Abstract
In order to find common genetic abnormalities that may identify loci of genes involved in the development of adenoid cystic carcinoma (ACC), we investigated DNA copy number changes in 24 of these tumors by comparative genomic hybridization (CGH). Our results indicate that unlike many carcinomas, ACCs have relatively few changes in DNA copy number overall. Twenty tumors had DNA copy number changes, which were mostly restricted to a few chromosomal arms. A frequent novel finding was the loss of DNA copy number in chromosome 12q (eight tumors, 33%) with the minimal common overlapping region at 12q12-q13. Deletion in this region has not been reported to be frequent in other types of cancer analyzed by CGH. In addition, deletions in 6q23-qter and 13q21-q22 and gains of chromosome 19 were observed in 25% to 38% of ACCs. Deletion of 19q, previously reported in a small series of ACC, was not identified in the current group of carcinomas. The current CGH results for chromosomes 12 and 19 were confirmed by microsatellite allelotyping. These results indicate that DNA copy number losses in 12q may be important in the oncogenesis of ACC and suggest that the 12q12-q13 region may harbor a new tumor-suppressor gene.
Keywords: comparative genomic hybridization, loss of heterozygosity, microsatellite markers, genetic deletion, adenoid cystic carcinoma
Introduction
Adenoid cystic carcinoma (ACC) is the most common malignant tumor of the submandibular and minor salivary glands [1,2], and also arises in other anatomic sites including the breast and vulva. Despite resection with and without postoperative irradiation, about 40% to 60% of patients with salivary ACC develop distant metastases [1]. The clinical course is often lengthy, however, and disease-specific mortality frequently occurs 10 to 20 years after therapy. ACC has a proclivity for invading nerves, a feature which, while certainly not unique, suggests specific molecular pathways leading to neurotropism. The neoplasm typically lacks the ability to metastasize to lymph nodes, preferring the hematogenous route. ACC characteristically shows features of myoepithelial differentiation and produces abundant basement membrane material and glycosaminoglycans, which may be linked to novel genetic mechanisms of differentiation.
Genetic changes that lead to the initiation and progression of ACC are poorly characterized. Cytogenetic studies have shown a relatively limited number of changes, the most frequent including alterations of 6q21-q24 (deletions or translocations), translocations of 9p13-p23, and, to a less extent, gains of chromosomes 7 and 8 [3-5]. After the detection of chromosome 17p13 deletions by fluorescent in situ hybridization [6], loss of heterozygosity (LOH) analysis in ACC has shown abnormalities of the p53 gene locus in up to half of ACCs [7,8]. In a low-resolution study of 10 ACCs, LOH analysis showed loss in 1p, 2p, 6q, and 17q in 20% to 30% and in 19q in 40% of cases [9]. High-resolution deletion mapping of 6q in six cases of ACC showed LOH in either 6q23 or 6q27 in two tumors [10]. In other cancers, comparative genomic hybridization (CGH) combined with LOH analysis has been shown to be a very powerful technique in identifying putative tumor-suppressor loci [11]. To date, however, there are no published surveys of global DNA copy number changes as detected by CGH for ACC.
To obtain a comprehensive profile of the DNA copy number changes in ACC, we screened 24 cases for DNA copy number alterations in whole tumor genomes by CGH, and further investigated chromosomes 19 and 12 for genetic alterations using microsatellite LOH analysis.
Materials and Methods
Tumors
This study was approved by the Institutional Review Board of the University of Virginia Health System. Twenty-four formalin-fixed, paraffin-embedded cases of ACC were obtained from the files of the Division of Surgical Pathology at the University of Virginia Health System. Sixteen patients were women and eight were men. The age range at diagnosis was 33 to 73 years (median 57 years). Ten tumors arose in the major salivary glands (lacrimal gland included), whereas 14 tumors developed in the minor salivary glands. The tumors were graded according to Szanto et al. [12] as lesions with no solid component (grade 1), <30% solid areas (grade 2), and >30% solid component (grade 3). Nine ACCs were grade 1, nine were grade 2, and six were grade 3. Pathologic stage was recorded based on the AJCC criteria [13]. Eight tumors were stage I, five were stage II, one was stage III, and 10 were stage IV. Patient follow-up data were obtained from the McIntire Tumor Registry at the University of Virginia Health System. Follow-up time ranged from 0.3 to 13.5 years (median 5.5 years) with 2 of 24 patients being diagnosed recently. Of the remaining patients, nine had developed metastases, while 13 had not. For the LOH study, five additional tumors collected from two women and three men were allelotyped using chromosomes 19 and 12 markers. Of these five tumors, one was grade 1, one was grade 2, and three were grade 3. Pathologic stage of these additional tumors included one stage I tumor, two stage IV tumors, and two tumors of unknown stage.
CGH
As the minimum sensitivity requirement for CGH is 50% of tumor material within a sample, paraffin-embedded tissue sections were dissected to obtain an estimated minimum of 70% tumor cells. DNA from paraffin-embedded tissue sections was extracted as previously described [14]. DNA extracted from peripheral blood cells of a healthy donor, along with DNA extracted from a gastric tumor with known DNA copy number changes, were, respectively, used as a negative and positive control in each CGH experiment. CGH was performed according to standard procedures with a modification using a mixture of fluorochromes conjugated to dCTP and dUTP nucleotides for nick translation [15]. Hybridizations, washings, and ISIS digital image analysis (MetaSystems, Altlussheim, Germany) were performed as described elsewhere [16]. Results from our controls and previous studies [16,17] indicated cut off levels of 1.17 and 0.85 for gains and losses, respectively. All CGH results were confirmed using a 99% confidence interval. Intra-experiment standard deviations for all positions in the CGH ratio profiles were calculated from the variation of the ratio values of all homologous chromosomes within the experiment. Confidence intervals for the ratio profiles were then computed by combining them with an empirical inter-experiment standard deviation and by estimating error probabilities based on the t-distribution.
Microsatellite LOH Assays
Chromosome 12 was selected for LOH analysis as CGH results of the present study showed a novel area of loss on this chromosome. Twenty-nine ACCs that were micro-dissected to 75% to 95% purity were chosen for LOH analysis. This group included 24 tumors that had been analyzed by CGH. Matched normal tissue was dissected for each cancer and DNA was extracted from both tissues as previously published [18]. MapPairs primers for microsatellite markers were obtained from Research Genetics (Huntsville, AL). Markers used on chromosome 12 were D12S391, D12S1301, and D12S1064 (Figure 2). The same panel of tumors was assayed for LOH using chromosome 19 markers D19S414, D19S220, D19S223, D19S246, D19S589, D19S254, and D19S210. Radiolabeled polymerase chain reaction (PCR) amplification, gel electrophoresis, autoradiography of microsatellite markers, and LOH determination were performed as previously described [19]. A tumor was determined to have undergone LOH at a particular locus only if the predominant band(s) associated with one allele showed a diminution in intensity of 50% or more in the tumor relative to normal [20], although most assays showed a complete or virtually complete absence of one allele in the tumor DNA in comparison to that of the matching normal DNA (Figure 3). All losses were confirmed in an independent PCR assay. In a subset of cases, duplex PCR was performed with primers to a portion of the β-2-microglobulin gene (B2M) to control for the presence of DNA of sufficient size in the genomic preparations to amplify the microsatellite alleles. The primers for this 264-bp product are: 5′-ATTCACCCCCACTGAAAAAG-3′ (F) and 5′-ACTAATCTGATCTTTACGAAC-3′ (R).
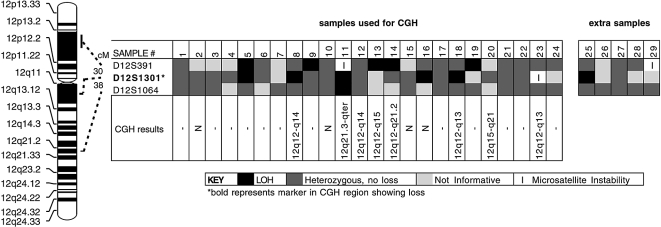
Figure 2.
LOH analysis using chromosome 12 markers in 29 ACCs. The microsatellite markers used are listed on the left, in order from pter (top) to qter (bottom). The results of the assays are shown as indicated in the key. The black squares indicate deleted loci. Deletions were found in 10 of 29 cancers (34%). The majority of losses were found at the D12S1301 marker, having loss in 6 of 10 cancers with deletions (60%) followed by the D12S391 marker having loss in 5 of 10 cancers with deletions (50%). CGH results are from Table 1.
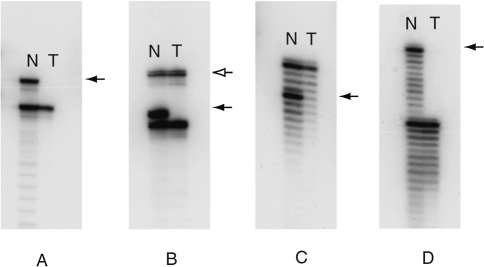
Figure 3.
Autoradiographs showing specific examples of LOH for each of four different ACCs on chromosome 12. (A) Marker D12S391 for case 13. (B) Duplex PCR with marker D12S391 and B2M for case 5. (C) Marker D12S1301 for case 16. (D) Marker D12S1301 for case 18. Left lanes show assays of non-neoplastic cells (N); right lanes show assays of neoplastic (tumor) cells (T). Solid arrows denote the microsatellite allele lost in the tumor samples. In Panel B, the open arrow points to the B2M duplex PCR product, which serves as a control for the presence of DNA fragments in the sample preparation of sufficient size to amplify the larger allele in the microsatellite PCR.
Statistics
The relationship between tumor site (major versus minor salivary glands), grade, stage, and the presence of metastasis was compared with the most frequent CGH gains or losses using Fisher's exact test (two-tailed). The relationship between the number of DNA copy alterations and tumor grade was examined using the unpaired t-test.
Results
CGH
Changes of DNA copy number were detected in 20 tumors. The average number of changes per tumor was 2.7. Losses of DNA sequences were more frequent than gains, and no high-level amplifications were seen. Grade 3 tumors had more changes (mean 5.7) than grades 1 and 2 neoplasms (mean 1.8, P<.001). The most frequent abnormalities included losses at 6q (25%), 12q (33%), and 13q (25%), and gains in chromosome 19 (38%). Table 1 shows the results in detail. Figure 1 presents a summary of the most common gains and losses of DNA sequences found in 20 ACCs with alterations in DNA copy number. There was no correlation between tumor site (major or minor salivary glands), grade, stage, or presence of metastasis with gain in 19 or loss in 6q, 12q, or 13.
Table 1.
Clinical, Histological, and CGH Karyotype Findings for 24 ACCs.
| Case | Age/Sex | Tumor Site | Grade/Stage | CGH Karyotype | |
| Losses | Gains | ||||
| 1 | 49/F | trachea | 1/I | 6q22.3-q23, X | - |
| 2 | 68/M | submandibular | 1/I | N | N |
| 3 | 54/F | trachea | 1/I | - | 15, 16, 17, 19 |
| 4 | 70/M | soft palate | 1/I | 6q23-qter, 9q31-qter, 13q14-q22 | 19 |
| 5 | 54/F | submandibular | 1/II | 8q11-q13, 9p | 3q13.3-q23 |
| 6 | 53/M | maxilla | 1/III | 6q23-qter | - |
| 7 | 65/M | parotid | 1/IV | 13q21-qter, 14 | - |
| 8 | 64/M | maxilla | 1/IV | 12q12-q14 | 16, 19 |
| 9 | 58/F | parotid | 1/IV | 9p21-pter | 19 |
| 10 | 57/F | soft palate | 2/I | N | N |
| 11 | 50/F | floor of mouth | 2/I | 12q21.3-qter | - |
| 12 | 36/F | bronchus | 2/IA | 6q11-q23, 12q12-q14, 13q14-q31 | 19 |
| 13 | 66/F | nasal septum | 2/II | 12q12-q15 | - |
| 14 | 64/M | parotid | 2/IV | 12q12-q21.2, 13q14-q31 | - |
| 15 | 33/F | parotid | 2/IV | N | N |
| 16 | 48/F | tongue base | 2/IVA | N | N |
| 17 | 43/F | trachea | 2/IIIB | 14 | 22 |
| 18 | 72/F | tongue | 2/unknown | 12q12-q13 | - |
| 19 | 69/M | soft palate | 3/II | - | 8 |
| 20 | 57/F | parotid | 3/IV | 6q11-q23, 9p21-pter, 12q15-q21, 13q13-q31 | 19 |
| 21 | 54/F | maxilla | 3/IVA | 1p35-pter, 1q41-qter, 5, 11p | 1p11-p34, 1q11-q32, 7, 9p, 11q12-q13 |
| 22 | 50/M | palate | 3/IVC | 6q23-qter, 9q, 11q21-q23 | 9p, 19, 22 |
| 23 | 73/F | lacrimal gland | 3/T2 N0 M0 | 1p33-pter, Xp21-pter, 6q22.2-q23, 7p13-p21.2, 12q12-q13, 13, 16q | 1p22-qter, 7q, 19 |
| 24 | 63/F | lacrimal gland | 3/T4a N0 M0 | 5q12-q21 | 19 |
M: male, F: female, N: normal.
Figure 1.
Summary of the most frequent gains and losses of DNA sequences in 20 ACCs with abnormal DNA copy number. Gains are on the right side of the chromosome ideogram and losses are on the left. A single line represents each tumor sample.
Microsatellite LOH Analysis
Chromosome 12 was selected for LOH analysis to confirm the finding of loss found by CGH at chromosome 12q12-q13. A microsatellite marker located within this cytogenetic location and two other markers bounding this region were used in a PCR amplification LOH assay (Figure 2). Results from LOH analysis showed a complete or virtually complete loss in all tumors with similar losses by CGH (Figure 3). LOH results showed an overall loss of 34% on chromosome 12, with the greatest loss occurring at the D12S1301 marker (30%) (Figures 2 and 3). The D12S1301 marker is located at the same chromosome 12q12-q13 cytogenetic region showing CGH loss (Figure 2). Marker D12S391, located on the p arm of chromosome 12, showed loss in 5 of 15 informative cases, while the D12S1064 marker, located on the q arm and distal to D12S1301, showed loss in only 1 of 22 informative cases (Figure 2). Of the 10 tumors demonstrating LOH, one of these was not analyzed by CGH (sample 25), and five had DNA copy number changes in 12q detected by CGH (samples 8, 11, 13, 14, 18), with one of these having 12q21-qter loss and distal to the 12q12-q13 region (sample 11). The other four tumors in Figure 2 did not show DNA copy number changes when analyzed by CGH (samples 5, 9, 16, 19). Sample 12 showed 12q12-q13 loss by CGH analysis, but did not show loss by LOH in any of the three markers examined. CGH analyses also showed DNA copy number changes at 12q in four tumors (samples 13, 14, 20, 23), but LOH analysis showed these to be not informative for 12q markers. Furthermore, two of these (samples 13 and 14), in addition to samples 9 and 19, had LOH at D12S391, as did case 5, which also had LOH at D12S1301.
Chromosome 19 was selected for LOH analysis because a previous study reporting deletion of this chromosome in ACC [9] was not consistent with our CGH findings of only gain of DNA copy number. Results from LOH analysis of chromosome 19 did not show loss for any of the seven markers, in agreement with our CGH results. The average of informative microsatellite assays per case was 3.9.
Discussion
The loss of chromosome 12q12-q13 is a novel finding in ACC. Such loss was the only detectable DNA copy number change in three tumors in this study, suggesting that a gene in this region plays a special role in ACC tumorigenesis. Further investigation of chromosome 12 by LOH analysis showed the overall loss on chromosome 12 to be similar to that found by CGH and that this loss likely includes a locus telomeric to D12S1301 since only 6 of 10 tumors with LOH included D12S1301. The finding of 12q loss by CGH analysis alone for sample 12 also leads us to believe that the loss may be telomeric to D12S1301. Interestingly, using the D12S391 marker, there was a high level of loss on the p arm of chromosome 12, suggesting that this loss may involve a relatively large area of chromosome 12 or perhaps, there is more than one consensus region residing on this chromosome — one at 12p11-13 and the other(s) at 12q. The fact, that LOH analysis alone showed loss on the p arm for four tumors (samples 9, 13, 14, and 19), suggests that this area may be involved by deletions too small for identification by CGH. The potential for loss on the q arm of chromosome 12 may perhaps extend further than that shown by LOH analyses because the two 12q markers are non-informative for a number of tested samples. The small microsatellite panel used in this study served only as an independent confirmation of the finding by CGH of deletions on chromosome 12. Further LOH studies using higher-density allelotyping of microsatellite markers are required to conclusively determine the number and localization of consensus regions on chromosome 12.
Although a recent literature review of 283 CGH studies showed that chromosome 12 deletions were not frequent in any of the 73 tumor entities analyzed [21], one study using microsatellite LOH assays found evidence of genetic deletion at 12q in salivary gland pleomorphic adenomas and in the adenoma component of carcinoma ex-pleomorphic adenoma [22]; the authors concluded that LOH at 12q may identify a subset of adenomas with the potential to progress to carcinoma. Although the majority of carcinomas arising in pleomorphic adenomas showed loss distal to the 12q12-q13 region, some of the deletions did extend to this locus, and in two cases, this region was the sole area of deletion. Interestingly, a translocation of 12q12-q13 in a case of myoepithelioma, a rare benign salivary gland neoplasm that shares ACC features of myoepithelial differentiation, has been reported [23]. A similar translocation has also been shown in renal oncocytoma [24]. These findings, together with our results, suggest that an unidentified gene with tumor-suppressor activity might be present at chromosome 12q12-q13. It is possible that this unknown gene might have functional importance for salivary gland tumors with myoepithelial differentiation. Further studies are required to determine the extent to which chromosome 12, and more specifically the region telomeric to the D12S1301 marker, may play in the development or progression of salivary gland tumors.
The CGH results of the present study, showing losses at 6q23-qter, are in agreement with the results of earlier cytogenetic and LOH findings in ACC [5,10], and further support the notion that deletions of genetic material at 6q may play an essential role in the development of this neoplasm. The chromosome 6q23-qter cytogenetic location includes the tumor-suppressor gene PLAGL1/LOT1/ZAC at 6q24.3. This gene was discovered to be downregulated during the transformation of a rat ovarian epithelial cell line [25], and was subsequently shown to be downregulated in primary human ovarian and breast carcinomas and cell lines [26,27]. In cell culture experiments, PLAGL1/LOT1/ZAC has been shown to have anti-proliferative and pro-apoptotic regulatory activities [28]. Therefore, further testing of PLAGL1/LOT1/ZAC as a potential candidate for tumor-specific deletion, mutation, and/or transcriptional down-regulation in ACC is required.
The frequent losses we observed in 13q21-q22 have not previously been reported in ACC. This genetic region is known to be deleted in several types of cancer, such as carcinomas of the lung, breast, prostate, and head and neck, as well as in tumors of bone (for review, see Ref. [29]). This genetic location is considerably distal to the RB1 locus (13q14), and may indicate the presence of a novel tumor-suppressor gene.
Gains in chromosome 19, frequently observed in the present collection of ACC, have been reported in several other types of carcinoma, including pancreatic [30,31], ovarian [32], and esophageal cancer [33]. In contrast to our findings, an earlier microsatellite LOH study by Johns et al. [9], using two markers on 19q — D19S210 and D19S246, reported a 40% rate of allelic loss in 10 ACCs. Our analysis of ACC using seven microsatellite markers (including the two 19q markers used in the previous study) failed to detect chromosomal loss, supporting the conclusions drawn from our CGH analysis. This discrepancy may be attributed to differences in interpretation of LOH assays, where allelic imbalances that occur in amplification events are interpreted as deletions.
The finding of a larger number of genetic changes in grade 3 tumors than in grades 1 and 2 neoplasms supports the hypothesis that more genetic alterations are required for tumor progression. Grade 3 ACC tumors are more poorly differentiated than grade 1 or 2 cancers in terms of tubule formation, and tend to be more aggressive clinically. The observation of a greater number of genetic abnormalities associated with more aggressive forms of neoplasia has been documented in other tumors [34].
Although specific DNA copy number abnormalities were detected in ACC, a much lower overall number of DNA copy number changes were seen compared with other epithelial neoplasms arising in the upper and lower aerodigestive tract, including nasopharyngeal carcinoma [35], head and neck squamous carcinoma [36,37], and small cell and non-small cell carcinomas of the lung [38–41]. Only a few ACCs (largely those that were grade 3) had the complexity of DNA copy number changes usually present in carcinomas of the aerodigestive tract. Our results showing limited areas of genetic loss in ACC are corroborated by a previous low-density allelotype analysis, which also did not detect many frequent deletion events [9]. The fact, that DNA copy number and cytogenetic abnormalities are less frequent in ACC, may indicate that a smaller number of genes are involved in the development of this neoplasm than is typical of most carcinomas. An alternative explanation for lack of DNA copy number abnormalities is the presence of the DNA replication error phenotype, for which an inverse correlation with number of cytogenetic abnormalities and genetic deletions has been found in colorectal carcinoma [42]. However, in our previous [43] and ongoing studies using microsatellite allelotyping of ACC, no widespread micro-satellite instability has been detected, as would be expected with the replication error phenotype. Hence, we favor the hypothesis that ACC develops as a result of fewer and more specific genetic alterations.
This study is the first global survey of DNA copy number changes in ACC, and points to the existence of possible novel tumor-suppressor genes on 6q, 12q, and 13q. Further LOH studies using higher-density allelotyping of micro-satellite markers are required for chromosome 12 as well as for chromosomes 6 and 13 to fine map regions that may contain, as yet, unidentified tumor-suppressor genes.
Acknowledgements
We thank Sharon Birdsall and Mark Clem for assistance in histologic sectioning and Craig Rumpel for technical assistance in microsatellite LOH analysis.
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Footnotes
This work was supported by grants from the Finnish Cancer Society and the National Organization for Rare Disorders (New Fairfield, CT). C. A. M. is also supported by grant no. 5K08CA74431-02 from the National Cancer Institute.
References
- 1.Ellis GL, Auclair PL. Tumors of the Salivary Glands. 3rd ed. Washington, DC: Armed Forces Institute of Pathology; 1996. [Google Scholar]
- 2.Vander Poorten VL, Balm AJ, Hilgers FJ, Tan IB, Loftus-Coll BM, Keus RB, Hart AA. Prognostic factors for long-term results of the treatment of patients with malignant submandibular gland tumors. Cancer. 1999;85:2255–2264. doi: 10.1002/(sici)1097-0142(19990515)85:10<2255::aid-cncr22>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Mark HF, Hanna I, Gnepp DR. Cytogenetic analysis of salivary gland type tumors. Oral Surg, Oral Med, Oral Pathol, Oral Radiol Endod. 1996;82:187–192. doi: 10.1016/s1079-2104(96)80223-x. [DOI] [PubMed] [Google Scholar]
- 4.Martins C, Fonseca I, Roque L, Pinto AE, Soares J. Malignant salivary gland neoplasms: a cytogenetic study of 19 cases. Eur J Cancer, Part B: Oral Oncol. 1996;32B:128–132. doi: 10.1016/0964-1955(95)00078-x. [DOI] [PubMed] [Google Scholar]
- 5.Nordkvist A, Mark J, Gustavsson H, Bang G, Stenman G. Nonrandom chromosome rearrangements in adenoid cystic carcinoma of the salivary glands. Genes, Chromosomes Cancer. 1994;10:115–121. doi: 10.1002/gcc.2870100206. [DOI] [PubMed] [Google Scholar]
- 6.Roijer E, Kas K, Klawitz I, Bullerdiek J, Van de Ven W, Stenman G. Identification of a yeast artificial chromosome spanning the 8q12 translocation breakpoint in pleomorphic adenomas with t(3;8)(p21;q12) Genes, Chromosomes Cancer. 1996;17:166–171. doi: 10.1002/(SICI)1098-2264(199611)17:3<166::AID-GCC4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto Y, Virmani AK, Wistuba II, McIntire D, Vuitch F, Albores-Saavedra J, Gazdar AF. Loss of heterozygosity and microsatellite alterations in p53 and RB genes in adenoid cystic carcinoma of the salivary glands. Hum Pathol. 1996;27:1204–1210. doi: 10.1016/s0046-8177(96)90316-0. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto Y, Wistuba II, Kishimoto Y, Virmani AK, Vuitch F, Albores-Saavedra J, Gazdar AF. DNA analysis at p53 locus in adenoid cystic carcinoma: comparison of molecular study and p53 immunostaining. Pathol Int. 1998;48:273–280. doi: 10.1111/j.1440-1827.1998.tb03905.x. [DOI] [PubMed] [Google Scholar]
- 9.Johns MM, Westra WH, Califano JA, Eisele D, Koch WM, Sidransky D. Allelotype of salivary gland tumors. Cancer Res. 1996;56:1151–1154. [PubMed] [Google Scholar]
- 10.Queimado L, Reis A, Fonseca I, Martins C, Lovett M, Soares J, Parreira L. A refined localization of two deleted regions in chromosome 6q associated with salivary gland carcinomas. Oncogene. 1998;16:83–88. doi: 10.1038/sj.onc.1201480. [DOI] [PubMed] [Google Scholar]
- 11.Hemminki A, Tomlinson I, Markie D, Jarvinen H, Sistonen P, Bjorkqvist AM, Knuutila S, Salovaara R, Bodmer W, Shibata D, de la Chapelle A, Aaltonen LA. Localization of a susceptibility locus for Peutz-Jeghers syndrome to 19p using comparative genomic hybridization and targeted linkage analysis. Nat Genet. 1997;15:87–90. doi: 10.1038/ng0197-87. [DOI] [PubMed] [Google Scholar]
- 12.Szanto PA, Luna MA, Tortoledo ME, White RA. Histologic grading of adenoid cystic carcinoma of the salivary glands. Cancer. 1984;54:1062–1069. doi: 10.1002/1097-0142(19840915)54:6<1062::aid-cncr2820540622>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 13.Fleming ID, Cooper JS, Henson DE, Hutter RVP, Kennedy BJ, Murphy GP, O'Sullivan B, Sobin LH, Yarbro JW. AJCC Cancer Staging Manual. 5th ed. Philadelphia: Lippincott-Raven Publishers; 1997. [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Rifai W, Larramendy ML, Bjorkqvist AM, Hemmer S, Knuutila S. Optimization of comparative genomic hybridization using fluorochrome conjugated to dCTP and dUTP nucleotides. Lab Invest. 1997;77:699–700. [PubMed] [Google Scholar]
- 16.El-Rifai W, Sarlomo-Rikala M, Miettinen M, Knuutila S, Andersson LC. DNA copy number losses in chromosome 14: an early change in gastrointestinal stromal tumors. Cancer Res. 1996;56:3230–3233. [PubMed] [Google Scholar]
- 17.El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res. 2000;60:3899–3903. [PubMed] [Google Scholar]
- 18.Moskaluk C, Kern S. Microdissection and PCR amplification of genomic DNA from histologic tissue sections. Am J Pathol. 1997;150:1547–1552. [PMC free article] [PubMed] [Google Scholar]
- 19.Rumpel CA, Powell SM, Moskaluk CA. Mapping of genetic deletions on the long arm of chromosome 4 in human esophageal adenocarcinomas. Am J Pathol. 1999;154:1329–1334. doi: 10.1016/S0002-9440(10)65386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emmert-Buck MR, Lubensky IA, Dong Q, Manickam P, Guru SC, Kester MB, Olufemi SE, Agarwal S, Burns AL, Spiegel AM, Collins FS, Marx SJ, Zhuang Z, Liotta LA, Chandrasekharappa SC, Debelenko LV. Localization of the multiple endocrine neoplasia type I (MEN1) gene based on tumor loss of heterozygosity analysis. Cancer Res. 1997;57:1855–1858. [PubMed] [Google Scholar]
- 21.Knuutila S, Autio K, Aalto Y. On-line access to CGH data of DNA sequence copy number changes. Am J Pathol. 2000;157:689. doi: 10.1016/S0002-9440(10)64579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Naggar AK, Callender D, Coombes MM, Hurr K, Luna MA, Batsakis JG. Molecular genetic alterations in carcinoma ex-pleomorphic adenoma: a putative progression model? Genes, Chromosomes Cancer. 2000;27:162–168. [PubMed] [Google Scholar]
- 23.El-Naggar AK, Lovell M, Callender DL, Ordonez NG, Killary AM. Cytogenetic analysis of a primary salivary gland myoepithelioma. Cancer Genet Cytogenet. 1999;113:49–53. doi: 10.1016/s0165-4608(98)00280-5. [DOI] [PubMed] [Google Scholar]
- 24.Dal Cin P, Van den Berghe H, Van Poppel H, Roskams T. Involvement of 12q12-13 is a nonrandom chromosome change in renal oncocytoma. Genes, Chromosomes Cancer. 1999;24:94. doi: 10.1002/(sici)1098-2264(199901)24:1<94::aid-gcc14>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Abdollahi A, Godwin AK, Miller PD, Getts LA, Schultz DC, Taguchi T, Testa JR, Hamilton TC. Identification of a gene containing zinc-finger motifs based on lost expression in malignantly transformed rat ovarian surface epithelial cells. Cancer Res. 1997;57:2029–2034. [PubMed] [Google Scholar]
- 26.Abdollahi A, Roberts D, Godwin AK, Schultz DC, Sonoda G, Testa JR, Hamilton TC. Identification of a zinc-finger gene at 6q25: a chromosomal region implicated in development of many solid tumors. Oncogene. 1997;14:1973–1979. doi: 10.1038/sj.onc.1201034. [DOI] [PubMed] [Google Scholar]
- 27.Bilanges B, Varrault A, Basyuk E, Rodriguez C, Mazumdar A, Pantaloni C, Bockaert J, Theillet C, Spengler D, Journot L. Loss of expression of the candidate tumor-suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene. 1999;18:3979–3988. doi: 10.1038/sj.onc.1202933. [DOI] [PubMed] [Google Scholar]
- 28.Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc-finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–2825. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis LJ, Li Y, Gerbault-Seureau M, Kuick R, Dutrillaux AM, Goubin G, Fawcett J, Cram S, Dutrillaux B, Hanash S, Muleris M. Amplification of DNA sequences from chromosome 19q13.1 in human pancreatic cell lines. Genomics. 1998;53:42–55. doi: 10.1006/geno.1998.5405. [DOI] [PubMed] [Google Scholar]
- 31.Hoglund M, Gorunova L, Andren-Sandberg A, Dawiskiba S, Mitelman F, Johansson B. Cytogenetic and fluorescence in situ hybridization analyses of chromosome 19 aberrations in pancreatic carcinomas: frequent loss of 19p13.3 and gain of 19q13.1-13.2. Genes Chromosomes Cancer. 1998;21:8–16. [PubMed] [Google Scholar]
- 32.Arnold N, Hagele L, Walz L, Schempp W, Pfisterer J, Bauknecht T, Kiechle M. Overrepresentation of 3q and 8q material and loss of 18q material are recurrent findings in advanced human ovarian cancer. Genes, Chromosomes Cancer. 1996;16:46–54. doi: 10.1002/(SICI)1098-2264(199605)16:1<46::AID-GCC7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Du Plessis L, Dietzsch E, Van Gele M, Van Roy N, Van Helden P, Parker MI, Mugwanya DK, De Groot M, Marx MP, Kotze MJ, Speleman F. Mapping of novel regions of DNA gain and loss by comparative genomic hybridization in esophageal carcinoma in the Black and colored populations of South Africa. Cancer Res. 1999;59:1877–1883. [PubMed] [Google Scholar]
- 34.Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. New York: McGraw-Hill; 1998. [Google Scholar]
- 35.Hui AB, Lo KW, Leung SF, Teo P, Fung MK, To KF, Wong N, Choi PH, Lee JC, Huang DP. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int J Cancer. 1999;82:498–503. doi: 10.1002/(sici)1097-0215(19990812)82:4<498::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 36.Bockmuhl U, Schwendel A, Dietel M, Petersen I. Distinct patterns of chromosomal alterations in high- and low-grade head and neck squamous cell carcinomas. Cancer Res. 1996;56:5325–5329. [PubMed] [Google Scholar]
- 37.Speicher MR, Howe C, Crotty P, du Manoir S, Costa J, Ward DC. Comparative genomic hybridization detects novel deletions and amplifications in head and neck squamous cell carcinomas. Cancer Res. 1995;55:1010–1013. [PubMed] [Google Scholar]
- 38.Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res. 1997;57:2116–2120. [PubMed] [Google Scholar]
- 39.Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, Schwendel A, Langreck H, Gellert K, Reichel M, Just K, du Manoir S, Cremer T, Dietel M, Ried T. Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res. 1997;57:2331–2335. [PubMed] [Google Scholar]
- 40.Petersen I, Langreck H, Wolf G, Schwendel A, Psille R, Vogt P, Reichel MB, Ried T, Dietel M. Small cell lung cancer is characterized by a high incidence of deletions on chromosomes 3p, 4q, 5q, 10q, 13q and 17p. Br J Cancer. 1997;75:79–86. doi: 10.1038/bjc.1997.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, Cremer T. Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res. 1994;54:1801–1806. [PubMed] [Google Scholar]
- 42.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–727. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 43.Cerilli LA, Swartzbaugh JR, Saadut R, Marshall CE, Rumpel CA, Moskaluk CA, Frierson HF., Jr Analysis of chromosome 9p21 deletion and p16 gene mutation in salivary gland carcinomas. Hum Pathol. 1999;30:1242–1246. doi: 10.1016/s0046-8177(99)90044-8. [DOI] [PubMed] [Google Scholar]