Abstract
We characterized rice cDNA sequences for OsDr1 and OsDrAp1, which encode structural homologs of the eukaryotic general repressors Dr1 and DrAp1, respectively. OsDr1 and OsDrAp1 are nuclear proteins that interact with each other and with the TATA binding protein/DNA complex. In vitro and in vivo functional analyses showed that OsDrAp1 functions as a repressor, unlike its role in other eukaryotic systems, in which DrAp1 is a corepressor. OsDr1 and OsDrAp1 functioned together as a much stronger repressor than either one alone. Functional dissections revealed that the N-terminal histone-fold domains of OsDr1 and OsDrAp1 were necessary and sufficient for their repression and protein–protein interaction with each other. The unique glutamine- and proline-rich domain of OsDr1 had no repression activity. The basic amino acid–rich region and an arginine and glycine repeat domain of OsDrAp1 enhanced its repression activity. Thus, although OsDr1 and OsDrAp1 function as repressors, the functions of the two components are reversed compared with those of their nonplant counterparts.
INTRODUCTION
Successful plant growth, development, and response to environmental cues require precise control of gene expression. A key control stage of differential gene expression is transcriptional regulation mediated by transcriptional factors. Studies of plant transcription factors have focused mainly on activators (Schwechheimer et al., 1998; Liu et al., 1999; Riechmann et al., 2000), whereas repressors have just begun to receive attention.
Several transcriptional repressors have been cloned from plants, and the elucidation of their functional mechanisms has begun. Transcriptional repressors have been found to mediate plant hormone signal transduction. For example, the soybean G box binding factor 2 (SGBF-2) is a basic leucine zipper (bZIP)–type factor that binds to the G box of the auxin response element of the GH3 promoter and represses both basal and auxin-induced transcription of a reporter gene (Liu et al., 1997). In Arabidopsis, auxin response factor 1 (ARF1) binds to the auxin response element TGTCTC and functions as a repressor (Ulmasov et al., 1999). Arabidopsis ethylene-responsive element binding factors 3 and 4 (AtERF3 and AtERF4) interact with the ethylene-responsive element (GCC box) and repress the basal activity of a reporter gene and the activity of the transcription activator AtERF5 (Fujimoto et al., 2000). Tobacco ERF3 (NtERF3) also functions as a GCC box–dependent transcriptional repressor. The C-terminal 35 amino acids of NtERF3, which contains the conserved L/FDLNL/F(X)P motif, are sufficient to confer the capacity for transcriptional repression on a heterologous DNA binding domain (Ohta et al., 2001). A development-specific transcriptional repressor gene, ROM2, has been cloned from French bean. ROM2 is a bZIP-type transcription factor that functions as a DNA binding site–dependent repressor. ROM2 represses PvALF-activated transcription of the promoter of phytohemagglutinin (DLEC2). The expression analysis indicates that the ROM2 repressor may play a role in silencing DLEC2 transcription during late embryogenesis (Chern et al., 1996). A Myb-type repressor gene, AtMYB4, involved in phenylpropanoid metabolism also has been cloned from Arabidopsis (Jin et al., 2000). AtMYB4 interacts directly with the promoter of the cinnamate 4-hydroxylase (C4H) gene and causes >20-fold repression of the expression of a C4H/GUS gene fusion in transfected protoplasts. The repression domain of AtMYB4 is thought to interact with the basal transcription machinery, thereby acting as a transcriptional repressor (Jin et al., 2000).
Transcriptional repressors associate with their target genes either directly through their DNA binding domains or indirectly by interacting with other DNA binding proteins or with the component of transcription machinery (Hanna-Rose and Hansen, 1996; Maldonado et al., 1999). All of the plant transcriptional repressors described above are gene specific and function by binding to specific DNA motifs. Transcriptional repressors that execute more general effects have not been described in plants. In other eukaryotic systems, several groups of general repressors have been identified to target components of the RNA polymerase II (RNAP II) core transcription machinery and inhibit the formation of the transcription initiation complex or inhibit transcriptional elongation (Maldonado et al., 1999). These general repressors globally affect RNAP II–mediated transcription.
The transcription of class II genes starts with the binding of TATA binding protein (TBP; the central component of TFIID) to the TATA box of a promoter. The binding of TBP to the promoter DNA is facilitated by TFIIA and is followed by TFIIB binding to give a more stable ternary complex that acts as the scaffold for subsequent recruiting of other general transcription factors and RNAP II (Orphanides et al., 1996). Several negative regulators have been identified to regulate the RNAP II–mediated transcriptional initiation in yeast, Drosophila, and mammalian systems. One of these negative regulators is the Dr1/DrAp1 complex. Dr1/DrAp1 was identified originally in human cells as a biochemical activity that inhibited TBP-dependent basal transcription in vitro (Inostroza et al., 1992). Dr1/DrAp1 globally represses RNAP II– and RNAP III–mediated transcription (Inostroza et al., 1992; White et al., 1994; Mermelstein et al., 1996; Gadbois et al., 1997; Kim et al., 1997; Prelich, 1997). Dr1 represses RNAP II transcription by precluding the entry of TFIIA and TFIIB into the preinitiation complex to prevent the formation of an active transcription complex (Kim et al., 1995; Goppelt et al., 1996; Cang et al., 1999). Biochemical analysis shows that Dr1 forms a heterotetramer with DrAp1 (for Dr1-associated protein1). DrAp1 itself cannot repress transcription. DrAp1 is a corepressor that enhances the repression activity of Dr1 (Mermelstein et al., 1996; Kim et al., 1997; Yeung et al., 1997). Drosophila Dr1/DrAp1 (dDr1/dDrAp1) is a bifunctional factor that represses the transcription of TATA-containing promoters and activates the transcription of a TATA-less promoter that contains a downstream element (Willy et al., 2000). Yeast Dr1/DrAp1 associates with the RNAP II preinitiation complex and selectively affects transcription both positively and negatively in vivo (Kim et al., 2000; Geisberg et al., 2001).
Here we report the characterization of two rice cDNAs, OsDr1 and OsDrAp1, which encode Dr1 and DrAp1 homologs, respectively. In vitro transcription analyses demonstrated that OsDrAp1 repressed the rice TATA binding protein (OsTBP)–enhanced transcription of a rice phenylalanine ammonia-lyase promoter/β-glucuronidase (GUS) gene fusion in rice whole-cell extracts. The combination of OsDr1 and OsDrAp1 eliminated the OsTBP-enhanced effects on the transcription of the reporter gene. This finding was confirmed by in vivo functional analysis, which showed that OsDrAp1 had strong repression activity, OsDr1 had weak repression activity, and OsDr1 and OsDrAp1 together functioned as a stronger repressor than either one alone. The histone-fold domains of OsDr1 and OsDrAp1 were necessary and sufficient for their functional interaction. Thus, although OsDr1/OsDrAp1, like its nonplant counterparts, functions as a repressor, it has a unique repression mechanism.
RESULTS
Isolation of cDNA Encoding Rice Dr1 and DrAp1
Expressed sequence tag (EST) clone r10n.PK0076.g1 was identified from the DuPont rice EST database (S. Tingey and G. Miao, unpublished data) to encode a truncated rice Dr1 homolog (OsDr1) with two unspliced introns (1082 and 84 bp). Using 5′ rapid amplification of cDNA ends and reverse transcriptase–mediated polymerase chain reaction techniques, we cloned the full-length cDNA of OsDr1. The coding region of OsDr1 is 891 bp; it encodes a polypeptide of 296 amino acids (molecular mass of 33.7 kD) that shows ∼34% identity to the human Dr1 (hDr1) and is much larger than that of Dr1 from other systems (Figure 1A). We also identified an EST (DuPont EST wre1n.pkoo37.g1) encoding a wheat Dr1 homolog (wDr1). The polypeptides of OsDr1 and wDr1 are ∼70% identical and are similar in size and structure (Figure 1A). Both OsDr1 and wDr1 have an extended C-terminal glutamine- and proline-rich domain (QP). The N-terminal 141 amino acids of OsDr1 is ∼68% identical to that of homologs from soybean (sDr1; DuPont EST ses2W.pk0043.b3) and Arabidopsis (aDr1; Kuromori and Yamamoto, 1994). Like Dr1 from nonplant systems, OsDr1 also has an N-terminal histone-fold domain that is ∼50 and 40% identical to the histone-fold domains of human Dr1 (hDr1) and yeast Dr1 (yDr1), respectively (Goppelt et al., 1996; Mermelstein et al., 1996; Yeung et al., 1997; Baxevanis and Landsman, 1998).
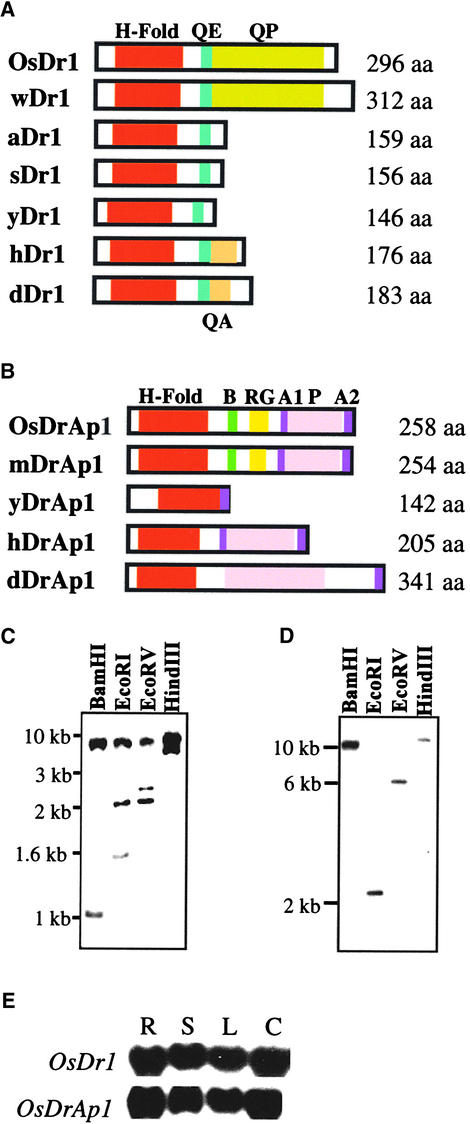
Figure 1.
Structure, Organization, and Expression of OsDr1 and OsDrAp1.
(A) OsDr1 was compared with Dr1 from wheat (wDr1), Arabidopsis (aDr1; Kuromori and Yamamoto, 1994), soybean (sDr1), yeast (yDr1; Kim et al., 1997), human (hDr1; Inostroza et al., 1992), and Drosophila (dDr1; Willy et al., 2000).
(B) OsDrAp1 was compared with DrAp1 from maize (mDrAp1), yeast (yDrAp1; Kim et al., 1997), human (hDrAp1; Goppelt et al., 1996), and Drosophila (dDrAp1; Willy et al., 2000).
H-Fold, histone-fold domain; QE, glutamine- and glutamate-rich domain; QA, glutamine- and alanine-rich domain; QP, glutamine- and proline-rich domain; aa, amino acids; B, basic amino acid stretch; RG, arginine-glycine repeat; A1 and A2, acidic amino acid–rich domain 1 and 2, respectively; P, proline-rich domain.
(C) and (D) DNA gel blot analysis of the organization of OsDr1 (C) and OsDrAp1 (D) in the rice genome.
(E) RNA gel blot analysis of the expression of OsDr1 and OsDrAp1 in rice. R, root; S, sheath and stem; L, leaf; C, suspension cells.
EST clone rls12.PK0015.e12 was identified from the DuPont EST database to encode a full-length rice DrAp1 homolog (OsDrAp1). The coding region of OsDrAp1 is 777 bp; it encodes a polypeptide of 259 amino acids (molecular mass of 28 kD), with ∼33% identity to DrAp1 from human (hDrAP1; Figure 1B) and ∼82% identity to a DrAp1 homolog from maize (mDrAp1; DuPont EST cs1.pk0049.a1). OsDrAp1 also has an N-terminal histone-fold domain that is ∼60 and 40% identical to the histone-fold domains of hDrAp1 and yeast DrAp1 (yDrAp1), respectively (Goppelt et al., 1996; Mermelstein et al., 1996; Kim et al., 1997; Baxevanis and Landsman, 1998). In OsDrAp1, there are two acidic amino acid–rich domains (residues 155 to 189 and 249 to 259) and one proline-rich domain (residues 192 to 238). Compared with nonplant DrAp1, OsDrAp1 has two unique domains, a basic amino acid–rich region (residues 102 to 119) and a six-arginine and six-glycine (RG) repeat region (residues 135 to 146).
Organization and Expression of OsDr1 and OsDrAp1
To estimate the number of OsDr1 and OsDrAp1 genes in the rice genome, we performed DNA gel blot analysis. Rice genomic DNA gel blots were hybridized separately with the full-length coding region of OsDr1 and OsDrAp1. OsDr1 probe hybridized to two to three restriction fragments of rice genomic DNA digested with BamHI, EcoRI, EcoRV, and HindIII (Figure 1C), suggesting that there is an OsDr1 gene family composed of no more than three genes in the rice genome. OsDrAp1 probe hybridized to one restriction fragment of rice genomic DNA digested with BamHI, EcoRI, EcoRV, and HindIII (Figure 1D), suggesting that there is only one OsDrAp1 gene in the rice genome.
To analyze the expression patterns of OsDr1 and OsDrAp1 in rice, we performed RNA gel blot analyses. RNA gel blots containing RNA samples from different tissues and suspension cells were hybridized with the same probes used for DNA gel blot analysis. The results showed that both OsDr1 and OsDrAp1 were expressed constitutively and abundantly in rice root, sheath, stem, leaf, and suspension cells (Figure 1E).
Molecular Interaction between OsDr1 and OsDrAp1
To determine whether OsDr1 and OsDrAp1 interact with each other, we first used the yeast two-hybrid system (Chien et al., 1991). The coding regions of OsDr1 and OsDrAp1 were inserted into the vectors pAD-Gal4 and pBD-Gal4 (Stratagene, San Diego, CA) to generate pAD::OsDr1 and pBD::OsDrAp1 constructs, respectively (Figure 2A). Yeast cotransformed with pAD::OsDr1 and pBD::OsDrAp1 could grow on −Trp, −Leu, and −His selection media (Figure 2B). Neither pAD::OsDr1 nor pBD::OsDrAp1 alone could support yeast growth on the triple selection media. To exclude the possibility that there was a mutation in yeast supporting growth on the triple selection media, we performed β-galactosidase assays. The results showed that only the yeast line containing both pAD::OsDr1 and pBD::OsDrAp1 constructs had β-galactosidase activity (Figure 2C); yeast lines containing either pAD::OsDr1 or pBD-OsDrAp1 alone had no β-galactosidase activity. These data provide evidence that there is a physical interaction between OsDr1 and OsDrAp1 in yeast.
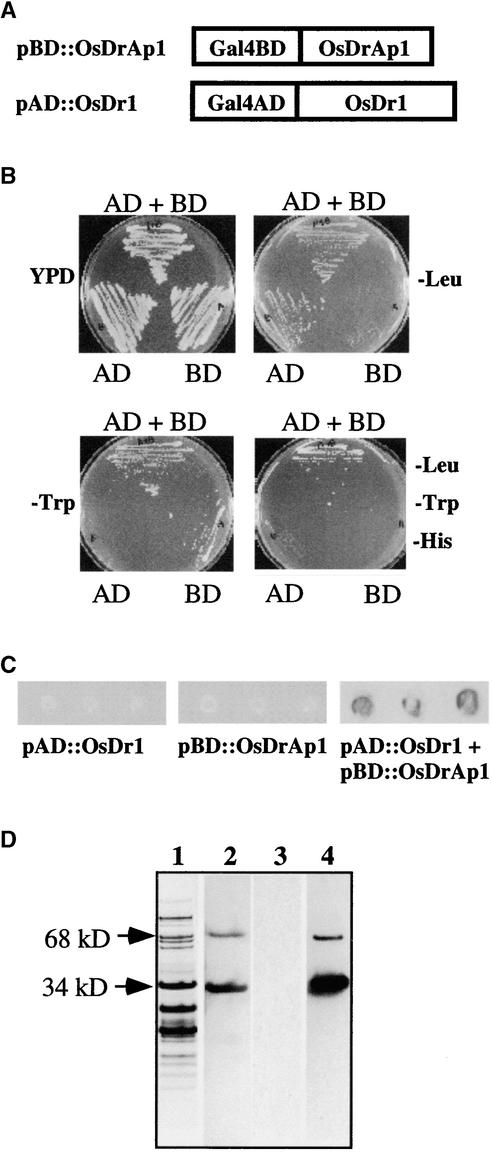
Figure 2.
Interaction of OsDr1 and OsDrAp1 in Yeast and in Vitro.
(A) Scheme of the fusion proteins in pBD::OsDrAp1 and pAD::OsDr1 constructs.
(B) Yeast growth on selection medium. AD, yeast strain transformed with pAD::OsDr1; BD, yeast strain transformed with pBD::OsDrAp1; AD + BD, yeast strain transformed with pAD::OsDr1 and pBD:: OsDrAp1 together. YPD, 20 g/L peptone, 10 g/L yeast extract, 20 g/L glucose, 40 mg/L adenine sulfate, 15 g/L agar, pH 5.8.
(C) β-Galactosidase activities in yeast cells transformed with pBD:: OsDrAp1, pAD::OsDr1, or pBD::OsDrAp1 and pAD::OsDr1 together.
(D) Interaction of recombinant OsDr1 and OsDrAp1 in vitro. Lane 1, the gel was stained with Coomassie blue; lanes 2 and 3, the proteins were blotted onto nitrocellulose membranes and incubated with His tag antibody (lane 2) and T7 tag antibody (lane 3); lane 4, the proteins blotted onto nitrocellulose membrane were denatured, renatured, and incubated with purified OsDr1 and with the T7 tag antibody.
Next, we performed protein overlay analysis (Harlow and Lane, 1988; Chen and Evans, 1995) using recombinant OsDr1 and OsDrAp1 that had 6 × His tags at their C termini; OsDr1 also had a T7 tag at its N terminus. Partially purified OsDrAp1 and BSA (as a control) were separated on four sets of 4 to 12% gradient polyacrylamide gels. One gel was stained with Coomassie blue to show the pattern of partially purified OsDrAp1 (Figure 2D, lane 1). The other three gels were blotted onto nitrocellulose membranes. The first membrane was incubated with His tag antibody to detect OsDrAp1. The results showed that there was a 34-kD OsDrAp1, which accounted for ∼15% of the total loaded proteins based on the Coomassie blue staining (Figure 2D, lanes 1 and 2). There also was a positive signal located at the 68-kD position, which might be a dimer of recombinant OsDrAp1. The proteins on the other two membranes were denatured and renatured. One of the membranes was incubated with the T7 tag antibody. The result showed no cross-reaction between the T7 tag antibody and OsDrAp1 (Figure 2D, lane 3). The other membrane was incubated with purified OsDr1 and then incubated with the T7 tag antibody. The signal pattern in the third membrane was exactly the same as in the first membrane (Figure 2D, lanes 2 and 4), suggesting that there was OsDr1/OsDrAp1 complex formation on the membrane that gave a positive signal with the T7 tag antibody. The signal detected at the 68-kD position (Figure 2D, lane 4) suggested that OsDr1 also interacted with the putative OsDrAp1 dimer. The other proteins in the partially purified OsDrAp1 preparations (Figure 2D, lane 1) and BSA (data not shown) did not interact with either the His tag or T7 tag antibodies.
OsDr1 and OsDrAp1 Are Nuclear Proteins Interacting with the TBP/DNA Complex
To study the localization of OsDr1 and OsDrAp1 in plant cells, we inserted the coding regions of OsDr1 and OsDrAp1 into the vector pRTL2-mGFP (von Arnim et al., 1998) to generate pOsDr1/mGFP and pOsDrAp1/mGFP constructs (Figure 3A), which contain OsDr1/mGFP and OsDrAp1/mGFP fusion genes, respectively. These constructs were introduced into tobacco NT-1 protoplasts, and the localization of the fusion proteins and the control mGFP were monitored by confocal microscopy in live transfected protoplasts. Figure 3B shows that the mGFP itself is localized in both cytoplasm and the nucleus. However, both OsDr1/mGFP and OsDrAp1/mGFP are localized nearly exclusively in the nucleus, excluding the nucleolus, of transfected protoplasts.
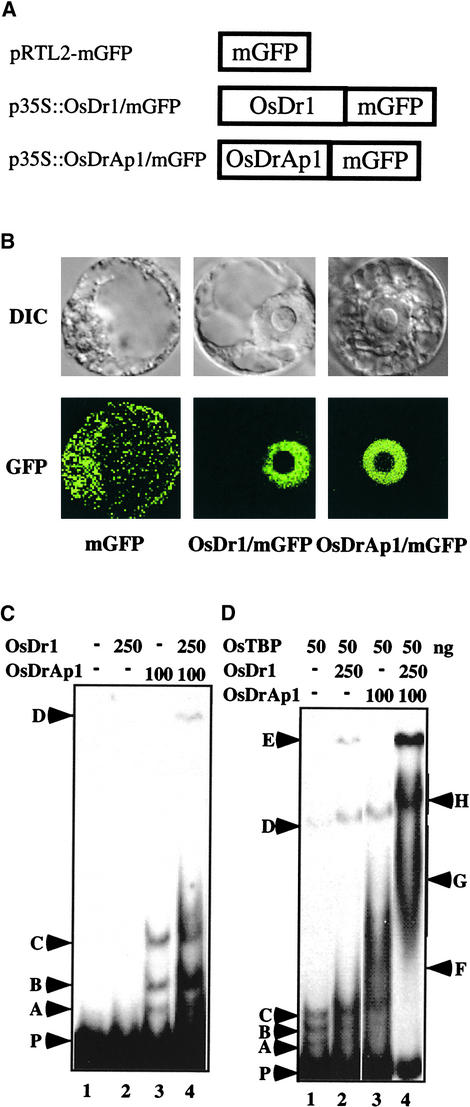
Figure 3.
Nucleus-Localized OsDr1 and OsDrAp1 Interact with the OsTBP/DNA Complex.
(A) Scheme of proteins in constructs pRTL2-mGFP, p35S::OsDr1/mGFP, and p35S::OsDrAp1/mGFP.
(B) Localization of mGFP, OsDr1/mGFP, and OsDrAp1/mGFP in tobacco NT-1 cells. DIC and GFP indicate differential interference contrast images and mGFP fluorescence images of the same cell transfected with the construct indicated.
(C) Recombinant OsDrAp1 binds to DNA. Gel shift assays with purified recombinant OsDr1 and OsDrAp1 are shown. Components and the positions of complexes are as indicated.
(D) Recombinant OsDr1 and OsDrAp1 interact with the OsTBP/DNA complex. Gel shift assays with purified recombinant OsDr1, OsDrAp1, and OsTBP are shown. Components and the positions of complexes are as indicated.
We then used gel shift assays to determine whether OsDr1 and OsDrAp1 interact with DNA. Using the DNA fragment of the rice pal gene from the −61 to +79 region (transcription start site as +1) as a probe (Zhu et al., 1995a) and purified recombinant OsDr1 and OsDrAp1 (data not shown), we found that OsDr1 alone had no DNA affinity. There was no detectable shifted band when 250 ng of purified OsDr1 was used in the reactions (Figure 3C, lane 2). Purified recombinant OsDrAp1 interacted with the DNA and formed three shifted bands, A, B, and C (Figure 3C, lane 3). One hundred nanograms of OsDrAp1 was enough to detect the DNA–protein interaction in the assays. OsDr1 and OsDrAp1 together produced stronger signals for the three shifted bands and also produced another shifted band (Figure 3C, lane 4, band D).
Next, we studied the interactions of OsDr1 and OsDrAp1 with the OsTBP/DNA complex. Purified recombinant OsTBP had weak affinity with the TATA element of the rice pal promoter (Q. Zhu, M.I. Ordiz, T. Dabi, R.N. Beachy, and C. Lamb, unpublished data) and produced several bands in gel shift assays (Figure 3D, lane 1, bands A through D). Upon addition of OsDr1 into the reaction of OsTBP and DNA, all of the OsTBP shifted bands were supershifted. More importantly, another shifted band also was produced at position E (Figure 3D, lane 2). The addition of OsDrAp1 to the reaction of OsTBP and DNA also caused OsTBP shifted bands A, B, and C to be supershifted to the region labeled F (Figure 3D, lane 3). The addition of OsDr1 and OsDrAp1 together not only supershifted the band from the F to the G region and the D band to H, it also produced a much stronger signal of the E band (Figure 3D, lane 4). These data showed that although either OsDr1 or OsDrAp1 alone interacted with the OsTBP/DNA complex, together they interacted strongly with the TBP/DNA complex to form larger complexes.
OsDr1 and OsDrAp1 Repress OsTBP-Enhanced pal Transcription in Vitro
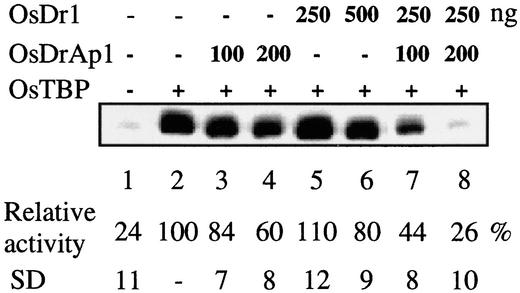
To study the function of OsDr1 and OsDrAp1, we used a plant in vitro transcription system using the rice pal promoter (−81 to +45)/GUS gene fusion as a reporter and rice whole-cell extracts as the source of the transcription machinery (Zhu et al., 1995b). Figure 4 shows that recombinant OsTBP enhanced pal basal transcription in rice whole-cell extracts (lanes 1 and 2), which confirmed our previous report (Q. Zhu, M.I. Ordiz, T. Dabi, R.N. Beachy, and C. Lamb, unpublished data). The addition of purified recombinant OsDrAp1 repressed transcription of the reporter gene in a dose-dependent manner. Two hundred nanograms of OsDrAp1 repressed transcription of the reporter gene by ∼40% (lane 4). However, the repression activity of OsDr1 was not as strong as that of OsDrAp1: 250 ng of OsDr1 exhibited no repression activity (lane 5), and 500 ng of OsDr1 repressed the transcription of the reporter gene by ∼20% (lane 6). However, a combination of 250 ng of OsDr1 with 200 ng of OsDrAp1 repressed the transcription of the reporter gene by ∼74%, which eliminated almost all of the enhanced effects of OsTBP (lanes 1, 2, and 8). This experiment was repeated four times, and the results were consistent.
Figure 4.
In Vitro Transcriptional Repression Activities of OsDr1 and OsDrAp1.
Recombinant OsTBP, OsDr1, and OsDrAp1 were added to the in vitro transcription mixture containing template and buffer at 10 min before the addition of rice whole-cell extracts. The transcript level of each reaction was measured by primer extension analysis. The products of primer extension reactions were separated on an 8% polyacrylamide denaturing gel.
OsDr1 and OsDrAp1 Function as Repressors in Vivo
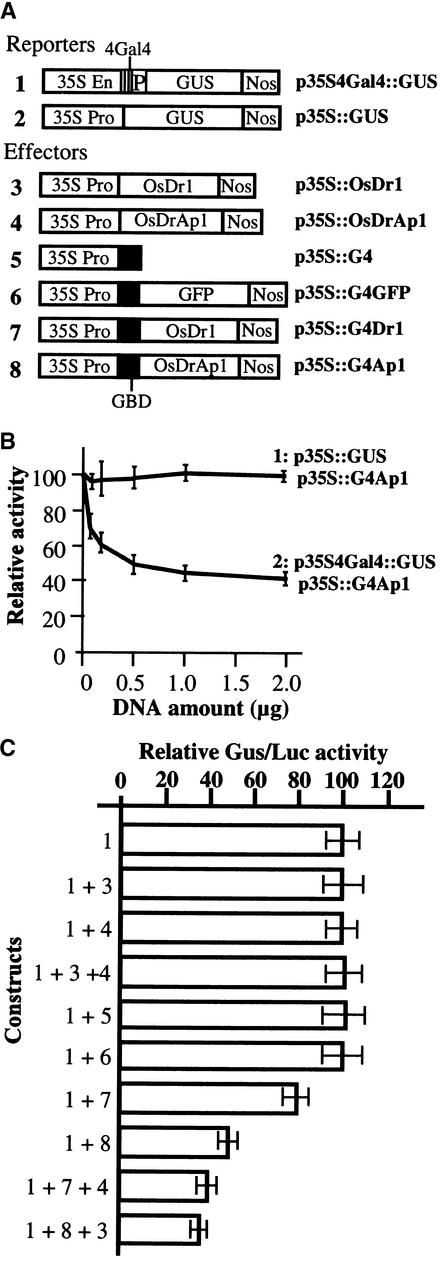
To elucidate the in vivo function of OsDr1 and OsDrAp1, we used the tobacco protoplast transfection assay (Liu et al., 1997). p35S4Gal4::GUS (Figure 5A) was used as the reporter, in which a chimeric promoter consisting of the 35S enhancer/tetramer of the GAL4 binding site/bean β-phaseolin storage protein gene minimal promoter (−65 to +80) was fused with the GUS open reading frame. p35S::LUC, which contained a 35S promoter/luciferase gene fusion, was used as an internal control for the transfection experiments. The ratio of GUS activity to luciferase activity was used as the measure of GUS gene expression. In NT-1 tobacco protoplasts, the activity of the chimeric promoter in the p35S4Gal4::GUS construct was as high as that in the wild-type 35S promoter (data not shown). p35S::G4 and p35S::G4GFP (Figure 5A), which contained the 35S promoter/GAL4 DNA binding domain gene fusion and the 35S promoter/GAL4 DNA binding domain fused in frame with the GFP coding region, respectively, were used as controls. p35S::G4Dr1 and p35S::G4Ap1 (Figure 5A), which contained the 35S promoter of Cauliflower mosaic virus/GAL4 DNA binding domain fused in frame with the OsDr1 and OsDrAp1 coding regions, respectively, were used as effectors. p35S::OsDr1 and p35S::OsDrAp1 (Figure 5A), which contained the 35S promoter of Cauliflower mosaic virus fused with OsDr1 and OsDrAp1, respectively, also were used in the transfection experiment.
Figure 5.
OsDr1/OsDrAp1 Functions as a Strong Repression Complex in Vivo.
(A) Scheme of the gene fusion structures in various constructs. 35S En, enhancer of the 35S promoter; 35S Pro, 35S promoter; 4Gal4, a tetramer of the Gal4 DNA binding site; P, minimal promoter (−65 to +80) of the bean β-phaseolin storage protein gene; GUS, β-glucuronidase coding region; Nos, nopaline synthase terminator; GBD, Gal4 DNA binding domain; GFP, green fluorescent protein.
(B) Titration of the Gal4/OsDrAp1 fusion protein (p35S::G4Ap1) on the repression of reporters. 1, GUS activities in tobacco protoplasts transfected with 2 μg of p35S::GUS and the indicated amounts of p35S::G4Ap1; 2, GUS activities of tobacco protoplasts transfected with 2 μg of p35S4Gal4::GUS and the indicated amounts of p35S::G4Ap1. Error bars indicate ±sd.
(C) Relative GUS/LUC activities in tobacco protoplasts transfected with the indicated constructs. Error bars indicate ±sd.
Cotransfection of the reporter p35S4Gal4::GUS with the effector p35S::G4Ap1 resulted in repression of the reporter gene, and the repression was a DNA dose-dependent response in the range of 0.1 to 2 μg of the effector; 2 μg of p35S::G4Ap1 repressed ∼55% of the expression of the p35S4Gal4::GUS reporter (Figure 5B, 2, and Figure 5C, 1 + 8). Two micrograms of effector DNA then was used for all further experiments. The repression by Gal4/OsDrAp1 also was GAL4 binding site specific, because there was no repression of GUS activity when p35S::GUS and p35S::G4Ap1 were used together for transfection (Figure 5B, 1).
Figure 5C shows that cotransfection of the reporter p35S4Gal4::GUS with the effector p35S::G4Dr1 also resulted in repression of the reporter gene expression (1 + 7). Overexpression of the GAL4 DNA binding domain alone did not repress the expression of the chimeric promoter (1 + 5), which was in agreement with other reports (Jin et al., 2000). Overexpression of the GAL4/GFP fusion protein did not repress the transcription of the chimeric promoter either (1 + 6). These results suggest that GAL4/Dr1- and GAL4/DrAp1-mediated transcriptional repression are not caused by steric hindrance of the GAL4 DNA binding domain. Overexpression of OsDr1 or OsDrAp1 alone or OsDr1 and OsDrAp1 together did not repress the expression of p35S4Gal4::GUS (1 + 3, 1 + 4, 1 + 3 + 4), which confirmed that OsDr1 and OsDrAp1 needed the GAL4 DNA binding domain to provide the GAL4 binding site–specific recognition in the Gal4/OsDr1 and Gal4/OsDrAp1 chimeric repressors to execute their functions (1 + 7, 1 + 8). The combination of p35S::G4Dr1 and p35S::OsDrAp1 exhibited stronger repression than did p35S::G4Dr1 alone (1 + 7 + 4, 1 + 7), whereas cotransfection of the reporter with p35S::OsDrAp1 had no effect on the reporter gene expression (1 + 4). Similarly, the combination of p35S::G4Ap1 and p35S::OsDr1 exhibited stronger repression than did p35S::G4Ap1 alone (1 + 8 + 3, 1 + 8), whereas cotransfection of the reporter with p35S::OsDr1 had no effect on the reporter gene expression (1 + 3). All of these transfection experiments were repeated at least three times with three to four replicates used each time, and the results were consistent.
Functional Dissection of OsDr1 and OsDrAp1
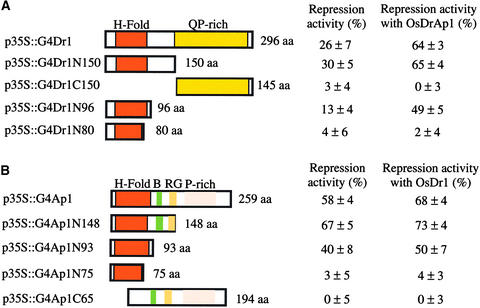
To dissect the functional domains of OsDr1, we made a series of N- and C-terminal deletion mutants and tested them for repression in vivo (Figure 6A). Construct p35S:: G4Dr1N150, which contained the GAL4 DNA binding domain fused in frame with the N-terminal 150 amino acids of OsDr1, had slightly stronger repression activity than full-length OsDr1. Construct p35S::G4Dr1C150, which contained the Gal4 DNA binding domain fused in frame with the OsDr1 C-terminal region from residue 151 to 296, had no repression activity. The N-terminal 96 amino acids of OsDr1 in p35S:: G4Dr1N96 still had repression activity. The N-terminal 80 amino acids of OsDr1 in p35S::G4Dr1N80, with a truncated histone-fold domain, had no repression activity. These results suggest that the histone-fold domain located in the N terminus of OsDr1 is necessary and sufficient for its in vivo repression activity.
Figure 6.
Histone-Fold Domains of OsDr1 and OsDrAp1 Are Necessary and Sufficient for Their Interaction and Repression Activities in Vivo.
Schemes of the mutants of OsDr1 (A) and OsDrAp1 (B) in the indicated constructs. Repression activity was measured as a percentage of the reduction of the GUS/LUC ratio ±sd. See Figure 1 for abbreviations.
(A) The repression activities of Gal4/OsDr1 and its derivatives with OsDrAp1 were analyzed by cotransfection of various constructs with p35::OsDrAp1.
(B) The repression activities of Gal4/OsDrAp1 and its derivatives with OsDr1 were analyzed by cotransfection of various constructs with p35::OsDr1.
Serial deletions of OsDrAp1 also were tested for repression in vivo (Figure 6B). Construct p35S::G4Ap1N148, which had a deletion of 111 amino acids at the C terminus of OsDrAp1, had stronger repression activity than full-length OsDrAp1. The N-terminal 93 amino acids of OsDrAp1 (p35S::G4Ap1N93) had repression activity. However, the N-terminal 75 amino acids (p35S::G4Ap1N75) had no repression activity. Deletion of the N-terminal 64 amino acids of OsDrAp1 (p35S::G4Ap1C65) also eliminated the repression activity of OsDrAp1. These data demonstrated that the N-terminal histone-fold domain of OsDrAp1 was necessary and sufficient for repression and that residues 93 to 148 were important for strong repression.
Combinations of various OsDr1 deletion mutants with p35S::OsDrAp1 led to stronger repression activity than the OsDr1 mutant alone. p35S::OsDrAp1 had no effect if the OsDr1 mutant had no repression activity by itself (Figure 6A). Similarly, combinations of various OsDrAp1 deletion mutants with p35S::OsDr1 showed stronger repression activity than the OsDrAp1 mutants alone, and p35S::OsDr1 had no effect if the OsDrAp1 mutant had no repression activity by itself (Figure 6B). Therefore, the histone-fold domain in both OsDr1 and OsDrAp1 is necessary for their functional interaction with each other to execute their combined repression activity.
DISCUSSION
We elucidated the functions of both components of the rice Dr1/DrAp1 complex through a combination of biochemical and molecular approaches. Our data demonstrate that although OsDr1 and OsDrAp1 are structural homologs of nonplant Dr1 and DrAp1, respectively, their functions are reversed compared with those of their nonplant counterparts (Inostroza et al., 1992; White et al., 1994; Gadbois et al., 1997; Kim et al., 1997; Prelich, 1997). Unlike in other systems, in which Dr1 is the repressor and DrAp1 functions as a regulatory subunit, OsDrAp1 has strong repression activity and OsDr1 has weak repression activity and plays the regulatory role of enhancing the repression activity of OsDrAp1.
Structure and Function Relationship of OsDr1 and OsDrAp1
OsDr1 is approximately twice as large as Dr1 from soybean, Arabidopsis (Kuromori and Yamamoto, 1994), and nonplant Dr1 (Figure 1A). The possibility that this increased length of the rice cDNA clone was attributable to a chimeric insert was excluded by the discovery of an EST from wheat encoding a Dr1 homolog of similar size and structure (Figure 1A). Analysis of the whole genome sequence of Arabidopsis shows that there are two genes encoding Dr1 (Riechmann et al., 2000). The significance of the size difference in Dr1 homologs from dicot and monocot plants is unknown. DNA gel blot results (Figure 1C) suggest that there is a small OsDr1 gene family in the rice genome. It will be interesting to determine whether rice has another Dr1 gene encoding a protein with a structure similar to that of dicot plant and nonplant Dr1.
Unlike hDr1 and dDr1, which have short C-terminal QA domains, OsDr1 has a long QP domain (Figure 1A). The QA domain of hDr1 had repression activity (Yeung et al., 1997; Goppelt et al., 1996). The QP domain of OsDr1 had no repression activity when tethered to the chimeric promoter (Figure 6A). Deletion of the QP domain in OsDr1 gave slightly stronger repression than did wild-type OsDr1 (Figure 6A). These results suggest that the QP domain of OsDr1 is not involved in repression or that it needs OsDrAp1 to execute its function.
In human and Drosophila, DrAp1 is larger than Dr1, whereas OsDrAp1 is smaller than OsDr1 (Figure 1B). Compared with nonplant DrAp1, OsDrAp1 has at least two extra motifs. The first one is a basic amino acid–rich motif (PRRRKAL) that the pSORT program recognizes as one of the three nuclear localization signal peptides. The second one is a six-RG repeat. These two domains are important for the strong repression activity of OsDrAp1 (Figure 6B). However the C-terminal 111 amino acids of OsDrAp1, which contains the two acidic domains and the proline-rich domain, is not involved in repression.
The N-terminal histone-fold domain is the most conserved region between plant and nonplant homologs of both Dr1 and DrAp1 (Figures 1A and 1B). In nucleosomes, the histone-fold motifs serve as dimerization and DNA binding domains of H2A-H2B and H3-H4 pairs (Baxevanis and Landsman, 1998). The octamic histone-fold domains of both H2A-H2B and H3-H4 in the nucleosome consist of three helices (Arents et al., 1991). The histone-fold domains of OsDr1 and OsDrAp1 resemble the same domains of H2B and H2A, respectively. Both contain one long middle helix and two short end helices in their predicted secondary structures (data not shown). The histone-fold domains of OsDr1 and OsDrAp1 are essential for their interaction and repression. The N-terminal 96 amino acids of OsDr1 and 93 amino acids of OsDrAp1, which contain the full-length histone-fold domain, are necessary and sufficient for their repression and functional interaction with the full-length OsDrAp1 and OsDr1 in transfected protoplasts, respectively. Deletions that affected the histone-fold domain in either OsDr1 or OsDrAp1 resulted in the total loss of their repression and interaction with OsDrAp1 or OsDr1, respectively (Figure 6). The histone-fold domains of OsDr1 and OsDrAp1, like those of H2A and H2B, may need to interact with each other and with DNA to execute their functions.
Molecular Interaction between OsDr1 and OsDrAp1
In nonplant systems, Dr1 and DrAp1 interact with each other to form a repression complex (Inostroza et al., 1992; Kim et al., 1995, 1997; Goppelt et al., 1996; Mermelstein et al., 1996; Prelich, 1997; Yeung et al., 1997). To study the possible interaction between OsDr1 and OsDrAp1, we used both in vivo and in vitro approaches. Yeast two-hybrid analysis showed that there was an interaction between OsDr1 and OsDrAp1 (Figures 2A to 2C). Interestingly, the interaction between OsDr1 and OsDrAp1 in yeast did not interfere with GAL4-based activation. The GAL4 activation domain might not be masked by the OsDr1/OsDrAp1 interaction. We also found that the N-terminal 96 amino acids, but not 80 amino acids of OsDr1, in the pAD::OsDr1 construct was sufficient to interact with pBD::OsDrAp1; likewise, the N-terminal 93 amino acids, but not 75 amino acids of OsDrAp1 in the pBD::OsDrAp1 construct, was sufficient to interact with pAD::OsDr1. Deletion of the N-terminal 64 amino acids of OsDrAp1 in the pBD::OsDrAp1 construct eliminated its ability to interact with pAD::OsDr1 (data not shown). These results suggest that the N-terminal histone-fold domains of both OsDr1 and OsDrAp1 are necessary and sufficient for their interaction with each other in yeast.
Protein overlay analysis showed that both the monomer and the putative dimer of OsDrAp1 interacted with OsDr1 on membranes (Figure 2D), which was in agreement with the reports that hDr1 and hDrAp1 could form heterotetramers (Mermelstein et al., 1996; Yeung et al., 1997). From these in vivo and in vitro data, we conclude that there is a physical interaction between OsDr1 and OsDrAp1.
OsDrAp1 Functions as a Repression Subunit Both in Vitro and in Vivo
We found that OsDrAp1 alone had repression activity both in vitro and in vivo, which was different from the regulatory function of the nonplant DrAp1 subunit. Recombinant OsTBP enhanced the pal basal transcription in a TATA box–dependent manner (Q. Zhu, M.I. Ordiz, T. Dabi, R.N. Beachy, and C. Lamb, unpublished data). OsDrAp1 effectively repressed OsTBP-enhanced pal transcription in rice whole- cell extracts (Figure 4, lanes 3 and 4), suggesting that OsDrAp1 functioned as a repressor in vitro.
Apparently, a greater amount of recombinant OsDr1 was needed for repression in rice whole-cell extract. Five hundred nanograms of OsDr1 gave ∼20% repression (Figure 4, lane 6). However, a combination of OsDr1 and OsDrAp1 gave much stronger repression than either one alone (Figure 4, lanes 7 and 8). In four repeated experiments, although the absolute signal among gels was different, the patterns were similar and the results were consistent. These data showed that OsDrAp1 functioned as a repression subunit and that OsDr1 and OsDrAp1 together functioned as a stronger repressor. We also used the in vitro transcription system to analyze the functions of rice transcription factor Rf2a (Yin et al., 1997) and Arabidopsis factor HY5 (Oyama et al., 1998) and found that both Rf2a and HY5 functioned as activators in vitro (Q. Zhu, M.I. Ordiz, T. Dabi, R.N. Beachy, and C. Lamb, unpublished data; Q. Zhu, R. Larkin, C. Fankhauser, M. Chatterjee, B. Maxwell, T. Oyama, K. Okada, C. Lamb, and J. Chory, unpublished data), in agreement with their functions in vivo. Therefore, the rice in vitro transcription system is a powerful approach to study the functions of both activators and repressors.
In agreement with our in vitro data, OsDrAp1 itself also functioned as a repressor in vivo when tethered to a chimeric promoter by the yeast GAL4 DNA binding domain (Figure 5). Overexpression of the GAL4 DNA binding domain alone or the GAL4/GFP fusion protein did not repress the transcription of the chimeric promoter (Figure 5C), in agreement with other reports (Jin et al., 2000). However, overexpression of the GAL4/OsDrAp1 fusion protein repressed transcription of the reporter (Figures 5B and 5C). Interestingly, overexpression of OsDrAp1 alone did not repress reporter gene expression (Figure 5C). We speculate that there are thousands of genes that are “on” in the transfected protoplasts and that the amount of OsDrAp1 is not enough to repress all of these genes. In the in vitro transcription analysis, there is only one promoter available to be repressed. When OsDrAp1 was tethered to a specific promoter, it showed strong repression activity. Similarly, overexpression of OsDr1 alone did not give any repression, whereas overexpression of GAL4/OsDr1 gave ∼26% repression (Figure 5C). These data showed that OsDrAp1 had stronger repression activity than did OsDr1 when tethered to a specific promoter. The combination of GAL4/OsDrAp1 with OsDr1, or GAL4/OsDr1 with OsDrAp1, gave stronger repression activity than did either GAL4/OsDrAp1 or GAL4/OsDr1 alone. The combination of OsDr1 and OsDrAp1 did not repress reporter gene expression (Figure 5C). Thus, we have defined the function of OsDrAp1 as a repression subunit and the function of OsDr1 as a regulatory subunit; these functions are opposite those of their structural counterparts in nonplant systems. The unique structural features of both OsDr1 and OsDrAp1 likely contribute to their different functions relative to their nonplant counterparts.
Repression Mechanism of the OsDr1/OsDrAp1 Complex
Transcriptional repression usually occurs during the assembly of the transcription initiation complex and early elongation (Hanna-Rose and Hansen, 1996; Maldonado et al., 1999). There are several transcriptional repression mechanisms. (1) A repressor can target transcription activators for degradation. For example, Arabidopsis COP1 and HY5 factors act antagonistically. HY5 is a bZIP transcription activator that binds to the promoters of light-inducible genes to activate gene expression and photomorphogenic development. COP1 is a RING finger protein with a WD-40 repeat that interacts directly with HY5 and may target it for proteasome-mediated degradation in the nucleus (Osterlund et al., 2000). In yeast, Srb10 is a cyclin-dependent kinase regulated by Srb11 cyclin. The Srb10/11 kinase is essential for a normal transcriptional response to galactose induction in vivo (Liao et al., 1995). Srb10 phosphorylates activator Gcn4, thereby marking it for recognition by SCF(Cdc4) ubiquitin ligase, which results in the degradation of Gcn4 (Chi et al., 2001). (2) A repressor can mask a transcriptional activator or the components of the transcription machinery. Yeast Srb10 also can phosphorylate the C-terminal domain of RNAP II to inhibit its participation in the transcription initiation complex, thereby inhibiting transcription (Hengartner et al., 1998). (3) A repressor can displace an activator to bind to a specific DNA element. MOT1, which is required for yeast viability, represses transcription by displacing TBP from DNA (Darst et al., 2001) and regulating the distribution of TBP between promoter and nonpromoter sites (Muldrow et al., 1999). (4) A repressor can block the interaction of an activator with other components of the transcription machinery or block the assembly of the transcription machinery. In human and yeast, Dr1/DrAp1-mediated repression requires Dr1 to interact with the TBP promoter complex. The Dr1/TBP/DNA complex precludes its interaction with TFIIB, thereby inhibiting the formation of the transcription machinery (Goppelt et al., 1996; Kim et al., 1995; Cang et al., 1999). Dr1 and DrAp1 are required for yeast viability (Kim et al., 1997). (5) A repressor can modify chromatin structure. Retinoblastoma (Rb) protein recruits the Sin3 histone deacetylase complex to repress transcription, presumably by altering local chromatin structure (Brehm et al., 1998). Rb also associates with histone methylase SUV39H1 and the methyl-lysine binding protein HP1 to repress the cyclin E promoter through the methylation of histone 3 (Nielsen et al., 2001).
Some repressors can repress a targeted gene through multiple mechanisms. Ssn6-Tup1, which represses >150 genes in yeast, represses transcription by interfering with transcription activators, recruiting histone deacetylases, and positioning nucleosomes at the promoters of its targeted genes (Smith and Johnson, 2000). Different repressors can use the same mediator to repress transcription. Dr1/DrAp1, MOT1, and NOT1 are different repressors that repress different genes. However, all of them interact with TBP directly, blocking its participation in the active transcription process (Maldonado et al., 1999). Transcriptional repression is an important mechanism involved in the regulation of gene expression. It is a dynamic and complex process.
To study the repression mechanisms of OsDr1/OsDrAp1, we used several approaches. First, we found that both OsDr1 and OsDrAp1 were nuclear proteins. The pSORT program predicted that OsDr1 and OsDrAp1 had ∼93 and 90% possibility, respectively, of being nuclear proteins. Using OsDr1/mGFP and OsDrAp1/mGFP gene fusions, we showed that these fusion proteins localized in the nuclei of transfected protoplasts (Figures 3A and 3B). Second, we found that OsDrAp1 was a DNA binding protein. Purified full-length recombinant OsDrAp1 produced several shifted bands in the gel shift assay (Figure 3C), suggesting that OsDrAp1 had weak affinity for DNA. Human DrAp1 itself had no affinity for DNA (Goppelt et al., 1996). The basic region, RG domain, and histone-fold domain of OsDrAp1 likely contribute its DNA binding activity. OsDr1 not only enhanced the DNA binding of OsDrAp1 but also produced a higher order complex with OsDrAp1 and DNA (Figure 3C). Third, we found that both OsDr1 and OsDrAp1 alone could interact with the OsTBP/DNA complex. The combination of OsDr1 and OsDrAp1 with OsTBP and DNA probe produced stronger and further-shifted bands, suggesting that OsDr1 and OsDrAp1 together interacted with the TBP/DNA complex (Figure 3D). In the case of human Dr1 and DrAp1, only the Dr1/DrAp1 complex can interact with the TBP/DNA complex (Goppelt et al., 1996). Finally, we found that recombinant OsDrAp1 itself repressed pal transcription in OsTBP-enhanced rice whole-cell extracts. OsDr1 and OsDrAp1 together eliminated the enhancement effect of OsTBP (Figure 4). These data suggest that OsDr1/OsDrAp1 interacts with the TBP/DNA complex to execute its repressor function.
In conclusion, we have discovered sharp differences in the repression mechanisms of the OsDr1/OsDrAp1 complex and their nonplant counterparts. Structural comparisons clearly show that OsDr1 and OsDrAp1 are from common ancestors, like Dr1 and DrAp1 from other systems. However, both OsDr1 and OsDrAp1 have acquired extra domains during evolution to execute their unique functions in rice. Comparative studies of Dr1 and DrAp1 genes from monocots and dicots will provide in-depth information regarding repression mechanisms and the evolutionary significance of the plant Dr1/DrAp1 complex. Because both OsDr1 and OsDrAp1 are expressed constitutively at high levels and they interact with the TBP/DNA complex to mediate their repression functions, it will be of interest to identify the factors and signals that regulate the repression functions of the plant Dr1/DrAp1 complex.
METHODS
Plant Materials
Rice (Oryza sativa cv IR72) suspension cells were cultured in N6 medium (Zhu et al., 1995a). Rice seed were germinated on wet Whatman paper in the dark at 27°C. Tobacco (Nicotiana tabacum) NT-1 suspension cells were cultured in Murashige and Skoog (1962) liquid medium (Allen et al., 1993).
Nucleic Acid Manipulation
General molecular cloning was performed according to standard methods (Sambrook et al., 1989). When polymerase chain reaction (PCR) or site-specific mutagenesis was involved in subcloning, the constructs were sequenced to confirm that no mistakes were generated. Site-directed mutagenesis and DNA and RNA gel blot hybridization were performed as described previously (Zhu et al., 1995a, 1995b).
5′ Rapid Amplification of cDNA Ends and Reverse Transcriptase–Mediated PCR Amplification of Full-Length cDNA of OsDr1
The 5′ end of OsDr1 cDNA was amplified according to the instructions of the 5′ rapid amplification of cDNA ends system (Gibco BRL, Rockville, MD). OsDr1 gene-specific oligonucleotide Q142 (5′-GTT-GCTCAGCTACACTTGTTCC-3′) was used as a primer for the first-strand cDNA synthesis, and Q143 (5′-TTGGAGAATCCAGGGTAT-CATGC-3′) and Q144 (5′-CAAGTGACGGCCGCTTGAACCTCT-TC-3′) were used as primers for first and second rounds of PCR amplification. The 350-bp DNA fragment from the second round of PCR was cloned into pGEM-T vector (Promega, Madison, WI). The DNA sequence confirmed that the 350-bp DNA fragment encoded the 5′ end of OsDr1.
To clone the full-length cDNA, OsDr1 gene-specific oligonucleotide Q148 (5′-CTTATACTGAGGCTACACAAC-3′) was used as a primer for the first-strand cDNA synthesis, and Q149 (5′-ATACCC-GGGTGAACTGTCCAAGCCATGTTC-3′) and Q150 (5′-TAAGAATTC-ATGGATCCGATGGATATCGTG-3′), designed to cover the translation stop and start codons, respectively, were used as primers for PCR amplification. The 900-bp PCR product was cloned into the pGEM-T vector to generate pGEM-OsDr1, which was confirmed by DNA sequencing.
Yeast Two-Hybrid Analysis
pBD-Gal4 and pAD-Gal4 have the Gal4 DNA binding domain and the activation domain, respectively, in the yeast two-hybrid system (Stratagene, San Diego, CA). The EcoRI-SmaI coding region fragment of pGEM-OsDr1 was cloned into pAD-Gal4 to generate pAD::OsDr1. The MfeI-PstI coding region fragment of OsDrAp1 was cloned into pBD-Gal4 to generate pBD::OsDrAp1. The pAD::OsDr1 and pBD::OsDrAp1 plasmids were transformed independently and cotransformed into the Stratagene yeast YRG-2 strain according to the method of Eible (1992). The pAD::OsDr1 transformants were selected on −Trp plates. The pBD::OsDrAp1 transformants were selected on −Leu plates. The interactions between pAD-OsDr1 and pBD::OsDrAp1 were selected on −Trp, −Leu, and −His plates. β-Galactosidase activities were analyzed as described by Jia et al. (2000).
A MfeI site was introduced into the cDNA of OsDrAp1 after residue 64 by site-directed mutagenesis. The MfeI-PstI fragments deleting the N-terminal 64 amino acids of OsDrAp1 were inserted into pBD-Gal4 to generate pBD::Ap1N65. A MscI site was introduced separately into the cDNA of OsDrAp1 at residues of 74, 92, and 148. The respective MfeI-MscI fragments of OsDrAp1 were inserted into the EcoRI-SmaI sites of pBD-Gal4 to generate the pBD::Ap1N75, pBD::Ap1N93, and pBD::Ap1N148 constructs respectively. The EcoRI-PmlI fragments of p35S::G4Dr1N96 and p35S::G4Dr1N80, which contained the N-terminal 96 amino acids and 80 amino acids of OsDr1, respectively, were used to replace the EcoRI-SmaI fragment of pAD::rDr1 to generate pAD::OsDr1N96 and pAD::OsDr1N80, respectively. The interactions of pAD::OsDr1 with pBD::Ap1N64, pBD::Ap1N75, pBD::Ap1N93, and pBD::Ap1N148 and the interactions of pBD::OsDrAp1 with pAD::OsDr1N96 and pAD::OsDr1N80 were analyzed separately using the same method described above.
Localization of mGFP Fusion Proteins
The EcoRI-NcoI coding region fragment of pGEM-OsDr1 was used to replace the EcoRI-NcoI fragment of pRTL2-mGFP (von Arnim et al., 1998) to generate pOsDr1/mGFP. The coding region of OsDrAp1 was amplified by PCR with oligonucleotides Q222 (5′-AACAATTGG-AGGAGCGGAGGCGGA-3′) and Q223 (5′-AACCATGGAATCCTC-GTTGTCGTAGTC-3′) as primers. The PCR product was inserted into pPCR-Script (Stratagene, San Diego, CA) to generate pST-OsDrAp1, which was digested completely with MfeI and digested partially with NcoI. The resulting 822-bp MfeI-NcoI fragment of OsDrAp1 was used to replace the EcoRI-NcoI fragment of pRTL-mGFP to generate pOsDrAp1/mGFP. Both the pOsDr1/mGFP and pOsDrAp1/mGFP constructs were confirmed by DNA sequencing. The localization of mGFP and its fusion proteins in living tobacco cells was monitored by confocal microscopy (Gindullis et al., 1999).
Expression and Purification of Recombinant OsDr1, OsDrAp1, and OsTBP in Escherichia coli
The EcoRI-BamHI fragment of pGEM-OsDr1 was inserted into pET33b to generate the pET33b-OsDr1 construct. The NcoI and XhoI sites were introduced into OsDrAp1 cDNAs around its translation start and stop codons, respectively, by in vitro mutagenesis. The 774-bp NcoI-XhoI fragment of OsDrAp1 was inserted into pET29b (Novagen, Madison, WI) to generate pET29-OsDrAp1. pET33b-OsDr1 and pET29-OsDrAp1 were transformed into E. coli BL21 (DE3) cells. Cells were grown at 30°C to an OD600 of 1.0, induced by 0.2 mM isopropyl-β-d-thiogalactopyranoside for 3 hr, and then harvested. The recombinant proteins were purified with nickel–nitrilotriacetic acid agarose beads (Qiagen, Valencia, CA) and purified further with T7 tag agarose beads for OsDr1 or S-protein agarose beads for OsDrAp1. The purified recombinant proteins were denatured with 2 M urea, dialyzed in renaturation buffer (20 mM Hepes-KOH, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, 20% glycerol, and 0.02% Nonidet P-40) overnight, and then concentrated with Centriprep10 (Amicon, Beverly, MA). Concentrated OsDr1 and OsDrAp1 were frozen in liquid N2 and stored at −70°C. Purification of recombinant OsTBP was as we reported previously (Q. Zhu, M.I. Ordiz, T. Dabi, R.N. Beachy, and C. Lamb, unpublished data).
Protein Overlay Analysis of the OsDr1–OsDrAp1 Interaction
Protein overlay analysis (Harlow and Lane, 1988; Chen and Evans, 1995; Jia et al., 2000) was performed with modifications. Briefly, partially purified recombinant OsDrAp1 protein and BSA were separated on 4 to 12% Bis-Tris gradient polyacrylamide gels and blotted onto a nitrocellulose membrane in NuPAGE Transfer Buffer (Novex, Carlsbad, CA) containing 20% methanol. After a brief rinse in binding buffer (20 mM Hepes, pH 7.0, 40 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 5% glycerol), the membrane was incubated for 45 min in binding buffer containing 6 M guanidine HCl. Then the membrane was incubated gradually in a binding buffer series with 3, 1.5, 0.75, and 0.375 M guanidine HCl for 15 min each. After a 15-min wash in binding buffer, the membrane was blocked for 3 hr in binding buffer containing 5% nonfat dry milk. After washing three times in binding buffer for a total of 30 min, the membrane was incubated with 50 μg/mL purified recombinant OsDr1 protein in binding buffer overnight. The membrane was rinsed once in binding buffer, washed three times in TBST buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween-20) for a total of 30 min, and then incubated with a 1:5000 dilution of horseradish peroxidase–conjugated T7 tag antibody (Novagen) for 1 hr in TBST buffer. After washing three times with TBST buffer for a total of 30 min, bound OsDr1 protein was detected with the enhanced chemiluminescence detection system (Pierce, Rockford, IL). Two replicas were detected with His tag antibody (Invitrogen, Carlsbad, CA) and T7 tag antibody immediately after the blotting to serve as controls. One gel was stained with Coomassie Brilliant Blue R 250.
Tobacco Protoplast Transfection Assay
Generation of Reporters and Effectors
The enhancer fragment (−832 to −50) of the 35S promoter was amplified by PCR and inserted into the SpeI-BamHI sites of pG4G vector (Liu and Odell, 1999) to generate the p35S4Gal4::β-glucuronidase (GUS) reporter construct. The coding region of the luciferase gene in pMAMneo-luc (Clontech, Palo Alto, CA) was amplified using primers LUC5′ (5′-GGCCATGGAAGACGCCAAAAAC-3′) and LUC3′ (5′-GGGGCCCGGTACCCGGGGATCC-3′) to produce a 1.8-kb fragment with 5′ NcoI and 3′ KpnI sites, which was used to replace the NcoI-KpnI fragment of pMH40 to generate the p35S::LUC construct. pMH40 was generated by insertion of the 35S promoter/GUS coding region/nopaline synthase terminator gene into pGEM9 (Promega).
All of the effector constructs were made using the vector p35SGal4VP16 (Liu and Odell, 1999) as the backbone. Thus, all of the effectors were in the cassette of the 35S promoter of Cauliflower mosaic virus and a 3′ octopine synthase terminator. All of the OsDr1 and OsDrAp1 full-length coding regions and deletion fragments were cloned into p35SGal4VP16 to replace the VP16 region. The GAL4 constructs contained the GAL4 DNA binding domain (amino acids 1 to 147) in frame with either full-length or deleted OsDr1 or OsDrAp1. The exact amino acid positions in each construct are listed in Figure 6.
Transfection Analysis
Tobacco NT-1 protoplasts were isolated using a modified procedure of Liu et al. (1997). Fifty milliliters of NT-1 suspension culture (4 to 5 days after subculture) were sedimented at 300g for 10 min in a Heraeus Megafuge (New York, NY). The suspension cells then were resuspended in 50 mL of enzyme solution containing 0.4 M mannitol, 20 mM Mes, pH 5.5, 1% Cellulase RS (Onozuka Yakult Honsha Co., Tokyo, Japan), and 0.1% Pectolyase Y-23 (Seishin Pharmaceutical, Tokyo, Japan). The cells were transferred into a 250-mL flask and shaken gently (60 rpm) at room temperature for 4 to 5 hr. Light microscopy was used to monitor the time course of protoplast preparation. The protoplasts were pelleted at 133g for 5 min, and the supernatant was removed by aspiration. The protoplasts were suspended gently in 30 mL of W5 solution (Goodall et al., 1990) and counted using a hemocytometer and light microscopy. The protoplasts were repelleted and suspended in W5 solution at 2 × 106 protoplasts per milliliter.
Freshly prepared protoplasts were pelleted and resuspended in MC buffer (5 mM Mes, 20 mM CaCl2, and 0.5 M mannitol, pH 5.7) at 2 × 106 protoplasts per milliliter. For each transfection analysis, 300 μL of tobacco protoplasts prepared as described above was mixed with up to 25 μL of supercoiled plasmid DNA. The plasmid DNA consisted of a mixture of 2 μg of the p35S4Gal4::GUS reporter construct, 0.1 μg of the p35S::LUC construct, and 2 μg of the effector construct. The p35S::LUC construct served as an internal standard for each transfection assay. The protoplasts and the DNAs were mixed with 0.3 mL of 40% (w/v) polyethylene glycol containing 0.1 M Ca(NO3)2 and 0.4 M mannitol, pH 10, at room temperature for 5 min. Four milliliters of Murashige and Skoog (1962) medium was added, and the protoplasts were incubated in the dark at room temperature for 40 to 48 hr. Transfections were performed at least three times, and at least three independent transfection experiments were performed with each effector construct.
Luciferase assays were performed by mixing 2 μL of extract with 100 μL of luciferase substrate (Promega) and measuring the emitted photons for 0.5 sec in a luminometer. Fluorometric GUS assays were performed as described (Jefferson, 1987). A fluorescence multiwell plate reader was used to measure GUS activity at 365 nm (excitation) and 455 nm (emission).
Gel Shift Assay
Oligonucleotides Q218 (5′-CCTCCGTCATCCGTCCTGCA-3′) and Q219 (5′-TTCTAGCTACTCGATTAGCT-3′) were used as primers to amplify a 140-bp DNA fragment from positions −69 to +71 of a rice pal promoter (Zhu et al., 1995a). The 140-bp fragment was purified, labeled with γ-32P-ATP, and used as a probe. DNA binding reactions were performed in a volume of 20 μL containing 0.5 × TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0), 10% glycerol, 1 mM DTT, 50 mM KCl, 30 ng of poly(dI-dC), and 0.1- to 0.25-ng probes. One hundred to 500 ng of purified recombinant proteins was added. The reaction products were separated on 5% native polyacrylamide gels in 0.5 × TBE buffer plus 2 mM DTT at 5 W for 1 hr at 4°C.
In Vitro Transcription Assays
Rice (cv IR72) cell suspension cultures were used for preparation of whole-cell extracts. In vitro transcription reactions were performed as described previously (Zhu et al., 1995b). In each experiment, the amounts of template and primer were the same in each reaction. The transcription reaction was initiated by the addition of different amounts of recombinant OsTBP, OsDr1, and OsDrAp1 to the reaction mixture 10 min before the addition of rice whole-cell extracts. Transcription products were analyzed by primer extension. The transcript levels were measured quantitatively by phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Accession Numbers
The GenBank accession numbers for the sequences described in this article are as follows: AF464902 (OsDr1), AF464903 (wDr1), AF464906 (sDr1), AF464904 (OsDrAp1), and AF464905 (mDrAp1).
Acknowledgments
We are grateful to Zhan-Bin Liu and Michele Smith-Jones for plasmids of pG4G, p35Gal4VP16, and p35S::LUC. We thank Steven Spiker for tobacco NT-1 cells. We thank Richard Howard, Liang Liang, and Timothy Bourett for their help with the confocal microscopy and Shawn Anderson, Zhan-Bin Liu, and Joan Odell for their critical reading of the manuscript. This research is part of the plant gene expression program of the Central Research and Development Department of the DuPont Company.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010320.
References
- Allen, G.C., Hall, G.E., Childs, L.C., Weisinger, A.K., Spiker, S., and Thompson, W.F. (1993). Scaffold attachment regions in-crease reporter gene expression in stably transformed plant cells. Plant Cell 5, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents, G., Burlingame, R.W., Wang, B.C., Love, W.E., and Moudrianakis, E.N. (1991). The nucleosomal core histone octamer at 3.1 Å resolution: A tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88, 10148–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis, A.D., and Landsman, D. (1998). Histone sequence database: New histone fold family members. Nucleic Acids Res. 26, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm, A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J., and Kouzarides, T. (1998). Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Cang, Y., Auble, D.T., and Prelish, G. (1999). A new regulatory domain on the TATA-binding protein. EMBO J. 18, 6662–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.J., and Evans, R.M. (1995). A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Chern, M.S., Bobb, A.J., and Bustos, M.M. (1996). The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 8, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y., Huddleston, M.J., Zhang, X., Young, R.A., Annan, R.S., Carr, S.A., and Deshaies, R.J. (2001). Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15, 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, C.-T., Bartel, P.L., Sternglanz, R., and Fields, S. (1991). The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88, 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst, R.P., Wang, D., and Auble, D.T. (2001). MOT1-catalyzed TBP-DNA disruption: Uncoupling DNA conformational change and role of upstream DNA. EMBO J. 20, 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eible, R. (1992). A simple and efficient procedure for transformation of yeast. Biotechniques 13, 18–20. [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadbois, E.L., Chao, D.M., Reese, J.C., Green, M.R., and Young, R.A. (1997). Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc. Natl. Acad. Sci. USA 94, 3145–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg, J.V., Holstege, F.C., Young, R.A., and Struhl, K. (2001). Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21, 2736–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis, F., Peffer, N.J., and Meier, I. (1999). MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall, G.J., Wiebauer, K., and Filipowicz, W. (1990). Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181, 148–161. [DOI] [PubMed] [Google Scholar]
- Goppelt, A., Stelzer, G., Lottspeich, F., and Meisterernst, M. (1996). A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 15, 3105–3116. [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose, W., and Hansen, U. (1996). Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 12, 229–234. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hengartner, C.J., Myer, V.E., Liao, S.M., Wilson, C.J., Koh, S.S., and Young, R.A. (1998). Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2, 43–53. [DOI] [PubMed] [Google Scholar]
- Inostroza, J., Mermelstein, F.H., Ha, I., Lane, W.S., and Reinberg, D. (1992). Dr1, a TATA-binding protein associated phosphoprotein and inhibitor of class II gene transcription. Cell 70, 477–489. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assay for chimeric genes in plants: The GUS fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Na, J.G., Hampsey, M., and Reinberg, D. (1997). The Dr1/DrAp1 heterodimer is a global repressor of transcription in vivo. Proc. Natl. Acad. Sci. USA 94, 820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Cabane, K., Hampsey, M., and Reinberg, D. (2000). Genetic analysis of the YDR1–BUR6 repressor complex reveals an intricate balance among transcriptional regulatory proteins in yeast. Mol. Cell. Biol. 20, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.K., Zhao, Y., Ge, H., Berstein, R., and Roder, R.G. (1995). TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J. Biol. Chem. 270, 10976–10981. [DOI] [PubMed] [Google Scholar]
- Kuromori, T., and Yamamoto, M. (1994). Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acids Res. 22, 5296–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, S.M., Zhang, J., Jeffery, D.A., Koleske, A.J., Thompson, C.M., Chao, D.M., Viljoen, M., van Vuuren, H.J., and Young, R.A. (1995). A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374, 193–196. [DOI] [PubMed] [Google Scholar]
- Liu, L., White, M.J., and MacRae, T.H. (1999). Transcription factors and their genes in higher plants: Functional domains, evolution and regulation. Eur. J. Biochem. 262, 247–257. [DOI] [PubMed] [Google Scholar]
- Liu, Z., and Odell, J.T. (1999). Specific gene activation by chimeric Gal4 transcription factors in stable transgenic plants. U.S. patent 5968793.
- Liu, Z., Hagen, G., and Guilfoyle, T.J. (1997). A G-box-binding protein from soybean binds to the E1 auxin-response element in the soybean GH3 promoter and contains a proline-rich repression domain. Plant Physiol. 115, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado, E., Hampsey, M., and Reinberg, D. (1999). Repression: Targeting the heart of the matter. Cell 99, 455–458. [DOI] [PubMed] [Google Scholar]
- Mermelstein, F., Yeung, K., Cao, J., Inostroza, J.A., Erdjument-Bromage, H., Eagelson, K., Landsman, G., Levitt, P., Tempst, P., and Reinberg, D. (1996). Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 10, 1033–1048. [DOI] [PubMed] [Google Scholar]
- Muldrow, T.A., Campbell, A.M., Weil, P.A., and Auble, D.T. (1999). MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol. Cell. Biol. 19, 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nielsen, S.J., Schneider, T., Bauer, U.M., Bannister, A.J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R.E., and Kouzarides, T. (2001). Rb targets histone H3 methylation and HP1 to promoters. Nature 412, 561–565. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides, G., Lagrange, T., and Reinberg, D. (1996). The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1998). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich, G. (1997). Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2a homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 17, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwechheimer, C., Zourelidou, M., and Bevan, M.W. (1998). Plant transcription factor studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 127–150. [DOI] [PubMed] [Google Scholar]
- Smith, R.L., and Johnson, A.D. (2000). Turning genes off by Ssn6-Tup1: A conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25, 325–330. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A., Deng, X.W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- White, R.J., Khoo, B.C.-E., Inostroza, J.A., Reinberg, D., and Jackson, S.P. (1994). Differential regulation of RNA polymerase I, II and III by the TBP-binding repressor Dr1. Science 266, 448–450. [DOI] [PubMed] [Google Scholar]
- Willy, P.J., Kobayashi, R., and Kadonaga, J.T. (2000). A basal transcription factor that activates or represses transcription. Science 290, 982–984. [DOI] [PubMed] [Google Scholar]
- Yeung, K., Kim, S., and Reinberg, A. (1997). Functional dissection of a human Dr1-DrAp1 repressor complex. Mol. Cell. Biol. 17, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Zhu, Q., Da, S., Lamb, C., and Beachy, R.N. (1997). RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 16, 5247–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q., Tsegaye, D., Beeche, A., Yamamoto, R., Lawton, M., and Lamb, C. (1995. a). Cloning and properties of a rice gene en-coding phenylalanine ammonia-lyase. Plant Mol. Biol. 29, 535–550. [DOI] [PubMed] [Google Scholar]
- Zhu, Q., Chappell, J., Hedrick, S., and Lamb, C. (1995. b). Accurate in vitro transcription by plant whole cell extracts from circularized plasmid templates. Plant J. 5, 1021–1030. [DOI] [PubMed] [Google Scholar]