Abstract
The deleted in colorectal carcinoma (DCC) gene is a potential tumor-suppressor gene on chromosome 18q21.3. The relatively high frequency of loss of heterozygosity (LOH) and loss of expression of this gene in neuroblastoma, especially in the advanced stages, imply the possibility of involvement of the DCC gene in progression of neuroblastoma. However, only few typical mutations have been identified in this gene, indicating that other possible mechanisms for the inactivation of this gene may exist. A polymorphic change (Arg to Gly) at DCC codon 201 is related to advanced colorectal carcinoma and increases in the tumors with absent DCC protein expression. In order to understand whether this change is associated with the development or progression of neuroblastoma, we investigated codon 201 polymorphism of the DCC gene in 102 primary neuroblastomas by polymerase chain reaction single-strand conformation polymorphism. We found no missense or nonsense mutations, but a polymorphic change from CGA (Arg) to GGA (Gly) at codon 201 resulting in three types of polymorphism: codon 201Gly type, codon 201Arg/Gly type, and codon 201Arg type. The codon 201Gly type occurred more frequently in disseminated (stages IV and IVs) neuroblastomas (72%) than in localized (stages I, II, and III) tumors (48%) (P=.035), and normal controls (38%) (P=.024). In addition, the codon 201Gly type was significantly more common in tumors found clinically (65%) than in those found by mass screening (35%) (P=.002). The results suggested that the codon 201Gly type of the DCC gene might be associated with a higher risk of disseminating neuroblastoma.
Keywords: tumor-suppressor gene, the DCC gene, PCR-SSCP, codon 201 polymorphism, neuroblastoma
Introduction
Inactivation of the tumor-suppressor gene has been shown to play an important role in the development of a variety of human cancers [1]. The deleted in colorectal carcinoma (DCC) gene, initially identified as a candidate tumor-suppressor gene on chromosome 18q21.3 in colorectal carcinoma [2], shows frequent allelic deletion and aberrant expression in many human tumors [3,4]. Alterations of the DCC gene are also associated with metastasis of various carcinomas [5–11]. The DCC gene encodes a neural cell adhesion molecule (NCAM)-like transmembrane protein. Its extracellular domain has four immunoglobulin (Ig)-like and six fibronectin type III-like domains, which have high homology to frazzled and UNC-40, the two transmembrane proteins of the Ig superfamily that are mainly expressed on motor neurons of the nervous system [12,13]. The cytoplasmic domain of the DCC gene shares similarity to neogenin. The expression of neogenin is dynamically regulated in the developing nervous system of chickens [14]. In addition, DCC has been identified as a receptor for netrin-1, a molecule involved in axon guidance and cell migration during development [15]. Thus, the DCC gene is a critical gene in regulating the growth and differentiation of the neural-origin cells.
Although there is no increase in the frequency of tumor formation in DCC hemizygous mice [16], reestablishment of the expression of DCC suppresses tumorigenicity [17,18]. Furthermore, DCC can induce apoptosis in cells outside the region of ligand availability (e.g., metastatic cells or locally invasive cells), suggesting that DCC may function as a tumor-suppressor and play a major role in suppressing invasion or metastasis of tumor cells [19].
Neuroblastoma is a malignant tumor of neural crest origin. It is very common in childhood neoplasm with an incidence of about 1 in 100,000 to 130,000 children under 15 years of age [20].We have reported the relatively high frequency of loss of heterozygosity (LOH) (31%) at DCC locus on chromosome 18q21 in neuroblastoma [21]. Reduced or missing expression of the DCC gene at the mRNA and protein levels in advanced stages of neuroblastoma has been observed both by us and by other groups [22–24], indicating that DCC is probably involved in the tumorigenesis of neuroblastoma. However, only a few mutations have been identified in this gene [23]. It is thus possible that mechanisms other than mutations are responsible for the inactivation of this gene. A polymorphic change (Arg to Gly) at DCC codon 201 was found to be related to advanced colorectal carcinomas and carcinomas lacking DCC protein expression [25,26]. In our previous study, this polymorphic change was observed in varieties of neuroblastoma cell lines and primary tumors [23]. Thus, we hypothesize that this polymorphic change might be associated with the development or progression of the neuroblastoma in the same way as it is in the colorectal carcinoma. In order to test this hypothesis, we investigated codon 201 polymorphism of the DCC gene in 102 primary tumors by polymerase chain reaction single-strand conformation polymorphism (PCR-SSCP), followed by sequencing.
Materials and Methods
Primary Tumor Specimens
One hundred and two tumor specimens were obtained from neuroblastoma patients. The samples were taken from the patients either at Saitama Children's Medical Center or at the University of Tokyo Hospital before chemotherapy. Tissue specimens were immediately frozen in liquid nitrogen after the surgical removal and stored at -80°C. Twelve noncancerous tissues including peripheral blood bone marrow were obtained from some of the above patients of stages I and II. The informed consent was obtained from the parents of the patients before the samples were taken. Twenty-one peripheral blood samples as normal controls for PCR-SSCP were obtained from healthy volunteers.
Patient Data
The stages of neuroblastoma were defined according to the classification of Evans et al. [27]. One hundred and two patients, aged from 18 days to 16 years with a median age of 12 months, were examined. These included 68 males and 34 females. Twenty-three were classified as stage I, 27 stage II, 16 stage III, 32 stage IV, and 4 stage IVs. Twenty-six patients including 13 stage I, 12 stage II, and 1 stage III were found by mass screening [28]. The status of the N-myc gene was examined in 59 patients including 12 patients in stage IV, five of whom had amplification. Patients in stages I and II were treated by either surgery alone or surgery plus chemotherapy that mainly consisted of vincristine and cyclophosphamide without radiotherapy. Patients in stages III and IV were administered with multidrug chemotherapy consisting of cyclophosphamide, doxorubicin, cisplatinum, and etoposide with or without surgery, radiotherapy, and hematopoietic stem cell transplantation [29].
PCR-SSCP Analysis of DCC Mutations
High-molecular-weight DNA was prepared from tumor tissues using the proteinase-K phenol-chloroform method and PCR-SSCP was performed as previously described [23]. The oligonucleotide primers for amplifying DCC codon 201 span a 353-fragment and were synthesized as described by Miyake et al. [5]. PCR for genomic DNA amplification proceeded in a 5-ml mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 100 mM of each dNTP, 5 pmol of each primer, 2.5 ml of [α-32P] dCTP (3000 Ci/mmol), and 0.25 units of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). The PCR consisted of an initial denaturing step: 2 minutes at 94°C; 35 cycles of amplification: 1 minute at 94°C for denaturation, 1 minute at 56°C for annealing, 1 minute at 72°C for extension, and 7 minutes at 72°C for the final extension step. After amplification, PCR mixtures were heated for 5 minutes at 80°C with 45 µL of formamide denaturing dye mixture (95% formamide, 20 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue), and then resolved by electrophoresis on a 5% nondenaturing polyacrylamide gel containing 45 mM Tris-borate (pH 8.3), 4 mM EDTA, and 1.6% Amp (ammonium peroxodisulfate) with and without 10% glycerol. Electrophoresis proceeded at 40 W for 2 to 3 hours at 26°C or 4°C. Gels were dried on filter paper and exposed to X-ray film at -80°C overnight.
Direct Sequencing of PCR-Amplified Fragments
Sequencing was performed as previously described [23]. Small gel slices containing the shift band detected by SSCP were excised, immersed in 20 µL of water, and placed at room temperature overnight. The extracts were centrifuged and PCR-amplified as described above. PCR products were purified using a Microcon 100 (Amicon, Beverly, MA) and sequenced by the dideoxy chain termination method.
Results
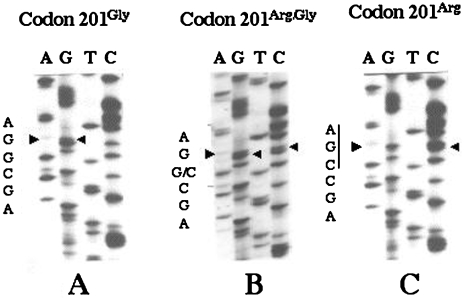
Using PCR-SSCP methodology, we investigated exon 3 of the DCC gene for codon 201 polymorphism in the 102 primary tumors, 12 noncancerous tissues, and 21 normal peripheral blood samples. Three patterns of band distribution were noted on the SSCP electrophoresis gels (Figure 1). DNA sequencing revealed that the polymorphism resulted from a CGA (Arg) to GGA (Gly) transversion in codon 201 at one or both alleles of the DCC gene (Figure 2). In tumor tissues, the most frequent transversion type was codon 201Gly (Figure 2A), which had one or two homozygous codon 201Gly alleles. The less frequent type was codon 201Arg/Gly type containing heterozygous codon 201Arg allele and codon 201Gly allele (Figure 2B). The least observed type was codon 201Arg type, which consisted of one or two homozygous alleles of wild type codon 201Arg (Figure 2C). In the 102 primary tumors, the frequencies of the codon 201Gly, codon 201Arg/Gly, and codon 201Arg types were 57% (58/102), 37% (38/102), and 6% (6/102), respectively. The types of codon 201 polymorphism were significantly correlated with the stages of neuroblastoma (P=.047, by Kruskal-Wallis test). The frequency of codon 201Gly type in stage IV+IVs tumors (26/36, 72%) was significantly higher than that in stage I+II+III tumors (32/66, 48%) (P=.035, by chi-square analysis) and that in normal controls (8/21, 38%) (P=.024, by chi-square analysis) (Table 1). Among the 26 tumors found by mass screening, 9 (35%) were codon 201Gly type, 12 (46%) were codon 201Arg/Gly, and 5 (19%) were codon 201Arg. In the 76 tumors found clinically, the frequencies of the three types were 49 (65%), 26 (34%), and 1 (1%), respectively. Codon 201Gly type of the DCC gene was significantly more frequent in the tumors found clinically (65%) than in those found by mass screening (35%) (P=.002, by chi-square analysis). Of the five tumors with N-myc amplification, codon 201Gly type was detected in two, codon 201Arg/Gly type in two, and codon 201Arg type in one tumor, respectively. The corresponding noncancerous tissues from stages I and II patients showed the same codon 201 alleles with the tumor tissues.
Figure 1.
Three types of codon 201 polymorphism within exon 3 of the DCC gene. Exon 3 was amplified from six primary tumors of neuroblastoma by PCR-SSCP and subjected to polyacrylamide gel electrophoresis. Three patterns of the bandshifts were noted on the electrophoresis gel and they are named as codon 201Arg type, codon 201Gly type, and codon 201Arg/Gly type according to the sequencing result.
Figure 2.
Position and nature of the codon 201 polymorphism. Small gel slices from the top second bands shown in Figure 1 were excised and applied to sequencing after PCR amplification. Variation of sequences at codon 201 was observed: codon 201Gly (A), a mutation of CGA (Arg) to GGA (Gly) at codon 201; codon 201Arg/Gly (B), both GGA (Gly) and CGA (Arg) present at codon 201; codon 201Arg (C), the wild type CGA (Arg) at codon 201. Arrow heads indicate the polymorphism nucleotides.
Table 1.
Correlation of Codon 201 Polymorphic Types of the DCC Gene with the Stages of Neuroblastoma.
| Stages | Number of total samples | Number (%) of codon 201Gly | Number (%) of codon 201Arg/Gly | Number (%) of codon 201Arg |
| I* | 23 | 10 (43%) | 11 (48%) | 2 (9%) |
| II* | 27 | 13 (48%) | 11 (47%) | 3 (11%) |
| III* | 16 | 9 (56%) | 6 (44%) | 0 |
| IV | 32 | 23 (72%) | 8 (25%) | 1 (3%) |
| IVs | 4 | 3 (75%) | 1 (25%) | 0 |
| Total | 102 | 58 (57%) | 38 (37%) | 6 (6%) |
| Normal control** | 21 | 8 (38%) | 10 (48%) | 3 (14%) |
The codon 201 polymorphic types of the DCC gene were significantly correlated with the stages of neuroblastoma (P=.047, by Kruskal-Wallis test). The frequency of codon 201Gly type was significantly higher in stages IV+IVs (26/36, 72%) than in stages I, II, and III (32/66, 48%) (*P=.035, by chi-square analysis) and in normal control (8/21, 38%) (**P=.024, by chi-square analysis).
Discussion
The DCC gene on chromosome 18q21 is considered to be one of the candidate tumor-suppressor genes in neuroblastoma because of the high frequency of LOH at DCC locus on chromosome 18 [21], the presence of a functional protein in primary tumors versus loss of expression in metastatic deposit, and the frequent reduced or loss of expression in advanced or disseminated tumors [22–24]. However, mechanisms for DCC inactivation are still unknown because only few mutations have been detected in neuroblastomas [23].
A polymorphic change from CGA (Arg) to GGA (Gly) at codon 201 of the DCC gene has been found to be associated with the progression of the colorectal carcinomas [25,26]. Minami et al. [25] demonstrated that codon 201Gly type was significantly increased in invasive colorectal carcinomas than adenomas, and 9 of 10 invasive cancer cases with LOH had remained only codon 201Gly allele. Another study by Schmitt et al. showed that 7 of 15 cases of the malignant colorectal tissues newly changed from codon 201Arg/Gly to codon 201Gly/Gly (both were codon 201Gly alleles) compared with their corresponding normal mucosa. The results from these two studies indicate that change from codon 201Arg/Gly to codon 201Gly type (remaining one codon 201Gly allele due to LOH [25] or two codon 201Gly alleles due to mutation [26]) is related to the malignant transformation. Previously, we found that the codon 201Gly type is frequently observed in neuroblastoma [23]; thus, we proposed that the polymorphic change from Arg to Gly at codon 201 of the DCC gene might be associated with neuroblastoma progression. Our results from the present study demonstrated that the codon 201Gly is more prevalent in disseminated neuroblastoma (stages IV and IVs) than in localized tumors (stages I, II, and III). In addition, the frequency of this abnormality is significantly higher in the patients found clinically than in those found by mass screening. This may be explained by the fact that most of the patients found by mass screening have stage I or II tumors. The data from this study suggest that the codon 201Gly type is likely related to a higher risk of dissemination of tumor cells. However, how codon 201Gly type becomes more prevalent in disseminated neuroblastoma remains unknown. Twelve noncancerous tissue specimens from stages I and II neuroblastomas (nondisseminated tumors) had the same codon 201 polymorphism types as the tumor tissues in the present study, indicating that the variation at codon 201 of the DCC gene is an inherited polymorphism. Thus, the possible explanations for the high incidence of codon 201Gly type in the disseminated neuroblastomas might be as follows. One possibility is that the codon 201Arg allele is preferentially lost in tumor cells due to LOH or mutation during progression of neuroblastoma similar to the scenario that happened in the advanced colorectal carcinoma [25,26]. The other possibility is that individuals with two codon 201Gly alleles are more susceptible to disseminated neuroblastoma than those without the codon 201Gly allele. We considered that the first mechanism is more possible in neuroblastoma based on the studies of others [25,26]; however, the second cannot be excluded until we compare the codon 201 polymorphism between tumors and the adjacent normal tissues from the patients with disseminated neuroblastomas in the future.
Not only unique somatic mutations in a gene could change the protein conformation and reduce or abolish the function of the gene, many functional polymorphisms intend to affect the expression of the protein and its activities [30–33]. The polymorphic change from CGA (Arg) to GGA (Gly) at codon 201 was also found to be associated with loss of the DCC protein in colorectal carcinomas [26]. Eight of nine colorectal tumors with both codon 201Gly alleles had no detectable DCC expression. Importantly, seven of them acquired a new codon 201Gly allele through mutation of Arg to Gly. Although we did not examine the DCC protein expression due to the limited amount of neuroblastoma samples, the polymorphic change from Arg to Gly at codon 201 resulting in codon 201Gly type documented in our study seems to have a good agreement with the loss of protein expression in neuroblastoma reported by Reale et al. [22]. First, our finding that codon 201Gly type was more prevalent in the disseminated tumors is consistent with Reale et al.'s result that loss of DCC protein expression was more frequently observed from patients with stage IV or IVs tumors. Second, stage IVs neuroblastomas occur in patients less than 1 year of age, and the majority of tumors spontaneously regress despite dissemination to distant sites [34]. Three of the four tumors from stage IVs patients showed the codon 201Gly type, implying that this mutation is probably related only to the higher risk of tumor cell dissemination, regardless of their true malignant potential. This result is consistent with the finding that DCC protein expression was undetectable in most stage IVs tumors [22]. Third, our study found that five neuroblastomas with N-myc amplification did not show a prediction to any codon 201 polymorphic type, indicating that the polymorphic change at codon 201 might be involved in the dissemination, not through the pathway regulated by N-myc. This phenomenon also agrees with the finding that there is no consistent relationship between loss of DCC protein and N-myc amplification [22]. However, because only a small number of cases in the second and third scenarios were investigated, further studies involving larger patient populations are required. In any case, it will be very interesting in the future to investigate the relationship between loss of DCC protein expression and codon 201 genotypes in disseminated neuroblastoma.
The association of the polymorphic change from Arg to Gly at codon 201 with the tumor progression or dissemination indicates that codon 201Gly type protein might have a weaker function than codon 201Arg type protein. Codon 201 is in the second of the four Ig-like domains of the DCC gene. This domain is in a highly conserved region that shares high similarity with adhesion molecule NCAM [2]. Developing neoplasia and metastasis is often associated with disruption of cell adhesion or cell-to-cell contacts [35,36]. The polymorphic change at codon 201 is from the amino acid Arg to Gly. The Arg is a basic, hydrophilic amino acid and has a big basic side chain, whereas Gly is a nonpolar amino acid with the least side chain volume. More importantly, occurrence of Gly residue would destabilize an α-helix structure in the second Ig-like domain because it has more conformational flexibility than other amino acids [37]. Taking together the changes in the side chain volume, hydropathy, and α-helix stability due to Gly substitution for Arg at codon 201 may alter the conformation of the DCC protein, resulting in the activity of the codon 201Gly type DCC protein much weaker than that of the wild type codon 201Arg DCC protein. This may diminish the tumor-suppressor function of DCC and contribute to the progression or dissemination of the tumors. Because codon 201Arg/Gly type is not shown to be related to the progression of the colorectal carcinomas [25,26] and the dissemination of neuroblastoma in this study, it is likely that the presence of one codon 201Arg allele is enough to produce functional DCC protein. Alternatively, the codon 201Arg allele is selectively transcribed. Loss of DCC expression and/or function activity may occur only when both of codon 201Arg alleles are deleted, for example, in codon 201Gly type tumors.
In summary, our study showed that the CGA (Arg) to GGA (Gly) substitution at codon 201 in both alleles is more frequent in disseminated neuroblastomas, suggesting that the polymorphism change from codon 201Arg type to codon 201Gly type might be involved in tumor dissemination. The high frequency of codon 201Gly type of the DCC gene in disseminated neuroblastomas also suggests that this variation might be a helpful genetic marker for these tumors.
Acknowledgements
We express appreciation to the biostatistician, Chikuma Hamada, Department of Pharmacoepidemiology, University of Tokyo, for his advice regarding statistical analysis. We thank Deborah Wilkinson for kindly reviewing the manuscript for English and Shouko Sohma for her skilled technical assistance. Our gratitude is also extended to the clinicians in Japan for generously providing samples and clinical data.
Abbreviations
- DCC
deleted in colorectal carcinoma gene
- LOH
loss of heterozygosity
- PCR-SSCP
polymerase chain reaction single-strand conformation polymorphism
Footnotes
This work was supported, in part, by a Grant-in-Aid for International Scientific Research, Scientific Research on Priority Areas, and Scientific Research from the Ministry of Education, Science, Sports, and Culture, Japan.
References
- 1.Weinberg RA. Tumor-suppressor gene. Science (Washington, DC) 1991;254:1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomus G, Kinzler KW, Vogelstein B. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science (Washington, DC) 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 3.Cho KR, Fearon ER. DCC: linking tumor-suppressor genes and altered cell surface interactions in cancer? Eur J Cancer. 1995;31A:1055–1060. doi: 10.1016/0959-8049(95)00128-6. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER. DCC: is there a connection between tumorigenesis and cell guidance molecules? Biochim Biophys Acta. 1996;1288:M17–M23. doi: 10.1016/0304-419x(96)00023-6. [DOI] [PubMed] [Google Scholar]
- 5.Miyake S, Nagai K, Yoshino K, Oto M, Endo M, Yuasa Y. Point mutations and allelic deletion of tumor-suppressor gene DCC in human esophageal squamous cell carcinomas and their relation to metastasis. Cancer Res. 1994;54:3007–3010. [PubMed] [Google Scholar]
- 6.Kikuchi-Yanoshita R, Konishi M, Fukunari H, Tanaka K, Miyaki M. Loss of expression of the DCC gene during progression of colorectal carcinomas in familial adenomatous polyposis and nonfamilial adenomatous polyposis patients. Cancer Res. 1992;52:3801–3803. [PubMed] [Google Scholar]
- 7.Ookawa K, Sakamoto M, Hirohashi S, Yoshida Y, Sugimura T, Terada M, Yokota J. Concordant p53 and DCC alterations and allelic losses on chromosomes 13q and 14q associated with liver metastases of colorectal carcinoma. Int J Cancer. 1993;53:382–387. doi: 10.1002/ijc.2910530307. [DOI] [PubMed] [Google Scholar]
- 8.Fang DC, Jass JR, Wang DX. Loss of heterozygosity and loss of expression of the DCC gene in gastric cancer. J Clin Pathol. 1998;51:593–596. doi: 10.1136/jcp.51.8.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida Y, Itoh F, Endo T, Hinoda Y, Imai K. Decreased DCC expression in human gastric cancers is clinicopathologically significant. Int J Pathol. 1998;79:634–639. doi: 10.1002/(sici)1097-0215(19981218)79:6<634::aid-ijc14>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 10.Saito M, Yamaguchi A, Goi T, Tsuchiyama T, Nakagawara G, Urano T, Shiku H, Furukawa K. Expression of DCC protein in colorectal tumors and its relationship to tumor progression and metastasis. Oncology. 1999;56:134–141. doi: 10.1159/000011954. [DOI] [PubMed] [Google Scholar]
- 11.Saegusa M, Hashimura M, Hara A, Okayasu I. Loss of expression of the gene deleted in colon carcinoma (DCC) is closely related to histologic differentiation and lymph node metastasis in endometrial carcinoma. Cancer. 1999;85:453–464. doi: 10.1002/(sici)1097-0142(19990115)85:2<453::aid-cncr25>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Kolodziej PA, Timpe LC, Mitchell KJ, Fried SR, Goodman CS, Jan LY, Jan YN. Frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell. 1996;87:197–204. doi: 10.1016/s0092-8674(00)81338-0. [DOI] [PubMed] [Google Scholar]
- 13.Chan SS-Y, Zheng H, Su M-W, Wilk R, Killeen MT, Hedgecock EM, Culotti JG. UNC-40, a C. elegans homolog of DCC (deleted in colorectal cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 1996;87:187–195. doi: 10.1016/s0092-8674(00)81337-9. [DOI] [PubMed] [Google Scholar]
- 14.Vielmetter J, Kayyem JF, Roman JM, Dreyer WJ. Neogenin, an avian cell surface protein expressed during terminal neuronal differentiation, is closely related to the human tumor-suppressor molecule deleted in colorectal cancer. J Cell Biol. 1994;127:2009–2020. doi: 10.1083/jcb.127.6.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in colorectal cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 16.Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, Simons J, Bronson RT, Gordon JI, Tessier-Lavigne M, Weinberg RA. Phenotype of mice lacking functional deleted in colorectal cancer (dcc) gene. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K, Oshimura M, Kikuchi R, Seki M, Hayashi T, Miyaki M. Suppression of tumorigenicity in human colon carcinoma cells by introduction of normal chromosome 5 or 18. Nature. 1991;349:340–342. doi: 10.1038/349340a0. [DOI] [PubMed] [Google Scholar]
- 18.Klingelhutz AJ, Hedrick L, Cho KR, McDougall JK. The DCC gene suppresses the malignant phenotype of transformed human epithelial cells. Oncogene. 1995;10:1581–1586. [PubMed] [Google Scholar]
- 19.Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Saivesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 20.Bessho F. Effects of mass screening on age-specific incidence of neuroblastoma. Int J Cancer. 1996;67:520–522. doi: 10.1002/(SICI)1097-0215(19960807)67:4<520::AID-IJC10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Takita J, Hayashi Y, Kohno T, Shiseki M, Yamaguchi N, Hanada R, Yamamoto K, Yokota J. Allelotype of neuroblastoma. Oncogene. 1995;11:1829–1834. [PubMed] [Google Scholar]
- 22.Reale MA, Reyes-Mugica M, Pierceall WE, Rubinstein MC, Hedrick L, Cohn SL, Nakagawara A, Brodeur GM, Fearon ER. Loss of DCC expression in neuroblastomas is associated with disease dissemination. Clin Cancer Res. 1996;2:1097–1102. [PubMed] [Google Scholar]
- 23.Kong XT, Choi SH, Inoue A, Xu F, Chen Tao, Takita J, Yokota J, Bessho F, Yanagisawa M, Hanada R, Yamamoto K, Hayashi Y. Expression and mutational analysis of the DCC, DPC4 and MADR2/JV18-1 genes in neuroblastoma. Cancer Res. 1997;57:3772–3778. [PubMed] [Google Scholar]
- 24.Reyes-Mugica M, Lin P, Yokota J, Reale MA. Status of deleted in colorectal cancer gene expression correlates with neuroblastoma metastasis. Lab Invest. 1998;78:669–675. [PubMed] [Google Scholar]
- 25.Minami R, Aoyama N, Honsako Y, Kasuga M, Fujimori T, Maeda S. Codon 201Arg/Gly polymorphism of DCC (deleted in colorectal carcinoma) gene in flat- and polypoid-type colorectal tumors. Dig Dis Sci. 1997;42:2446–2452. doi: 10.1023/a:1018839907159. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt CA, Thaler KR, Witting BM, Kaulen H, Meyer Zum Buschenfelde K-H, Dippold WG. Detection of the DCC gene product in normal and malignant colorectal tissues and its relation to a codon 201 mutation. Br J Cancer. 1998;77:588–594. doi: 10.1038/bjc.1998.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans AE, D'Angio GJ, Randolph J. A proposed staging for children with neuroblastoma. A Children's Cancer Study Group. Cancer. 1971;27:374–378. doi: 10.1002/1097-0142(197102)27:2<374::aid-cncr2820270221>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi Y, Hanada R, Yamamoto K. Biology of neuroblastoma in Japan found by screening. Am J Pediatr Hematol/Oncol. 1992;14:342–347. [PubMed] [Google Scholar]
- 29.Sawaguchi S, Kaneko M, Uchino J, Takeda T, Iwafuchi M, Matsuyama S, Takahashi H, Nakajo T, Hoshi Y, Okabe I, Yokoyama J, Nishihira H, Sakaki S, Sakurai M, Sawada T, Nagahara N, Tsuchida Y. Treatment of advanced neuroblastoma with emphasis on intensive induction chemotherapy: a report from the Study Group of Japan. Cancer. 1990;66:1879–1887. doi: 10.1002/1097-0142(19901101)66:9<1879::aid-cncr2820660905>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 30.Facher EA, Becich MJ, Deka A, Law JC. Association between human cancer and two polymorphisms occurring together in the p21Waf1/Clp1 cyclin-dependent kinase inhibitor gene. Cancer. 1997;79:2424–2429. doi: 10.1002/(sici)1097-0142(19970615)79:12<2424::aid-cncr19>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 31.Frazier ML, O'Donnell FT, Kong S, Gu X, Campos I, Luthra R, Lynch PM, Amos CI. Age-associated risk of cancer among individuals with N-acetyltransferase 2 (NAT2) mutations and mutations in DNA mismatch repair genes. Cancer Res. 2001;61:1269–1271. [PubMed] [Google Scholar]
- 32.Yu Y, Okayasu R, Weil MM, Silver A, McCarthy M, Ryan Z, Long S, Cox R, Ullrich RL. Elevated breast cancer risk in irradiated BALB/c mice associated with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- 33.Hu YJ, Korotkov KV, Mehta R, Hatfield DL, Rotimi CN, Luke A, Prewitt TE, Cooper RS, Stock W, Vokes WS, Dolan ME, Gladyshev VN, Diamond AM. Distribution and functional consequences of nucleotide polymorphisms in the 3′-untranslated region of the human Sep15 gene. Cancer Res. 2001;61:2307–2310. [PubMed] [Google Scholar]
- 34.Hayashi Y, Inaba T, Hanada R, Yamada I, Nakagome Y, Yamamoto K. Similar chromosomal patterns and lack of N-myc gene amplification localized and IV-S stage neuroblastoma in infants. Med Pediatr Oncol. 1989;17:111–115. doi: 10.1002/mpo.2950170208. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface, modulation, and alternative RNA splicing. Science (Washington, DC) 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- 36.Edelman GM, Crossin KL. Cell adhesion molecules: amplifications for a molecular histology. Annu Rev Biochem. 1991;60:155–190. doi: 10.1146/annurev.bi.60.070191.001103. [DOI] [PubMed] [Google Scholar]
- 37.Nelson DL, Cox MM. The three-dimensional structure of proteins. In: Nelson DL, Cox MM, editors. Lehninger Principles of Biochemistry. NY: Worth Publishers; 2000. pp. 159–202. [Google Scholar]