Abstract
We show here that, although genes constitute only a small percentage of the maize genome, it is possible to identify them phenotypically as Ac receptor sites. Simple and efficient Ac transposition assays based on the well-studied endosperm markers bz and wx were used to generate a collection of >1300 independent Ac transposants. The majority of transposed Ac elements are linked to either the bz or the wx donor loci on chromosome 9. A few of the insertions produce obvious visible phenotypes, but most of them do not, suggesting that these populations will be more useful for reverse genetics than for forward transposon mutagenesis. An inverse polymerase chain reaction method was adapted for the isolation of DNA adjacent to the transposed Ac elements (tac sites). Most Ac insertions were into unique DNA. By sequencing tac sites and comparing the sequences to existing databases, insertions were identified in a number of putative maize genes. The expression of most of these genes was confirmed by RNA gel blot analysis. We report here the isolation and characterization of the first 46 tac sites from the two insertion libraries.
INTRODUCTION
The maize genome is made up largely of repetitive DNA (Hake and Walbot, 1980), of which a significant fraction consists of methylated retrotransposons arranged in clusters (SanMiguel et al., 1996). Genes likely make up <5% of the total nuclear genome and are found in hypomethylated CpG islands (Antequera and Bird, 1988; Bennetzen et al., 1994). This also is the genomic component into which the transposon Activator (Ac) inserts (Dellaporta and Chomet, 1985; Chen et al., 1987). One can take advantage of this property of Ac to identify genes phenotypically as Ac receptor sites, hereafter termed tac sites (for transposed Ac sites). Genes then would be isolated and sequenced as the DNA adjacent to Ac. An advantage of this approach to maize functional genomics is that, in addition to a sequence that can be compared with the existing databases, it generates an insertion library that can be screened for subtle mutant phenotypes, particularly after learning where the genes are expressed. Many genes are expected to have minor effects or to be induced only under special conditions and would be missed from a conventional transposon mutagenesis screen designed to identify gross changes in phenotype (Thatcher et al., 1998).
There is ample evidence that other maize transposons, such as Mu and Spm/En, also insert preferentially into hypomethylated DNA (Chandler and Walbot, 1986; Cone et al., 1986; Schmidt et al., 1987; Bennetzen et al., 1988). Furthermore, maize genes and transposons occupy an isochore of DNA with an extremely narrow G-C content that constitutes only 10 to 20% of the genome (Carels et al., 1995). Thus, in general, transposons can be used to search for genes within the bulk of repetitive DNA. This concept served as the basis for projects that generated large pools of genomic DNAs from tens of thousands of plants carrying Mutator elements at multiple locations in the genome (Das and Martienssen, 1995; Meeley and Briggs, 1995). From these populations, popularly known as Mu gene machines, several mutations have been identified in genes for which a DNA sequence was available (Bensen et al., 1995; Frey et al., 1997). This resource has enabled investigators to proceed from sequence to mutant very rapidly. Most mutations recovered from the Mu gene machines, as well as from regular Mu transposon mutagenesis experiments, carry the nonautonomous, but highly abundant, Mu1 element (Bennetzen, 1996).
Ac insertion libraries represent a complementary approach to the Mu gene machine. As an insertion, Ac has several advantages: (1) Ac transpositions can be selected phenotypically. Thus, it is possible to use convenient seed phenotypes to assemble a large collection of independent genomic transpositions of Ac (trAcs) with little effort. (2) Ac has a dominant trans-acting phenotype, namely, the capacity to trans-activate the excision of a defective Ds (Dissociation) element from a reporter allele located elsewhere. Therefore, trAcs can be identified in heterozygous conditions and then rendered homozygous to screen for mutant phenotypes. (3) Ac's dominant phenotype permits the quick and simple analysis of cosegregation between a trAc and a new mutation, information that is useful to the investigator in deciding on the next experimental step. (4) Ac's visible phenotype allows the mapping of a trAc insertion in the genome independently of the molecular mapping of its adjacent sequence. (5) Ac tends to transpose to sites that are linked closely to the donor site (Van Schaik and Brink, 1959; Greenblatt, 1984; Dooner and Belachew, 1989). This property of Ac can be used to “saturate” defined regions of the genome with insertions. Placing Ac in launching pads at multiple regularly spaced locations would enable selective targeting of the entire genome, as was done in Arabidopsis in the work that led to the directed targeting of the FAE1 gene (James et al., 1995). (6) The number of sequences in the genome that are homologous with the central part of the Ac element is low (four to six) (Fedoroff et al., 1983). Furthermore, most of those sequences are methylated, whereas Ac is not (Schwartz and Dennis, 1986; Chomet et al., 1987). These two features make it feasible to devise polymerase chain reaction (PCR) strategies for the isolation of DNA adjacent to Ac using Ac primers.
This discussion should serve to illustrate why the Mu and Ac approaches to functional genomic analysis in maize are complementary. The power of both strategies lies in their ability to define gene function through gene disruption. In the former, mutagenesis relies on the mobilization of high-copy-number, nonautonomous Mu1 elements that do not transpose locally (Lisch et al., 1995), and in the latter, it relies on the regional mobilization of the single-copy, autonomous Ac element that targets nearby sites preferentially.
We have generated a large Ac insertion library consisting of 1225 independent transpositions from one launching platform, the wx-m7(Ac) mutable allele in 9S. Only 61 of the 1225 Ac transposants yielded clear mutant phenotypes, of which 6 cosegregated with Ac. Hence, factors other than Ac were responsible for most of the mutations, suggesting that these populations will be more useful for reverse genetics than for forward mutagenesis. As expected, most transposed elements were linked to the wx donor locus. We have begun to isolate and sequence the DNA adjacent to the trAc elements from this and an additional, smaller insertion library generated previously from the bz-m2(Ac) allele (Dooner and Belachew, 1989; Dooner et al., 1994). We report here the characterization of the first 46 tac sites isolated from these libraries. All Ac insertion sites isolated to date correspond to low-copy DNA, and more than half have homology with either known genes or expressed sequence tags (ESTs) in the sequence databases. The expression of a subset of tac sites, including some lacking homology with any known sequence, was confirmed by RNA gel blot analysis.
RESULTS
Generation and Genetic Characterization of an Ac Insertion Library from wx-m7(Ac)
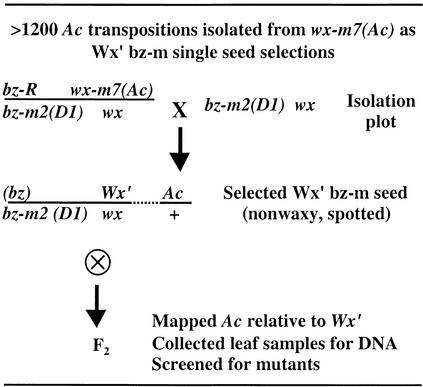
We have developed an efficient single-seed selection scheme to identify germinal transpositions of Ac from the wx-m7(Ac) mutable allele. Figure 1 outlines the genetic derivation of the transposants. The system relies on the coupling of two easily scored endosperm markers, wx and bz, to monitor Ac transposition in the female germline. wx-m7(Ac) is a highly mutable allele described by McClintock (1964). It reverts to Wx′ in the germline, with frequencies on the order of a few percent. The 4.6-kb Ac element is inserted upstream of the Wx coding sequence (Muller-Neumann et al., 1984; Klosgen et al., 1986), in a region of the Wx gene in which excision footprints do not seem to interfere with gene function. The reporter allele bz-m2(D1) produces a heavily spotted aleurone phenotype in the presence of Ac and a solid-bronze phenotype in its absence. The other two alleles, bz-R and wx, are stable mutations. In ears of bz-R wx-m7(Ac)/bz-m2(D1) wx heterozygotes pollinated with bz-m2(D1) wx, germinal transpositions of Ac can be selected as single exceptional kernels with a nonwaxy, spotted (Wx′ bz-m) phenotype among a majority of kernels with a waxy or waxy mutable phenotype.
Figure 1.
Derivation of an Ac Insertion Library from the wx-m7(Ac) Mutable Allele.
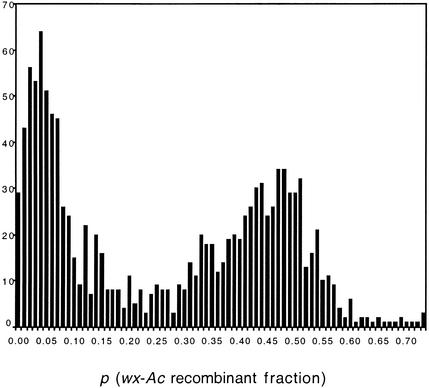
A collection of 1225 independent Ac transposants was generated, and all of the trAc sites were mapped relative to the wx donor locus by two-point crosses. The distribution of Ac receptor sites is shown in Figure 2. As has been observed at other maize loci (Greenblatt, 1984; Dooner and Belachew, 1989), the majority of trAc elements (63%) are linked to the donor locus. Of the 1225 families produced by selfing the primary transposants, 61 (or ∼5%) segregated for obviously visible mutants. Table 1 lists the obvious phenotypes identified either in the ear of the primary transposants or in field screens of their progeny conducted at different stages of development. The mutations were typical of those recovered in other mutagenized populations and affected various parts of the plant. Segregating mutant seed phenotypes included defective kernels, dull endosperms, and empty pericarps. Several F2 families in the nursery segregated seedling phenotypes, such as pale green, luteus, and albino, which often were lethal. Some of the adult plant phenotypes observed were zebra cross-bands, wrinkled leaves, yellow green, and wilty. Cosegregation analysis of the trAc and the new mutant revealed that only a minority (six, or ∼10%) may be tagged by Ac. At least two of them are tagged by a Ds element, judging from the somatic and germinal reversions that occur only in the presence of Ac.
Figure 2.
Distribution of 1225 trAcs among Wx′ Revertants from wx-m7(Ac).
The genetic distance between the wx donor locus and a trAc is plotted against the number of trAcs for each genetic distance. Genetic distance is expressed as p, the fraction of recombinants between wx and Ac. The distribution of p values is bimodal. The left cluster corresponds to receptor sites closely linked to wx on 9S, and the right cluster corresponds to unlinked receptor sites.
Table 1.
Segregating Mutant Phenotypes
| Mutant Phenotype | No. | Cosegregation with Ac |
|---|---|---|
| Seed | ||
| Dull | 2 | |
| Defective kernel (small endosperm, germless) |
15 | |
| Anthocyanin low | 1 | |
| Abnormal kernel, low anthocyanin | 1 | |
| Abnormal kernel (small kernel, with embryo) |
4 | |
| Lethal embryo (normal endosperm, germless) |
3 | 2 |
| Inverted kernel | 1 | |
| Paper-thin kernel | 3 | |
| New Wx-m | 1 | 1 |
| Sugary | 1 | |
| Seedling | ||
| Pale green lethal | 4 | |
| Pale green | 2 | |
| Luteus | 3 | |
| Plant | ||
| Short, pale green, wilty | 2 | 1 |
| Adherent upper whorls | 1 | 1 |
| Ear semisterility | 3 | |
| Pale green, shorter | 6 | |
| Yellow green | 1 | |
| Ramosa tassel | 1 | |
| Abnormal leaf blades | 1 | |
| Stunted, necrotic | 1 | |
| Wrinkled leaf | 1 | |
| White stripes | 1 | |
| Zebra | 1 | |
| Pollen | ||
| Reduced male transmission | 1 | 1 |
| Total | 61 | 6 |
Although this collection of 1225 transposants is of potential utility for gene isolation by conventional transposon tagging, our primary objective is to isolate the DNA adjacent to the trAc element (tac sites), which in most cases will correspond to genes, independent of whether the trAc confers a mutant phenotype. We also are isolating tac sites from a smaller collection of 102 trAcs derived from the bz-m2(Ac) mutable allele (Dooner and Belachew, 1989). All of the trAcs in this set were mapped relative to the bz donor locus in chromosome 9 (Dooner and Belachew, 1989; Dooner et al., 1994), and those that were unlinked to bz were assigned subsequently to chromosomes on the basis of their linkage to reciprocal translocation break points (Dooner et al., 1994). The latter group of trAc stocks could serve as starting points for localized transposon mutagenesis from chromosomes other than 9 (Dooner, 1995). All of the stocks described in this report have been deposited in the Maize Stock Center (Urbana, IL).
Isolation of tac Sites
The potential utility of a maize Ac insertion library for functional genomics will depend on the ease with which tac sites can be isolated. In transgenic dicots, in which the introduced Ac usually is present in one or two copies, tac sites can be isolated readily by PCR approaches, such as inverse PCR (IPCR) (Whitham et al., 1994; James et al., 1995) and thermal asymmetric interlaced PCR (TAIL-PCR) (Tsugeki et al., 1996). Maize, on the other hand, has several endogenous Ac-homologous sequences (Fedoroff et al., 1983), which render tac site isolation by PCR-based methods much more difficult. Although IPCR has been used to isolate an Spm-adjacent sequence in maize (Earp et al., 1990), this method has not been used widely because usually too many nonspecific sequences are amplified. To minimize the amplification of unwanted sequences, we have introduced a gel-purification step before performing IPCR. Although somewhat laborious, the method works well and, to date, it has enabled us to clone 42 tac sites corresponding to germinal transpositions of Ac. This is the largest number of DNA sequences adjacent to confirmed germinal transpositions of an autonomous and therefore phenotypically traceable transposable element yet isolated in maize.
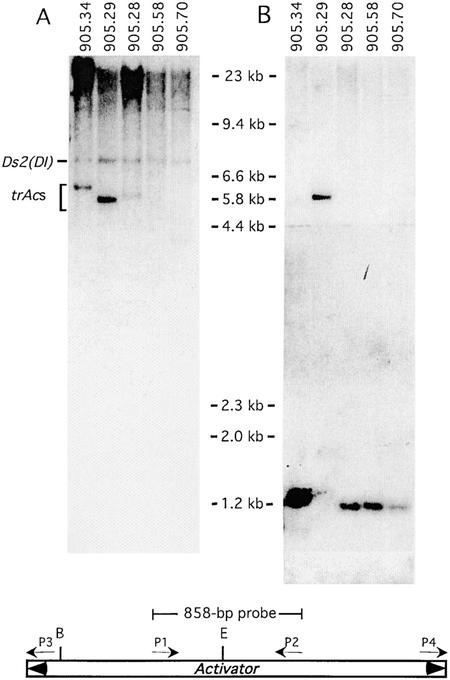
First, we examined diagnostic DNA gel blots of primary transposants or their progeny to identify unique Ac-hybridizing bands of a size appropriate for IPCR amplification of the DNA flanking Ac (5 to 9 kb). An internal control in these blots is provided by the bz-m2(D1) allele, which, like trAc, is present in heterozygous condition and hybridizes to the Ac probe. An example of such a blot is shown in Figure 3A. Then, we cut out the corresponding fragment from a preparative gel and performed IPCR to amplify the tac site. We digested genomic DNA with either methylation-sensitive (PstI, SalI) or methylation-insensitive (BglII, SstI, NcoI, EcoRI) enzymes. Except for EcoRI, none of these enzymes cut within Ac, so our IPCR products should contain sequences adjacent to both sides of Ac. Thus, we can determine immediately after sequencing if the Ac ends are flanked by the 8-bp duplication expected of a new Ac insertion. Most of the cloned tac sites were chosen on the basis of the feasibility of amplifying them by IPCR.
Figure 3.
DNA Gel Blot Analysis of Ac Insertion Sites.
Each lane on the gel corresponds to DNA from an individual transposant digested with PstI.
(A) Diagnostic hybridization with the 858-bp Ac probe shown below. New Ac-hybridizing bands are detected in transposants 905.34, 905.29, and 905.28.
(B) Confirmation hybridization of the same membrane with the IPCR product from transposant 905.29 amplified with primers P3 and P4 (putative tac905.29 site). The probe detects a 4.6-kb larger band only in the lane containing DNA from transposant 905.29.
B, BamHI; E, EcoRI.
To confirm that we had isolated a true tac site, we compared the DNAs of transposants. If a true tac site has been isolated, then the transposant contributing the amplified sequence that is used as a probe should have a band that is 4.6 kb larger than the band in the other transposants. That is precisely the pattern seen in Figure 3B, where transposant 905.29 contributed both the probe and the DNA in the second lane. Confirmation DNA gel blots probed with the putative tac sites sometimes failed to reveal the expected polymorphism, suggesting that the isolated DNA most likely corresponds to either a preexisting Ds insertion or a somatic Ac transposition. The repeated recovery of clones with the same sequences from different transposants suggests that most of these sequences represent preexisting Ds insertions in our stocks. Remarkably, almost all of the probes tested detected only one or two bands, showing that our IPCR strategy leads to the amplification of single- or low-copy DNA.
Characterization of tac Sites
Table 2 lists the 46 tac sites that have been sequenced and confirmed by DNA gel blot analysis to date. Most (41 of 46) detected either one or two bands in genomic DNA gel blots, and 54% (25 of 46) had homology with sequences in the public databases (BLAST search of GenBank databases). Ac elements in the 900 series (sites 1 to 39) are derived from wx-m7(Ac). They were mapped relative to the wx donor locus and, in the case of Ac902.33, by the recombinant inbred method (Burr et al., 1988). Ac elements with a four-digit designation (sites 40 to 46) are derived from bz-m2(Ac). They were mapped relative to the bz donor locus (Dooner and Belachew, 1989; Dooner et al., 1994) and, in the case of Ac8193 and Ac8194, by the wx reciprocal translocation method (Dooner et al., 1994). Reflecting the location of the wx and bz donor loci, a large number of the cloned tac sites reside on 9S.
Table 2.
Characterization of Isolated tac Sites
| No. | tac Site | Enzyme a | Sequence Homology | E Value | Expression Data | Location | Copy No. |
|---|---|---|---|---|---|---|---|
| 1 | tac901.04 | SstI | Arabidopsis cytochrome P450 | 5e−40 | Undetected | 9: 13 cM from wxb | 1 |
| 2 | tac902.05 | SalI | tRNA-gly (GCG) | 2e−33 | Constitutive | 9: 29 cM from wx | 1 |
| 3 | tac902.07 | SalI | Arabidopsis ring zinc finger | 1e−17 | Seedling; tassel; 3-week-old embryo |
9: 18 cM from wx | 1 |
| 4 | tac902.33 | SacI | Rice plastid S9 ribosomal protein | 6e−86 | Endosperm; ear; tassel | 1L, close to telo | 1 |
| 5 | tac903.05 | SstI | No homology | Wounded or salt-treated seedlings |
Unlinked to wx | 1 | |
| 6 | tac903.06 | SstI | Arabidopsis auxin-inducible protein |
4e−6 | Etiolated seedlings; light repressed |
Unlinked to wx | 1 |
| 7 | tac905.05 | EcoRII | No homology | Undetected | 9S: 7 cM from wx | 1 | |
| 8 | tac905.29 | PstI | Arabidopsis Ser Thr kinase | 7e−11 | Tassel; roots | 9S: 9 cM from wx | 1 |
| 9 | tac907.25 | PstI | Arabidopsis sugar transporter | 1e−8 | Roots | Unlinked to wx | 1 |
| 10 | tac907.49 | PstI | No homology | Undetected | Unlinked to wx | 1 | |
| 11 | tac907.51 | NcoI | Arabidopsis short-chain ADHc | 3e−33 | Higher in older tissues | Unlinked to wx | 1 |
| 12 | tac907.54 | PstI | Arabidopsis hypothetical protein | 7e−25 | Immature ear; 2-week-old seed | Unlinked to wx | 1 |
| 13 | tac909.06 | EcoRI | Tobacco hypothetical protein | 1e−5 | Undetected | Unlinked to wx | 4–5 |
| 14 | tac911.14 | EcoRI | Arabidopsis DNA- 3-methyladenine glycosidase |
3e−27 | Mature leaves; silk | Unlinked to wx | 1 |
| 15 | tac911.16 | PstI | No homology | Undetected | Unlinked to wx | 1 | |
| 16 | tac911.17 | PstI | No homology | Undetected | Unlinked to wx | 1 | |
| 17 | tac911.22 | PstI | Maize early embryo EST | 5e−10 | Tassel (3 to 8 cm); stem | Unlinked to wx | 1 |
| 18 | tac911.28 | PstI | Arabidopsis Ser Thr kinase | 2e−7 | Endosperm; seedlings | Unlinked to wx | 3 |
| 19 | tac911.30 | PstI | No homology | Undetected | Unlinked to wx | 1 | |
| 20 | tac913.01 | PstI | No homology | Roots: salt induced; seed | 9S: 8 cM from wx | 1 | |
| 21 | tac913.09 | PstI | Arabidopsis hypothetical protein | 8e−6 | Tassel (3 to 8 cm); seedling shoot |
9S: 10 cM from wx | 2 |
| 22 | tac913.14 | PstI | No homology | Endosperm; seedling shoot; root |
9S: 12 cM from wx | 1 | |
| 23 | tac914.07 | PstI | No homology | Undetected | Unlinked to wx | 1 | |
| 24 | tac914.11 | PstI | No homology | Undetected | 9S: 16 cM from wx | 1 | |
| 25 | tac914.13 | BglII | No homology | Undetected | 9S: 21 cM from wx | 1 | |
| 26 | tac914.23 | PstI | Maize wx:Ac to intron 4 | 7e−82 | Endosperm; pollen | 9S: wx | 1 |
| 27 | tac917.09 | NcoI | No homology | Immature ear | 9S: 4 cM from wx | 4 | |
| 28 | tac917.12 | EcoRI | Arabidopsis NAM-like protein | 3e−5 | Etiolated seedlings | 9S: 14 cM from wx | 1 |
| 29 | tac917.27 | PstI | Arabidopsis auxin efflux carrier protein |
1e−5 | Tassel; seedling; silk | 9S: 12 cM from wx | 1 |
| 30 | tac917.29 | PstI | No homology | Seedling shoot and root; 3-week-old embryo |
9S: 8 cM from wx | 1 | |
| 31 | tac918.02 | PstI | No homology | Undetected | Unlinked to wx | 1 | |
| 32 | tac919.27 | PstI | Arabidopsis Pro-rich protein | 2e−5 | Tassel; stem; silk | 9: 35 cM from wx | 1 |
| 33 | tac919.39 | BglII | Barley SERK1 Leu-rich repeat protein |
1e−14 | Undetected | 9: 37 cM from wx | 1 |
| 34 | tac919.41 | BglII | No homology | Etiolated seedlings | 9: 32 cM from wx | 1 | |
| 35 | tac921.20 | EcoRI | Rice Arg decarboxylase | 4e−5 | Tassel; leaf | 9S: 3 cM from wx | 1 |
| 36 | tac923.08 | EcoRI | No homology | 3-week-old embryo | Unlinked to wx | 2 | |
| 37 | tac923.13 | EcoRI | No homology | Undetected | Unlinked to wx | 1 | |
| 38 | tac923.17 | EcoRI | No homology | Immature seed; tassel | Unlinked to wx | 1 | |
| 39 | tac929.20 | EcoRI | Arabidopsis unknown protein | 2e−15 | Undetected | 9S: 9 cM from wx | 1 |
| 40 | tac2094 | λ | Arabidopsis MAPK kinased | e−113 | Root; ear | 9S: <1 cM from bz | 4 |
| 41 | tac6067 | λ | Maize sesquiterpene cyclase | 0 | Leaves, volicitin induced | 9S: <1 cM from bz | 1 |
| 42 | tac6087 | λ | Maize sesquiterpene cyclase | 0 | Leaves, volicitin induced | 9S: <1 cM from bz | 1 |
| 43 | tac6058 | PCR | No homology | Endosperm; tassel | 9S: <1 cM from bz | 1 | |
| 44 | tac7077 | EcoRII | No homology | Constitutive | 9S: <1 cM from bz | 2 | |
| 45 | tac8193 | PstI | No homology | Undetected | 5: 4 cM from T5-9c | 1 | |
| 46 | tac8194 | PstI | Rice zinc finger protein | 2e−12 | Undetected | 7: 3 cM from T7-9a | 1 |
Restriction enzyme used to digest genomic DNA; four tac sites were isolated by either cloning in λ or direct PCR.
cM, centimorgan.
ADH, alcohol dehydrogenase.
MAPK, mitogen-activated protein kinase.
Several tac sites have been identified that have homology with other plant genes, mostly from Arabidopsis and rice. Among them are genes encoding proteins of predicted function, such as ring zinc finger proteins, Ser/Thr kinases, and a DNA-3-methyladenine glycosidase, as well as hypothetical proteins of unknown function. As indicated above, although several mutants have been identified, only a minority of them appear to be tagged by Ac on the basis of genetic cosegregation data. Two interesting examples of novel mutations apparently created by trAcs are the lethal embryo mutations that resulted from the insertion of Ac902.33 (Table 2, site 5) in a gene homologous with the rice chloroplast ribosomal protein S9 gene and Ac921.20 (Table 2, site 35) in a gene homologous with the rice Arg decarboxylase gene.
Five of the tac sites isolated from the collection of trAcs from bz-m2(Ac) are very close to bz on 9S (Table 2, sites 40 to 44). These tac sites have been studied in greater detail because they have helped to identify genes in the gene-rich bz region (Fu et al., 2001). Two trAcs (Ac6087 and Ac6067) landed in stc1, a sesquiterpene cyclase gene that is located immediately distal to bz (Table 2, sites 41 and 42). This gene is involved in the tritrophic interaction, a fascinating system of indirect defense that enables maize to protect itself from herbivorous insects by attracting parasites of the herbivore (Turlings et al., 1990; Farmer, 1997). stc1 is induced by volicitin, a compound produced in the salivary glands of beet army worms that forage on maize leaves (Shen et al., 2000). Both Ac insertions are null: the stc1 gene fails to be induced in response to volicitin and, as a consequence, a specific sesquiterpene is missing from the mixture of volicitin-elicited volatiles in the mutant seedlings. These are good examples of Ac insertions whose nonobvious phenotype could be investigated in detail after isolating the corresponding tac sites and identifying the gene in which Ac had inserted. Two other trAcs (Ac6058 and Ac7077) have tagged sequences with no homology in the public sequence databases (Table 2, sites 43 and 44). We have shown that these are genes by isolating cDNA clones for both tac sites from a tassel cDNA library (Fu et al., 2001; W. Park and H.K. Dooner, unpublished data). Thus, tac sites can be used to define plant genes that are not found in the sequence databases even after the Arabidopsis genome sequencing project has been largely completed (Arabidopsis Genome Initiative, 2000).
Expression of tac Sites
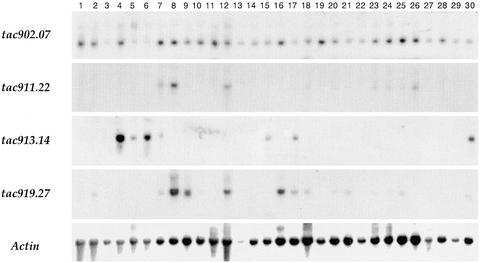
The expression of all tac sites listed in Table 2 was analyzed by RNA gel blotting. RNA from the following maize tissues was included in the analysis: developing endosperms and embryos (2 to 5 weeks); seedling shoots (1 week); juvenile roots (4 weeks); adult roots, leaves, and stems (10 weeks); immature tassels from four developmental stages (4 to 15 cm, 20 to 27 cm, 29 to 42 cm, and 42 to 50 cm); and immature ears from two developmental stages (3 and 8 cm). RNA samples from special tissue sources also were included in some analyses, for example, from seedling leaves treated with the insect elicitor volicitin (Shen et al., 2000) and from juvenile roots treated with NaCl. Figure 4 shows representative RNA gel blots for four different tac sites. The results from all RNA gel blots are summarized in Table 2 (Expression Data), which lists the tissue(s) giving the strongest signals for each tac site.
Figure 4.
RNA Gel Blot Analysis of Several tac Sites.
Total RNA from a variety of tissues was hybridized with the tac site probes indicated at left. Lanes 1 to 4, seed at various developmental stages (1, 1 DAP; 2, 5 DAP; 3, 7 DAP; 4, 21 DAP); lane 5, 14-DAP endosperm; lane 6, 35-DAP endosperm; lane 7, 21-DAP embryo; lanes 8 to 11, tassels at different developmental stages (lane 8, 3.8 cm; lane 9, 20 to 27 cm; lane 10, 30 to 33 cm; lane 11, 44 cm) at anthesis; lane 12, adult stem; lane 13, adult leaf; lane 14, adult root; lane 15, juvenile root; lane 16, silk from 8-cm ear; lane 17, 6-day-old seedling shoot; lane 18, 8-day-old seedling shoot; lane 19, 14-day-old seedling shoot, cut and incubated for 2 hr in water; lanes 20 to 24, 14-day-old seedling shoots incubated in different solutions (lane 20, 0.2 M NaCl for 48 hr; lane 21, 2% Suc for 48 hr; lane 22, water for 48 hr; lane 23, 20 mM KCl for 8 hr; lane 24, 20 mM KCl for 4 hr, followed by water for 4 hr); lane 25, 10-day-old etiolated seedling shoot after 8-hr light exposure; lane 26, 10-day-old etiolated seedling shoot in continuous darkness; lanes 27 to 29, 2-cm leaf segments from 14-day-old seedlings incubated for 7 hr under different treatments (lane 27, water in light; lane 28, water in darkness; lane 29, 7% hydrogen peroxide in light); and lane 30, 6-day-old seedling root.
A critical issue in this type of expression analysis is whether the signal detected by the tac site probe can be attributed specifically to the putative gene defined by the Ac insertion. This is particularly relevant when the gene in question encodes a protein belonging to a family of related proteins. Therefore, we distinguish two types of tac sites: those that give single bands in high-stringency confirmation DNA gel blots, and those that give more than one band. Among the 46 tac sites, 31 (67%) gave a positive signal with RNA collected from at least one tissue. Of these 31, 6 gave more than one band in DNA gel blots, making the results of the RNA gel blot analysis ambiguous. For one of these six (tac7077; see above), we have obtained cDNA evidence of expression. The remaining expressed tac sites gave single bands in high-stringency DNA gel blots, indicating that the signal detected in RNA gel blots is specific. Therefore, at least half of the sequences tagged by Ac are expressed.
One way to confirm the specificity of the signal detected by a tac site is to use RNA from a deletion mutation as a negative control. We have done so for the five tac sites located in the vicinity of bz (tac2094, tac6067, tac6087, tac6058, and tac7077) and have verified that the signal detected in RNA gel blots by probes that gave single bands in DNA gel blot analysis was, in fact, tac site specific (Fu et al., 2001; B. Shen, D. Lu, and H.K. Dooner, unpublished data). The only tac site in the bz region that hybridized to RNA from both the wild type and the sh-bz-X2 deletion control was tac7077, which consistently detected two bands in DNA gel blots. Although this is a powerful genetic approach, it is limited to those regions of the genome that are homozygous viable when deleted, an infrequent occurrence in maize. A second way to verify signal specificity is to monitor gene expression in Ac homozygous individuals, of which most presumably are mutated for the gene in question. If the signal is specific to the tac site being tested, such individuals should show either highly reduced transcript levels or, occasionally, an altered transcript size from Ac-mediated improper splicing (Wessler, 1988). Because the survey nature of our project precludes this type of exhaustive RNA analysis, we have not performed tests with mutant RNAs.
An alternative means to determine that a particular tac site is expressed is to establish that the predicted 3′ untranslated region of a cDNA or an EST matches the tac site genomic sequence. In addition to the tac6058 and tac7077 cDNAs described above, we have isolated cDNAs corresponding to Ac902.33 and Ac921.20, the two Acs that apparently resulted in embryo-lethal mutations. Again, because of the nature of our work, we have limited this type of analysis to only a few tac sites.
Overall, the data on DNA homology, copy number, and expression presented in Table 2 show that Ac inserts preferentially into genes and that Ac is, in fact, an excellent gene-searching engine in the highly repetitive maize genome. There are two interesting correlates in Table 2 that are worth noting. Most tac sites having homology with sequences in the databases are expressed in some tissue (20 of 25). On the other hand, 52% of tac sites lacking homology with sequences in the databases failed to give signals in RNA gel blots (11 of 21). These may represent Ac insertions in genes that are expressed at very low levels or only under special conditions. Alternately, they could represent Ac insertions in intergenic regions or long introns.
DISCUSSION
Single-Kernel Selection of Ac Transposants
Many schemes that monitor the transposition of Ac using Ds reporters have been described in maize and other plants. The most efficient schemes combine Ac and Ds mutable alleles that are expressed in the kernel (e.g., in the pericarp and aleurone; Dellaporta and Moreno, 1994) or seedling (Jones et al., 1989; Sundaresan et al., 1995), permitting the ready screen of a large number of individuals. In this study, we have taken advantage of the excellent endosperm genetics of maize to develop a simple assay that detects the germinal transpositions of Ac. The system uses the two easily scored endosperm markers wx and bz to monitor Ac transposition in either the female or the male germline. We use the Ac parent as female because this makes it possible to identify premeiotic transpositions as clonal sectors in the ear. If the Ac parent is used as male, premeiotic sectors are randomized upon pollen shedding, making clonal information much more difficult to obtain. As shown in Figure 1, transpositions of Ac that occur at or near meiosis in the Ac female parent can be selected as individual kernels having a unique nonparental phenotype. No attempt was made in this study to select for either linked or unlinked transpositions. However, stocks carrying appropriate Ac and Ds mutable alleles flanked by endosperm markers also can be used to identify transpositions of Ac specifically to unlinked sites as exceptional single kernels in an ear (Dooner et al., 1994).
Ac as a Gene Searcher
We have shown here that in maize the transposon Ac inserts preferentially into genes. This was suspected from previous work that had established that maize genes reside in hypomethylated CpG islands (Antequera and Bird, 1988; Bennetzen et al., 1994) and that Ac transposes preferentially into hypomethylated DNA (Chen et al., 1987). By isolating and sequencing the DNA adjacent to transposed Ac elements (tac sites), comparing their sequences to the nucleic acid databases, examining their copy numbers in DNA gel blots, and analyzing their expression by RNA gel blot analysis, we have confirmed that most Ac receptor sites in maize correspond to genes. Significantly, no matches to retrotransposons—sequences that make up a large percentage of the repetitive DNA in the maize genome (SanMiguel et al., 1996)—have been detected, supporting the notion that sites of Ac visitation tend to correspond to unique DNA. Another maize transposon, the miniature inverted repeat transposable element Heartbreaker, displays a similar insertion site preference (Zhang et al., 2000), in agreement with the finding that miniature inverted repeat transposable elements and retrotransposons occupy different domains in the maize genome (Tikhonov et al., 1999).
Our observations that all tac sites isolated to date are present in one or a few copies in the genome and that many tac sites are homologous with known genes and show tissue-specific patterns of expression suggest that Ac is a reliable gene searcher in the highly repetitive maize genome. In fact, we have used Ac to identify new putative genes. Some of the tac sites have no homology with any sequence in the databases, yet they are transcribed in specific tissues, suggesting that they are genes (Table 2, tac913.01, tac913.14, tac6058, and tac7077). For the latter two sites, we have isolated full-length cDNAs and defined exon-intron junctions (Fu et al., 2001; W. Park, unpublished data). Therefore, homology with a tac site sequence may turn out to be a useful gene-recognition criterion in maize.
Maize class II transposons (i.e., those that excise during transposition) also are being used to locate genes in Arabidopsis and rice, species with much smaller genomes and a much lower abundance of retrotransposons than maize (Arabidopsis Genome Initiative, 2000; Le et al., 2000; Turcotte et al., 2001). Two types of transposant populations have been generated: those containing selected transpositions of either Ds or dSpm to a site unlinked to the donor locus (Parinov et al., 1999; Tissier et al., 1999), and those containing unselected transpositions of Ac, Ds, or dSpm/I, a majority of which will be linked to the donor locus (Dubois et al., 1998; Wisman et al., 1998; Chin et al., 1999; Enoki et al., 1999; Ito et al., 1999; Speulman et al., 1999). Many sequences adjacent to the transposed elements have been isolated from these populations, and a large number of them show similarity to gene sequences in the databases. Thus, in these species, maize transposons also transpose preferentially into genes and can be used as efficient tools for functional genomics (Martienssen, 1998). Yet, it is in maize, with its high content of repetitive DNA, that the value of excisive transposons as gene searchers is clearest. Maize functional genomics projects using transposons are being pursued by several groups of investigators (summaries are available at http://plantgenome.sdsc.edu/projects.html). The transposons being used in these projects are either Ac or Mutator, but all of the strategies rely directly or indirectly on the propensity of maize class II transposons to insert preferentially into genes.
We do not know at present if Ac occasionally transposes into repetitive retrotransposon DNA in maize. Because these sequences are highly methylated (Burr et al., 1988; Bennetzen et al., 1994; Rabinowicz et al., 1999), it is likely that if such a transposition occurred, Ac would become methylated and inactivated (Schwartz and Dennis, 1986; Chomet et al., 1987). In fact, a methylated and inactive “cryptic” Ac element has been isolated from repetitive, methylated DNA (Leu et al., 1992). Yet, this element was present at the same position in several unrelated inbred lines, indicating that an old transposition event placed the cryptic Ac in its present location in the genome. Transposed Ac elements identified by virtue of their activity may represent a selected set, namely, transpositions into genes. No tac sites with homology with retrotransposons were recovered in the present study, although both methylation-sensitive and methylation-insensitive enzymes were used to identify IPCR-clonable tac sites. The fraction of tac sites having homology with sequences in the databases was similar whether the genomic DNA was digested with a methylation-sensitive (0.48) or a methylation-insensitive (0.58) enzyme. This suggests that there is no bias in using methylation-sensitive enzymes to identify IPCR-clonable bands. Digesting genomic DNA with methylation-sensitive enzymes is preferable because the number of Ac-hybridizing bands is reduced considerably (Chen et al., 1987). In heterologous systems, the insertion of class II maize elements in repetitive DNA occurs rarely. In the one study that reported that dSpm could insert in repetitive DNA, including retrotransposons, the methylation status of those sequences was not analyzed (Tissier et al., 1999).
Positioning of Ac Insertions within Genes
A recent analysis of Ds-insertion lines in Arabidopsis revealed a preference for mobilized Ds elements to reinsert close to the 5′ ends of genes (Parinov et al., 1999). When the distance between the site of insertion and the start codon was plotted for 232 Ds transpositions, a strong preference was found for insertion close to the 5′ end of the gene. A similar study in Arabidopsis analyzed transposed En/Spm elements in 48,000 plant lines, of which ∼80% result from independent transpositions of a dSpm element (Tissier et al., 1999). The authors reported no preference for the 5′ ends of genes, although they did notice a preference for genic DNA. Specifically, they analyzed the flanking sequences from 1200 independent insertions and found that 70% of the insertions were into coding regions.
We have performed a similar analysis with our independent Ac insertions. However, we are limited by the current state of maize sequencing. In cases in which maize genomic and either cDNA or EST sequences were available, we used that information to determine the position of the Ac insertion in the gene. We were able to do this for Ac902.33, Ac914.23, Ac921.20, Ac2094, Ac6058, Ac6067, Ac6087, and Ac7077. Otherwise, we used the most similar sequence in the database and made the assumption that exon size and number would be conserved across plant species. Given the sequence information available at present, we determined either the codon position of the Ac insertion in the coding region or the number of nucleotides away from the beginning or the end of translation. Flanking sequences that had no homology with sequences in the public databases or had homology with an EST for which no genomic sequence is available were ignored. Ac6058 and Ac7077 were considered because, although they have no homology with database sequences, we have complete genomic and cDNA sequences as well as RNA gel blot expression data for both of them.
Our data, summarized in Table 3, show that there is no preference for Ac to insert close to the ATG start codon. In fact, the majority of our insertions appear to occur in the middle of genes. Of the insertions for which a genic position was determined, 75% were in exonic DNA. Interestingly, the insertions tend to occur near the ends of exons, because 50% were within one of the two terminal fifths of the exon.
Table 3.
Ac Location within Genes
| trAc | Gene Homology | Ac Location | Position in Gene |
|---|---|---|---|
| Ac2094 | Maize MAPK kinasea | 5′ UTRb | 35 nucleotides upstream of ATG |
| Ac6058 | Maize cDNA, no database homology | Codon 87 | Middle of 2nd exon |
| Ac6067 | Maize sesquiterpene cyclase | Codon 45 | Middle of 1st exon |
| Ac6087 | Maize sesquiterpene cyclase | 5′ UTR | 104 nucleotides upstream of ATG |
| Ac7077 | Maize cDNA, no database homology | 5′ UTR | 175 nucleotides upstream of ATG |
| Ac8194 | Arabidopsis zinc finger protein | Codon 314 | 5′ end of 3rd exon |
| Ac901.04 | Arabidopsis cytochrome P450 | Codon 102 | Middle of last exon |
| Ac902.05 | Arabidopsis tRNA-gly (GCG) | Intergenic space | 50 nucleotides 3′ of terminator |
| Ac902.07 | Arabidopsis ring zinc finger protein | Codon 171, | Middle of 1st exon |
| Ac902.33 | Maize plastid S9 ribosomal protein | Codon 184 | 3′ end of last exon |
| Ac905.29 | Arabidopsis Ser Thr kinase | Codon 540 | 3′ end of 3rd exon |
| Ac907.25 | Arabidopsis sugar transporter | Codon 14 | 5′ end of 1st exon |
| Ac907.51 | Arabidopsis short-chain ADHc | Codon 80 | 5′ end of 1st exon |
| Ac907.54 | Arabidopsis hypothetical protein | 5′ UTR | 59 nucleotides upstream of ATG |
| Ac911.14 | Arabidopsis DNA-3-methyladenine glycosidase | Codon 266 | 3′ end of 4th exon |
| Ac911.28 | Arabidopsis Ser Thr kinase | Codon 434 | Middle of only exon |
| Ac913.09 | Arabidopsis hypothetical protein | Codon 544 | 3′ end of last exon (4th) |
| Ac914.23 | Maize starch synthase | Codon 301 | 3′ end of 4th exon |
| Ac917.12 | Arabidopsis NAM1-like protein | Codon 191 | Middle of last exon |
| Ac917.27 | Arabidopsis PIN1 auxin efflux carrier protein | Codon 305 | 3′ end of 1st exon |
| Ac919.27 | Arabidopsis Pro-rich protein | Intergenic space | 366 nucleotides after stop codon |
| Ac919.39 | Barley Leu-rich repeat protein | Codon 220 | 3′ end of only exon |
| Ac921.20 | Rice Arg decarboxylase | Codon 580 | 3′ end of only exon |
| Ac929.20 | Arabidopsis unknown protein | Codon 2229 | Middle of 11th exon |
MAPK, mitogen-activated protein kinase.
UTR, untranslated region.
ADH, alcohol dehydrogenase.
Mutants in Ac Stocks
New mutants with obvious visible phenotypes segregated in ∼5% of the F2 families from the primary Ac transposants. The dominant trans-acting effect of Ac on the excision of Ds from the reporter allele bz-m2(D1) allowed us to test for cosegregation between trAc and the new mutation in each F2 family. We found that Ac cosegregated with a mutation in just a handful of cases. The tac sites in the two Ac-cosegregating mutations with the most dramatic phenotype, embryo lethality, have been isolated. They correspond to Ac insertions in genes encoding proteins with homology with the rice chloroplast ribosomal protein S9 and Arg decarboxylase. Currently, we are attempting to obtain germinal revertants of these mutants to confirm that they are, in fact, tagged by Ac. Isolating revertants from lethal mutants is not a trivial task, but it is facilitated by our ability to identify heterozygotes on the basis of Ac's dominant phenotype.
Transposed Ds elements represent a likely source of the new mutations that do not cosegregate with Ac. In total, there are ∼40 to 50 copies of the short 0.4-kb Ds1 elements and the larger Ds elements in the maize genome (Dennis et al., 1988). The short Ds1 elements have internal sequences completely unrelated to Ac, whereas the larger Ds elements arise from internal deletions of Ac. Possibly, both types of elements contribute to the spectrum of mutants recovered in Ac stocks. Mutants tagged by both classes of Ds elements have been reported at a diversity of loci (summarized by Neuffer et al., 1997). If new mutations are caused by Ds elements, they should be unstable in the presence of Ac. However, somatic instability is not always easy to detect, particularly if the size of the somatic sectors is small. We have detected somatic instability of a pale green seedling mutant, a sugary endosperm mutant, and a zebra-striped plant mutant and have shown that the instability of the latter two is dependent on an Ac element located elsewhere in the genome. On the other hand, we have not seen somatic instability of mutant phenotypes, such as luteus and albino seedlings, in which somatic reversion should have been readily detectable. It is possible, therefore, that some of the mutants recovered are not tagged by either Ac or Ds.
Some of the mutations could be spontaneous point mutations, but the overall mutation frequency for obvious visible phenotypes (5%) is much higher than has been observed in similar experiments. For instance, Neuffer (1978) reported a spontaneous overall mutation frequency of 0.7%, which is much lower than what we observed. Insertions that do not belong to the Ac-Ds family are other possible sources of mutations. The recovery of mutants harboring insertions other than those known to be present in a particular stock is common in transposon mutagenesis experiments (Schmidt et al., 1987; Schnable et al., 1989; Johal and Briggs, 1992; Michel et al., 1995; Tacke et al., 1995). However, such mutants usually show somatic instability. Finally, there exists the possibility that some of the stable mutants arise from two-step transpositions of Ac that create a transposon footprint at the first site of insertion. Although this possibility often is raised as a concern by researchers who espouse two-element systems for transposon mutagenesis, there are no data to support it. Furthermore, given the preference of Ac to transpose to closely linked sites, one would expect that the mutations in our study frequently would be linked to the trAc element, but they are not. Thus, we do not believe that the latter mechanism plays a major role in the generation of mutations that are not tagged by Ac.
Given the fact that Ac transposes preferentially into genes, it might seem paradoxical that so few of the trAcs cause obvious mutant phenotypes. The cumulative evidence for the large-scale duplication, triplication, and even greater amplification of segments of the maize genome (Helentjaris et al., 1988; Gaut and Doebley, 1994; Gaut, 2001) suggests that a possible reason for the dearth of mutant phenotypes is genetic redundancy. However, redundancy in maize rarely is complete. The evolved differences in function between duplicate genes can range from slight quantitative differences of expression in certain tissues (orp1 and orp2; Wright et al., 1992) to completely nonoverlapping spatial expression patterns (sh2 and agp1; Giroux and Hannah, 1994). A collection of lines carrying Ac insertions in different, partially sequenced genes will help investigators to screen for subtle mutant phenotypes, particularly after obtaining information on where in the plant the mutated gene is expressed. It also will make it possible to identify mutations lacking obvious visible phenotypes (Shen et al., 2000) or mutations in genes with minor quantitative effects that would be missed from conventional transposon mutagenesis screens designed to identify gross changes in phenotype.
METHODS
Description of Maize Stocks
All of the stocks used in this study were in the common genetic background of the maize (Zea mays) inbred line W22. wx-m7(Ac) is an unstable wx allele described by McClintock (1964). It arose by insertion of the 4.6-kb Ac element in the 5′ untranslated region of the Wx gene (Muller-Neumann et al., 1984; Klosgen et al., 1986). bz-m2(Ac) is an unstable bz mutation that arose from the insertion of Ac in the second exon of the bz gene (McClintock, 1955; Ralston et al., 1988). bz-m2(D1) is the first derivative from bz-m2(Ac) isolated by McClintock (1962). It harbors a 3.3-kb internally deleted Ds element at the same position as Ac in bz-m2(Ac) (Dooner et al., 1986). bz-R and wx are stable recessive mutations.
Derivation of an Ac Insertion Library from wx-m7(Ac)
bz-R wx-m7(Ac)/bz-m2(D1) wx heterozygotes were pollinated with bz-m2(D1) wx in a large hand-pollinated isolation plot. In the resulting test cross ears, which will have predominantly waxy, bronze kernels, germinal transpositions of Ac can be selected as exceptional kernels that appear shiny and spotted (Wx′ bz-m). From 1185 ears, more than 2500 independent Wx′ bz-m kernels were selected. These occurred as separate kernels in the ears, indicative of meiotic or postmeiotic Ac transposition events. Very few ear sectors contained multiple adjacent Wx′ bz-m kernels, which are indicative of premeiotic events. Of the single-kernel selections, approximately half were heritable; that is, they had resulted from Ac transposition events that were included in both endosperm and embryo. The other half had noncorresponding genotypes in the endosperm and embryo and, most likely, resulted from gametophytic Ac transposition events that were recovered only in the endosperm. Kernels with nonconcordant embryos and endosperms are a common occurrence in selection schemes for germinal transpositions of other transposable elements.
Selfed ears were harvested from 1225 confirmed primary transposants, and rough estimates of the wx-Ac linkage were obtained for each transposant by counting the number of spotted and nonspotted kernels among the waxy class, the majority of which would have received at least one copy of the bz-m2(D1) reporter allele. The recombinant fraction p was estimated as 1−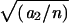 where a1 is the number of bz-m wx kernels, a2 is the number of bz wx kernels, and n = a1 + a2. Errors in the estimate of p in such two-point mapping arise from excisions of Ac without reinsertion and from transpositions of Ac to unlinked sites. The latter source is significant only for very closely linked receptor sites and results in a slight displacement of the modal p value away from 0. Ac was found to be unlinked to the wx donor locus (the null hypothesis tested) in ∼37% of transposants and was found to be linked in the remaining transposants. This crude distribution is not very different from what has been observed at other loci (Greenblatt, 1984; Dooner and Belachew, 1989).
where a1 is the number of bz-m wx kernels, a2 is the number of bz wx kernels, and n = a1 + a2. Errors in the estimate of p in such two-point mapping arise from excisions of Ac without reinsertion and from transpositions of Ac to unlinked sites. The latter source is significant only for very closely linked receptor sites and results in a slight displacement of the modal p value away from 0. Ac was found to be unlinked to the wx donor locus (the null hypothesis tested) in ∼37% of transposants and was found to be linked in the remaining transposants. This crude distribution is not very different from what has been observed at other loci (Greenblatt, 1984; Dooner and Belachew, 1989).
Nucleic Acid Extraction, Blotting, and Hybridization
Leaf DNA was isolated by a urea extraction procedure (Greene et al., 1994). Total RNA was extracted using TRIzol reagent according to the manufacturer's protocol (Life Technologies, Rockville, MD). DNA concentration was established by comparing aliquots of sample DNA with 1 μg of λDNA on an agarose gel stained with ethidium bromide. RNA concentration was determined by comparing samples with standards on ethidium bromide–stained gels and UV light spectrophotometry. Restriction enzyme–digested DNA (10 μg for methylation sensitive, 8 μg for methylation insensitive) and total RNA (30 μg) were resolved on 0.8 and 1.2% agarose gels, respectively, and then transferred to Hybond N+ membranes (Amersham Pharmacia, Piscataway, NJ). 32P-labeled probes were generated with Ready-To-Go DNA labeling beads (Amersham Pharmacia). The probe used to detect Ac-hybridizing bands was a polymerase chain reaction (PCR)-generated fragment from the middle of Ac. The primers used to generate this probe were Ac2047 and Ac2905R (P1 and P2, respectively, in Figure 3). The probe used for RNA gel blot analysis and tac site confirmation was a PCR-generated fragment corresponding to the complete inverse PCR (IPCR) product with minimal Ac ends. The primers used to create this probe were Ac24R and Ac4542 (P3 and P4, respectively, in Figure 3). These primers generate a probe containing 48 nucleotides of Ac, which are insufficient to reveal any additional bands in blots. Hybridization and membrane washing followed established protocols (Church and Gilbert, 1984). The sequences of the primers described above are as follows: Ac2047 (P1), 5′-GGATTGATGATGATTGGTGTCTCC-3′; Ac2905R (P2), 5′-TTGTACCTAGGGTCAAGGAAGCAT-3′; Ac24R (P3), 5′-TTTCCCATCCTA-CTTTCATCCCTG-3′; and Ac4542 (P4), 5′-CGTTACCGACCGTTT-TCATCCCTA-3′.
tac Site Amplification and Sequencing
Independent tac sites were identified in diagnostic DNA gel blots as restriction fragment length polymorphisms when hybridized with the internal Ac probe. Fragments <9 kb were selected as likely substrates for IPCR amplification. Thirty micrograms of genomic DNA then was digested with the appropriate enzyme and separated by gel electrophoresis as described above. The region containing the band of interest was excised and purified using a QiaQuick Gel Extraction Column (Qiagen, Valencia, CA). The purified fragment was diluted to 2 ng/μL and circularized using T4 DNA Ligase (New England Biolabs, Beverly, MA). The ligation reaction was extracted once with phenol:chloroform (1:1) and precipitated with 0.1 vol NaOAc and 2 vol ethanol. The precipitation reaction was centrifuged for 20 min at 14,000g, and then the pellet was washed with 70% ethanol. The pellet was vacuum dried and resuspended in 50 μL of deionized water.
IPCR was performed according to the protocol of QiaTaq (Qiagen). An aliquot of the ligated DNA sample recovered from fragment purification was used as a template for IPCR. The reaction consisted of 1 × PCR buffer, 1 × Q buffer (Qiagen), 1 mM total deoxynucleotide triphosphates, 400 nmol of each primer, and 2 units of rTth polymerase (Perkin-Elmer). The reaction was denatured at 95°C for 3 min, cooled to 80°C, and held for the addition of polymerase. The reaction continued with 37 cycles of 30 sec of denaturation at 94°C, 30 sec of annealing at 62°C, and 4 min of extension at 72°C. The last 20 cycles autoextended the extension time by 20 sec per cycle. The first pair of primers, designated Ac2578R and Ac4372, often produced a very faint band when electrophoresed on an agarose gel. The products from the first reaction were diluted 10-fold, and then 1 μL was used for the nested reaction. This produced a total 500-fold dilution of template DNA for the nested amplification.
The nested amplification was performed as described above with the following changes. The polymerase used was QiaTaq polymerase (Qiagen) to facilitate T/A cloning. The primers used were Ac100R and Ac4473. The sequences of the four oligonucleotide primers used in the nested PCR were as follows: Ac2578R, 5′-GACAAACATACC-TGCGAGGATCAC-3′; Ac4372, 5′-ACCGAACAAAAATACCGGTTCCCG-3′; Ac100R, 5′-ATATCCCGTTTCCGTTCCGTTTTC-3′; and Ac4473, 5′-TCCCGTTTTCGTTTCCGTC-3′. After the nested reaction, a clear, sharp band was visible. The band of interest was excised from a gel and purified with a BIO101 glass bead kit (Q-Bio, Carlsbad, CA). The PCR product then was cloned into a pGEM-EasyT vector (Promega, Madison, WI) and sequenced on an ABI377 DNA sequencer (Applied Biosystems, Foster City, CA). Fragments were analyzed using the Lasergene99 software suite (DNAStar, Madison, WI). Ac genomic DNA junctions were identified, and the flanking genomic DNA was used as a query against the various GenBank databases.
Accession Numbers
The tac sites listed in Table 2 have been submitted to GenBank under the following accession numbers: tac901.04, AY065586; tac902.05, AH011332, AY065594, AY065595; tac902.07, AY065576; tac902.33, AH011329, AY065572, AY065573; tac903.05, AH011338, AY065606, AY065607; tac903.06, AH011337, AY065604, AY065605; tac905.5, AF338121; tac905.29, AH011322, AY065560, AY065561; tac907.25, AH011327, AY065568, AY065569; tac907.49, AH011328, AY065570, AY065571; tac907.51, AH011334, AY065598, AY065599; tac907.54, AH011343, AY065616, AY065617; tac909.06, AF338125; tac911.14, AY065584; tac911.16, AY065587; tac911.17, AY065589; tac911.22, AH011336, AY065602, AY065603; tac911.28, AH011323, AY065558, AY065559; tac911.30, AH011325, AY065564, AY065565; tac913.01, AH011330, AY065590, AY065591; tac913.09, AH011340, AY065610, AY065611; tac913.14, AH011333, AY065596, AY065597; tac914.07, AH011324, AY065562, AY065563; tac914.11, AY065577; tac914.13, AY065585; tac914.23, AH011342, AY065614, AY065615; tac917.09, AH011331, AY065592, AY065593; tac917.12, AY065578; tac917.27, AH011326, AY065566, AY065567; tac917.29, AY065588; tac918.02, AH011335, AY065600, AY065601; tac919.27, AY065579; tac919.39, AH011339, AY065608, AY065609; tac919.41, AH011341, AY065612, AY065613; tac921.20, AF338139; tac923.08, AY065582; tac923.13, AY065580; tac923.17, AY065581; tac929.20, AY065583; tac2094, AF338110; tac6067, AF338112; tac6087, AF338113; tac6058, AF338111; tac7077, AF338114; tac8193, AY065574; tac8194, AY065575.
Acknowledgments
We thank Huihua Fu and Binzhang Shen for comments on the manuscript, Huihua Fu, Zhengrong Ma, Binzhang Shen, and Zhenwei Zheng for sharing unpublished sequence data, María I. Cruz for laboratory assistance, and Krystyna Dooner for assistance in generating the Ac mapping data. This research was supported by National Science Foundation Plant Genome Program Grant No. DBI 98-13364.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010468.
References
- Antequera, F., and Bird, A.P. (1988). Unmethylated CpG islands associated with genes in higher plant DNA. EMBO J. 7, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (1996). The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204, 195–229. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., Brown, W.E., and Springer, P.S. (1988). The state of DNA modification within and flanking maize transposable elements. In Plant Transposable Elements, O.E. Nelson, ed (New York: Plenum Press), pp. 237–250.
- Bennetzen, J.L., Schrick, K., Springer, P.S., Brown, W.E., and SanMiguel, P. (1994). Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37, 565–576. [DOI] [PubMed] [Google Scholar]
- Bensen, R.J., Johal, G.S., Crane, V.C., Tossberg, J.T., Schnable, P.S., Meeley, R.B., and Briggs, S.P. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, B., Burr, F.A., Thompson, K.H., Albertson, M.C., and Stuber, C.W. (1988). Gene mapping with recombinant inbreds in maize. Genetics 118, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels, N., Barakat, A., and Bernardi, G. (1995). The gene distribution of the maize genome. Proc. Natl. Acad. Sci. USA 92, 11057–11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L., and Walbot, V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Greenblatt, I.M., and Dellaporta, S.L. (1987). Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, H.G., Choe, M.S., Lee, S.H., Park, S.H., Koo, J.C., Kim, N.Y., Lee, J.J., Oh, B.G., Yi, G.H., Kim, S.C., Choi, H.C., Cho, M.J., and Han, C.D. (1999). Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 19, 615–623. [DOI] [PubMed] [Google Scholar]
- Chomet, P.S., Wessler, S., and Dellaporta, S.L. (1987). Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K., Burr, F., and Burr, B. (1986). Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83, 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, L., and Martienssen, R. (1995). Site-selected transposon mutagenesis at the hcf106 locus in maize. Plant Cell 7, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., and Chomet, P.S. (1985). The activation of maize controlling elements. In Genetic Flux in Plants, B. Hohn and E. Dennis, eds (New York: Springer-Verlag), pp. 169–216.
- Dellaporta, S.L., and Moreno, M.A. (1994). Gene tagging with Ac/Ds elements in maize. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 219–233.
- Dennis, E.S., Finnegan, E.J., Taylor, B.H., Peterson, T.A., Walker, A.R., and Peacock, W.J. (1988). Maize transposable elements: Structure, function, and regulation. In Plant Transposable Elements, O.E. Nelson, ed (New York: Plenum Press), pp. 101–113.
- Dooner, H.K. (1995). Stocks carrying transposed Ac elements available from the Maize Genetics Co-op. Maize Genet. Coop. Newsl. 69, 115–116. [Google Scholar]
- Dooner, H.K., and Belachew, A. (1989). Transposition pattern of the maize element Ac from the bz-m2(Ac) allele. Genetics 122, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., English, J., Ralston, E., and Weck, E. (1986). A single genetic unit specifies two transposition functions in the maize element Activator. Science 234, 210–211. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K., Belachew, A., Burgess, D., Harding, S., Ralston, M., and Ralston, E. (1994). Distribution of unlinked receptor sites for transposed Ac elements from the bz-m2(Ac) allele in maize. Genetics 136, 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, P., Cutler, S., and Belzile, F.J. (1998). Regional insertional mutagenesis on chromosome III of Arabidopsis thaliana using the maize Ac element. Plant J. 13, 141–151. [DOI] [PubMed] [Google Scholar]
- Earp, D.J., Lowe, B., and Baker, B. (1990). Amplification of genomic sequences flanking transposable elements in host and heterologous plants: A tool for transposon tagging and genome characterization. Nucleic Acids Res. 18, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki, H., Izawa, T., Kawahara, M., Komatsu, M., Koh, S., Kyozuka, J., and Shimamoto, K. (1999). Ac as a tool for the functional genomics of rice. Plant J. 19, 605–613. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E. (1997). New fatty acid base signals: A lesson from the plant world. Science 276, 912–913. [Google Scholar]
- Fedoroff, N.V., Wessler, S., and Shure, M. (1983). Isolation of the transposable maize controlling elements Ac and Ds. Cell 35, 235–242. [DOI] [PubMed] [Google Scholar]
- Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grun, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R.B., Briggs, S.P., Simcox, K., and Gierl, A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277, 696–699. [DOI] [PubMed] [Google Scholar]
- Fu, H., Park, W., Yan, X., Zheng, Z., Shen, B., and Dooner, H.K. (2001). The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98, 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. (2001). Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genet. Res. 11, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J. (1994). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux, M.J., and Hannah, L.C. (1994). ADP-glucose pyrophosphorylase in shrunken2 and brittle2 mutants of maize. Mol. Gen. Genet. 243, 400–408. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I.M. (1984). A chromosome replication pattern deduced from pericarp phenotypes resulting from movement of the transposable element Modulator in maize. Genetics 108, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, B., Walko, R., and Hake, S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138, 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake, S., and Walbot, V. (1980). The genome of Zea mays, its or-ganization and homology to related grasses. Chromosoma 79, 251–270. [Google Scholar]
- Helentjaris, T., Weber, D., and Wright, S. (1988). Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Seki, M., Hayashida, N., Shibata, D., and Shinozaki, K. (1999). Regional insertional mutagenesis of genes on Arabidopsis thaliana chromosome V using the Ac/Ds transposon in combination with a cDNA scanning method. Plant J. 17, 433–444. [DOI] [PubMed] [Google Scholar]
- James, D.W., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H.K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION 1 (FAE1) gene with the maize transposon Activator. Plant Cell 7, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal, G.S., and Briggs, S.P. (1992). Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258, 985–987. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Carland, F., Maliga, P., and Dooner, H.K. (1989). Visual detection of transposition of the maize element Activator (Ac) in tobacco seedlings. Science 244, 204–207. [DOI] [PubMed] [Google Scholar]
- Klosgen, R.B., Gierl, A., Schwarz-Sommer, S., and Saedler, H. (1986). Molecular analysis of the waxy locus of Zea mays. Mol. Gen. Genet. 203, 237–244. [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, J.Y., Sun, Y.H., Lai, Y.K., and Chen, J. (1992). A maize cryptic Ac-homologous sequence derived from an Activator transposable element does not transpose. Mol. Gen. Genet. 233, 411–418. [DOI] [PubMed] [Google Scholar]
- Lisch, D., Chomet, P., and Freeling, M. (1995). Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics 139, 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1955). Controlled mutation in maize. Carnegie Inst. Washington Year Book 54, 245–255. [PubMed] [Google Scholar]
- McClintock, B. (1962). Topographical relations between elements of control systems in maize. Carnegie Inst. Washington Year Book 61, 448–461. [Google Scholar]
- McClintock, B. (1964). Aspects of gene regulation in maize. Carnegie Inst. Washington Year Book 63, 592–602. [Google Scholar]
- Meeley, R., and Briggs, S. (1995). Reverse genetics for maize. Maize Genet. Coop. Newsl. 69, 67–82. [Google Scholar]
- Michel, D., Hartings, H., Lanzini, S., Michel, M., Motto, M., Riboldi, G.R., Salamini, F., and Doring, H.-P. (1995). Insertion mutations at the maize Opaque2 locus induced by transposable element families Ac, En/Spm and Bg. Mol. Gen. Genet. 248, 287–292. [DOI] [PubMed] [Google Scholar]
- Muller-Neumann, M., Yoder, J., and Starlinger, P. (1984). The DNA sequence of the transposable element Ac of Zea mays L. Mol. Gen. Genet. 198, 19–24. [Google Scholar]
- Neuffer, M.G. (1978). Induction of genetic variability. In Maize Breeding and Genetics, D.B. Walden, ed (New York: John Wiley and Sons), pp. 579–600.
- Neuffer, M.G., Coe, E.H., and Wessler, S. (1997). The Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Parinov, S., Sevugan, M., De, Y., Yang, W.C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R., and Martienssen, R.A. (1999). Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 23, 305–308. [DOI] [PubMed] [Google Scholar]
- Ralston, E.J., English, J., and Dooner, H.K. (1988). Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics 119, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Burr, F.A., and Burr, B. (1987). Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science 238, 960–963. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., Peterson, P.A., and Saedler, H. (1989). The bz-rcy allele of the Cy transposable element system of Zea mays contains a Mu-like element insertion. Mol. Gen. Genet. 217, 459–463. [DOI] [PubMed] [Google Scholar]
- Schwartz, D., and Dennis, E. (1986). Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol. Gen. Genet. 205, 476–482. [Google Scholar]
- Shen, B., Zheng, Z., and Dooner, H.K. (2000). A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: Characterization of wild-type and mutant alleles. Proc. Natl. Acad. Sci. USA 97, 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speulman, E., Metz, P.L., van Arkel, G., te Lintel Hekkert, B., Stiekema, W.J., and Pereira, A. (1999). A two-component enhancer-inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell 11, 1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Tacke, E., Korfhage, C., Michel, D., Maddaloni, M., Motto, M., Lanzini, S., Salamini, F., and Doring, H.-P. (1995). Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J. 8, 907–917. [DOI] [PubMed] [Google Scholar]
- Thatcher, J.W., Shaw, J.M., and Dickinson, W.J. (1998). Marginal fitness contributions of nonessential genes in yeast. Proc. Natl. Acad. Sci. USA 95, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki, R., Kochieva, E.Z., and Fedoroff, N.V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10, 479–489. [DOI] [PubMed] [Google Scholar]
- Turcotte, K., Srinivasan, S., and Bureau, T. (2001). Survey of transposable elements from rice genomic sequences. Plant J. 25, 169–179. [DOI] [PubMed] [Google Scholar]
- Turlings, T.C.J., Tumlinson, J.H., and Lewis, W.J. (1990). Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 250, 1251–1253. [DOI] [PubMed] [Google Scholar]
- Van Schaik, N., and Brink, R.A. (1959). Transposition of Modulator, a component of the variegated pericarp in maize. Genetics 44, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S. (1988). Phenotypic diversity mediated by the maize transposable elements Ac and Ds. Science 242, 399–405. [DOI] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wisman, E., Hartmann, U., Sagasser, M., Baumann, E., Palme, K., Hahlbrock, K., Saedler, H., and Weisshaar, B. (1998). Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA 95, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, A.D., Moehlenkamp, C.A., Perrot, G.H., Neuffer, M.G., and Cone, K.C. (1992). The maize auxotrophic mutant orange pericarp is defective in duplicate genes for tryptophan synthase beta. Plant Cell 4, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Arbuckle, J., and Wessler, S.R. (2000). Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc. Natl. Acad. Sci. USA 97, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]