Abstract
The epitope specificities and functional activities of monoclonal antibodies (MAbs) specific for the murine leukemia virus (MuLV) SU envelope protein subunit were determined. Neutralizing antibodies were directed towards two distinct sites in MuLV SU: one overlapping the major receptor-binding pocket in the N-terminal domain and the other involving a region that includes the most C-terminal disulfide-bonded loop. Two other groups of MAbs, reactive with distinct sites in the N-terminal domain or in the proline-rich region (PRR), did not neutralize MuLV infectivity. Only the neutralizing MAbs specific for the receptor-binding pocket were able to block binding of purified SU and MuLV virions to cells expressing the ecotropic MuLV receptor, mCAT-1. Whereas the neutralizing MAbs specific for the C-terminal domain did not interfere with the SU-mCAT-1 interaction, they efficiently inhibited cell-to-cell fusion mediated by MuLV Env, indicating that they interfered with a postattachment event necessary for fusion. The C-terminal domain MAbs displayed the highest neutralization titers and binding activities. However, the nonneutralizing PRR-specific MAbs bound to intact virions with affinities similar to those of the neutralizing receptor-binding pocket-specific MAbs, indicating that epitope exposure, while necessary, is not sufficient for viral neutralization by MAbs. These results identify two separate neutralization domains in MuLV SU and suggest a role for the C-terminal domain in a postattachment step necessary for viral fusion.
The murine leukemia virus (MuLV) envelope proteins consist of SU (gp70) and TM (transmembrane [p15E]), two subunits that exist on the virion surface as trimeric complexes (22, 50) of disulfide-linked heterodimers (56). The SU subunit is responsible for binding to the cell surface receptor (10, 14), which for ecotropic MuLV is the cationic amino acid transporter, mCAT-1 (1, 26). Receptor binding by ecotropic SU has been mapped to the amino-terminal 236 amino acids, and this region is therefore called the “receptor binding domain” (RBD) (19). While much of this amino-terminal domain is well conserved among all MuLVs regardless of receptor usage, the RBD contains three variable regions (VRA, VRB, and VRC) that are relatively conserved only among MuLV envs that use the same receptor (13, 27, 46, 63). Structural and mutational studies of the ecotropic RBD have identified a putative receptor-binding pocket in VRA that includes the five residues S84, D86, R85, R97, and W102 (2, 8, 13, 36), consistent with observations that VRA plays a dominant role in receptor choice (41).
Despite considerable effort spent generating MuLV-specific monoclonal antibodies (MAbs), relatively few such MAbs have been well characterized, and these studies have not provided detailed information about the structure and function of MuLV Env (7, 34, 37, 43, 57, 58, 62). Two rat MAbs have been described that have potent neutralization activity against MuLV. 35/56 was isolated from a W/Fu rat immunized with AKR ecotropic MuLV and was type specific for a class of endogenous MuLVs related to Akv (44, 49). Its reactivity strongly correlated with the GIX epitope, originally defined as an inherited Mendelian marker present on thymocytes of certain strains of mice and subsequently shown to be present on endogenously expressed MuLV Env proteins (48, 64). The 35/56 epitope was roughly mapped to the C-terminal domain of gp70 by biochemical fragmentation analysis (55). An independently isolated rat MAb, 83A25, was broadly reactive with a C-terminal epitope present on the envelope glycoproteins of many ecotropic, polytropic, xenotropic, and amphotropic MuLVs (12), but absent from both the Rauscher and Friend isolates.
The present study describes new MAbs specific for sites in the RBD or proline-rich region (PRR) of Friend SU, isolated from mice immunized with a recombinant fusion protein consisting of the first 263 residues of Friend SU joined to the V1/V2 domain of human immunodeficiency virus type 1 (HIV-1) gp120. The transgenic XenoMouse G2 strain, which produces fully human antibodies (20, 39), was used to isolate most of the new MAbs described in this study. These mice have been engineered by functionally inactivating the murine heavy chain and kappa light chain immunoglobulin loci and incorporating megabase-size inserts of human DNA carrying immunoglobulin heavy chain and kappa light chain loci that express the majority of the human antibody repertoire. Although the original impetus of the experiments described here was to generate human MAbs against the HIV-1 domains of these proteins, most of the MAbs generated were directed against epitopes within native MuLV SU. The epitope specificity and functional activity of a number of these novel MuLV SU-specific MAbs, including two directed against a neutralization site in the RBD, are described. In addition, the C-terminal domain epitopes recognized by the neutralizing rat MAbs 35/56 and 83A25 are defined more precisely, and the mechanisms by which these MAbs neutralize MuLV are addressed.
MATERIALS AND METHODS
Purification of recombinant proteins and MAbs.
The recombinant MuLV SU truncation protein and MuLV-HIV-1 fusion proteins were expressed from the human cytomegalovirus promoter as described previously (52). The truncation protein contained the first 263 residues of Friend clone 57 MuLV SU. The MuLV-HIV-1 fusion proteins joined a 96-amino-acid fragment encompassing the V1/V2 domain of the CaseA2 (65) or SF162 (6) isolate of HIV-1 SU to the C terminus of the 263-residue N-terminal fragment of MuLV SU. These recombinant proteins contained a polyhistidine affinity tag that was used to purify these proteins on Ni+2-nitrilotriacetic acid resin, as described previously (52). The purity of the fusion proteins was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining, and their concentration was determined by A280, assuming an extinction coefficient of 1.0.
MAbs were purified from hybridoma cell supernatants with GammaBind G Sepharose or protein A Sepharose Fast Flow (Amersham Pharmacia Biotech AB) according to the manufacturer's recommendations. The purity and concentration of purified MAbs were assayed as described above, except that an extinction coefficient of 1.35 (17) was used. The MuLV SU-specific rat MAbs 35/56 (44) and 83A25 (12) have been described previously. The HIV-1 SU-specific human MAb 5145a (53) and the MuLV p12gag-specific rat MAb 10BA10 (23) were produced in this laboratory. The MuLV TM-specific MAbs 42/114 (16) and 10CE11 (33) have been described previously. Goat polyclonal sera raised to Rauscher MuLV SU or CA were purchased from Quality Biotech, Camden, N.J.
The Friend, Friend S84I, PVC211 (21, 60), and PVC211 D86A RBD proteins were provided by J. Cunningham (Harvard Medical School). Friend S84I has the Friend RBD sequence with an isoleucine substitution for serine at residue 84, and PVC211 D86A is the RBD of the Friend-derived PVC211 clone containing an alanine substitution for aspartate at residue 86 (8). Full-length soluble MuLV SU was purified from virus particles by lectin affinity chromatography essentially as described previously (59). The purity and concentration of this MuLV SU were determined as described above.
Cell lines.
XC rat sarcoma cells, Chinese hamster ovary (CHO), mouse NIH 3T3 fibroblasts, human 293 fibroblasts, mouse Sp2/0 myelomas, and all hybridomas were cultured in cDMEM (Dulbecco's modified Eagle's medium [Gibco] supplemented with 10% fetal bovine serum [Sigma, St. Louis, Mo.] and penicillin-streptomycin [Gibco] at 100 μg/ml) unless otherwise indicated. 293 cells expressing mCAT-1t (293.mCAT) (9) were provided by James Cunningham and were cultured as described above under G418 selection.
Viruses.
NIH 3T3 cells chronically infected with MuLVs of either ecotropic Friend clone 57 (15), Moloney clone 1 (40), AKR.623 (35), or an infectious mutant of Friend clone 57 containing a seven-residue insertion at position 243 of SU (24) were prepared by transfection of cDNA clones of these viruses into NIH 3T3 cells and passage of the cells until infection approached 90% as assayed by immunofluorescence. Culture supernatants from these cell lines, clarified by low-speed (3,000 rpm) centrifugation and filtration through a 0.8-μm-pore-diameter filter (Nalgene), were used as the source for virions. For assays using concentrated virus (enzyme-linked immunosorbent assay [ELISA] and Western blot), the viral supernatant was concentrated by centrifugation at 22,000 × g at 4°C for 2 h followed by resuspension of the pellet in phosphate-buffered saline (PBS).
To produce MuLV-luciferase pseudotypes, 293 cells (2.5 × 106 in a 100-mm-diameter plate) were transfected with a mixture of three plasmids by using Fugene-6 reagent (Roche Biochemicals, Indianapolis, Ind.) as described previously (61). The plasmids were (i) an MuLV gag/pol expression plasmid constructed by cloning an AflII-SacI fragment (nucleotides 642 to 5820) from Friend FB29 MuLV (25) into pcDNA3.1/Zeo (−) (Invitrogen); (ii) an MuLV env expression plasmid consisting of the MuLV env gene (a 2,349-bp XbaI-EcoRV fragment) of interest in pcDNA3.1/Zeo (−); and (iii) a pBABE-luciferase plasmid prepared by moving the luciferase gene from pSP-luc+ (Promega) into pBABE-Puro (42). Forty-eight hours posttransfection, the culture supernatant was harvested and clarified by filtration (0.45-μm pore diameter).
MuLVs bearing Friend 57/AKR.623 chimeric Envs were prepared by utilizing existing or newly created convenient restriction sites in both sequences. Silent mutations were introduced into the AKR.623 sequence via PCR mutagenesis in order to introduce sites present in the Friend 57 sequence. These included modification of an existing AKR BstEII site (G to C at nucleotide 1350) and creation of PstI (T to C at nucleotide 1615 and A to G at nucleotide 1617) and BsaAI (C to G at nucleotide 1254) sites (nucleotide numbering from the A of the initiation codon of AKR.623 env). The following restriction fragments were cloned from this AKR.623 sequence into an EcoRV-PstI fragment of Friend clone 57 env in pSP72 (Promega, Madison, Wis.): (i) AvrII-PstI (amino acids 328 to 514), (ii) AvrII-BsaAI (amino acids 328 to 393), (iii) AvrII-BstEII (amino acids 328 to 426), (iv) BsaAI-BstEII (amino acids 393 to 426), and (v) BstEII-PstI (amino acids 426 to 514). The amino acid numbering begins at the N-terminal alanine of Friend 57 SU, and the chimeras are illustrated in Fig. 1. The resulting plasmids were verified by sequencing. The chimeric fragments were cloned into a two-long terminal repeat MuLV genomic plasmid (24) in pSP72. The resultant proviral plasmids were transfected into NIH 3T3 cells, and MuLV infection was monitored by immunofluorescence.
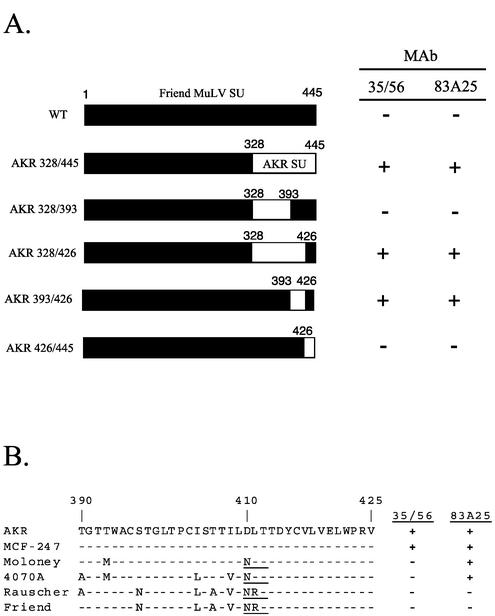
FIG. 1.
Mapping of the 35/56 and 83A25 epitopes. (A) Black rectangles indicate sequences from Friend clone 57 MuLV SU, and white rectangles indicate sequences from AKR.623 MuLV SU. Residue numbers (derived from the Friend sequence) are indicated above the bars. Only the SU sequence is shown. NIH 3T3 cells infected with MuLVs bearing these chimeras were assayed for 35/56 and 83A25 reactivity by immunofluorescence. +, positive reaction between antibody and infected cells; −, no reaction. (B) Sequence alignment for various MuLV SUs from residues 390 to 425. The primary amino acid sequence for AKR.623 is shown at the top. A dash indicates conservation; polymorphisms are indicated by the corresponding residue letter. Reactivity with 35/56 or 83A25, based on data taken from references 12 and 55, is indicated on the right. An N-linked glycosylation site present in some sequences is underlined.
MuLV neutralization assays.
Neutralization of infectious MuLV was quantitated with an immunofluorescence-based strategy essentially as described in reference 23. Percent neutralization was calculated as [1 − (% infection/% infection of control wells)] × 100. The 50% neutralization dose (ND50) is defined as the concentration of MAb that reduced the number of infected cells by 50%.
For luciferase assays, MuLV-luciferase pseudotypes were used to infect NIH 3T3 cells plated the day before at 2 × 104 cells per well in 24-well plates. The NIH 3T3 cells were treated with Polybrene (Sigma; final concentration, 4 μg/ml) prior to infection. A dilution of the MuLV-luciferase pseudotype previously determined to yield ∼20,000 relative light units (RLU) was incubated with serial dilutions of antibody for 1 h at 37°C. The MuLV-luciferase-antibody mixture was added to the cells, incubated overnight, and removed the next day by aspiration and replaced with cDMEM. Three days postinfection, the cells were harvested for measurement of luciferase as previously described (28). All samples were tested in triplicate. Percent neutralization is calculated as [1 − (RLU of treated wells/RLU of control wells)] × 100.
Immunizations and production of MAbs.
For immunizations, the recombinant MuLV(1-263)/HIV-V1/V2 SU proteins were formulated in RAS monophosphoryl lipid A-Squalene adjuvant (RIBI Immunochemicals, Inc.), and animals were primed by subcutaneous injection at a dose of 25 μg/kg of body weight. Booster immunizations consisting of 5-μg/kg doses of the same antigen and adjuvant were administered subcutaneously at 3-week intervals. Animals were bled 7 to 8 days postboost, and the titers were determined by ELISA against the recombinant SU protein used for the immunogen.
Four days following a final intraperitoneal booster immunization with the antigen in PBS, animals were sacrificed, and splenocytes collected by gentle disruption of lymphoid tissue in DMEM. The splenocytes were combined with Sp2/0 mouse myeloma cells and fused by stirring in a solution of 50% polyethylene glycol 1450-10% dimethyl sulfoxide (Sigma) in PBS. The fused cells were washed with DMEM and resuspended in a rich selection medium—HAT selection media supplemented with recombinant interleukin 6 (Roche Biochemicals) and OPI (Sigma) (17)—and then transferred to 96-well culture plates (typically 30 plates per fusion). When the colonies were visible by eye, culture supernatants were screened for immunogen-specific antibodies by ELISA. Positive hybridomas were passaged and rescreened. Hybridomas that remained positive were cloned by limiting dilution.
Immunoassays.
ELISAs (53) and Western blot assays (54) were performed essentially as described previously. Fluorescence-activated cell sorting (FACS) assays to detect MuLV binding were based on a previously published protocol (68). 293 or 293.mCAT cells were detached with trypsin-EDTA (Celltech), and a total of 5 × 105 cells were incubated with 1 ml of pseudotyped MuLV particles at 4°C for 2 h with gentle agitation. Cells were washed with 1 ml of ice-cold 10% fetal bovine serum in PBS (FACS buffer) and resuspended in polyclonal goat anti-Rauscher SU serum diluted 1:250 in FACS buffer. Cells were incubated for 1 h at 4°C, washed again, and incubated with fluorescein isothiocyanate (FITC)-conjugated anti-goat immunoglobulin G (Zymed) at 5 μg/ml in FACS buffer. After a half-hour incubation at 4°C, cells were washed again, fixed in 4% paraformaldehyde in PBS, and analyzed by flow cytometry with a FacsCalibur flow cytometer (Becton Dickson) and associated software. For MuLV binding inhibition, MuLV pseudotypes were incubated with anti-SU antibodies for 1 h at 37°C prior to addition to receptor-expressing cells.
Assay of MuLV-induced fusion.
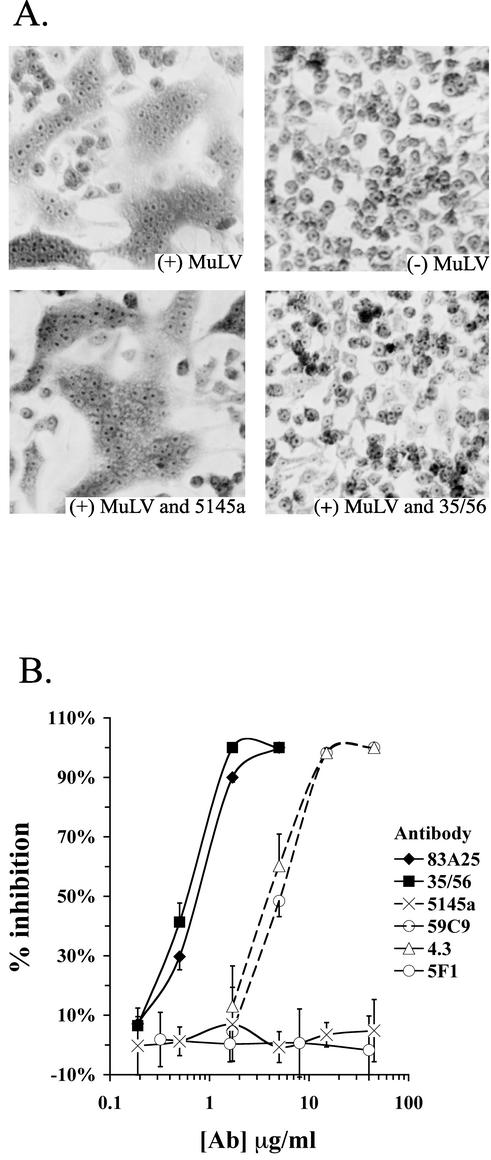
Syncytium formation of XC cells was assayed essentially as described previously (49). Briefly, the MuLV supernatant was incubated with various concentrations of antibody for 1 h at 37°C prior to addition to XC cell monolayers. After 2.5 h, the cells were washed, fixed with methanol, and stained with cresyl violet. Syncytia were quantitated by counting the number of fused cells containing more than four nuclei in four equal areas, representing ∼1,500 cells for each data point.
MuLV SU binding assays.
Purified MuLV SU (50 μg of total protein) was labeled with 125I by using a Bolton-Hunter reagent kit (ICN). A typical labeling reaction resulted in SU with a specific activity of 103 cpm/ng. For binding analysis, 30 ng of labeled SU was added to 5 × 105 293.mCAT cells and incubated for 1.5 h at room temperature. The cells were pelleted and washed three times with DMEM plus 5% fetal bovine serum to remove unbound label. Bound SU was then quantitated with a gamma counter.
Immunoprecipitation.
Radiolabeled immunoprecipitation and SDS-PAGE assays were performed essentially as described previously (51). For virion precipitation assays, supernatant medium containing labeled virions was first applied to a Sephacryl S-1000 Superfine (Pharmacia) column to remove any soluble SU from intact virions. Virions were collected in the void volume and used in the precipitation assay, with or without prior solubilization with 0.5% Nonidet P-40 (NP-40). Quantitation of labeled MuLV CA was performed with a Storm 850 PhosphorImager and ImageQuant software (Molecular Dynamics).
RESULTS
Epitope mapping of C-terminal MAbs 35/56 and 83A25.
Two potently neutralizing rat MAbs, 35/56 and 83A25, have previously been shown to recognize epitopes in the C-terminal domain of SU (12, 55). The disulfide dependence of their epitopes was demonstrated by Western blotting, and the reactivity of these MAbs with SU from AKR.623 and Friend clone 57 virions was analyzed by ELISA (Table 1). As previously reported, both 35/56 and 83A25 reacted with ecotropic AKR.623 SU, while neither MAb reacted with Friend clone 57. Taking advantage of this specificity, chimeras between Friend and AKR SUs were used to map determinants of the 35/56 and 83A25 epitopes. Segments of the Friend SU C-terminal domain were replaced with homologous sequences from AKR.623, and the resultant chimeric SUs were assayed for biological function and immunoreactivity. Each of the chimeric Envs was able to mediate MuLV infection as efficiently as wild-type Friend Env (4). Substitution of residues 393 to 426 from AKR.623 was sufficient for expression of both the 35/56 and 83A25 epitopes (Fig. 1A), indicating that their type specificities were determined by sequences in this region and suggesting that both MAbs were directed against sites in this region of SU. A sequence comparison of this region from a number of SUs that had been typed for these two epitopes revealed several polymorphisms that may account for the differential expression of these epitopes (Fig. 1B). The closest sequence to AKR.623 and MCF-247, the only isolates reactive with 35/56, was Moloney ecotropic clone 1. There were only two differences among these sequences in this region: a T-to-M change at 393 and a D-to-N change at 410, which generated an N-linked glycosylation site in Moloney SU that was absent in the AKR protein. The T393 residue of AKR.623 was present in two of the nonreactive sequences (Friend and Rauscher), while the D residue at 410 was replaced by N for all of the nonreactive sequences. This suggests either that D410 was necessary for the 35/56 epitope or that the creation of the N-linked glycosylation site by the D-to-N substitution at this position resulted in blocking of this epitope. For 83A25, three consistent differences were found between the reactive and nonreactive sequences, at positions 397 (S versus N), 406 (T versus A), and 411 (L versus R). More complete mapping of these epitopes will be presented elsewhere.
TABLE 1.
Characterization of SU-specific MAbs in this study
| MAb | MuLV recognized by MAba | Reactive with reduced SUb | Neutralization activityc |
|---|---|---|---|
| 35/56 | AKR, MCF 247 | No | + |
| 83A25 | Broadly reactive | No | + |
| 4.3 | Friend 57 | No | + |
| 59C9 | Friend 57 | No | + |
| 3.3 | Friend 57 | Yes | − |
| 2.1 | Friend 57 | Yes | − |
| 2.4 | Friend 57 | Yes | − |
| 4A1 | Friend 57 | Yes | − |
| 11B6 | Friend 57 | Yes | − |
| 13B3 | Friend 57 | Yes | − |
| 5F1 | Friend 57 | Yes | − |
| 28G12 | Friend 57 | Yes | − |
| J16 | Friend 57 | Yes | − |
| M19 | Friend 57 | Yes | − |
| 3.2 | Friend 57, Moloney clone 1 | Yes | − |
| 3.5 | Friend 57, Moloney clone 1 | Yes | − |
| 3.6 | Friend 57, Moloney clone 1 | Yes | − |
35/56 reactivity was taken from reference 55, and 83A25 reactivity was taken from reference 12. Reactivity of new MAbs was assayed against pelleted Friend clone 57, Moloney clone 1, AKR.623, and amphotropic (4070A) and polytropic MCF.247 MuLVs by ELISA.
All MAbs were assayed against reduced and nonreduced Friend clone 57 SU (novel MAbs) or AKR.623 SU (35/56 and 83A25) by Western blotting.
As determined by immunofluorescence-based MuLV neutralization assay.
Generation of new MuLV SU-specific MAbs.
A recombinant protein consisting of the N-terminal 263 amino acids of the Friend MuLV SU joined to the V1/V2 domain of the HIV-1 SU protein, MuLV(1-263)/HIV-V1/V2 (25), was used to immunize rats and XenoMouse G2 animals. The resulting immune response was monitored by ELISA versus the SU fusion protein used as the immunogen. Typically, serum titers plateaued after three to four booster immunizations (data not shown). Hybridomas were generated and screened by ELISA initially against MuLV(1-263)/HIV-V1/V2 to identify hybridomas producing MAbs against the immunogen and then against MuLV(1-263) and MuLV(1-33)/HIV-V1/V2 proteins to map the epitopes to either the MuLV or HIV SU sequences.
The MuLV-derived region of the immunogen was immunodominant: of 38 hybridomas isolated, thirty two (84%) were MuLV specific (4). The six MAbs specific for the HIV-V1/V2 sequence did not react with native HIV-1 SU by ELISA (data not shown), indicating that they were specific for epitopes unique to the immunogen or for nonnative structures of the HIV-1 V1/V2 domain. In contrast with these results, immunization of XenoMouse G2 animals with a recombinant HIV-1 SU did yield a large number of HIV-1 SU-specific MAbs, including MAbs against native V1/V2 epitopes (18).
Characterization of MuLV SU-specific MAbs.
The MuLV(1-263)-specific MAbs all reacted with native SU protein purified from Friend MuLV virions (4). All of the MAbs reacted with intact Friend MuLV by ELISA, and three antibodies (3.2, 3.5, and 3.6) also recognized Moloney ecotropic virions (Table 1). None of the antibodies reacted with AKR.623, amphotropic 4070A, or polytropic MCF-247 virions, nor did they react with control material from uninfected NIH 3T3 cells. All of the antibodies reacted with SU in Western blot assays, as did a commercial polyclonal serum raised to MuLV SU (24). Two antibodies, 4.3 and 59C9, recognized epitopes sensitive to reduction of disulfide bonds by dithiothreitol (DTT), while the reactivity of the remaining XenoMouse MAbs was not affected by DTT treatment (Table 1).
Epitope mapping of novel SU-specific antibodies.
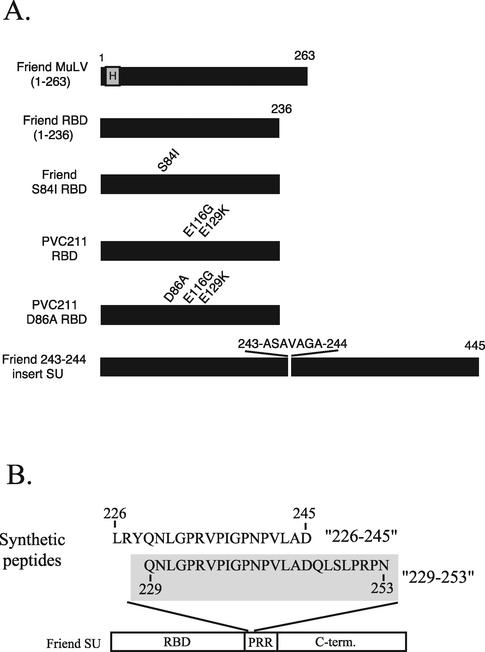
The epitopes recognized by the MAbs generated in this study were mapped against a panel of recombinant proteins and peptides based on Friend clone 57 SU and a functional mutant Friend SU protein with a seven-residue insert at position 243-244 (Fig. 2 and Table 2).
FIG. 2.
Antigens used for ELISAs to map SU-specific MAbs. (A) Black rectangles represent sequences derived from MuLV SU, and the letter H represents a six-histidine affinity tag. Amino acid numbers are given above the bars, and single-amino-acid differences from the Friend clone 57 sequence are indicated. The Friend SU bearing an insert between residues 243 and 244 has been described previously (24); viruses bearing this mutation are infectious. (B) Two synthetic peptides based on the Friend clone 57 MuLV SU PRR are shown: a 20-mer, 226-245, and a 25-mer, 229-253, which is shaded. The sequences are aligned, and the corresponding residue numbers of Friend SU are indicated. A schematic of Friend SU below the peptides shows the domain organization of SU. C-term., C-terminal domain.
TABLE 2.
Mapping the epitopes of the N-terminal anti-SU MAbs
| Antibody classification | OD405
|
||||||
|---|---|---|---|---|---|---|---|
| Friend MuLV (1-263)a | Friend RBD (1-236)a | Friend S84I RBDa | PVC211 D86A RBDa | 229-253 peptidea | 226-245 peptidea | 243-244 insert Envb | |
| RBD specific, neutralizing | |||||||
| 4.3 | 1.1 | 1.5 | 0.0 | 1.3 | 0.0 | 0.0 | + |
| 59C9 | 1.4 | 2.1 | 0.0 | 1.7 | 0.0 | 0.0 | + |
| RBD specific, nonneutralizing | |||||||
| 2.1 | 0.9 | 1.8 | 1.6 | 1.9 | 0.1 | 0.0 | + |
| 3.2 | 1.1 | 1.4 | 1.1 | 1.3 | 0.0 | 0.0 | + |
| 3.5 | 0.5 | 0.9 | 0.8 | 1.0 | 0.2 | 0.0 | + |
| 3.6 | 0.8 | 1.2 | 1.1 | 1.3 | 0.0 | 0.0 | + |
| J16 | 1.8 | 1.3 | 1.0 | 1.4 | 0.0 | 0.0 | + |
| PRR specific, nonneutralizing | |||||||
| 2.4 | 1.8 | 0.2 | 0.2 | 0.2 | 1.4 | 0.0 | − |
| 3.3 | 2.9 | 0.1 | 0.1 | 0.2 | 2.8 | 0.0 | − |
| 4A1 | 2.8 | 0.0 | 0.0 | 0.0 | 1.8 | 0.0 | − |
| 5F1 | 2.5 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | − |
| 28G12 | 2.3 | 0.0 | 0.0 | 0.0 | 2.3 | 0.0 | − |
| 11B6 | 2.4 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | − |
| 13B3 | 2.2 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | − |
| Polyclonal anti-SU serum | 1.7 | 1.6 | NDc | 1.3 | 0.8 | 1.8 | + |
Antibodies were used in an ELISA at 1 μg/ml. Values (optical density at 405 nm [OD405]) are the averages of triplicate samples.
Antibody reactivities with 3T3 cells infected with MuLV bearing the 243-ASAVAGA-244 insertion were assayed via immunofluorescence.
ND, not determined.
By virtue of their reactivity with these antigens, the new MAbs could be classified into three groups (Table 2). The two disulfide-dependent MAbs, 4.3 and 59C9, were both sensitive to the S84I substitution in Friend RBD, but not to the D86A substitution in PVC211 RBD, suggesting that S84 is a contact site for these antibodies. The S84 and D86 residues have been identified as components of the binding pocket for the mCAT-1 receptor (8). It is possible that the S84I mutation caused gross misfolding of Env and that this accounted for the lack of recognition by these MAbs. However, it has been reported that this mutant Env was processed normally and incorporated into virions at wild-type levels, suggesting that the S84I mutant was properly folded (8). These data indicated that the 59C9 and 4.3 antibodies recognized the same or highly related discontinuous epitopes, which possibly included at least one residue involved in the binding of SU to its receptor. Because these two MAbs had neutralizing activity against Friend clone 57 MuLV (see below), this group was termed “RBD specific, neutralizing.”
Five other MAbs were also RBD specific, in that their reactivities were not affected by truncation at residue 236 or by the 7-amino-acid insertion at residue 243 (Table 2). Unlike the first group, the reactivities of these antibodies were not affected by the S84I substitution, and they all reacted with reduced SU protein (Table 1). These MAbs did not neutralize MuLV (see below), and thus this group was termed “RBD specific, nonneutralizing.”
Reactivity by the third class of MAbs was abolished by the truncation of the N-terminal domain of SU at residue 236 and by the insertion between residues 243 and 244 (Table 2). These data indicated that sequences located in the PRR between residues 236 and 263 contributed to these epitopes and suggested that they spanned residues 243 and 244. The MAb panel was tested for reactivity against two overlapping synthetic peptides based on the Friend clone 57 sequence (Fig. 2B). As expected, none of the RBD-specific MAbs reacted with either peptide. The remaining seven MAbs all recognized the 229-253 peptide (Table 2), corresponding to the N-terminal portion of the PRR, but not the overlapping 226-245 peptide, indicating an important role for residues 246 to 253 in the structure of this epitope. Because none of the MAbs that recognized the 229-253 peptide neutralized MuLV, this group of MAbs was termed “PRR specific, nonneutralizing” (Table 2).
Neutralization of Friend MuLV by SU-specific antibodies.
The complete panel of novel MAbs was assayed for neutralization against Friend clone 57 MuLV by using a previously described immunofluorescence assay (4). Only two of these MAbs, 59C9 and 4.3, had neutralizing activity, both with ND50s of approximately 1 μg/ml. These two MAbs were both specific for disulfide-dependent epitopes in the RBD and dependent on the S84 residue.
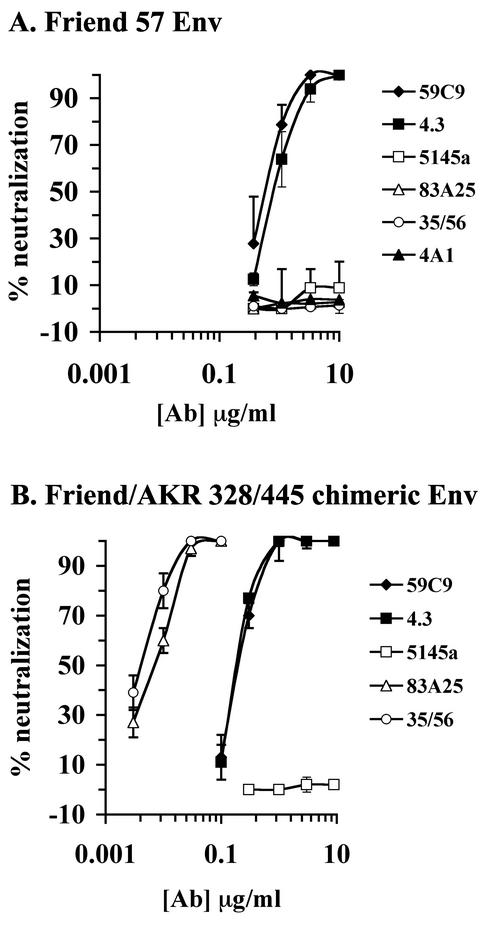
Neutralization by these antibodies was also assayed with MuLV luciferase reporter viruses bearing either wild-type Friend clone 57 Env or a chimeric Env comprising the first 328 residues from Friend clone 57 with the remaining residues from AKR.623 MuLV (Friend/AKR 328/445). This chimera expressed both the N-terminal epitopes recognized by the panel of new MAbs and the C-terminal epitopes recognized by 35/56 and 83A25, allowing direct comparison of the neutralization potencies of these MAbs. In the luciferase assay, 59C9 and 4.3 neutralized the Friend clone 57-pseudotyped MuLV with ND50s of 0.6 and 0.8 μg/ml, respectively (Fig. 3A). These MAbs also neutralized virus pseudotyped with the Friend/AKR 328/445 Env, which was approximately fourfold more sensitive to 59C9 and 4.3 than the Friend pseudotype (ND50s of 0.2 μg/ml) (Fig. 3B). 35/56 and 83A25 did not neutralize the Friend clone 57 pseudotype (consistent with the lack of reactivity of these MAbs for this Env), but neutralized the Friend/AKR 328/445 pseudotype very potently, with ND50s of 0.005 and 0.008 μg/ml, respectively. This was 25- to 40-fold-less antibody than required for an equivalent level of neutralization by the 59C9 and 4.3 MAbs (Fig. 3B). No neutralization was observed with a PRR-specific MAb (4A1) or an irrelevant HIV-1-specific MAb (5145a).
FIG. 3.
Neutralization of MuLV by SU-specific MAbs. Neutralization assays for the indicated MAbs were performed with MuLV-luciferase-encoding virions pseudotyped with Friend clone 57 (A) or Friend/AKR 328/445 chimeric Env (B). Values are averages of triplicate samples.
Mechanisms of neutralization: inhibition of SU binding to receptor.
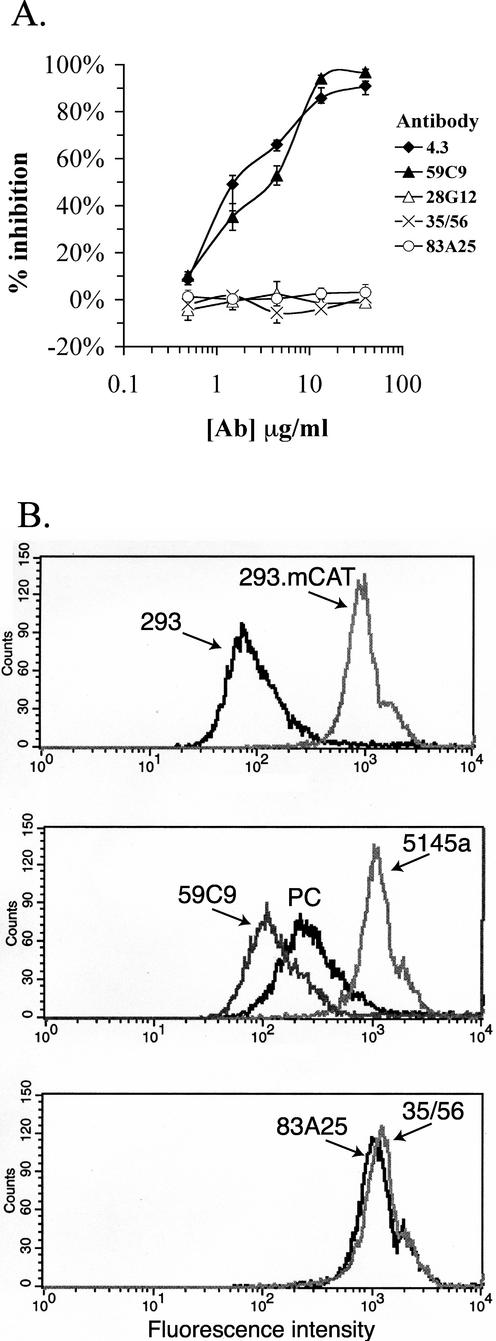
The involvement of a residue within the receptor-binding pocket in the reactivity of the novel neutralizing MAbs suggested that these antibodies might interfere with the interaction between SU and the mCAT-1 receptor. To test this hypothesis, a binding assay utilizing soluble SU was established. Friend/AKR 328/445 SU purified from MuLV virions by lectin affinity chromatography was labeled with 125I. The labeled SU was reactive with all MAbs used in the binding inhibition assay and bound specifically to 293 cells expressing mCAT-1 (data not shown). 59C9 and 4.3 effectively blocked binding of SU to 293.mCAT cells (50% inhibitory doses [ID50s] of 2 to 4 μg/ml; Fig. 4A), while the C-terminal-neutralizing MAbs and the PRR-specific MAb 28G12 did not block binding at concentrations as high as 40 μg/ml.
FIG. 4.
Inhibition of MuLV SU binding to mCAT-1 by MAbs. (A) Purified Friend/AKR 328/445 chimeric SU labeled with 125I was incubated with the MAbs at the indicated concentrations for 1 h at 37°C and then assayed for the ability to bind 293.mCAT cells. The results are presented as percent inhibition of the binding level in the absence of MAb. Values are averages of triplicate samples. (B) MuLV pseudotyped with the Friend/AKR 393/426 chimeric Env was incubated with 293 or 293.mCAT cells (top panel) for 1 h at 37°C, and virion binding was detected by FACS. The MuLV particles were pretreated with 59C9, 5145a, or polyclonal anti-SU serum (PC) (middle panel) or 83A25 or 35/56 (bottom panel) for 1 h at 37°C and then assayed for binding as described above. The MAbs were used at 20 μg/ml, and the serum was used at a 1:100 dilution.
A FACS-based virion-binding assay was used in order to determine whether the MAbs that inhibited binding of SU to mCAT-1 also blocked binding of intact virions to cells. The pseudotyped MuLV used in these experiments bound specifically to mCAT-1-expressing cells (Fig. 4B). Pretreatment of the virus with SU-specific polyclonal serum reduced binding moderately, while treatment with 59C9 at 20 μg/ml reduced binding of MuLV to near the background level obtained for 293 cells not expressing mCAT-1. However, pretreatment of virions with the C-terminal MAbs 35/56 and 83A25 or a negative control MAb, 5145a, at the same concentration had no inhibitory effect in this assay. These data indicated that inhibition of receptor binding accounted for neutralization by the binding pocket-specific MAbs 59C9 and 4.3, but not for neutralization by the C-terminal domain-specific MAbs 35/56 and 83A25.
Mechanisms of neutralization: inhibition of MuLV-induced fusion.
The inability of the C-terminal MAbs to block the binding of SU or virions to receptor suggested that their neutralizing activity was due to interference with a later step in Env function. Their ability to inhibit membrane fusion induced by MuLV was therefore tested with a syncytium formation assay. When MuLV particles are added to monolayers of rat XC cells, syncytia are formed (Fig. 5A). This fusion was not dependent on viral replication, since syncytia were counted shortly after addition of virus, and thus this assay measured fusion from without. Both virus-cell and cell-cell fusions are required for syncytium formation. Titration of the neutralizing MAbs in this assay revealed that the C-terminal MAbs 35/56 and 83A25 inhibited syncytium formation considerably more potently than the N-terminal-neutralizing MAbs (Fig. 5B), mirroring their performance in neutralization assays. Neither a PRR-reactive MAb (5F1) nor an anti-HIV-1 MAb (5145a) showed inhibition in this assay. The fact that the C-terminal MAbs 35/56 and 83A25 blocked fusion but not binding of SU or virions to cells expressing mCAT-1 indicated that their efficient neutralization involved the blocking of a step between virion attachment and membrane fusion.
FIG. 5.
Inhibition of syncytium formation by MAbs. (A) Viral supernatant from NIH 3T3 cells infected with MuLV bearing the Friend/AKR 393/426 Env was incubated with or without MAbs for 1 h at 37°C. The supernatant was then used to overlay XC cells, and syncytium formation was analyzed after a 2.5-h incubation at 37°C. The MAbs were used at 15 μg/ml. (B) The indicated MAbs were titrated for their ability to inhibit syncytium formation as described in panel A. After fixation and staining of syncytia, fused nuclei were counted in four equal areas for each data point, and this number was used to calculate a percent inhibition relative to no antibody treatment.
Nonneutralizing SU-specific MAbs can also bind MuLV virions.
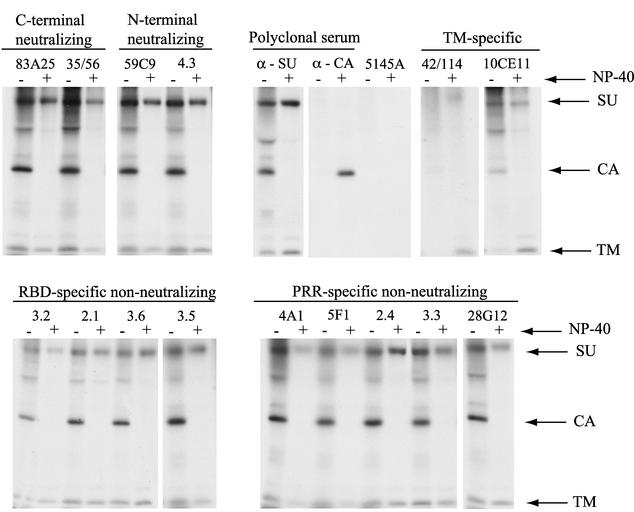
The MAbs isolated in this study were raised to and assayed against soluble SU proteins. The lack of neutralization activity found for the majority of these MAbs might therefore have been due to weak affinity for the native Env oligomers present on MuLV virions. This has been proposed as the explanation for MAbs that fail to neutralize HIV-1 (5, 47). To test this hypothesis, the ability of anti-SU MAbs to bind intact MuLV virions was examined in an intact virion precipitation assay. This assay is based on the detection of the internal viral capsid protein (CA [p30]) after immunoprecipitation of intact, purified virions by anti-Env MAbs (Fig. 6).
FIG. 6.
Precipitation of intact virions with Env-specific MAbs. Culture supernatants from NIH 3T3 cells infected with MuLV bearing the chimeric Friend/AKR 393/426 Env labeled with [35S]methionine and cysteine were immunoprecipitated in the presence (+) or absence (−) of NP-40 with the MAbs (20 μg/ml) or polyclonal sera indicated. Precipitates were analyzed by SDS-PAGE and autoradiography. TM refers to the transmembrane subunit of Env, p15E, and CA refers to the MuLV capsid protein, p30. The presence of a CA band is indicative of binding of the SU-specific MAbs to intact virions.
All of the SU-specific MAbs precipitated intact virions to some degree, while a negative control MAb (5145a) did not (Fig. 6). When virions were first lysed with detergent, no CA was precipitated by any of the anti-Env reagents tested, demonstrating that CA precipitation in the absence of detergent was dependent on the binding of the MAbs to the surface of intact virions. The strongest virion precipitation activity was seen for the neutralizing MAbs and the PRR-specific nonneutralizing MAbs. The RBD-specific nonneutralizing group of MAbs precipitated virions less efficiently. Two TM-specific MAbs that do not neutralize MuLV precipitated intact virions very poorly (10CE11) or not at all (42/114), indicating that their respective epitopes were at best slightly exposed on virions. After disruption of the viral membrane with detergent, all of the SU-specific MAbs efficiently precipitated SU, and the TM-specific MAbs precipitated TM (Fig. 6). These data indicated that the inability of the PRR-specific, nonneutralizing MAbs to neutralize MuLV was not due to an inability of these antibodies to bind to virions.
Relative binding activities of MAbs for intact virions and monomeric SU.
The relative affinities of the MAbs used in this study for monomeric MuLV SU were then assayed by titration against purified MuLV SU by ELISA (4). With the exception of 83A25 (whose affinity was four- to sevenfold higher than those of the other MAbs), MAbs in all groups possessed similar affinities (within twofold) for monomeric MuLV SU, defined as the MAb concentration that gave half-maximal binding (Table 3). This indicated that the low affinity of the RBD-specific, nonneutralizing MAbs for virions was due to poor exposure of their epitopes on native Env multimers, not to an inherent low affinity of these MAbs for SU.
TABLE 3.
Relative affinities and neutralization titers of anti-SU MAbs
| MAb and classification | MAb concn (nM) yielding half-maximal binding to:
|
ND50c | |
|---|---|---|---|
| MuLV virionsa | MuLV SUb | ||
| RBD specific, neutralizing | |||
| 59C9 | 1.3 | 0.8 | 1.1 |
| RBD specific, nonneutralizing | |||
| 2.1 | 11.0 | 1.5 | >230 |
| 3.6 | 5.7 | 0.9 | >230 |
| PRR specific, nonneutralizing | |||
| 3.3 | NDd | 1.1 | >230 |
| 5F1 | 1.7 | 0.9 | >230 |
| C terminal, neutralizing | |||
| 35/56 | 0.3 | 0.8 | 0.03 |
| 83A25 | 0.1 | 0.2 | 0.04 |
Determined by PhosphorImager analysis of virion precipitation.
Determined by ELISA against purified SU.
Concentration of MAb that reduced infectivity by 50% (determined inMuLV-luciferase neutralization assays).
ND, not determined.
The relative affinities of the various MAbs for intact virions were then examined more quantitatively by titrating MAbs from each group for their ability to precipitate CA and determining the concentration that yielded half-maximal precipitation (Table 3). The affinities of the C-terminal-specific MAbs for virions were roughly 5- to 10-fold greater than that of the RBD-specific, neutralizing MAb, 59C9. This was consistent with, and may account for, the more potent neutralizing activities of the C-terminal-specific MAbs. The affinity of the PRR-specific, nonneutralizing MAb 5F1 was only slightly lower than that of 59C9, confirming that the inability of this antibody to neutralize Friend MuLV was not due to low affinity of this MAb for intact virions. The affinities of MAbs 2.1 and 3.6 for virions were 5- to 10-fold less than that of 59C9, suggesting that low affinity accounted for the lack of detectable neutralization by MAbs from the RBD-specific, nonneutralizing group.
DISCUSSION
This study describes a number of new MAbs, isolated from a transgenic strain of mice engineered to produce human antibodies, that were directed against defined sites in the RBD or PRR of MuLV SU. The neutralizing activities and neutralization mechanisms of these antibodies and those of two previously described rat MAbs directed against sites in the C-terminal domain of SU were studied. The two RBD-specific, neutralizing MAbs, 4.3 and 59C9, mapped to a disulfide-dependent epitope that included residue 84, an element of the binding pocket for the mCAT-1 receptor. Binding studies performed with both soluble SU (4.3 and 59C9) and intact viral particles (59C9) indicated that these MAbs neutralized MuLV by blocking the binding interaction between SU and the mCAT-1 receptor. Critical residues for the binding of two C-terminal-specific rat MAbs, 35/56 and 83A25, were mapped to one or more of six polymorphisms in a 15-residue region including the C-terminal disulfide-bonded loop of SU. These MAbs neutralized MuLV with approximately 25-fold-greater potency than the RBD-specific MAbs; this increased potency may be related to a higher affinity of these MAbs for intact virions. Binding studies demonstrated that this potent neutralization was not mediated by interference with receptor binding, but by blockage of an undefined postattachment step necessary for fusion between the viral and cellular membranes.
These experiments also identified at least three other SU epitopes not involved in neutralization of MuLV. Two epitopes were broadly mapped to the RBD, whose respective MAbs were termed the “RBD-specific, nonneutralizing group” (Table 1). One epitope was represented by MAbs 2.1 and J16, which recognized Friend clone 57, but not Moloney or AKR.623 MuLV (Table 1). The other RBD epitope was recognized by MAbs 3.2, 3.5, and 3.6, which reacted with the Friend and Moloney MuLV sequences, but not AKR (Table 1). The data presented here did not exclude the possibility that these RBD-specific, nonneutralizing MAbs recognized a larger number of distinct epitopes not resolved by these experiments. The lack of neutralizing activity by these MAbs correlated with their low affinity for intact virions, which presumably was due to poor exposure of their epitopes on the surface of intact virions (Table 3).
A third distinct epitope defined by the novel MAbs was a linear sequence centered around residues 246 to 253 in the PRR of SU. Despite good exposure of this region on the surface of intact virions, these MAbs did not possess detectable neutralizing activity. This demonstrates that surface exposure is not a sufficient criterion for a site on SU to act as a neutralization epitope. This region of the PRR is the most variable region of SU, even among viral isolates of the same receptor class, and tolerates large insertions and deletions with little or no deleterious effects on SU function (24, 66). About 50% of the MAbs isolated in these experiments mapped to this epitope cluster, suggesting that this region was highly immunogenic. These results suggest that one function of these sequences may be to act as an immune decoy to divert the humoral response from conserved sites that are critical for function.
The dependence of both 35/56 and 83A25 binding on a small C-terminal region from AKR.623 MuLV mapped determinants of the epitopes of these MAbs to the same small region. This result was unanticipated because of the strikingly different strain distributions of these two epitopes. The similar potencies and mechanisms of neutralization by these two MAbs were consistent with their recognition of a common functional domain.
The mechanistic studies described in this paper clearly demonstrated that the two C-terminal-specific MAbs did not block binding of virions to the mCAT-1 receptor, but nonetheless interfered with the subsequent fusion reaction. Although there is a view that virus neutralization is mediated only by steric interference of attachment (5), postattachment neutralization has previously been reported for other enveloped viruses, including HIV-1, bovine leukemia virus, and influenza virus (11, 38, 45), and thus is not unique to MuLV.
The C-terminal region that determines the 35/56 and 83A25 epitopes is not particularly hydrophobic and does not possess any other obvious structural feature that would account for a direct role in fusion. It is possible that these epitopes require additional nonpolymorphic residues that, considering the conformational nature of these epitopes, might lie outside of the identified region. These other residues might be directly involved in the fusion reaction. Alternatively, the region involved in fusion may be a site distinct from these epitopes that is sterically or allosterically affected by binding of these MAbs to their targets. Rather than directly interfering with the fusion reaction, binding of these MAbs might disrupt a step in the transduction of a signal from the RBD through the C-terminal domain of SU that activates TM for fusion. Relevant to this are recent studies suggesting that complex interactions between the N-terminal region, PRR, and C-terminal domains of SU are involved in regulating the fusogenicity of the Env complex (3, 30-32). Another possibility is that binding of these antibodies interferes with the interaction between SU and an unidentified cell surface component that functions as a second receptor.
Additional information about the structure of these epitopes and that of the overall protein domain surrounding these epitopes is needed to further determine their roles in viral infection and neutralization. Particularly useful for addressing these questions would be the determination of the three-dimensional structure of the entire SU protein and its associated TM regions. Crystallization of membrane proteins is often facilitated by forming complexes with specific antibody fragments that stabilize individual domains and interdomain interactions (29, 67). The availability of well-characterized MAbs against defined conformational epitopes located at distinct domains of SU could provide useful tools for crystallizing MuLV SU. In addition to determining the structure of SU, such crystals would also provide the precise definition of the molecular features of these epitopes, which should help elucidate their roles in neutralization.
Acknowledgments
We thank the laboratories of James Cunningham for providing RBD proteins and the 293.mCAT-1 cell line; Frank Malik and Ulrich Hammerling for providing the 83A25 and 35/56 hybridoma cell lines, respectively; and Jose Corvalan, Abgenix, for providing some of the MAbs used for this study.
These studies were aided by grant AI46283 from the Public Health Service to A.P.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhart, M. D. 2002. A study of retroviral neutralization by monoclonal antibodies specific for the murine leukemia virus envelope subunit SU. Ph.D. dissertation. New York University, New York, N.Y.
- 5.Burton, D., E. Saphire, and P. Parren. 2001. A model for neutralization of viruses based on antibody coating of the virion surface, p. 109-143. In D. Burton (ed.), Antibodies in viral infection. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 6.Cheng-Mayer, C., C. Weiss, D. Seto, and J. A. Levy. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. USA 86:8575-8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro, B., K. Wehrly, M. Cloyd, W. Britt, J. Portis, J. Collins, and J. Nishio. 1981. Characterization of mouse monoclonal antibodies specific for Friend murine leukemia virus-induced erythroleukemia cells: Friend-specific and FMR-specific antigens. Virology 112:131-144. [DOI] [PubMed] [Google Scholar]
- 8.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol. 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLarco, J. E., and G. J. Todaro. 1976. Membrane receptors for murine leukemia viruses: characterization using the purified viral envelope glycoprotein gp71. Cell 8:365-371. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, M. J., and N. J. Dimmock. 2001. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. J. Virol. 75:10208-10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, L. H., R. P. Morrison, F. G. Malik, J. Portis, and W. J. Britt. 1990. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J. Virol. 64:6176-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 14.Fowler, A. K., D. R. Twardzik, C. D. Reed, O. S. Weislow, and A. Hellman. 1977. Binding characteristics of Rauscher leukemia virus envelope glycoprotein gp71 to murine lymphoid cells. J. Virol. 24:729-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friend, C. 1957. Cell-free transmission in adult Swiss mice of a disease having the character of a leukemia. J. Exp. Med. 105:302-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerling, U., A. Pinter, and P. V. O'Donnell. 1980. Monoclonal antibodies as a tool in research on oncogenic viruses, p. 255-256. In G. A. Beth (ed.), The role of viruses in human cancer. Elsevier, Amsterdam, The Netherlands.
- 17.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 139-242. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.He, Y., W. J. Honnen, C. P. Krachmarov, M. Burkhart, S. C. Kayman, J. Corvalan, and A. Pinter. 2002. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human IgG loci. J. Immunol. 169:595-605. [DOI] [PubMed] [Google Scholar]
- 19.Heard, J. M., and O. Danos. 1991. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 65:4026-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakobovits, A. 1995. Production of fully human antibodies by transgenic mice. Curr. Opin. Biotechnol. 6:561-566. [DOI] [PubMed] [Google Scholar]
- 21.Kai, K., K. Mitsuno, N. Goto, Y. Ami, S. Ando, and M. Kanoe. 1996. Factors affecting induction of neurological disorders in mice by paralysis-inducing Friend-related PVC viruses. J. Vet. Med. Sci. 58:285-290. [DOI] [PubMed] [Google Scholar]
- 22.Kamps, C., Y. C. Lin, and P. K. Wong. 1991. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology 184:687-694. [DOI] [PubMed] [Google Scholar]
- 23.Kayman, S. C., R. Kopelman, S. Projan, D. M. Kinney, and A. Pinter. 1991. Mutational analysis of N-linked glycosylation sites of Friend murine leukemia virus envelope protein. J. Virol. 65:5323-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kayman, S. C., H. Park, M. Saxon, and A. Pinter. 1999. The hypervariable domain of the murine leukemia virus surface protein tolerates large insertions and deletions, enabling development of a retroviral particle display system. J. Virol. 73:1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J., E. I. Closs, L. M. Albritton, and J. M. Cunningham. 1991. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature 352:725-758. [DOI] [PubMed] [Google Scholar]
- 27.Koch, W., G. Hunsmann, and R. Friedrich. 1983. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J. Virol. 45:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krachmarov, C. P., S. C. Kayman, W. J. Honnen, O. Trochev, and A. Pinter. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1737-1748. [DOI] [PubMed] [Google Scholar]
- 29.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavillette, D., B. Boson, S. J. Russell, and F.-L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavillette, D., A. Ruggieri, B. Boson, M. Maurice, and F.-L. Cosset. 2002. Relationship between SU subdomains that regulate the receptor-mediated transition from the native (fusion-inhibited) to the fusion-active conformation of the murine leukemia virus glycoprotein. J. Virol. 76:9673-9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavillette, D., A. Ruggieri, S. J. Russell, and F.-L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Z., A. Pinter, and S. C. Kayman. 1997. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex. J. Virol. 71:7012-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lostrom, M. E., S. R. Stone, M. Tam, W. N. Burnette, A. Pinter, and R. C. Nowinski. 1979. Monoclonal antibodies against murine leukemia viruses: identification of six antigenic determinants of the p15(E) and gp70 envelope proteins. Virology 98:336-350. [DOI] [PubMed] [Google Scholar]
- 35.Lowy, D. R., E. Rands, S. K. Chattopadhyay, C. F. Garon, and G. L. Hager. 1980. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc. Natl. Acad. Sci. USA 77:614-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAtee, F. J., and J. L. Portis. 1985. Monoclonal antibodies specific for wild mouse neurotropic retrovirus: detection of comparable levels of virus replication in mouse strains susceptible and resistant to paralytic disease. J. Virol. 56:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McInerney, T., L. McLain, S. J. Armstrong, and N. J. Dimmock. 1997. A human IgG1 (b12) specific for the CD4 binding site of HIV-1 neutralizes by inhibiting the virus fusion entry process, but b12 Fab neutralizes by inhibiting a postfusion event. Virology 233:313-326. [DOI] [PubMed] [Google Scholar]
- 39.Mendez, M. J., L. L. Green, J. R. Corvalan, X. C. Jia, C. E. Maynard-Currie, X. D. Yang, M. L. Gallo, D. M. Louie, D. V. Lee, K. L. Erickson, J. Luna, C. M. Roy, H. Abderrahim, F. Kirschenbaum, M. Noguchi, D. H. Smith, A. Fukushima, J. F. Hales, S. Klapholz, M. H. Finer, et al. 1997. Functional transplant of megabase human immunoglobulin loci recapitulates human antibody response in mice. Nat. Genet. 15:146-156. [DOI] [PubMed] [Google Scholar]
- 40.Moloney, J. 1960. Biological studies on a lymphoid leukemia virus extracted from sarcoma 37. I. Origin and introductory observations. J. Natl. Cancer Inst. Monogr. 24:933. [PubMed] [Google Scholar]
- 41.Morgan, R. A., O. Nussbaum, D. D. Muenchau, L. Shu, L. Couture, and W. F. Anderson. 1993. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J. Virol. 67:4712-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niman, H. L., and J. H. Elder. 1982. Structural analysis of Rauscher virus gp70 using monoclonal antibodies: sites of antigenicity and P15(E) linkage. Virology 123:187-205. [DOI] [PubMed] [Google Scholar]
- 44.Nowinski, R. C., R. Pickering, P. V. O'Donnell, A. Pinter, and U. Hammerling. 1981. Selective neutralization of ecotropic murine leukemia virus by monoclonal antibodies: localization of a site on the gp70 protein associated with ecotropism. Virology 111:84-92. [DOI] [PubMed] [Google Scholar]
- 45.Orlik, O., J. Ban, J. Hlavaty, C. Altaner, R. Kettmann, D. Portetelle, and G. A. Splitter. 1997. Polyclonal bovine sera but not virus-neutralizing monoclonal antibodies block bovine leukemia virus (BLV) gp51 binding to recombinant BLV receptor BLVRcp1. J. Virol. 71:3263-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ott, D., R. Friedrich, and A. Rein. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J. Virol. 64:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren, P. W. H. I., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierotti, M., A. B. D. Leo, A. Pinter, P. V. O'Donnell, U. Hammerling, and E. Fleissner. 1981. The GIX antigen of murine leukemia virus: an analysis with monoclonal antibodies. Virology 112:450-460. [DOI] [PubMed] [Google Scholar]
- 49.Pinter, A., T.-E. Chen, A. Lowy, N. G. Cortez, and S. Silagi. 1986. Ecotropic murine leukemia virus-induced fusion of murine cells. J. Virol. 57:1048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinter, A., and E. Fleissner. 1979. Characterization of oligomeric complexes of murine and feline leukemia virus envelope and core components formed upon cross-linking. J. Virol. 30:157-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinter, A., and W. J. Honnen. 1988. O-linked glycosylation of retroviral envelope gene products. J. Virol. 62:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinter, A., W. J. Honnen, S. C. Kayman, O. Troshev, and Z. Wu. 1998. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine 16:1803-1811. [DOI] [PubMed] [Google Scholar]
- 53.Pinter, A., W. J. Honnen, M. E. Racho, and S. A. Tilley. 1993. A potent, neutralizing human monoclonal antibody against a unique epitope overlapping the CD4-binding site of HIV-1 gp120 that is broadly conserved across North American and African virus isolates. AIDS Res. Hum. Retrovir. 9:985-996. [DOI] [PubMed] [Google Scholar]
- 54.Pinter, A., W. J. Honnen, S. A. Tilley, C. Bona, H. Zaghouani, M. K. Gorny, and S. Zolla-Pazner. 1989. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J. Virol. 63:2674-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinter, A., W. J. Honnen, J.-S. Tung, P. V. O'Donnell, and U. Hammerling. 1982. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology 116:499-516. [DOI] [PubMed] [Google Scholar]
- 56.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Portis, J. L., and F. J. McAtee. 1983. Monoclonal antibodies derived during graft-versus-host reaction. II. Antibodies detect unique determinants common to many MCF viruses. Virology 126:96-105. [DOI] [PubMed] [Google Scholar]
- 58.Portis, J. L., F. J. McAtee, and M. W. Cloyd. 1982. Monoclonal antibodies to xenotropic and MCF murine leukemia viruses derived during the graft-versus-host reaction. Virology 118:181-190. [DOI] [PubMed] [Google Scholar]
- 59.Pyle, S., D. J. Chabot, T. L. Miller, S. A. Serabyn, J. W. Bess, Jr., and L. O. Arthur. 1991. Large-scale purification of gp70 from Moloney murine leukemia virus. J. Virol. Methods 32:303-315. [DOI] [PubMed] [Google Scholar]
- 60.Shibamura, S., K. Kai, T. Akatsuka, and T. Odaka. 1989. Biochemical and immunological characterization of murine leukemia viruses that are paralysis-inducing in rats. Jpn. J. Cancer Res. 80:24-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stone, M. R., and R. C. Nowinski. 1980. Topological mapping of murine leukemia virus proteins by competition-binding assays with monoclonal antibodies. Virology 100:370-381. [DOI] [PubMed] [Google Scholar]
- 63.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 61:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tung, J. S., E. S. Vitetta, E. Fleissner, and E. A. Boyse. 1975. Biochemical evidence linking the GIX thymocyte surface antigen to the gp69/71 envelope glycoprotein of murine leukemia virus. J. Exp. Med. 198:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, N., T. Zhu, and D. D. Ho. 1995. Sequence diversity of V1 and V2 domains of gp120 from human immunodeficiency virus type 1: lack of correlation with viral phenotype. J. Virol. 69:2708-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, B. W., P. M. Cannon, E. M. Gordon, F. L. Hall, and W. F. Anderson. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J. Virol. 72:5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 68.Yu, H., N. Soong, and W. F. Anderson. 1995. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J. Virol. 69:6557-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]