Abstract
The tonoplast was proposed as a default destination of membrane-bound proteins without specific targeting signals. To investigate the nature of this targeting, we created type I fusion proteins with green fluorescent protein followed by the transmembrane domain of the human lysosomal protein LAMP1. We varied the length of the transmembrane domain from 23 to either 20 or 17 amino acids by deletion within the hydrophobic domain. The resulting chimeras, called TM23, TM20, and TM17, were expressed either transiently or stably in tobacco. TM23 clearly accumulated in the plasmalemma, as confirmed by immunoelectron microscopy. In contrast, TM17 clearly was retained in the endoplasmic reticulum, and TM20 accumulated in small mobile structures. The nature of the TM20-labeled compartments was investigated by coexpression with a marker localized mainly in the Golgi apparatus, AtERD2, fused to a yellow fluorescent protein. The strict colocalization of both fluorescent proteins indicated that TM20 accumulated in the Golgi apparatus. To further test the default destination of type I membrane proteins, green fluorescent protein was fused to the 19–amino acid transmembrane domain of the plant vacuolar sorting receptor BP-80. The resulting chimera also accumulated in the Golgi instead of in post-Golgi compartments, where native BP-80 localized. Additionally, when the transmembrane domain of BP-80 was lengthened to 22 amino acids, the reporter escaped the Golgi and accumulated in the plasma membrane. Thus, the tonoplast apparently is not a favored default destination for type I membrane proteins in plants. Moreover, the target membrane where the chimera concentrates is not unique and depends at least in part on the length of the membrane-spanning domain.
INTRODUCTION
The sorting of integral proteins in plants is not well understood. Nevertheless, it is accepted that peptidic signals exposed in the cytosol are responsible for targeting to the correct subcellular location in plant cells. This signal-mediated sorting is opposed to a default transport that is believed to happen when no signal is present on a protein. Although the default destination within the secretory pathway for a soluble protein is secretion, the default membrane is unclear. The most informative results about the location where membrane proteins would accumulate by default were provided by a study of α-TIP (Höfte and Chrispeels, 1992). In this experiment, the last 48 amino acids of α-TIP, which contains the sixth transmembrane domain, were sufficient to target a reporter protein to the tonoplast. Because the deletion of the cytosolic C-terminal 15 amino acids from the α-TIP sequence did not prevent the tonoplast accumulation of the truncated protein, the authors indirectly deduced a role for the sixth membrane-spanning domain. Either this α-TIP transmembrane domain would be sufficient for vacuolar location or the tonoplast would be the default destination for membrane proteins.
More recently, the same sixth transmembrane domain of α-TIP was used in a chimeric construct. In this different context, it appeared unable to send a reporter protein to a lytic compartment and did not reach the Golgi (Jiang and Rogers, 1998). Another study used a chimeric protein with the transmembrane domain as well as the C-terminal domain of the yeast Wbp1 (dolichyl-diphosphooligosaccharide protein glycosyltransferase β-subunit precursor) in which the endoplasmic reticulum (ER) retention motif was destroyed (Barrieu and Chrispeels, 1999). Because a release of the soluble reporter to the vacuole occurred, the authors proposed that the presence of the transmembrane domain may send an invertase reporter to the vacuole by a default process. The problem of these approaches is that the location of the membrane protein itself is not addressed directly by analyzing the location of membrane-bound reporters. Instead, conclusions are suggested indirectly from the location of a soluble reporter released from the membrane chimera.
As proposed by Barrieu and Chrispeels (1999), it is possible that, while reaching a lytic compartment on its way to its default destination, a chimeric membrane protein releases its reporter. The released soluble protein then can be transported farther down the secretory pathway while the membrane portion remains in a different location. In our opinion, the destination for a membrane protein should be defined strictly as the membrane where the integral protein finally resides. The role of the transmembrane domain itself in transport through the secretory pathway also was investigated for type II (Munro, 1995a) and type IV (Pedrazzini et al., 1996; Rayner and Pelham, 1997; Yang et al., 1997) proteins in animal and yeast cells. In these studies, the length of the transmembrane domain had a dominant role over its single signals and its overall hydrophobic nature. The most frequently identified potential signals are four Phe residues found in the transmembrane domain of sialyl transferase that play a role in Golgi retention. Nevertheless, when all four Phe residues were replaced by Ile, the accumulation at the plasma membrane was threefold higher than in the native form but barely reached 4% of that of a positive control. This finding suggested that single residues have a somewhat limited role in subcellular location.
We wanted to determine if the tonoplast is the default destination in plants and if the length of the transmembrane domain has any effect on this subcellular location. For this purpose, we designed constructs in which the reporter green fluorescent protein (GFP) was fused to a transmembrane domain from a human lysosomal protein (LAMP1). We also included a few charged amino acids from the original cytosolic domain to ensure an efficient anchoring in the membrane. We then modified the length of the hydrophobic domain by removing either three or six internal amino acids. This should shorten the hydrophobic region without disturbing the flanking sequences that are in contact with the hydrophilic heads of the lipid bilayer. We ensured that our reporter was not released from the chimera and therefore addressed the membrane protein location in whole cells using the fluorescence emitted by the GFP reporter. We also used a transmembrane domain originating from a plant protein, the plant vacuolar sorting receptor BP-80. Because this receptor normally resides in compartments very close to the vacuole, it was a good candidate to reach the tonoplast upon mutagenesis.
RESULTS
Three Chimeric Proteins, TM23, TM20, and TM17, Accumulate in Different Membranes of the Secretory Pathway
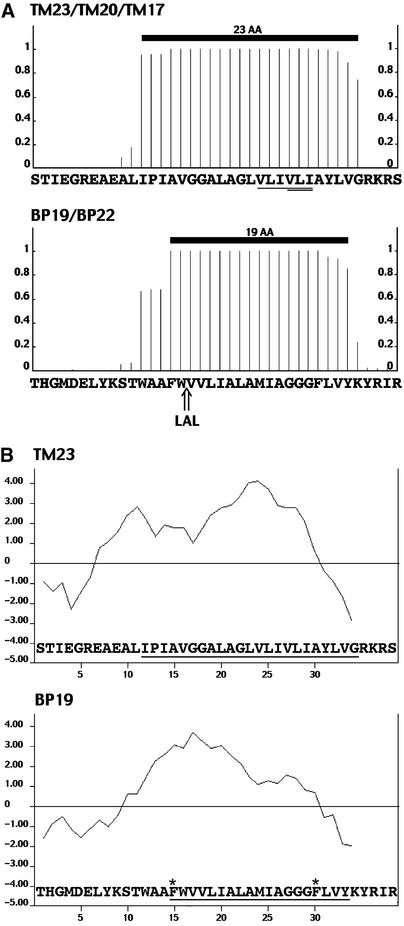
Figure 1A shows the three fusion proteins, TM23, TM20, and TM17, with the detailed sequence of the C-terminal region. We used the TMHMM program (Sonnhammer et al., 1998) to estimate the length of the transmembrane domain and accordingly named the constructs with the corresponding length in amino acids for the transmembrane domain. TM23 clearly harbors 23 hydrophobic amino acids, all of them having the highest chance to be integral in a membrane (minimum of 78%). We chose this sequence for two reasons: (1) the limits between the membrane part and the flanking regions are clear, and (2) the use of a human sequence should decrease the chance of having amino acids with potential selective retention information for any membrane in plants. As shown in Figure 1A, the amino acids chosen for deletions are fully inside the expected transmembrane domain and therefore should affect only its length. Each of the three constructs then was expressed transiently in tobacco leaves by infiltration with transformed Agrobacterium tumefaciens.
Figure 1.
Estimated Membrane-Spanning Domains and Hydrophobic Plots for the Membrane Fusion Chimeras with GFP.
Detailed are the most C-terminal amino acids of the chimeras (x axes). AA, amino acid.
(A) Sequences from the chimeras were submitted to the TMHMM program to estimate their probability of being inside a membrane (y axes). Thick black lines represent the position of the transmembrane domain as estimated by TMHMM. The sequence at top shows the TM23 transmembrane domain and the derived TM20 and TM17. The amino acids removed from the original LAMP1 transmembrane domain from TM23 to produce TM20 are single underlined. The three amino acids removed from TM20 to produce TM17 are double underlined. The sequence at bottom shows the BP19 chimera, including the transmembrane domain of the vacuolar sorting receptor BP-80 and a corresponding derivative, BP22, with the added tripeptide LAL in the transmembrane domain.
(B) Hydropathy plots of TM23 and BP19 using the program of Kyte and Doolittle (1982). The hydropathy index of each residue is represented on the y axes. The position of the transmembrane domain estimated by the TMHMM program is underlined. Two Phe residues internal to the membrane-spanning domain of BP-80 are indicated with stars.
Twenty-four hours after transformation, we always found dominant ER labeling for all three constructs (data not shown). This means that all of the fusion proteins had entered the ER as expected. After 48 hr, the labeling had reached stable patterns similar to those found at 72 hr and shown in Figures 2A to 2C. Importantly, GFP accumulation patterns also remained unaffected when protein synthesis was blocked for several hours using cycloheximide and protease inhibitors (data not shown), suggesting that the GFP accumulation we observed is representative of the final location of the reporter within the secretory pathway. We used a quantitative color mode to collect and present these images because it allowed us to (1) set the microscope in a nonpixel saturating mode, and subsequently (2) appreciate the relative quantity of fluorescence found in the various labeled structures. The color reflects the amount of GFP fluorescence collected by the confocal microscope, with a gradient of red to white from the weakest to the strongest signal, respectively.
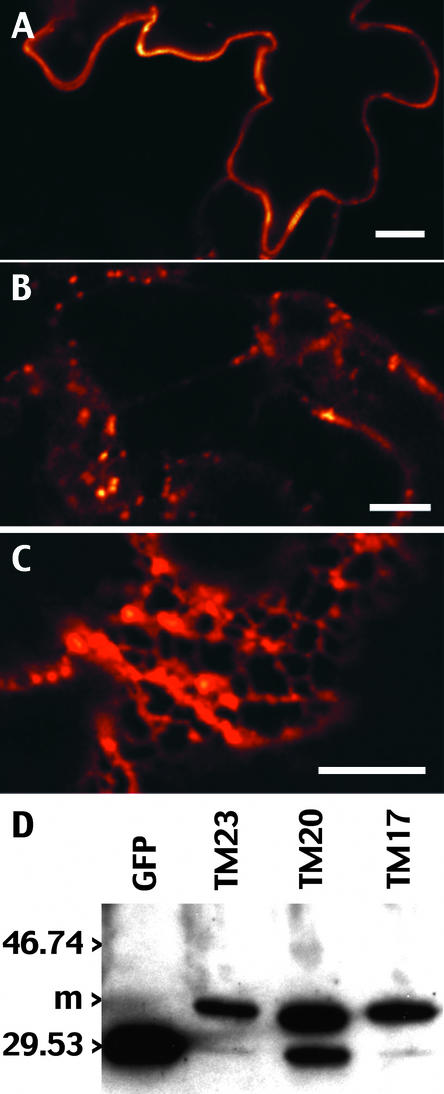
Figure 2.
Differential Accumulation Pattern of Transiently Expressed TM23, TM20, and TM17 Reflects the Accumulation of a Membrane-Bound Reporter.
(A) to (C) Tobacco lower leaf epidermis expressing the transmembrane chimeras at 72 hr after infiltration. The GFP accumulation pattern was observed with a confocal microscope from cells expressing TM23 (A), TM20 (B), or TM17 (C). The scale of colors from red to white represents the amount of GFP accumulation from the lowest to the highest GFP signal, respectively. Images were collected in a linear mode of GFP fluorescence. Bars = 10 μm.
(D) Immunoblot with anti-GFP antibodies of a crude extract of leaves transiently expressing TM23, TM20, or TM17. A soluble GFP was used as a control (GFP lane). m, positions of the membrane forms of the transmembrane chimeras. Positions of molecular mass markers (in kilodaltons) are shown at left.
As shown in Figures 2A to 2C, the accumulation patterns were very different depending on the construct expressed. GFP accumulated in a continuous line at the periphery of the epidermal cells for TM23 (Figure 2A), in dots that were extremely mobile for TM20 (Figure 2B), or in an ER-like pattern for TM17 (Figure 2C). Because it was necessary to determine that the GFP fluorescence detected in the living tissue originated from a membrane-bound form of the reporter, we performed immunoblot analysis using anti-GFP antibodies on a crude protein extract from leaf tissue expressing TM23, TM20, or TM17 (Figure 2D). From the three plant extracts, we detected a major form at 35 kD that clearly is larger than the apparent molecular mass of the reporter by itself. This observed molecular mass is in accordance with the expected size of the fusion proteins, which should be 29 kD for GFP plus 4.5 kD for the 32 to 38 amino acids from LAMP1. In addition to the membrane form, we detected a minor band that was almost invisible for TM23 and TM17 but was clear for TM20. By comparison with soluble GFP, we concluded that this band corresponds to GFP that was clipped off the chimeras.
Interestingly, we found in several separate extractions that construct TM20 always gave a greater amount of released GFP than the two other constructs. In fact, for the TM20 construct, we occasionally detected a slight fluorescence from GFP inside the vacuole in highly expressing cells. In addition, we noticed that the percentage of clipped GFP from each of the three chimeras increased drastically with longer extraction procedures and during storage of the extract, even in the presence of a wide range of protease inhibitors. Therefore, the cleavage suggests a high sensibility of the fusion proteins to proteases at the membrane level. For this reason, any extensive subcellular fractionation of the protein extracts was very delicate. Still, by immunoblot analysis of the three constructs, the 35-kD form was found exclusively in the membrane and not in the soluble fraction (100,000g supernatant), confirming its anchoring within membranes (data not shown).
The immunoblot shown here is representative of what was obtained from several independent transient expression and extraction procedures. Therefore, it reflects the forms of GFP present in vivo (Figure 2D). We estimated the relative amounts of the soluble/membrane form of GFP from the blot and found 4% soluble form for TM23 and 6% for TM17. Even for TM20, the large majority of the chimera was in the membrane form, with 28% in the soluble form. To conclude, the pattern of GFP accumulation represents the subcellular location where the membrane-bound reporter concentrates for each of the three transmembrane constructs. Therefore, the length of the transmembrane domain clearly affects the subcellular destination of type I membrane proteins.
GFP Fused to a 23–Amino Acid Transmembrane Domain Accumulates in the Plasma Membrane
To identify further the membrane at which the fusion proteins accumulated, we produced transgenic plants expressing each of the transmembrane reporters (Figure 1A). As shown in Figures 3A to 3C, we observed a tobacco leaf from a plant stably transformed with TM23 using confocal microscopy. The clear and sharp fluorescent line found previously after transient expression (Figure 2A) also was clearly visible in stably transformed plants and was found in every epidermal and guard cell (Figure 3A). As shown in a more detailed view (Figure 3B), the labeling clearly was concentrated on two separate lines from two adjacent cells, leaving the cell wall space unstained. In addition, small structures often were associated with the line type of labeling (Figure 3A). The location of some spots in the middle of the cell (Figure 3A, arrowheads) did not correspond to internal labeling but was attributable instead to the fact that the optical section was at the level of the surface of the leaf epidermis.
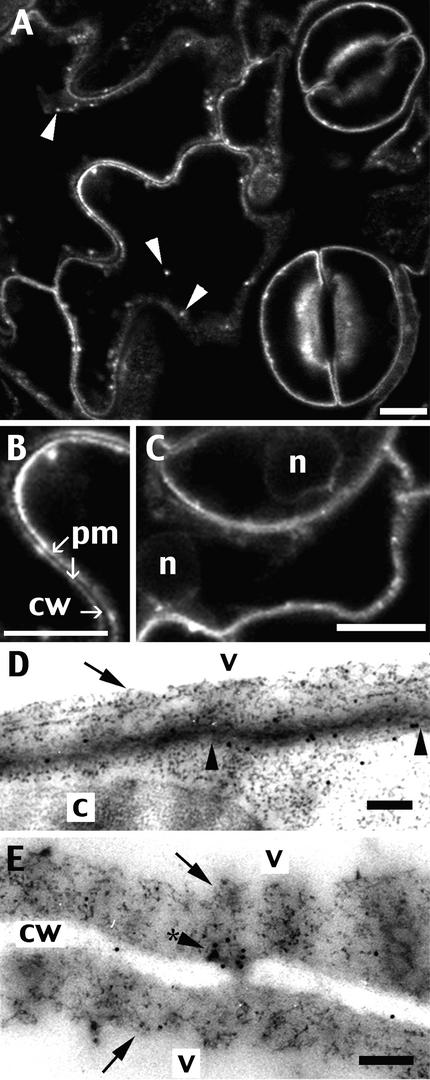
Figure 3.
Tobacco Lower Leaf Epidermis Stably Expressing TM23 Accumulates GFP in the Plasma Membrane.
Small pieces of a leaf stably expressing the GFP chimera were cut off and either observed using a confocal microscope ([A] to [C]) or fixed and immunolabeled with anti-GFP antibodies for electron microscopy ([D] and [E]). In addition to the dominant line-type of labeling, dots are visible (arrowheads, [A]) that are equivalent to structures associated with the plasma membrane (arrowhead with a star [E]). The plasma membrane-specific location of TM23 is especially visible within a 900-nm-long portion of membrane delimited by arrowheads (D). No labeling was found on the tonoplast, marked by arrows ([D] and [E]). c, chloroplast; cw, cell wall; n, nucleus; pm, plasma membrane; v, vacuole. Bars = 10 μm for (A) to (C) and 200 nm for (D) and (E).
Because epidermal cells have a very small relative volume of cytosol, one could argue that this line type of labeling corresponds to the tonoplast. We examined more carefully guard cells within an optical section through the nucleus (Figure 3C) and found that the fluorescent line remained at the periphery of the cell and did not follow the shape of the nucleus, as would be the case for tonoplast staining. Although this finding strongly suggests that the reporter TM23 accumulated in the plasma membrane, we performed immunolabeling at the electron microscopic level using anti-GFP antibodies (Figures 3D and 3E). The gold particles were found almost exclusively on the plasma membrane (13 of the 17 visible in Figure 3D) and not on the tonoplast (arrows). This result is attributable to anti-GFP antibodies, because the negative control gave very little nonspecific labeling (data not shown).
The gold particles often aligned clearly along the plasma membrane, as shown in the small portion of membrane between the two arrowheads in Figure 3D. Within this ∼900-nm-long portion of membrane, nine aligned gold particles were found. In Figure 3E, an accumulation of gold particles (7 of 11) also could be seen in a structure of ∼100 nm (arrowhead with star) associated with the plasma membrane and clearly separated from the tonoplast (arrows). These structures often were found by electron microscopy and probably are equivalent to the fluorescent dots detected by confocal microscopy (Figure 3A, arrowheads). We conclude that TM23 accumulates along the plasma membrane and not in the tonoplast of the large central vacuole.
GFP Fused to a Transmembrane Domain of 17 Amino Acids Accumulates in the ER
Figure 4 shows the leaf epidermis from tobacco plants stably transformed with TM17. We found in every cell a clear fluorescent network and a ring around the nucleus. This network was particularly visible in an optical section at the level of the cortical ER, as shown in Figure 4B. This very distinctive pattern can be identified as an ER labeled with GFP based on published data (Boevink et al., 1996, 1998). In medial cross-sections of the cells, the labeled strands appeared as a discontinuous line at the periphery of the cell (Figure 4C). This typical ER accumulation pattern of GFP, added to the fact that the reporter is membrane bound (and therefore not free in the cytosol), leads to the conclusion that TM17 accumulates in the ER. This means that changing the length of the transmembrane domain from 23 to 17 amino acids leads to a displacement of the chimera accumulation from the plasma membrane to the ER. As the images of single confocal sections suggest, the surfaces of the epidermal cells clearly were not flat. This can be visualized more clearly in the three-dimensional reconstruction shown in Figure 4D that was made from consecutive confocal images, each representing 1 μm of the tissue thickness.
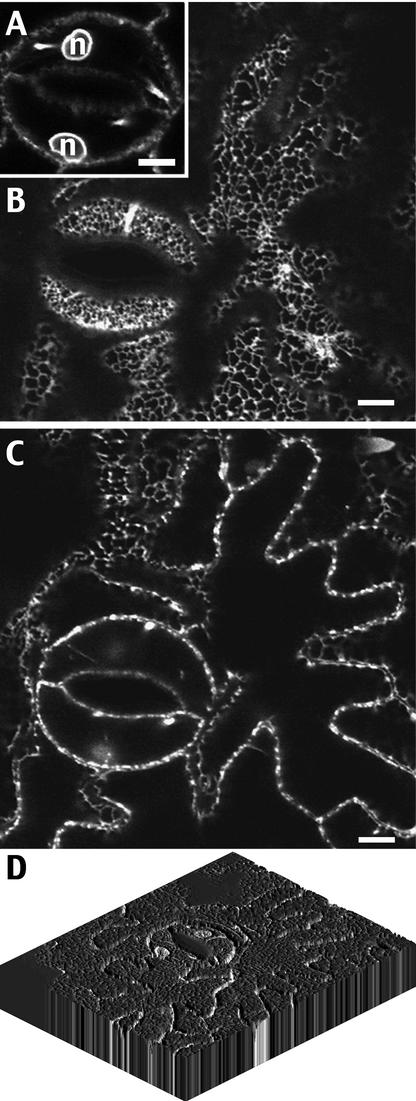
Figure 4.
Tobacco Lower Leaf Epidermal Cells Stably Transformed by TM17 Accumulate GFP in the Endoplasmic Reticulum.
(A) to (C) Small pieces of a leaf stably expressing the GFP chimera were cut off and observed using a confocal microscope. The same area of cells is presented in various confocal planes at the level of the nucleus of stomata cells (A), at the level of the cortical ER (B), or on a median plane of the epidermal cell layer (C). n, nucleus. Bars = 10 μm.
(D) Three-dimensional reconstruction of labeled epidermal cells using the program NIH Image 1.62/3DV1.01. Fourteen individual confocal sections were taken from the same labeled area, each corresponding to 1 μm of thickness, and were treated to rebuild the three-dimensional view.
GFP Fused to a Transmembrane Domain of 20 Amino Acids Accumulates in the Golgi
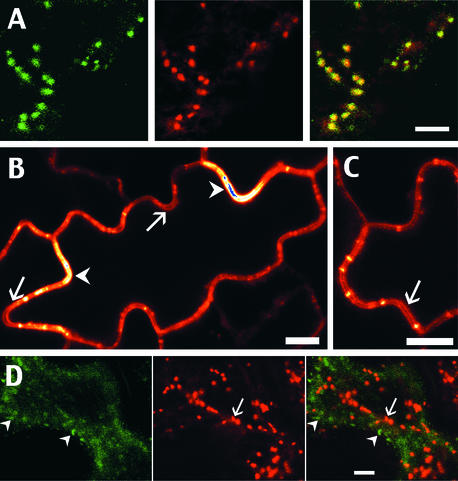
As shown in Figures 5A to 5F, tobacco plants stably expressing TM20 (which is intermediate in length between TM23 and TM17) gave a drastically different GFP accumulation pattern that resembled neither plasma membrane nor ER labeling. For TM20, GFP was found predominantly in small bodies that were extremely mobile in the cells and often were found to follow cytoplasmic strands traversing the vacuole (data not shown). These cytoplasmic strands sometimes were detected using confocal microscopy, although they showed very weak fluorescence compared with the dots (Figures 2B and 5A to 5F). The fact that these structures have an internal localization is clearer in guard cells, in which the cytoplasm is slightly thicker than in epidermal cells. In Figures 5B to 5F, successive optical sections are shown of the same guard cell. Especially in Figures 5E and 5F, the GFP-labeled compartments appear between the vacuole and the nucleus. Based on size, number, and dynamics, the TM20 structures are very similar to Golgi bodies (Boevink et al., 1998). On the other hand, the fact that, in a transient expression approach, some GFP was found in the vacuole could be taken as an indication that these structures are prevacuoles.
Figure 5.
A Single Transmembrane Domain of 20 Amino Acids Leads to the Accumulation of a GFP Chimera in the Golgi.
Small pieces of a leaf, stably ([A] to [F]) or transiently ([G] and [H]) expressing the GFP chimera TM20, were cut off and observed using a confocal microscope. In (G) and (H), epidermal cells were double transformed with the GFP chimera TM20 (green) and the AtERD2-YFP marker for the Golgi apparatus and the ER (red). The two images collected from the same optical section were merged to show the colocalization of the two markers (right panels, yellow). n, nucleus. Bars = 10 μm for (A) to (F) and 5 μm for (G) and (H).
To determine the nature of the TM20 compartments, we performed transient coexpression of TM20 and a ER-Golgi marker, each of them carrying color variants of GFP. To label the Golgi, we used yellow fluorescent protein (YFP) fused to the sequence of AtERD2, similar to the previously published AtERD2-GFP (Boevink et al., 1998). This chimeric protein was shown previously to accumulate selectively in the cis-Golgi and in the ER when overexpressed in tobacco cells, as demonstrated by a GFP accumulation pattern and immunoelectron microscopy (Boevink et al., 1998; Saint-Jore et al., 2002). Simultaneously, we infiltrated the same leaf tissue with the ER-Golgi marker and TM20 and then selected epidermal cells that were doubly transformed, based on the presence of both fluorescent markers. The results of this double transformation are shown in Figures 5G and 5H.
Within the same optical section, the TM20 compartments labeled with GFP are shown in green and the Golgi labeled with the AtERD2-YFP chimera are shown in red. As expected, AtERD2-YFP accumulated in small bodies, which are Golgi and associated with the ER (Figure 5G, red). The merged images from the GFP and YFP channels (Figures 5G and 5H, right) demonstrate, by the yellow color of every dot, that each of the TM20 structures colocalized with a Golgi compartment. In the greater magnification view shown in Figure 5H, the overlap between the two types of labeling appears total. The confocal mode used to collect these images was controlled by ensuring that any possible communication between the YFP and GFP channels could not be detected.
Therefore, these images are representative of doubly labeled cells, and both markers are always colocalized. The fact that the two types of labeling never appeared separate, even in highly mobile Golgi, is taken as an additional indication that these compartments are identical (data not shown). The movement of these labeled Golgi also indicates that the cell is alive; therefore, the colocalization of both markers reflects normal cell activity. These results clearly show that TM20 accumulates in the Golgi apparatus and not in a prevacuolar compartment, which is separated physically from the Golgi, as shown by previous electron microscopy studies (Conceiçao et al., 1997; Sanderfoot et al., 1998).
Elongation of the BP-80 Transmembrane Domain Modifies the Accumulation Site of the Reporter from the Golgi to the Plasma Membrane
Because we used the transmembrane domain of a human protein for our studies, we wanted to determine where the GFP would be sent when fused to a hydrophobic domain from a plant protein. As shown in Figure 1A, we fused the GFP sequence to the transmembrane domain and a few charged amino acids from the cytosolic domain of the pea vacuolar sorting receptor BP-80, also called VSRPS-1 (Paris et al., 1997). The length of the transmembrane domain for the resulting chimera, BP19, was estimated using the TMHMM program described previously and was found to be 19 amino acids (Figure 1A). We transiently transformed tobacco leaves with this construct and observed the cells by confocal microscopy.
As shown in Figure 6A, the typical pattern of GFP accumulation observed for BP19 corresponds to small dots, very similar to TM20/Golgi structures in terms of size and mobility. By contrast with TM20 expression, we found no labeling inside the vacuole, even in transiently transformed cells expressing a very high level of BP19. As shown in Figure 6A, we performed a double transformation using two color variants of fluorescent proteins to address the localization of BP19. We coexpressed the ER-Golgi marker fused to YFP (AtERD2-YFP) and BP19 (GFP). The fluorescence from BP19 structures is represented in green and the YFP-labeled Golgi, from the same optical section, is represented in red. As shown by the yellow dots on the merged channels, the BP19 compartments colocalize strictly with the Golgi, demonstrating that the transmembrane domain from a plant protein also led to an accumulation of the chimera in the Golgi.
Figure 6.
Lengthening the Plant Transmembrane Domain of the Vacuolar Sorting Receptor BP-80 from 19 to 22 Amino Acids Affects the Localization of the Reporter from the Golgi to the Plasma Membrane.
Small pieces of leaves transiently expressing the GFP chimeras BP19 and BP22 were observed using a confocal microscope.
(A) Cells were double transformed with the GFP chimera BP19 (green) and the AtERD2-YFP marker for the Golgi apparatus and the ER (red). The two images collected from the same optical section were merged to show the colocalization of the two markers (right panels, yellow).
(B) and (C) Epidermal cells expressing the BP22 construct were observed using the quantitative mode of the microscope. The scale of colors from red to white represents the amount of GFP accumulation from the lowest to the highest GFP signal, respectively. Blue represents the saturating emission of fluorescence (arrowheads). Plasma membrane labeling of two adjacent cells is clearly visible in some areas of the cells (arrows).
(D) Cells were double transformed with the GFP chimera BP22 (green) and the AtERD2-YFP marker for the Golgi apparatus and the ER (red). The two images collected from the same optical section were merged to show that none of the BP22 bodies (arrowheads) localize together with the Golgi (arrow).
Bars = 5 μm for (A) and (D) and 10 μm for (B) and (C).
Because the native BP-80 normally accumulates either in the trans-Golgi network or in the prevacuole to the lytic vacuole, its transmembrane domain should be compatible with subcellular locations farther down the secretory pathway, on the route to the vacuole (Humair et al., 2001). Nevertheless, we detected no accumulation of the reporter anywhere downstream of the Golgi, including in the tonoplast. Compared with the transmembrane chimeras, the BP19 protein with a predicted transmembrane domain length of 19 amino acids was expected to accumulate in the same location as TM20, and this is what we observed.
As shown in Figure 1A, we then added three hydrophobic residues, LAL, within the BP19 construct to create BP22. The introduced amino acids are in the expected transmembrane domain of BP-80 and should lengthen it to a 22–amino acid version. BP22 was expressed transiently in tobacco leaves, and the accumulation pattern of GFP was visualized with a confocal microscope. Figures 6B and 6C show BP22 accumulation in a quantitative mode for the typical labeling. Most cells were labeled with the BP22 construct, and the highly homogenous pattern obtained is a line at the periphery of the cells. This labeling is distinct from that of cell wall accumulation because two adjacent lines often were seen when two neighboring cells expressed the construct (Figures 6B and 6C, arrows). This is identical to what was found for a plasma membrane GFP pattern (TM23; Figures 2A and 3B) and is a strong indication that BP22 also accumulated in the plasma membrane.
As for TM23 (Figure 2A), some membrane areas contained more GFP fluorescence (Figure 6B, arrowheads), with occasional saturation of the detection system (Figure 6B, blue areas). By contrast with TM23, BP22 also showed some dots along the membrane that appeared to bridge over two adjacent cells (Figure 6C) and that were not mobile (data not shown). To exclude the possibility that these dots were Golgi, we coexpressed BP22 together with AtERD2-YFP. As shown in Figure 6D, BP22 accumulating dots (green, arrowheads) were clearly distinct from Golgi accumulating AtERD2-YFP (red, arrows). Because of the convex shape of the epidermal cells, BP22 always appeared slightly more peripheral than Golgi. Both structures were found in the same focal plane that simultaneously crossed the plasma membrane at the cell periphery and the cytosol in the central zone of the cell.
To conclude, lengthening the transmembrane domain of BP-80 to 22 amino acids also led to an accumulation of the reporter protein farther down the secretory pathway, from the Golgi to the plasma membrane. Interestingly, the BP22 chimera concentrated neither in compartments where the native BP-80 resides (the trans-Golgi network and the prevacuole) nor in the tonoplast. Therefore, the lengthening of the transmembrane domain allowed the chimera to exit the Golgi but apparently not to take a vacuolar route.
DISCUSSION
In mammalian cells, the default destination for membrane proteins in the secretory pathway was first believed to be the plasma membrane (Pfeffer and Rothman, 1987). Lysosomal membrane proteins are transported from the Golgi via two pathways that are mediated by sorting motifs exposed on their cytosolic portions (Hopkins, 1992). These proteins either may transit only by an endosomal/prelysosomal compartment or may first reach the plasma membrane and then be endocytosed in a signal-dependent manner. Then, the length of the transmembrane domain was shown to play an important role by allowing the exit from the Golgi apparatus, the membrane of which is thinner than compartments farther along the secretory pathway. A synthetic hydrophobic sequence of 17 Leu residues was sufficient for Golgi retention, whereas the protein was accumulated in the plasmalemma with 23 Leu residues (Munro, 1995b).
In yeast cells, the membrane of the vacuole can be reached by a default process (Roberts et al., 1992). More precisely, the prevacuolar compartment is the destination where the protein can be transported with no signal requirement, and further transport to the vacuole occurs via a maturation process of the prevacuole. In plant cells, the default destination for membrane proteins is far less clear and is complicated by the existence of a more complex vacuolar system. Several results indicated that the tonoplast could be reached by default (Höfte and Chrispeels, 1992; Jiang and Rogers, 1998; Barrieu and Chrispeels, 1999), although this conclusion was based indirectly on the vacuolar location of released soluble reporters.
We wanted to address the question of default transport for membrane proteins and study the role of the length of the transmembrane domain for protein location in plants. For this purpose, we created several chimeric proteins in which a reporter, GFP, was fused to various transmembrane domains with lengths of 23, 22, 20, 19, or 17 amino acids. We then expressed these constructs in tobacco using either a transient or a stable expression approach and characterized the subcellular location of membrane reporter accumulation. We found that GFP chimeras do not always accumulate in the same membrane but are distributed throughout the secretory pathway in a transmembrane length–dependent manner. We also found no accumulation in the tonoplast and additional indications that the vacuolar route cannot be taken efficiently without positive sorting signals.
The Role of the Length of the Transmembrane Domain in Subcellular Location
We used two sources of transmembrane domains for this study. One originated from the human protein LAMP1, and the other was derived from the pea vacuolar sorting receptor BP-80. In both cases, the chimeras clearly accumulated in the plant plasma membrane, with a 23– or 22–amino acid membrane-spanning domain, or in the Golgi, with a 20– or 19–amino acid domain. A shorter transmembrane domain of 17 amino acids led to ER retention in tobacco cells. By contrast with LAMP1, the limits of the transmembrane domain for BP-80 were not so obvious. When we started to record our results, TMHMM version 1.0 estimated the BP-80 transmembrane domain to be 19 amino acids long, because the N-terminal three residues (WAA) were excluded from the membrane-spanning domain (Figure 1A). Meanwhile, we have used version 2.0 of the program several times on the same peptide sequence and found slight differences in the results for the interface-located residues.
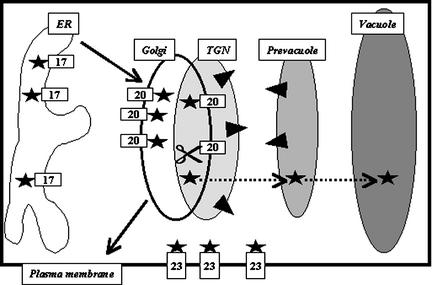
The latest profile indicates a >80% chance for the WAA residues to be part of the transmembrane domain and a 55% chance for the Tyr residue situated at the other end of transmembrane domain. This gives a novel estimated length for the BP-80 transmembrane domain of 21 amino acids. This predicted length of the BP-80 transmembrane domain does not seem to be influenced by the neighboring GFP, because the same value is predicted by the TMHMM program for the native BP-80 protein and BP19. These findings illustrate the limits in determining transmembrane domain length with an estimation program. Because the LAMP1 transmembrane domain is always found to be 23 amino acids by TMHMM, we propose to use the obtained values of 23, 20, and 17 amino acids for transmembrane constructs as references for the locations in the plasma membrane, the Golgi, and the ER, respectively, as shown on Figure 7.
Figure 7.
Model for the Distribution of Membrane Proteins within the Plant Secretory Pathway.
Membrane-bound chimeras to GFP (stars) are found at various locations in living tobacco cells. The membrane corresponds to the ER, the Golgi, or the plasma membrane for a transmembrane domain length of 17, 20, and 23 amino acids, respectively. This length-dependent distribution also is found when using the transmembrane domain of BP-80, a vacuolar sorting receptor (black triangles) normally localized in the trans-Golgi network (TGN) and the prevacuole. In some cases, reporter accumulated in the Golgi may be in contact with a lytic environment that is responsible for the cleavage (scissors) of the linker between the GFP and the membrane. The soluble released reporter then could reach the vacuole in a process that remains to be characterized (dotted arrows). Importantly, no tonoplast accumulation of the reporters was detected. This finding suggests that some positive information is required for membrane proteins in plants to efficiently reach the compartments located beyond the Golgi on the vacuolar route (gray compartments).
In seed tissue, the native protein BP-80 (Figure 7, triangles) has been shown to localize in the trans-Golgi and the prevacuole (Paris et al., 1997). This vacuolar receptor is the prototype of a large family of homologs with few known functional informations (Shimada et al., 1997; Ahmed et al., 2000). Nevertheless, these variants are found mostly in the same subcellular locations as BP-80 (Ahmed et al., 1997; Sanderfoot et al., 1998). When comparing all seven variants identified in Arabidopsis, the length of their transmembrane domains (residues with a >80% chance to be in the membrane) is estimated to be 21 amino acids, with one exception at 22 and another at 23 amino acids (atbp80f and atbp80d, respectively; Hadlington and Denecke, 2000). Unfortunately, this type of comparison is difficult to generalize to the entire secretory pathway because very few resident proteins with single membrane-spanning domains have been characterized and localized clearly.
We cannot use proteins with multispanning domains for this comparison because it is proposed that their helices often adopt a slightly tilted conformation compared with the membrane surface and often present polar residues that may interact with each other in a multiple–α-helices membrane-spanning domain conformation. For the Golgi, all of the single-span resident proteins identified to date are of type II. Using the TMHMM program, the length of the transmembrane domain was 16 residues for the Arabidopsis mannosyl-oligosaccharide 1,2-α-mannosidase, 17 residues for α-glucosidase-1, and 18 residues for xylosyl transferase. For the ER-resident Arabidopsis calnexins, the transmembrane domain is expected to be either 21 or 22 amino acids.
The membrane type I protein Cf-9 (Jones et al., 1994) is predicted to have a 19-residue transmembrane domain. First proposed to be a plasma membrane resident protein, it is in fact accumulated in the ER (Benghezal et al., 2000). Finally, the type II plasma membrane–residing endo-1,4-β-glucanase family carries a 23–amino acid transmembrane domain (Mølhoj et al., 2001). Our results suggest that the membrane thickness increases in plants from the ER to the Golgi and finally to the plasma membrane, as demonstrated previously in animal cells (Munro, 1995b). Whether this variation in thickness will be reflected in the lengths of the resident proteins remains to be determined when a larger pool of type I proteins is available from plants. In the examples studied here, the estimated length obtained with TMHMM was correct in assessing the subcellular location of the membrane protein.
The lipid composition of membranes of the secretory pathway in animal cells differs markedly from the ER to the plasma membrane. Two main components of the lipidic bilayer are believed to play a role in generating the thickness gradient of the membranes, cholesterol and the glycosphingolipids. These two elements would associate in a stable platform in the Golgi and ultimately would form a lipid raft at the plasma membrane level (Brown and London, 2000). Therefore, the plasma membrane is enriched in cholesterol compared with the ER, and the Golgi presents an intermediate composition. This lipid selection results in differences in the physical properties of the various membranes, with critical effects on their specific functions. Therefore, the plasma membrane, as a physical barrier for the cell, should be more impermeable, whereas the ER should be highly flexible, allowing extensive formation of tubules and vesicles.
The lipid differences also affect the membrane protein distribution, favoring the presence of proteins with a transmembrane domain length matching the membrane thickness (Killian, 1998). In plant cells, the lipid composition of the membrane in the secretory pathway also shows a gradient of sterols, with increasing amounts from the ER to the Golgi and finally the plasma membrane. In leek seedlings, the amount of sterol compared with phospholipids increases 2.6 times from the ER to the Golgi and 2.7 times from the Golgi to the plasma membrane (Moreau et al., 1998). Additionally, the transport of these sterols is blocked by monensin and brefeldin A and follows a kinetic that is compatible with a vesicle-mediated pathway. The glycosphingolipids also are abundant in the plasma membrane and concentrated in the outer leaflet. The membrane heterogeneity in a plant system, therefore, supports a similar transport of the domains of sterols and sphingolipids from the ER to the plasma membrane together with associated proteins.
Additional support for this hypothesis comes from the recent demonstration that the plant ER can select a subclass of lipids in vesicles formed in vitro using a cell-free system from leek cells (Sturbois-Balcerzak et al., 1999). One of the selected lipids is the phosphatidylserine carrying very-long-chain fatty acids (>18 carbons). Compared with the plasma membrane, the tonoplast has almost the same amount of glycosphingolipids and also presents a similar ratio of sterol to glycerolipids. In contrast, the amount of proteins is 2.7 times lower in the tonoplast than in the plasma membrane. These findings in lipids fit well with a variation in membrane thickness from the ER to the plasma membrane in plant cells, as in animal system. In contrast, the similarity in lipid composition between the tonoplast and the plasma membrane suggests that features other than the physical properties of the transmembrane domains are used to sort membrane proteins between the vacuolar and secretion routes.
How to Distinguish between Membrane-Bound and Soluble Released Reporters
A small proportion of GFP was occasionally detected in the vacuole of cells highly expressing the Golgi-localized TM20. Because the large majority of the reporter was bound to the membrane, this indicated that GFP was detectable when released in the vacuole. Several studies support the hypothesis that the reporter can be released from the membrane in a compartment upstream of the vacuole and later can reach the vacuole in its soluble form.
First, the vacuolar GFP released from TM20 was found exclusively after transient expression in cells that were much more fluorescent than average and that showed symptoms of stress, such as the aggregation and immobility of Golgi compartments and nonspecific labeling. This result indicates that a state of stress can affect the localization of membrane proteins. In a study by Barrieu and Chrispeels (1999), the reporter used was a yeast invertase fused to 111 amino acids of the yeast ER protein Wbp1p, including a single transmembrane segment and the C-terminal cytosolic tail, in which the ER retention signal had been inactivated by mutagenesis. In transgenic tobacco, some soluble active invertase was found to accumulate in the vacuole. Interestingly, the leaf tissue used for this study was under strong stress, although probably because of the presence of active invertase rather than because of an overload of the secretory pathway. This stressed phenotype, characterized by the appearance of yellow and brown sectors on leaf tissue, was found occasionally after our infiltration procedure and always correlated with the highly disturbed GFP accumulation patterns described previously.
Another study with GFP fused to the transmembrane segment and the cytosolic tail of PV72, a pumpkin homolog of pea BP-80, also indicated a cell stage–dependent accumulation of fluorescence in BY-2 tobacco cells (Mitsuhashi et al., 2000). The GFP fluorescence patterns changed from small dots to vacuolar staining depending on whether the cells originated from a 3-day-old culture or from 10-day-old calli. Based on the known subcellular location of BP-80, the nature of the dots could be either the trans-Golgi network and/or the prevacuole. Therefore, the fact that all of the membrane-bound GFP was released and accumulated in the vacuole in callus cells suggests drastic changes within those compartments, including in their lytic properties. Prolonged retention in a compartment containing some proteases will be necessary to release the reporter from the membrane, and this would explain why there is no soluble GFP in the vacuole for plasma membrane–accumulating chimeras.
The yeast trans-Golgi–residing enzyme Kex2p apparently has at least an equivalent in tobacco cells that functions on a typical Kex2p site (Jiang and Rogers, 1998). This homolog, therefore, is a potential candidate for the clipping of Golgi-accumulated reporters. In fact, the nature of the linker used to attach the reporter to the membrane also directly influenced this Golgi-associated lytic effect. This linker is very short for BP19 but nine amino acids longer (IEGREAEAL) for TM20. Both are much shorter that the ∼70–amino acid invertase-Wbp1p linker that contains a potential cleavage site for Kex2p (Barrieu and Chrispeels, 1999). With a transmembrane domain length estimated to be 20 amino acids, the Wbp1 chimera is expected to localize in the Golgi and should be in close contact with a lytic compartment such as the trans-Golgi network.
Finally, the linker also could play a role in diverting the reporter from being secreted after its release from the membrane (Figure 7, dotted line). It has been established that many C-terminal sequences may function as vacuolar sorting determinants with little sequence specificity (for review, see Matsuoka and Neuhaus, 1999). The potential role of the TM20 linker as a vacuolar signal also may explain the difference between TM20 and BP19 in terms of soluble GFP detected in the vacuole.
Our results and previously published data suggest that a membrane protein accumulating in the Golgi is in contact with a lytic environment that can release the reporter from the membrane depending on the linker and on the state of the cell. Although the “clipping off” process of the reporter for some membrane chimeras (Figure 7, scissors) remains to be characterized, it is clear that the vacuolar accumulation of soluble reporter proteins is not the only evidence for the accumulation of the membrane-bound chimera in the tonoplast. Proteolytic release in a compartment upstream of the site of sorting for soluble proteins also can lead to the same result (Figure 7, dotted line).
Can the Tonoplast Be Reached by Default in Plant Cells?
Interestingly, none of the chimeras we tested led to an accumulation of the reporter on the tonoplast or to a total release of soluble GFP in the vacuole, although we know that fluorescence is detectable in this location using our experimental procedures. This is especially relevant for the chimera made with the transmembrane domain of a receptor involved in transporting proteins to the lytic vacuole, BP-80. Even when the BP-80–based chimera was allowed to exit the Golgi by lengthening the transmembrane domain, the reporter was found in the plasma membrane but not in the tonoplast. By contrast with previous data, we used living cells and observed membrane-bound reporters. Apparently, the length is not the limiting feature that prevents tonoplast localization of the membrane reporter.
All of these results strongly suggest that positive information is required in the plant secretory pathway for an efficient accumulation of a membrane protein in compartments downstream of the Golgi (Figure 7, gray) on the vacuolar route. This contradicts the previously published hypothesis that the tonoplast can be reached by default in plant cells. Höfte and Chrispeels (1992) fused the sixth transmembrane domain and the C terminus of α-TIP to the reporter protein phosphinotricin acetyltransferase (PAT). The whole chimera PAT-TIP accumulated predominantly in the vacuole. To interpret their results, the authors postulated either that the tonoplast can be reached by default or that some vacuolar targeting information is present within the transmembrane domain of α-TIP. Our results support this second hypothesis, because, by contrast with α-TIP, neither LAMP1 nor BP-80 should carry any vacuolar targeting information within their transmembrane domains.
As mentioned above, Barrieu and Chrispeels (1999) also proposed the tonoplast as a default destination in plants using a chimera developed to address the same question in yeast (Gaynor et al., 1994). In transgenic tobacco cells expressing the Wbp1p chimera, cell fractionation indicated that as much as half of the chimera produced was bound to the membrane. Unfortunately, the exact location of this membrane form could not be addressed in this study. If the whole chimera had reached the tonoplast, it is unlikely that only half of it would have released its soluble marker into the lytic environment of the vacuole. Our results reinforce the second explanation of Barrieu and Chrispeels for their findings. That is, the reporter Wbp1 could remain in a compartment upstream of the vacuole, where it would be clipped from the membrane and transported to the vacuole in its soluble form.
Signals within the Transmembrane Domains
The first membrane-spanning domain described in this article originated from a human lysosomal resident protein, and to date, only the cytosolic portion of this protein has been shown to play a role in lysosome targeting (Rohrer et al., 1996). The native factors that control the localization of the full-length BP-80 in its compartments, the trans-Golgi network and the prevacuole, are unknown. Our results support indirectly the crucial role of BP-80 cytosolic domain in traffic (Jiang and Rogers, 1998). Searches for retention signals within transmembrane domains have been made in the animal and yeast systems. To date, only Phe residues were suggested to play a role in Golgi retention in mammalian cells. The mammalian sialyl transferase, a type II trans-Golgi–resident protein, contains four Phe residues in its transmembrane domain. These four amino acids often are found in animal Golgi-resident proteins and are proposed to play a role in the kin recognition model for Golgi localization. Mutation of the four Phe residues to Ile leads to a threefold increase in plasma membrane accumulation (Munro, 1995b). Nevertheless, the Phe effect is very weak, because this accumulation represents only 4% of a positive control protein for plasma membrane transport.
For the Arabidopsis type II Golgi-resident proteins, the number of Phe residues also is highly variable, from none in α-glucosidase-1, to one in α-mannosidase-1, to 4 in xylosyl transferase. In the BP constructs used here, the transmembrane domain of BP-80 contains two Phe residues (Figure 1B, stars). Neither the position nor the number of Phe residues is conserved within the BP-80 family. Of the 20 variants analyzed, nine have no Phe residues, five have only one Phe residue, five have two Phe residues, like BP-80 (although the positions are not always conserved), and one maize variant has four Phe residues. These results can be taken as an indication that these residues are not essential for the location of this receptor family. On the other hand, one of the Phe residues in BP-80 is replaced by a highly conserved Tyr in most homologs. Additionally, the Phe residues of BP-80 do not prevent the accumulation of the BP reporter protein in the plasma membrane when the transmembrane domain is lengthened to 22 amino acids. This suggests a minor role, if any, of these residues compared with the length (as demonstrated previously in mammalian cells; Munro, 1995b).
To investigate the role of Phe in the Golgi retention of chimera BP19, we replaced these residues with Leu and transiently expressed the resulting chimera in tobacco leaves. Unfortunately, we detected almost no fluorescent cells for this mutant construct. We repeated the assay four times with two independent clones of transformed Agrobacterium. Occasionally, using a laser power stronger than usual, we detected one positive cell with weak ER-like labeling (data not shown). Treatments with protease inhibitors or brefeldin A did not improve the overall GFP fluorescence of the leaf tissue. Whether this result can be taken as an indication of a role of Phe in Golgi retention or as a reflection of some indirect effect such as nonefficient membrane anchoring at the ER level remains to be determined. TMHMM analysis of the mutated construct did not reveal any difference in the estimated transmembrane domain compared with BP19. The role of Phe in membrane retention, therefore, remains to be investigated and would require another complete study with additional quantitative approaches.
The hydrophobic features of the transmembrane domain also play a role in retention within membranes of the secretory pathway, as shown using SNAREs, which are C-terminally anchored proteins, in yeast (Rayner and Pelham, 1997). Both Sft1p, a Golgi SNARE, and Ufe1p, an ER SNARE, possess transmembrane domains of the same length. Neither key amino acids nor the overall hydropathy of the transmembrane domain is responsible for membrane retention. Nevertheless, the location of the residues relative to the α-helix axis is an important feature. For Ufe1p, the presence of a slightly polar face within the α-helix forming the transmembrane domain plays a role in ER retention.
The hydropathy plots for TM23 and BP19 revealed small differences in terms of residue distribution (Figure 2B). Although both transmembrane and BP constructs accumulated in the same type of membrane, for an equivalent transmembrane domain length, the most hydrophobic residues were within the luminal half for BP-80 or the cytosolic half for LAMP1. This finding suggests that this apparent heterogeneity in hydropathy has no effect on subcellular location. In terms of the distribution of residues along the α-helix axis, no obvious polarity appeared in the constructs used when constructing α-helix wheel plots (data not shown). Because we removed or added amino acids in groups of three, we may have interfered only slightly with this α-helix–based distribution.
Conclusion
We present here the location of a membrane-bound chimera in apparently unstressed living cells. This finding clearly demonstrates that the membrane where the reporter accumulates can be the ER, the Golgi, or the plasma membrane (Figure 7). In addition, there is no indication that the tonoplast can be reached by default for type I membrane proteins in nonstressed cells. In contrast, our results suggest that some positive information is required for membrane proteins to reach post-Golgi compartments on the vacuolar route efficiently (Figure 7, gray). The need for vacuolar targeting information would fit with the possible existence of two or three different vacuolar destinations in plant cells (Paris et al., 1996) and therefore different pathways (Matsuoka et al., 1995; Jiang and Rogers, 1998), a situation unique to plant cells.
It is reasonable, therefore, to suppose that positive information is required for membrane proteins to take the proper vacuolar route from the Golgi. This information apparently is not present in the transmembrane portion of the proteins, because BP-80–based constructs never reached the post-Golgi compartments where the full-length receptor resides (Figure 7, triangles). Also, note that the TGN and the prevacuole are distinguishable from the Golgi by confocal microscopy when using a combination of fluorescent proteins (Kim et al., 2001). It should be remembered that the fluorescence approach used here allows us to detect the main location where the chimera is concentrated. We cannot exclude the possibility that a small portion of the chimera also may be present in a different subcellular location that could be detectable by more sensitive biochemical approaches.
METHODS
Constructs
We used the previously described sequence of green fluorescent protein (GFP) modified for plants (Haseloff et al., 1997; Di Sansebastiano et al., 1998) with the difference that we used a more stable variant, called GFP6, with the mutations F64L and S65T (Cormack et al., 1996). The enhanced yellow variant of GFP (YFP) was purchased from Clontech (Palo Alto, CA). Both sequences were fused in frame with the signal peptide from the Arabidopsis thaliana chitinase. The fluorescent protein constructs were inserted into plant binary vectors, either pBIN (Haseloff et al., 1997) for both stable transformation and leaf infiltration of transmembrane constructs or pVKH18-EN6 (Batoko et al., 2000) for transient expression of BP19 or the YFP fused to AtERD2. This leads in both cases to the expression of all fusion proteins under the control of an enhanced promoter of Cauliflower mosaic virus.
For TM23, a secreted form of GFP6 was fused in frame to a potential cleavage site for factor X followed by the original sequence for the human LAMP1 transmembrane domain (lysosome-associated membrane protein-1) and by the four amino acids RKRS from the original cytosolic domain of human LAMP1. Overlapping oligonucleotides were used to reconstitute the double-stranded DNA with (1) a SalI cohesive end in 5′, (2) the sequence encoding the peptide IEGREAE (the factor X cleavage site), (3) amino acids 382 to 409 from the human LAMP1 sequence, and (4) a cohesive end for PstI. The resulting SalI-PstI fragment was cloned at the corresponding sites of pSGFP6K (Di Sansebastiano et al., 1998) in place of the sequence encoding the ER retention motif KDEL. For TM23, the resulting amino acid sequence added to GFP was 5′-STIEGREAEALIPIAVGGALAGLVLIVLIAYLVGRKRS-3′.
TM20 and TM17 were made in a similar way, but oligonucleotides were designed to make two modified forms in which the amino acids underlined in the sequence shown above were removed. TM20 was made by removing one block of three amino acids, VLI, whereas TM17 was made by removing all six amino acids. TM20 lacks the original amino acids 395 to 397, whereas TM17 lacks amino acids 395 to 400, from the human LAMP1 sequence. The size of the hydrophobic transmembrane domain was calculated using the prediction program for transmembrane helices based on a hidden Markov model (TMHMM version 1.0), which was replaced recently by TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0), that was developed by Anders Krogh and Erik Sonnhammer (Sonnhammer et al., 1998). Using this program, the predicted transmembrane domain of TM23 is 23 amino acids long. The size of the truncated transmembrane domain was 20 and 17 amino acids for TM20 and TM17, respectively.
The BP19 construct was made by polymerase chain reaction to generate a SalI site at amino acids 563 and 564 and a stop codon plus a SacI site after amino acid 592 from the original BP-80 sequence (Paris et al., 1997). The resulting fragment was fused to the sequence of GFP6 from TM23 into binary vector pVKH18-EN6 using a triple ligation approach. The BP22 construct was made from BP19 by polymerase chain reaction using an antisense oligonucleotide (with a SacI site), a sense primer with a SalI site in 5′, and the addition of the 5′-CTTGCTCTC-3′ coding sequence. The fragment SalI-SacI then was cloned to replace its equivalent in BP19 to generate BP22, which has the additional tripeptide LAL between residues 569 and 570 of the original BP-80 sequence.
To generate the AtERD2-YFP construct, we used the AtERD2-GFP construct pVKH18-EN6-AtERD2-GFP (Saint-Jore et al., 2002) and the enhanced YFP sequence. We created HindIII and SalI sites at the 5′ end of YFP and added a C-myc tag and a SacI site at the 3′ end. The resulting fragment was cloned into the HindIII and SacI sites of pVKH18-EN6-ERD2-GFP. The sequence for AtERD2 was subcloned upstream of the YFP sequence in the expression cassette of the binary vector pVKH18-EN6.
Immunolabeling
Rabbit antiserum directed against GFP isolated from Aequorea victoria was purchased from Molecular Probes (A-6455; Eugene, OR). The dilutions used were either 1:50 for immunogold labeling or 1:5000 for protein gel blot analysis. The secondary anti-rabbit antibodies conjugated to alkaline phosphatase were purchased from Sigma (A-3687; St. Louis, MO) and were used for protein gel blot analysis at a dilution of 1:30,000. Secondary anti-rabbit antibodies for immunogold labeling, coupled to 15-nm gold particles, were purchased from Biocell TEBU (goat anti-rabbit IgG [H+L], EM.GAR 15; Le Perray en Yvelines, France) and were used at a dilution of 1:40.
Immunoblots were revealed with alkaline phosphatase using the Immun-Star chemiluminescent detection system from Bio-Rad (170-5012; Hercules, CA). The reference used for the soluble GFP (10 ng/lane) was provided by Clontech (rEGFP, 8365-1).
Transient and Stable Expression in Tobacco
For stable expression, the three transmembrane constructs in pBIN were used to transform Agrobacterium tumefaciens strain GV3101 p2260 (a gift from Barbara Hohn, Friedrich Miescher-Institut, Basel, Switzerland) using a classic triparental mating protocol (Ditta et al., 1980) with the helper strain pRK2013 (Ditta et al., 1980; Spielman and Simpson, 1986) or by heat shock (Koncz and Schell, 1986). Plants then were transformed using a previously described technique (Horsch et al., 1985) starting from leaves of tobacco (Nicotiana tabacum cv SR1).
For transient expression, leaves of 3- to 4-week-old tobacco plants growing at 20 to 22°C were used. Each expression vector was introduced into Agrobacterium strain GV3101 p2260 as described above. A single colony from the transformants was inoculated in 5 mL of YEB medium (5 g/L beef extract, 1 g/L yeast extract, 5 g/L Suc, and 0.5 g/L MgSO4·7H2O) supplemented with 100 μg/mL kanamycin. The bacterial culture was incubated at 30°C with agitation for 20 hr. One milliliter of bacterial culture then was pelleted by centrifugation at 2200g for 5 min at room temperature. The pellet was washed once with 1 mL of infiltration medium (Batoko et al., 2000) and then resuspended in 1 mL of the same buffer. For experiments requiring coexpression of two different constructs, 0.5 mL of each bacterial culture was mixed before the washing step. The bacterial suspension was inoculated through the stomata using a 1-mL plastic syringe and by gentle pressure on the lower epidermal surface. Transformed plants then were incubated under normal growth conditions.
Confocal Microscopy
We observed the fluorescence emitted by the fluorescent proteins at 36 to 72 hr after transient expression or from leaf pieces of stably transformed tobacco. Briefly, segments of transformed leaves (∼1 cm2) were cut using a razor blade and mounted in water for imaging. We used either (1) a confocal system from Leica (TCS 4D; Wetzlar, Germany) coupled to a DM RBE microscope from Leica with a fluorescein isothiocyanate setting for GFP fluorescence, or (2) an inverted Zeiss LSM 510 laser scanning microscope (Jena, Germany), both with a ×40 oil immersion objective. For imaging of the coexpression of YFP and GFP constructs, excitation lines of an argon ion laser of 488 nm for GFP and 514 nm for YFP were used alternately with line switching on the multitrack facility of the microscope. GFP fluorescence was detected using a 515-nm dichroic beam splitter and a 475- to 525-nm bandpass filter, and YFP fluorescence was detected with a 515-nm dichroic filter and a 535- to 590-nm bypass filter. In this way, any communication and bleeding of fluorescence were eliminated. Enhanced-quality images were acquired with Zeiss LSM 510 imaging system software, and postacquisition image processing was performed with LSM 5 Image Browser (Zeiss) and Adobe Photoshop F1 4.0 software (Mountain View, CA).
Electron Microscopy
Chemical fixation and immunogold staining were performed as described previously (Fleurat-Lessard et al., 1997). Young tobacco leaf tissues were cut into 1- to 2-mm pieces and fixed for 45 min in a mixture of 1.5% (w/v) paraformaldehyde and 0.5% glutaraldehyde in 0.05 M phosphate buffer, pH 7.2. The tissue then was washed abundantly (at least six baths for a total of 1 hr) in the same buffer and was submitted to postfixation of 3 min in 1% (v/v) OsO4. After gradual dehydration in ethanol, the leaf pieces were embedded overnight in London White Resin (L012; TAAB Laboratories, St. Germain en Laye, France). Resin polymerization was performed in a capsule of gelatin at 55°C for 24 hr. Thin sections were spread carefully with toluene vapor and were collected on parlodion-coated gold grids. The sections were incubated overnight with a 1:50 dilution of antibodies against GFP. Secondary antibodies labeled with 15-nm gold particles then were applied for 2 hr. The samples were washed and treated subsequently for contrast in a saturated uranyl acetate solution (8 min) and in lead citrate (3 min). A control experiment omitted the primary antibodies. Observations were made at 60 kV with a CM 100 Philips microscope (Eindhoven, The Netherlands).
Accession Numbers
The accession numbers for the sequences described in this article are AY039536 (Arabidopsis mannosyl-oligosaccharide 1,2-α-mannosidase), AY055858 (Arabidopsis α-glucosidase-1), AF272852 (Arabidopsis xylosyl transferase), AT5 g61790 and AT5 g07340 (the ER-resident Arabidopsis calnexins), P11279 (human LAMP1), and U79958 (the original BP-80 sequence).
Acknowledgments
Special thanks are given to Claude Saint-Jore (Oxford Brookes University) and Ian Moore (Oxford University) for providing the AtERD2-GFP construct. We thank V. Gomord (Centre National de la Recherche Scientifique, Unité Mixte de Recherche 6037, Rouen, France) for critical reading of the manuscript. This work was supported by a European Molecular Biology Organization long-term fellowship to N.P. (57-1996), a Swiss National Foundation grant (3100-46926.96), and a Biotechnology and Biological Sciences Research Council grant to C.H. (PO8503).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.000620.
References
- Ahmed, S.U., Bar-Peled, M., and Raikhel, N.V. (1997). Cloning and subcellular location of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol. 114, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S.U., Rojo, E., Kovaleva, V., Venkataraman, S., Dombrowski, J.E., Matsuoka, K., and Raikhel, N.V. (2000). The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 149, 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrieu, F., and Chrispeels, M.J. (1999). Delivery of a secreted soluble protein to the vacuole via a membrane anchor. Plant Physiol. 120, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H.-Q., Hawes, C., and Moore, I. (2000). A Rab1 GTPase is required for transport between endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal, M., Wasteneys, G.O., and Jones, D.A. (2000). The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12, 1179–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P., Cruz, S.S., Hawes, C., Harris, N., and Oparka, K.J. (1996). Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J. 10, 935–941. [Google Scholar]
- Boevink, P., Oparka, K., Cruz, S.S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Brown, D.A., and London, E. (2000). Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224. [DOI] [PubMed] [Google Scholar]
- Conceiçao, A.D.S., Marty-Mazars, D., Bassham, D.C., Sanderfoot, A.A., Marty, F., and Raikhel, N.V. (1997). The syntaxin homolog atPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9, 571–582. [PMC free article] [PubMed] [Google Scholar]
- Cormack, B.P., Valdivia, R.H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (gfp). Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Di Sansebastiano, G.-P., Paris, N., Marc-Martin, S., and Neuhaus, J.-M. (1998). Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J. 15, 449–457. [DOI] [PubMed] [Google Scholar]
- Ditta, G., Stanfield, S., Corbid, D., and Helsinki, D.R. (1980). Broad host range DNA cloning system for gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurat-Lessard, P., Frangne, N., Maeshima, M., Ratajczak, R., Bonnemain, J.L., and Martinoia, E. (1997). Increased expression of vacuolar aquaporin and H+-ATPase related to motor cell function in Mimosa pudica L. Plant Physiol. 114, 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, E.C., Heesen, S.T., Graham, T.R., Aebi, M., and Emr, S.D. (1994). Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J. Cell Biol. 127, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlington, J.L., and Denecke, J. (2000). Sorting of soluble proteins in the secretory pathway of plants. Curr. Opin. Plant Biol. 3, 461–468. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, R.K., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfte, H., and Chrispeels, M.J. (1992). Protein sorting to the vacuolar membrane. Plant Cell 4, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, C.R. (1992). Selective membrane protein trafficking: Vectorial flow and filter. Trends Biochem. Sci. 17, 27–32. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, J.E., Hoffmann, N.L., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Humair, D., Hernández Felipe, D., Neuhaus, J.-M., and Paris, N. (2001). Demonstration in yeast of the function of BP-80, a putative plant vacuolar sorting receptor. Plant Cell 13, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L.W., and Rogers, J.C. (1998). Integral membrane protein sorting to vacuoles in plant cells: Evidence for two pathways. J. Cell Biol. 143, 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D.G. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Killian, J.A. (1998). Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta 1376, 401–416. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.-W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., and Neuhaus, J.-M. (1999). Cis-elements of protein transport to the plant vacuoles. J. Exp. Bot. 50, 165–174. [Google Scholar]
- Matsuoka, K., Bassham, D.C., Raikhel, N., and Nakamura, K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130, 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi, N., Shimada, T., Mano, S., Nishimura, M., and Hara-Nishimura, I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY2 cells. Plant Cell Physiol. 41, 993–1001. [DOI] [PubMed] [Google Scholar]
- Mølhoj, M., Ulvskov, P., and Dal Degan, F. (2001). Characterization of a functional soluble form of a Brassica napus membrane-anchored endo-1,4-β-glucanase heterologously expressed in Pichia pastoris. J. Plant Physiol. 127, 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, P., Hartmann, M.-A., Perret, A.-M., Sturbois-Balcerzak, B., and Cassagne, C. (1998). Transport of sterols to the plasma membrane of leek seedlings. Plant Physiol. 117, 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. (1995. a). A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem. Soc. Trans. 23, 527–530. [DOI] [PubMed] [Google Scholar]
- Munro, S. (1995. b). An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14, 4695–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, N., Stanley, C.M., Jones, R.L., and Rogers, J.C. (1996). Plant cells contain two functionally distinct vacuolar compartments. Cell 85, 563–572. [DOI] [PubMed] [Google Scholar]
- Paris, N., Rogers, S.W., Jiang, L., Kirsch, T., Beevers, L., Phillips, T.E., and Rogers, J.C. (1997). Molecular cloning and further characterization of a probable plant vacuolar sorting receptor. Plant Physiol. 115, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini, E., Villa, A., and Borgese, N. (1996). A mutant cytochrome b(5) with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc. Natl. Acad. Sci. USA 93, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S.R., and Rothman, J.E. (1987). Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu. Rev. Biochem. 56, 828–852. [DOI] [PubMed] [Google Scholar]
- Rayner, J.C., and Pelham, H.R.B. (1997). Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16, 1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., Nothwehr, S.F., and Stevens, T.H. (1992). Membrane protein sorting in the yeast secretory pathway: Evidence that the vacuole may be the default compartment. J. Cell Biol. 119, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer, J., Schweizer, A., Russell, D., and Kornfeld, S. (1996). The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J. Cell Biol. 132, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore, C.M., Evins, J., Brandizzi, F., Batoko, H., Moore, I., and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29, 661–678. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Ahmed, S.U., Marty-Mazars, D., Rapoport, I., Kirchhausen, T., Marty, F., and Raikhel, N.V. (1998). A putative vacuolar cargo receptor partially colocalizes with atPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 95, 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T., Kuroyanagi, M., Nishimura, M., and Hara-Nishimura, I. (1997). A pumpkin 72-kDa membrane protein of precursor-accumulating vesicles has characteristics of a vacuolar sorting receptor. Plant Cell Physiol. 38, 1414–1420. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen, eds (Menlo Park, CA: American Association for Artificial Intelligence Press), pp. 175–182. [PubMed]
- Spielman, A., and Simpson, R.B. (1986). T-DNA structure in transgenic tobacco plants with multiple independent integration sites. Mol. Gen. Genet. 205, 34–41. [Google Scholar]
- Sturbois-Balcerzak, B., Vincent, P., Maneta-Peyret, L., Duvert, M., Satiat-Jeunemaitre, B., Cassagne, C., and Moreau, P. (1999). ATP-dependent formation of phosphatidylserine-rich vesicles from the endoplasmic reticulum of leek cells. Plant Physiol. 120, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M., Ellenberg, J., Bonifacino, J.S., and Weissman, A.M. (1997). The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272, 1970–1975. [DOI] [PubMed] [Google Scholar]