Rhizobium lipooligosaccharides rescue a carrot somatic embryo mutant. Plant Cell 5, 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, A.J., Hendriks, T., Meijer, E.A., Penning, M., Lo Schiavo, F., Terzi, M., Van Kammen, A., and De Vries, S.C. (1995). Transient reduction in secreted 32 kD chitinase prevents somatic embryogenesis in the carrot (Daucus carota L.) variant ts11. Dev. Genet. 16, 332–343. [Google Scholar]
- Domon, J.-M., Neutelings, G., Roger, D., David, A., and David, H. (2000). A basic chitinase-like protein secreted by embryogenic tissues of Pinus caribaea acts on arabinogalactan proteins extracted from the same cell lines. J. Plant Physiol. 156, 33–39. [Google Scholar]
- Goormachtig, S., Lievens, S., Van De Velde, W., Van Montagu, M., and Holsters, M. (1998). Srchi13, a novel early nodulin from Sesbania rostrata, is related to acidic class III chitinases. Plant Cell 10, 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, L.S., and Sticklen, M.B. (1994). Plant chitinases. Can. J. Bot. 72, 1057–1083. [Google Scholar]
- Hoson, T., and Wada, S. (1986). N-Acetylglucosamine-containing glycopeptide released from rice coleoptile cell walls by protease treatment. Plant Cell Physiol. 27, 303–309. [Google Scholar]
- Jach, G., Görnhardt, B., Mundy, J., Logemann, J., Pinsdorf, E., Leah, R., Schell, J., and Maas, C. (1995). Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8, 97–109. [DOI] [PubMed] [Google Scholar]
- John, M., Röhrig, H., Schmidt, J., Wieneke, U., and Schell, J. (1993). Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 90, 625–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, P.R., and Ecker, J.R. (1998). The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 32, 227–254. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Levorson, J., and Chlan, C.A. (1997). Plant chitinase consensus sequences. Plant Mol. Biol. Rep. 15, 122–133. [Google Scholar]
- Meins, F., Fritig, B., Linthorst, H.J.M., Mikkelsen, J.D., Neuhaus, J.-M., and Ryals, J. (1994). Plant chitinase genes. Plant Mol. Biol. Rep. 12, 22–28. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nühse, T.S., Peck, S.C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275, 7521–7526. [DOI] [PubMed] [Google Scholar]
- Passarinho, P.A., Van Hengel, A.J., Fransz, P.F., and De Vries, S.C. (2001). Expression pattern of the Arabidopsis thaliana AtEP3/AtchiIV endochitinase gene. Planta 212, 556–567. [DOI] [PubMed] [Google Scholar]
- Prasad, T.K., and Cline, M.G. (1987). Shoot inversion inhibition of stem elongation in Pharbitis nil. Plant Physiol. 85, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado, A.P., Pinheiro, C., Vidal, S., Chaves, I., Ricardo, C.P.P., and Rodrigues-Pousada, C. (2000). The Lupinus albus class-III chitinase gene, IF-3, is constitutively expressed in vegetative organs and developing seeds. Planta 210, 543–550. [DOI] [PubMed] [Google Scholar]
- Samac, D.A., and Shah, D.M. (1991). Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac, D.A., Hironaka, C.M., Yallaly, P.E., and Shah, D.M. (1990). Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 93, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J., Röhrig, H., John, M., Wieneke, U., Stacey, G., Koncz, C., and Schell, J. (1993). Alteration of plant growth and development by Rhizobium nodA and nodB genes involved in the synthesis of oligosaccharide signal molecules. Plant J. 4, 651–658. [Google Scholar]
- Semino, C.E., and Allende, M.L. (2000). Chitin oligosaccharides as candidate patterning agents in zebrafish embryogenesis. Int. J. Dev. Biol. 44, 183–193. [PubMed] [Google Scholar]
- Semino, C.E., and Robbins, P.W. (1995). Synthesis of “Nod”-like chitin oligosaccharides by the Xenopus developmental protein DG42. Proc. Natl. Acad. Sci. USA 92, 3498–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino, C.E., Specht, C.A., Raimondi, A., and Robbins, P.W. (1996). Homologs of the Xenopus developmental gene DG42 are present in zebrafish and mouse and are involved in the synthesis of Nod-like chitin oligosaccharides during early embryogenesis. Proc. Natl. Acad. Sci. USA 93, 4548–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink, H.P., Wifjes, A.H.M., Van Vliet, T.B., Kijne, J.W., and Lugtenberg, B.J.J. (1993). Rhizobial lipo-oligosaccharide signals and their role in plant morphogenesis: Are analogous lipophilic chitin derivatives produced by the plant? Aust. J. Plant Physiol. 20, 380–392. [Google Scholar]
- Takakura, Y., Ito, T., Saito, H., Inoue, T., Komari, T., and Kuwata, S. (2000). Flower-predominant expression of a gene encoding a novel class I chitinase in rice (Oryza sativa L.). Plant Mol. Biol. 42, 883–897. [DOI] [PubMed] [Google Scholar]
- Tanimoto, M., Roberts, K., and Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943–948. [DOI] [PubMed] [Google Scholar]
- Van Hengel, A.J., Guzzo, F., Van Kammen, A., and De Vries, S.C. (1998). Expression pattern of the carrot EP3 endochitinase genes in suspension culture and in developing seeds. Plant Physiol. 117, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Spronsen, P.C., Van Brussel, A.A.N., and Kijne, J.W. (1995). Nod factors produced by Rhizobium leguminosarum biovar viciae induce ethylene-related changes in root cortical cells of Vicia sativa ssp. nigra. Eur. J. Cell Biol. 68, 463–469. [PubMed] [Google Scholar]
- Walter, M.H. (1992). Regulation of lignification in defense. In Genes Involved in Plant Defense, T. Boller and F. Meins, eds (New York: Springer-Verlag), pp. 327–352.
- Wemmer, T., Kaufmann, H., Kirch, H.-H., Schneider, K., Lottspeich, F., and Thompson, R.D. (1994). The most abundant soluble basic protein of the stylar transmitting tract in potato (Solanum tuberosum L.) is an endochitinase. Planta 194, 264–273. [PubMed] [Google Scholar]
- Zaat, S.A.J., Van Brussel, A.A.N., Tak, T., Lugtenberg, B.J.J., and Kijne, J.W. (1989). The ethylene-inhibitor aminoethoxyvinylglycine restores normal nodulation by Rhizobium leguminosarum biovar. viciae on Vicia sativa subsp. nigra by suppressing the ‘thick and short roots’ phenotype. Planta 177, 141–150. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Ripperger, A., and Ye, Z.-H. (2000). Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 123, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Maher, E.A., Masoud, S., Dixon, R.A., and Lamb, C.J. (1994). Enhanced protection against fungal attack by constitutive coexpression of chitinase and glucanase genes in transgenic tobacco. Bio/Technology 12, 807–812. [Google Scholar]