Abstract
Mammalian telomeres consist of TTAGGG repeats, telomeric repeat binding factor (TRF), and other proteins, resulting in a protective structure at chromosome ends. Although structure and function of the somatic telomeric complex has been elucidated in some detail, the protein composition of mammalian meiotic telomeres is undetermined. Here we show, by indirect immunofluorescence (IF), that the meiotic telomere complex is similar to its somatic counterpart and contains significant amounts of TRF1, TRF2, and hRap1, while tankyrase, a poly-(ADP-ribose)polymerase at somatic telomeres and nuclear pores, forms small signals at ends of human meiotic chromosome cores. Analysis of rodent spermatocytes reveals Trf1 at mouse, TRF2 at rat, and mammalian Rap1 at meiotic telomeres of both rodents. Moreover, we demonstrate that telomere repositioning during meiotic prophase occurs in sectors of the nuclear envelope that are distinct from nuclear pore-dense areas. The latter form during preleptotene/leptotene and are present during entire prophase I.
INTRODUCTION
Telomeres are essential for chromosome stability and regulate the replicative life-span of somatic cells (Muller 1938, McClintock 1941; for reviews see, Blackburn and Greider 1995, de Lange 1998a, Dandjinou et al., 1999). They also play an important role in meiosis, in particular in the chromosome pairing process, which takes place during first meiotic prophase (for reviews see von Wettstein et al., 1984, Dernburg et al., 1995, de Lange 1998b, Zickler and Kleckner 1998). Meiosis is a specialized cell division that contributes to sexual reproduction by reducing the diploid chromosome number to the haploid. As a precondition, the reductional division requires the lengthwise alignment, synaptic pairing, and recombination of homologous chromosomes (von Wettstein et al., 1984, Loidl 1990, Kleckner 1996, Roeder 1997). Linear chromosomes with telomeres are indispensable for faithful homologue pairing and segregation (Naito et al., 1998, Rockmill and Roeder 1998, Ishikawa and Naito 1999).
In contrast to the situation in Saccharomyces cerevisiae, where telomeres aggregate in a few clusters at the nuclear periphery (Gilson et al., 1993), mammalian telomeres are scattered throughout the nuclear volume in most somatic and premeiotic cells (Vourc'h et al., 1993, Scherthan et al., 1996; Luderus et al., 1996). At the onset of first meiotic prophase, mammalian telomeres relocate to the nuclear envelope and then transiently congregate at a limited sector of the nuclear membrane in the vicinity of the centrosome (bouquet formation) (Rasmussen and Holm 1980, Loidl 1990, Dernburg et al., 1995, Trelles-Sticken et al., 1999). In certain plant and grasshopper species, bouquet formation has been associated with the concentration of nuclear pores close to the membrane-attached telomeres (Loidl 1990, Zickler and Kleckner 1998). Pore clustering is also known from human spermatogenesis (Fawcett and Chemes 1979), but its relation to meiotic telomere clustering has not been determined.
In the asynaptic meiosis of Schizosaccharomyces pombe, it has been shown that telomere clustering is a response to mating pheromone, and it contributes to homologue pairing (Chikashige et al., 1997). Bouquet formation depends on the presence of the Taz1 protein (Cooper et al., 1998, Nimmo et al., 1998), which is related to the mammalian telomeric repeat binding factors, TRF1 and TRF2, with which it shares both a C-terminal Myb-type DNA binding domain and a large central TRF homology domain (TRFH) (Cooper et al., 1997; Bilaud et al., 1996; Li et al., 2000).
Compared with the detailed knowledge on telomere clustering and bouquet formation in the asynaptic meiosis of fission yeast (Cooper 2000), literally nothing is known about the components of the telomeric complex during mammalian meiosis. Somatic mammalian telomeres consist of 5 to 50 kb of TTAGGG repeats (Moyzis et al., 1988, de Lange et al., 1990, Kipling and Cooke 1990), that associate with specific proteins to form functional telomeres that protect chromosome ends and regulate telomere maintenance by telomerase, the reverse transcriptase responsible for addition of TTAGGG repeats to chromosome ends (Blackburn and Greider, 1995; Nugent and Lundblad, 1998). The mammalian telomeric complex consists of two related telomeric repeat binding factors (TRF1 and TRF2) and their interacting partners. Both TRF1 and TRF2 bind duplex telomere repeat arrays as homodimers, using a C-terminal Myb domain (Chong et al., 1995; Bianchi et al., 1997; Broccoli et al., 1997a; Bianchi et al., 1999). TRF1 and its interacting protein TIN2 are important regulators of telomere length (van Steensel and de Lange 1997; Kim et al., 1999), possibly by modulating the access of telomerase. Furthermore, TRF1 has been shown to mediate parallel pairing of telomere sequence tracks in vitro (Griffith et al., 1998), raising the possibility that this protein could be involved in associations of closely spaced telomeres during the bouquet stage (Scherthan et al., 1996; Bass et al. 1997) and in telomere associations frequently seen in spermatid nuclei (Zalenski et al., 1997; Meyer-Ficca et al., 1998) and mitotic cells (Wagenaar 1969).
TRF2 shares considerable homology with TRF1, but its N-terminus is very basic rather than acidic in TRF1 (Bilaud et al., 1997; Broccoli et al., 1997a). This structural distinction between TRF1 and TRF2 is reflected in functional difference. Although TRF2, like TRF1, has been implicated in telomere length control (Smogorzewska et al., 2000), TRF2 also plays a key role in telomere protection. TRF2 is required to maintain telomere integrity and to prevent chromosome end fusions (van Steensel et al., 1998) and p53-dependent apoptosis, which ensues when TRF2-depleted chromosome termini are detected by DNA damage checkpoints (Karlseder et al., 1999). TRF2 has been proposed to mediate its protective tasks by hiding the end of the double helix in a t-loop structure (Griffith et al., 1999).
Recently, the human ortholog of the budding yeast duplex telomere repeat binding protein scRap1 (Shore and Nasmyth, 1987; Klein et al., 1992) has been isolated by a two-hybrid screen with TRF2 as bait (Li et al., 2000). Human (h)Rap1 shows considerable sequence homology to scRap1, however, it binds the telomeric complex through protein interaction with TRF2 (Li et al., 2000), in contrast with the direct duplex repeat binding properties of scRap1 (Longtine et al., 1989).
In addition to their interaction with telomerase, mammalian telomeres associate with a second enzyme, tankyrase (Smith et al., 1998). Tankyrase is an ankyrin-related protein with a C-terminal poly-(ADP-ribose)-polymerase (PARP) domain that locates to telomeres through its interaction with TRF1 (Smith et al., 1998; Smith and de Lange, 1999). In vitro, tankyrase can poly(ADP) ribosylate itself and TRF1, and this modification results in the loss of telomeric DNA binding activity of TRF1 (Smith et al., 1998). In vivo, targeting of tankyrase to the nucleus results in removal of TRF1 from telomeres and inappropriate growth of telomeres in telomerase-expressing cells (Smith and de Lange, 2000). In addition to its presence at the telomere, tankyrase locates to nuclear pore complexes in interphase cells and relocates to centrosomes in mitosis (Smith and de Lange, 1999). The function of tankyrase at these nontelomeric sites is not known.
Indirect immunofluorescence (IF) has demonstrated that TRF1, TRF2, and hRap1 are present at telomeres throughout the division cycle of somatic cells (Chong et al., 1995; van Steensel and de Lange, 1997; van Steensel et al., 1998; Li et al., 2000). In interphase, these factors are detectable as discrete dispersed nuclear dots, and for TRF1 it was demonstrated that these sites coincide with TTAGGG repeat loci. The telomeric localization of TRF2 and hRap1 in interphase was inferred from colocalization with TRF1. In condensed mitotic chromosomes, each of these proteins can be visualized at terminal sites, in prophase, metaphase, anaphase, and telophase. Therefore, it is generally assumed that the TTAGGG repeats are always complexed with this set of proteins. Indeed, TRF2 can also be observed at TTAGGG repeat sequences at chromosome-internal sites, a situation frequently encountered in Chinese hamster chromosomes (Smogorzewska et al., 2000). Despite the obvious interest in the role of the telomeric complex in meiosis (see above), there is no information on the expression and localization of telomeric proteins in mammalian meiosis.
Here we report on the protein composition of the mammalian telomeric complex at meiosis. We demonstrate that TRF1 and -2, hRAP1 and tankyrase are expressed in male meiocytes, and that these factors are located at telomeres throughout prophase I. Furthermore, our studies confirm the previous description of the movement of meiotic telomeres, and they now reveal that this movement is associated with an independent redistribution of nuclear pores. Unexpectedly, we find that nuclear pore complexes and telomeres migrate in separate, generally nonoverlapping territories of the nuclear envelope, indicating at least two migrations that contribute to the altered organization of the meiotic nuclear envelope. These results indicate that the components of somatic telomeres also reside at chromosome ends during mammalian meiosis and redistribute independently from meiotic nuclear pore complexes.
MATERIALS AND METHODS
Testicular Specimens
Mouse spermatocytes were obtained from C57Bl6/129 mice and from several randombred adult Mus musculus males, obtained at the local zoo supply. Animals were killed by cervical dislocation, and the testes were immediately resected and processed, as given below. A human testis sample was obtained from a 37-year-old male of proven fertility, in the course of a reverse vasectomy, via needle biopsy under local anesthesia. A second tissue sample was obtained from an 80-year-old male of proven fertility, by open incisional biopsy, in association with an orchitectomy under general anesthesia . All tissues were shock-frozen for 5 min in isopentane at −70°C, transferred to liquid nitrogen for another 5 min, and finally stored at −70°C until further use.
Testicular Preparations
Structurally preserved nuclei for simultaneous synaptonemal complex (SC)-immunostaining and FISH were prepared by mincing fresh or frozen testicular tissue in MEM medium (Life Technologies, GIBCO, Ann Arbor, MI)/0.5% mammalian protease inhibitor (Sigma, St. Louis, MO) at 4°C. After removal of tissue pieces, a drop of the suspension was placed on clean aminosilane-coated glass slides and immediately mixed with two drops of fixative (3.7% formaldehyde, 0.1 M sucrose, pH 7.4). After air drying at room temperature, slides were stored at −20°C until further use. Swab preparations were obtained by touching the surface of an ethanol-cleaned aminosilane-coated glass slide with a freshly thawed small testis tissue piece, which was lifted off after a few seconds. The adhering cells were then fixed for 10 min in 3.7% formaldehyde/PBS at room temperature. Finally, aldehyde groups were quenched by a 5 min wash in PBS, 0.5% Glycin (wt/vol). Preparations were then subjected to IF as described below.
Detergent Spreading
Surface spreading was performed as follows: About 50 μl of a testicular suspension was placed on a glass slide and mixed either with 50 μl of ionic detergent solution 1% Lipsol (Sci. Lab. Suppl., Nottingham, United Kingdom; Albini and Jones, 1984) or with 250 μl of a 1% solution of the nonionic detergent Triton X-100 (Serva, Heidelberg, Germany). Swelling and spreading of spermatocytes was monitored by phase contrast microscopy. When cells obtained an opaque appearance, 300 μl of fixative (3.7% acid free formaldehyde [Merck, Darmstadt, Germany], 0.1 M Sucrose, pH 7.4) were added to the slide and gently mixed by tilting. Slides were then air-dried at 37°C and stored at -20°C until further use.
Antisera
The following affinity-purified antibodies were used in the immunostaining experiments: TRF1: rabbit anti-human TRF1 (#371; van Steensel and de Lange, 1997), mouse anti-human full length TRF1 (Smith and de Lange, 1999), and rabbit anti-mouse-Trf1 antisera (#644, J. Karlseder and T. deLange, unpublished data) (Broccoli et al., 1997b). TRF2: rabbit anti-TRF2 antibodies #508; #647 (van Steensel et al., 1998; Zhu et al., 2000) were used as a mixture (diluted 1/1000 in PBS). Anti-TRF2 mAbs15 and 8 (Imgenex, San Diego, CA) were used in single reactions (diluted 1/1000). Tankyrase: the following affinity purified rabbit antitankyrase sera were mixed in equal volumes: #465 (Smith et al., 1998) and #762 to aa 973-1149 of tankyrase, and #763 to full-length tankyrase (Smith and de Lange, unpublished data). A rabbit anti-human-Rap1 antiserum (#765; Li et al., 2000) was used to detect mammalian Rap1. All antisera were used at 1/1000 dilution in PBS/0.1%Tween 20/0.2% BSA/0.1% gelatin (PBTG). All antibodies were tested in individual staining reactions for their specificity and performance. Controls without primary antibodies were all negative (our unpublished results).
A polyclonal rabbit anti-SCP3 antiserum was used to detect axial cores and complete SCs (Lammers et al., 1994). mAb414 (BABCO, Berkeley, CA), which is a universal monoclonal antibody to nuclear pore complex proteins like nucleoporin p62 and related glycosylated nucleoporins (Davis and Blobel 1987), was used to stain nuclear pores.
IF Staining
Fresh or frozen preparations were rinsed in water and then in PBS/0.1% glycin to remove sucrose and formaldehyde. IF staining of telomere proteins and SCP3 of the axial/lateral element of the SC (Lammers et al., 1994), was performed as described earlier (Scherthan et al., 1996). The conditions of immunostaining applied are known to reveal TRFs and hRap1 in somatic cells (Chong et al., 1995, Smith and de Lange 1999, Li et al. 2000). Briefly, preparations were extracted for 30 min with 0.5% Triton-X-100/PBS, washed with PBS, incubated for 10 min in PBTG, followed by incubation with antibody solution containing the primary anti-telomere protein antisera (diluted 1/1000 in PBTG) in a moist chamber at 4°C, overnight. Subsequently, preparations were rinsed 3x3 min with PBS/Tween 20 and incubated for 30 min at 37°C with a biotinylated secondary goat anti-rabbit antibody (Vector Labs, diluted 1/500 in PBS), which was visualized with Avidin-Cy3 (1/1000; Sigma). For double labeling, preparations obtained by this procedure were further subjected to immunostaining with anti-SCP3 or nuclear pore antibodies, which were applied for 60 min at 37°C, followed by three 3-min washes in PBTG, and detection with appropriate secondary FITC-conjugated antibodies (Vector Labs, Burlingame, CA) (see Pandita et al., 1999). Finally, preparations were mounted in antifade solution (Vectashield, Vector Labs) containing 0.5 μg/ml DAPI (Sigma) to reveal nuclear DNA. In mouse testicular suspension preparations, the stage-specific distribution of SCP3 proteins and/or DAPI-bright heterochromatin clusters (Scherthan et al., 1996) was utilized to identify spermatocytes at various stages of prophase I.
Because SC and antitelomere protein antisera used were in many cases derived in rabbits, all costaining reactions were performed consecutively, and with and without SCP3 detection. An identical distribution of telomere protein signals was obtained in all cases (our unpublished results; compare e.g., Figures 2 and 3). Furthermore, all IF staining reactions were carried out in parallel on 1) structurally preserved testicular nuclei, 2) on mild nuclear spreads obtained with the nonionic detergent Triton-X 100, and 3) on spreads obtained with the ionic detergent mixture Lipsol. The latter has been shown to produce extensively spread nucleoids that are particularly amenable to SC analysis (Albini and Jones, 1984). Dependent on the preparation method and detergent used, some antigens were only revealed in one particular type of preparation (see RESULTS).
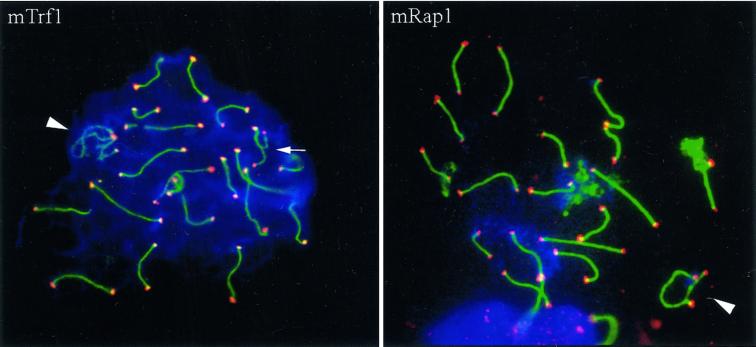
Figure 2.
(mTrf1) Spread pachytene spermatocyte I of the mouse (Mus musculus) stained with anti-SCP3 antiserum (FITC, green) and anti-mouse Trf1 (Cy3; red). Strong Trf1 signals are present at ends of SCs, unpaired axes (arrow), and the telomeres of the sex chromosomes (arrowhead). One strong Trf1 signal is seen at the association site of the distal ends of the XY pair. (mRap1) IF staining of mammalian Rap1 with anti-hRap1 Abs (red, Cy3) in a spread mouse pachytene nucleus. The ends of the SCs are capped by strong, large mRap1 signals, which sometimes extend beyond the SC ends. The XY pair displays mRap1 signals at the ends of the paired and the two unpaired meiotic chromosome cores (arrowhead). The fuzzy green material at three bivalents results from SCP3 fluorescence at remnants of nucleolar material carried by three autosomes. DNA is shown in blue (DAPI).
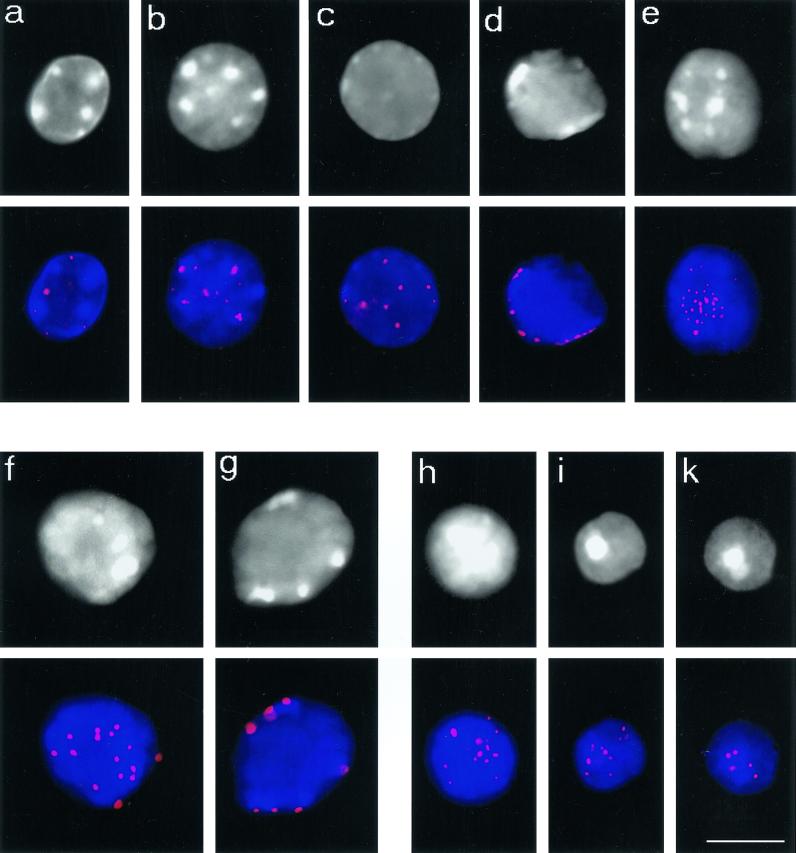
Figure 3.
Telomere redistribution during mouse spermatogenesis as displayed by Trf1 immunofluorescence (Cy3, red). Staging was deduced from stage-specific heterochromatin and telomere distribution patterns (see Scherthan et al., 1996). DAPI staining of nuclear DNA is given at the top (gray). DAPI-bright heterochromatin is seen as whitish clusters. (a) Nucleus of a spermatogonium with dispersed telomere distribution. (b) Premeiotic nucleus with prominent intranuclear heterochromatin clusters and dispersed telomeric Trf1 signals. (c) Preleptotene nucleus with faint peripheral heterochromatin and scattered Trf1 telomere signals (d) Late preleptotene nucleus displaying exclusively perinuclear telomere distribution. Note the small telomere signals, compared with f and g. (e) Top of bouquet nucleus (corresponding to leptotene/zygotene) with clustered telomeres. Small Trf1 signal diameter indicates that most telomeres have not yet paired. (f and g) Pachytene nuclei as identified by prominent peripheral heterochromatin clusters and large peripheral telomere signals. (f) Top of pachytene nucleus with numerous signals. (g) Focal plane at maximum diameter of a pachytene nucleus displaying Trf1 signals at the nuclear envelope. (h-k) Haploid round spermatids showing Trf1 telomere signals at the DAPI-bright heterochromatin chromocenters over the central region of the spermatid nuclei. Bar: 10 μm, it applies to all details.
To exclude potential artifacts induced by air drying and/or detergent extraction protocols, control nuclear pore/telomere IF experiments were performed to undisrupted suspension and swab preparation nuclei that had been obtained on aminosilane-coated glass slides without allowing the suspension to dry out completely. To this end, suspension nuclei were allowed to settle on the slides for 30–45 min at room temperature, followed by a 10-min fixation in 3.7% formaldehyde/0.1 M sucrose. The still moist cross-linking fixative was then washed off as described above, and the nuclei were subjected to IF staining without detergent extraction.
IF-FISH
Combinatorial immunostaining and telomere FISH was carried out as described earlier (Scherthan et al., 1996). Briefly, TRF1 was first immunostained using the biotin/avidin-Cy3 system. Preparations were then denatured in the presence of an FITC-labeled PNA telomere repeat probe (DAKO, Carpinteria, CA) and hybridized for 12 h at 37°C, followed by three 5-min washes in 0.05xSSC. Preparations were finally embedded in antifade (Vector Labs) containing 0.5 μg/ml DAPI as DNA counterstain and were analyzed in the microscope.
Microscopic Evaluation
Preparations were evaluated using a Zeiss Axioskop epifluorescence microscope (Carl Zeiss, Jena, Germany) equipped with single- and double-band pass filters for excitation of green, red, and blue, and for simultaneous excitation of red and green fluorescence (Chroma Technologies, Brattleboro, VT) and 63x and 100x plan-neofluoar lenses. Three-dimensional evaluation of immunostained nuclei was performed in some experiments by carefully focusing through the nuclei using a 100x plan neofluoar lens. Digital black-and-white images were recorded with a cooled CCD camera (Hamamatsu Photonics, Bridgewater, NY) and merged to RGB-images by the ISIS fluorescence image analysis system (MetaSystems, Altlussheim, Germany). Fluorescence profiles were derived from the gray-scale values of the pixels along interactively determined polygon tracks in digital-RGB images using the “profile” measurement option of the ISIS image analysis package. The program determines the relative gray values in each color-channel from three pixels laying perpendicular across the selected line and displays the relative variations in fluorescence intensity along this line as a profile graph in the image. Signals, e.g. in red and green fluorescent channels, which colocalize create peaks at identical positions. Please note that the profiles displayed are calibrated at the strongest signal encountered in a line polygon.
RESULTS
TRF1 Is a Component of Meiotic Telomeres
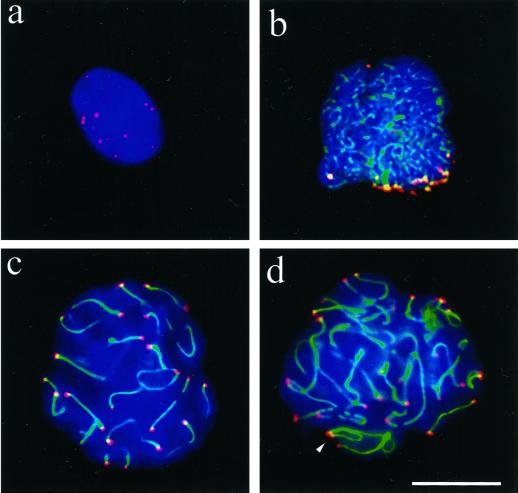
We immunostained ionic and nonionic detergent spread testicular preparations of several mice and two human donors with antibodies directed against human TRF1 with Abs #371 (van Steensel and de Lange, 1997) and its rather divergent mouse ortholog Trf1 with Abs #644 (Broccoli et al., 1997b; Karlseder and de Lange, unpublished data). IF disclosed that TRF1 is present at telomeres scattered throughout somatic nuclei (our unpublished results), consistent with the pattern reported previously (Luderus et al., 1996). Combinatorial TRF1 IF and telomere FISH demonstrated colocalization of TTAGGG repeat signals and TRF1 in all cell types (our unpublished results). In spread spermatocytes, TRF1 IF generated distinct signal spots that, in a few favorably spread preleptotene nuclei, reached the theoretical 92 signals that are expected if all human telomeres were detected (Figure 1a). At leptotene, TRF1 was detected at the ends of the developing axes of the not yet paired homologues (Figure 1b). Pachytene nuclei generally displayed strong signals at SCs ends which often extended between some nonhomologous telomeres (Figure 1, c and d). Undisrupted or mildly spread nuclei generally showed more compact TRF1 signals (see Figure 9).
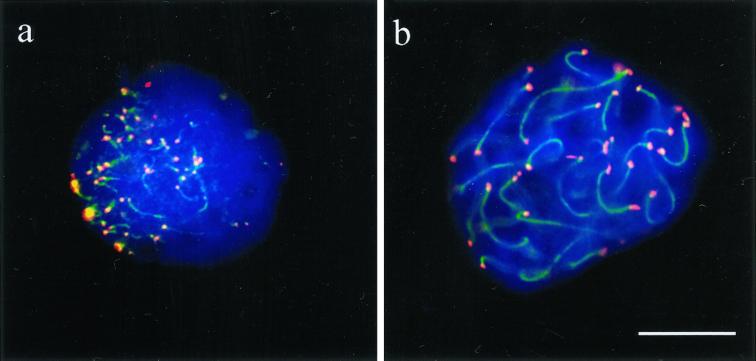
Figure 1.
Spreads of human spermatocytes I immunostained with anti-TRF1 (#371; Cy3, red) and anti-SCP3 synaptonemal complex protein (FITC, green). (a) Spread preleptotene nucleus (identified by its extensive diameter and week patchy SCP3 deposits to the right), which exhibits distinct TRF1 signal spots that equal the diploid number of individual telomeres (92). (b) Mildly spread leptotene nucleus (focal plane on top of nucleus) with TRF1 signals at the ends of developing axial elements (green threads), which are accumulated at a limited region of the nucleus (bouquet arrangement). (c) Pachytene spread. Distinct TRF1 signals are seen at both ends of the SCs. Note that TRF1 forms thread-like connections between several nonhomologous telomeres. The enlarged inset displays two nonhomologous SC ends that are connected by a TRF1 stretch. (d) Late pachytene spread: prominent TRF1 signals cap the SC ends. The sex chromosome cores form a condensed XY bivalent (arrow) that also contains TRF1 at its telomeres. DNA is counterstained with DAPI (blue).
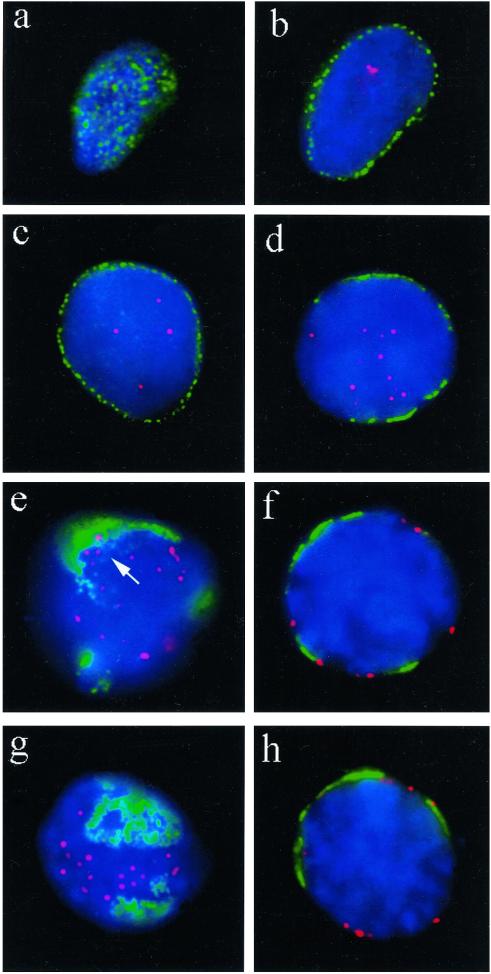
Figure 9.
Distribution of TRF1-tagged telomeres (red, Cy3) in relation to nuclear pore (NUP) positioning (green, FITC; detected with mAb414) in undisrupted human premeiotic and meiotic testicular suspension nuclei. (a) Focal plane at the top of a premeiotic nucleus reveals the typical, more or less even distribution of nuclear pores. (b) Nuclear pore (NUP) IF at the equatorial plane of the same nucleus creates a rim-like signal distribution, while a few telomere signals are present in the interior. (c) NUP rim-staining at the equator of another premeiotic nucleus. Telomere signals are seen in the nuclear interior. (d) Patchy perinuclear distribution of NUPs of an early meiotic nucleus (as deduced by its increased DAPI intensity) with internal telomeres (preleptotene). (e) Focal plane at the top of a spermatocyte I nucleus that displays pronounced accumulation of NUPs. Telomeres are at the nuclear periphery. The immediate vicinity of a few telomeres that are close to NUP clusters is devoid of NUP fluorescence (arrow). (f) Center view of a pachytene nucleus (note the thread-like DAPI-stained chromatin). Telomeres locate adjacent to NUP patches or at NUP-free regions of the nuclear envelope. (g and h) Top and equatorial focal plane, respectively, of an early pachytene nucleus (deduced from locally accumulated telomeres and thread-like chromatin) with telomeres remote from NPC-dense areas. In the equatorial plane, a telomere spot colocalizes with NUP signal stretch (h, to the right). Nuclear DNA is counterstained in blue (DAPI).
As in the human situation, immunostaining with antibodies #644 to mouse Trf1 revealed distinct signal spots in premeiotic and meiotic nuclei of spreads and structurally preserved nuclei (Figures 2, 3). As in human meiocytes, Trf1 was found in abundance at the ends of paired and unpaired meiotic chromosome cores (Figure 2). To investigate telomere proteins during the dynamic redistribution of meiotic chromosome ends, we immunostained structurally preserved nuclei in testicular suspensions, since detergent spreading disrupts three-dimensional nuclear architecture (see Pandita et al., 1999). Undisrupted testicular nuclei were staged according to the stage-specific distribution patterns of DAPI-bright mouse heterochromatin and telomeres (Scherthan et al., 1996). Mouse spermatogonia and somatic nuclei displayed Trf1 signals scattered throughout the nuclear lumen (Figure 3, a and b), which is consistent with the 3D telomere distribution seen by FISH (Vourc'h et al., 1993, Scherthan et al., 1996) and TTAGGG repeat EM-in situ hybridization (Luderus et al., 1996). Preleptotene nuclei, as identified by diffuse perinuclear heterochromatin distribution, showed mTrf1 telomere signals scattered throughout the nuclear lumen (Figure 3c). Late preleptotene nuclei, which display distinct perinuclear heterochromatin clusters, exhibited exclusively peripheral and small Trf1 signals (Figure 3d). Spermatocytes I with a bouquet topology, which is representative of the leptotene/zygotene transition stage (Scherthan et al., 1996), displayed most Trf1 signals accumulated in a limited region of the nucleus (Figure 3e). Mouse pachytene spermatocytes, as identified by prominent peripheral heterochromatin clusters and an increased nuclear diameter, exhibited large peripherally distributed telomere Trf1 signals (Figure 3, f and g), which is consistent with FISH analysis (Scherthan et al., 1996). Trf1 telomere signals increased in size when chromosome ends were synaptically paired. This was noted for all telomere-specific proteins and types of preparation (see below), irrespective of the species investigated (Figures 1, 3, 4, 7, 9). Haploid round spermatids exhibited small Tfr1 signals in the vicinity of the central DAPI-bright chromocenter (Figure 3, h–k), which is in agreement with the telomere distribution observed by FISH in this cell type (Zalenski et al., 1997).
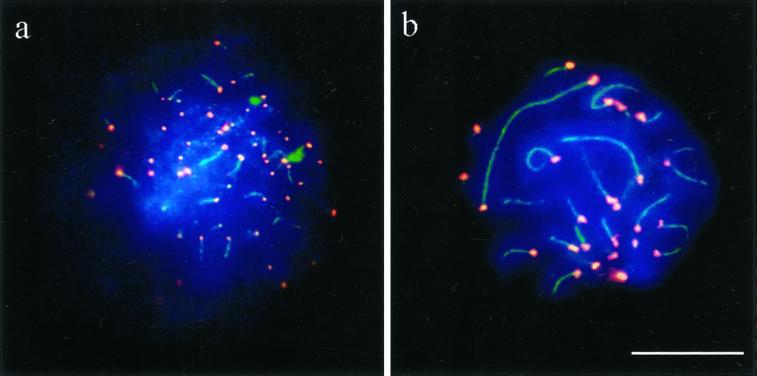
Figure 4.
Spread human spermatocytes I after immunostaining with anti-TRF2 (Cy3; red) and anti-SCP3 (FITC, green). (a) Top region of a mildly spread leptotene nucleus, which shows distinct TRF2 signal spots at the ends of developing axial elements (faint green threads). Ends of cores and TRF2 signals overlap (yellow) (b) Focal plane at the top of a spread pachytene nucleus. Distinct TRF2 signals are seen at the ends of the SCs. Most of the SCs extend beyond the focal plane. DNA is stained with DAPI (blue). Bar represents 10 μm.
Figure 7.
Immunofluorescent staining of hRap1 (red, Cy3) in mildly spread human spermatocytes I. Focal planes at top of nuclei. (a) A human leptotene spermatocyte displays small but distinct hRap1 signals at the ends of unpaired chromosome axes (green, FITC). (b) Large hRap1 signals are seen at the ends of SCs of a mildly spread pachytene nucleus. DNA is counterstained in blue (DAPI). Bar: 10 μm.
TRF1 was revealed in all types of preparations and was found to be abundant at the ends of mammalian meiotic chromosome cores during all substages of prophase I and haploid spermatids. Furthermore, TRF1 has also been detected at the ends of female meiotic chromosomes (C. Tease and M. Hultén, unpublished data). Altogether, it appears that it may be a major component of the meiotic telomere complex.
TRF2
Reminiscent of the labeling obtained for TRF1, in human spermatocytes TRF2 formed a dispersed, dot-like IF signal pattern in premeiotic nuclei (our unpublished results) and localized to the ends of unpaired and paired meiotic chromosome cores as distinct signal spots (Figure 4). In spreads obtained with the nonionic detergent Triton-X100 and swab preparations of rat testes, only mouse anti-hTRF2 Abs produced distinct signals at meiotic chromosome ends (our unpublished results). Failure to reveal detectable IF signals by the other anti-TRF2 sera tested in rat and the absence of TRF2 IF signals in all types of mouse preparations could be due to masked or altered epitopes in the homologous rodent proteins, since the antibodies used were raised against the human protein. In any case, the results obtained in human and rat testicular suspensions suggest that TRF2 is also a discrete component of the mammalian meiotic telomere.
To test for the relative positions of the two TRF proteins at meiotic telomeres, we performed colocalization experiments with mouse anti-hTRF1 full-length protein antibodies and rabbit anti-hTRF2 antibodies (#508, #647). In human spreads, the differential IF detection of both proteins resulted in colocalized telomeric signals with a mixed color (Figure 5a). The colocalization was verified at the fluorescence microscope level using the profile measurement option of the ISIS software (see MATERIALS AND METHODS). Polygon lines drawn across telomeric signals of spermatocytes at different prophase stages showed peaks of signal intensities at identical locations in both channels (Figure 5a). Of 223 telomeres investigated, 220 were found to contain both proteins in similar concentrations as judged from the fluorescence signal intensities and profile analysis (our unpublished results). Only one signal spot showed TRF1, and two spots showed TRF2 immunofluorescence only. In conclusion, it appears that TRF1 and TRF2 colocalize to the meiotic telomere throughout all stages of prophase I.
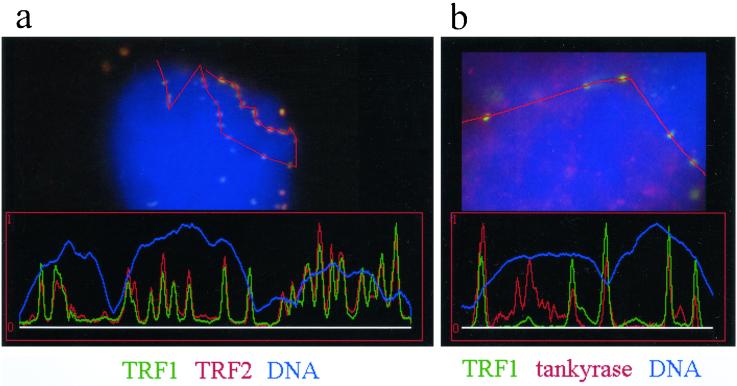
Figure 5.
Analysis of colocalization of fluorescent signals of telomeric proteins in human testes nuclei. Fluorescence profile analysis is conducted along polygon lines drawn across telomeric signals and adjacent chromatin in raw digital images (see methods). Colocalization is indicated by signal peaks in the green and red channel at identical positions. (a) A fluorescence profile derived from a line polygon drawn across 19 telomeres at the top of a bouquet nucleus (leptotene/zygotene) costained with TRF1 (green, FITC) and TRF2 (red, Cy3). Perfect colocalization of similar sized signals is seen. The DAPI profile (DNA, blue) appears uneven since it runs along the nuclear periphery. (b) Colocalization of tankyrase (red) and five TRF1 signals in a sector of a spread spermatocyte nucleus. Each TRF1 signal peak is colocalized with a distinct tankyrase signal peak. Additional tankyrase signals which create additional peaks in the red channel likely result from reminders of nontelomeric tankyrase e.g., at nuclear pore remnants (see text).
Tankyrase Locates to Meiotic Telomeres
Of all types of preparations tested, tankyrase was revealed only in ionic detergent-spreads, where it produced numerous granular IF signals throughout the chromatin of premeiotic nuclei (Figure 6a). It can be anticipated that the spreading-induced collapse of membrane structures may lead to a dispersion of cytoplasmic and nuclear pore-bound tankyrase throughout the nuclei. In meiocytes, distinct tankyrase signals were obtained at the ends of developing axial cores of leptotene chromosomes (Figure 6b) and at the ends of the SCs (Figure 6, c and d). Confluent signals were sometimes observed between several chromosomes ends, which resemble the observations in the TRF1 experiments and may result from TRF1-dependent tankyrase localization to the telomeric complex. Compared with the signals created by the other telomere binding proteins investigated, most tankyrase signals obtained were relatively small and of variable size. We further confirmed the colocalization of tankyrase and TRF1 by fluorescence profile analysis across telomeric signals. Profile analysis of 145 TRF1 tagged meiotic telomeres (Figure 5b) disclosed distinct tankyrase signals at 125 of these telomeres. While TRF1 localized exclusively to the telomeres, extratelomeric tankyrase signals were noted as a fine granular background scattered over the chromatin of spermatocyte nuclei (Figure 6, b and c). These extratelomeric signals might represent remnants of nontelomeric tankyrase dispersed by spreading-induced disintegration of the nuclear membrane –detergent extraction of nonfixed, isolated meiocyte nuclei has been shown to disintegrate cytoplasm and nuclear envelope (Stick and Schwarz 1983). This may also explain why we failed to detect a nuclear pore-associated tankyrase distribution, which has been previously observed in cultured cross-linking fixed somatic cells (Smith and de Lange 1999). Additionally, tankyrase may redistribute at meiotic prophase when pore distribution is significantly altered (Fawcett and Chemes 1979, see below).
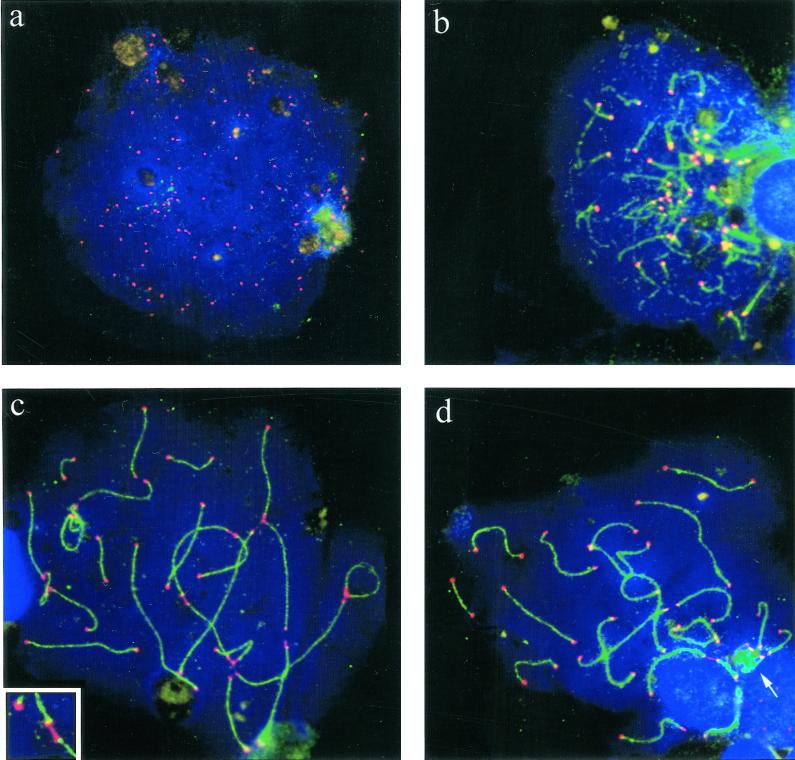
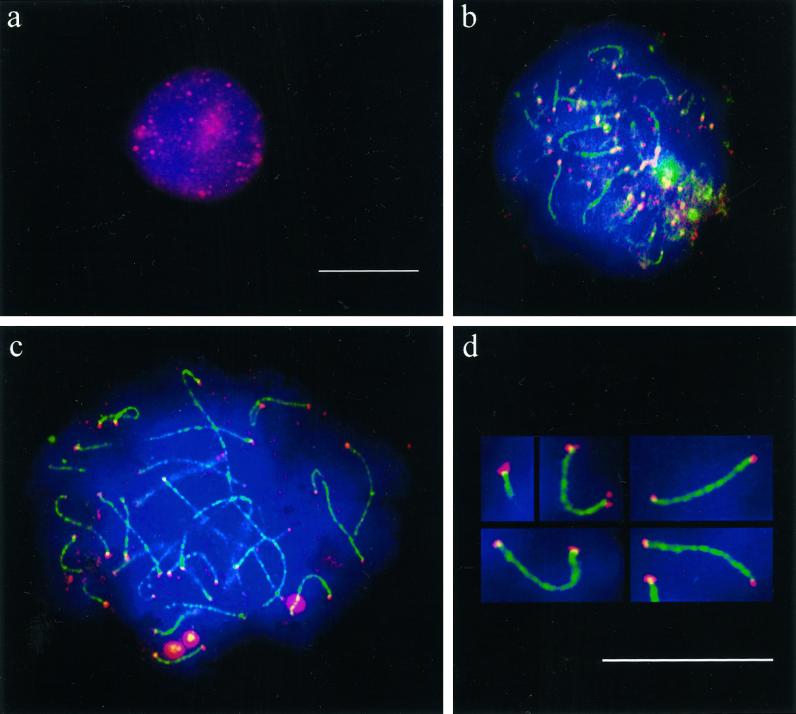
Figure 6.
Ionic detergent spread human testicular nuclei after immunostaining with anti-tankyrase (Cy3; red) and anti-SCP3 (FITC, green) antibodies. DNA is stained with DAPI (blue). (a) Premeiotic cell displaying numerous tankyrase signals scattered throughout the nucleus and its periphery. This pattern agrees with the localization of tankyrase at telomeres and nuclear pores (Smith and de Lange 1999). (b) Top of a mildly spread leptotene nucleus at the bouquet stage (accumulated telomeres). The axial elements (green threads) are capped with tankyrase signals (reddish). Surplus small red signal dots are scattered between the clustered chromosome ends and possibly represent remnants of extratelomeric tankyrase. (c) Pachytene spread showing small but distinct tankyrase signals at the ends of the SCs (green). The bright red dots are background grains often seen in this testes sample. (d) Magnified details of SCs showing distinct tankyrase signals at their termini. Sometimes, the tankyrase signals were split in two and seen to extend beyond the SC ends. This is particularly evident in the first two details. In the upper left detail the axial core extends beyond the focal plane. Bars in a and d represent 10 μm. The bar in a applies also to b and c.
Usually, telomeric tankyrase signals were confined to a fraction of spermatocytes in a preparation, suggesting that there could be lower amounts of tankyrase at meiotic telomeres compared with the other telomere proteins investigated. The requirement for ionic detergent spreading to reveal tankyrase signals suggests that most of the protein may be masked in the telomere complex or attachment plaque and/or that protein may be lost during disruption of the nuclear membranes. Investigation of rodent telomeres failed to reveal tankyrase signals with all preparation types and antitankyrase sera tested (# 465; # 762; # 763), which could be due to limited access of antibodies in undisrupted nuclei and insufficient retention of tankyrase during the spreading procedures applied. Because tankyrase is highly conserved between mammalian species (S. Smith, A. Himelblau, and TdL, unpublished data) and because tankyrase was detected by Western analysis in rat testes nuclear extract (Smith et al. 1998), a diminished affinity of the anti-human tankyrase sera to homologous rodent proteins seems not to be a likely cause for the observed variation. In any case, the results in human spermatogenesis unequivocally suggest that tankyrase is also a distinct component of meiotic telomeres.
Mammalian Rap1 Is Found at Meiotic Telomeres
In all types of human testicular preparations, immunostaining with antibody # 765 to full length mammalian mRap1 (Li et al., 2000) revealed scattered small distinct mRap1 signals in somatic nuclei (our unpublished results) and distinct signal spots at the ends of unpaired meiotic chromosomes cores (Figure 7a). As in the case of TRF1 and TRF2, the signals seen at SC ends of human pachytene spermatocytes were generally strong and large (Figure 7b). Mouse and rat meiotic chromosome cores also displayed mRap1 at the ends of paired and unpaired meiotic chromosome cores obtained in mild nonionic detergent spreads and swab preparations (Figures 2b, 8), thereby demonstrating the conserved nature of this protein and its localization to the meiotic telomeric complex.
Figure 8.
Mildly spread nuclei from testicular suspensions of the rat (Rattus norvegicus) immunostained with anti-hRap1 Abs (Cy3; red) and anti-SCP3 Abs (FITC, green). DNA is shown in blue (DAPI). (a) Premeiotic nucleus with numerous mRap1 signal spots. SC proteins are absent. (b) Mildly spread leptotene/early zygotene nucleus. Numerous thin axial cores are seen with synapsis in progress between two chromosome ends (thick green signal stretch) near the bouquet basis. The latter contains numerous closely spaced chromosome ends, capped with strong mRap1 signals. (c) Mildly spread late pachytene nucleus with distinct mRap1 signals at SC ends. Some SCs extend beyond the focal plane, which is at the top of this nucleus. (d) Mildly spread pachytene nucleus with strong mRap1 signals at SC ends. The closely spaced ends of the backfolded cores of the XY bivalent exhibit distinct mRap1 signals (arrowhead). Top of nucleus shown. Bar in c: 10 μm.
Telomere Redistribution at Meiosis Leads to Congregation of Nuclear Pore Complexes in Telomere-Free Areas
During plant and insect meiosis telomeres and nuclear pores accumulate in limited areas of the nuclear envelope (e.g., Gillies 1975, Church 1976, Holm 1977). The present investigation showed that tankyrase, which is found at somatic telomeres and nuclear pore complexes (NPC) (Smith and de Lange 1999), is present in low but detectable amounts at meiotic telomeres. Furthermore, a recent observation in yeast suggests a possible link between telomeres and NPCs in vegetative cells (Galy et al., 2000), and ends of spread mammalian pachytene chromosomes have been found to be occasionally attached to pore-rich fragments of the nuclear envelope (see Moses 1977). This prompted us to test for a potential association of meiotic telomeres with NPCs during mammalian meiosis. To this end, we obtained cross-linking fixed human testis suspension nuclei and immunostained these with mAb414, which binds to p62 and related nucleoporins (Davis and Blobel 1987, Aris and Blobel 1989). NPCs were found to be evenly distributed across the envelope of human somatic nuclei, giving rise to a typical rim pattern at the equatorial focal plane of nuclei, while TRF1-tagged somatic telomeres, in contrast, were distributed throughout the nuclear lumen (Figure 9, a–c). The more or less even somatic NPC distribution was changed to a patchy pattern in early meiotic nuclei that displayed telomere signals in the nuclear interior (preleptotene; Figure 9d). More advanced spermatocytes (leptotene–pachytene, as identified by threadlike DAPI staining of the chromosomes; Figure 9, f and h) exhibited a dramatic accumulation of NPCs in a few restricted areas of the nuclear envelope (NE), while the NE-attached telomeres were generally distributed in pore-free areas (Figure 9, f and h). In several cases where telomeres were seen near NPC accumulations, the immediate vicinity of these telomeres appeared free of NPC fluorescence signals (Figure 9e). These observations mirror earlier 3D-EM observations in grasshopper meiosis (see Church 1976). To exclude potential negative influence of the detergent extraction used in the standard IF protocol, we also performed immunostaining without Triton extraction of suspension nuclei. However, the results obtained were similar to the ones with detergent extraction (our unpublished results). This agrees with Alsheimer et al. (1999), who showed that short Triton extraction of cross-linking fixed meiocytes leaves the nuclear periphery intact. When we determined the fraction of TRF1 telomere signals colocalizing or partially overlapping with NPC fluorescence signals in nonextracted spermatocyte I nuclei by profile measurements, it appeared that only 17% of TRF1 signals showed an association with NPC fluorescence, while 83% of TRF1 telomere signal spots (n = 217) were devoid of NPC signals. Similar values were obtained in detergent-extracted spermatocyte I nuclei. Furthermore, the largely mutual exclusive distribution properties of telomeres and NPCs were observed from leptotene–diplotene as deduced from SCP3 and NPC costaining experiments (our unpublished results). Altogether, it appears that the nuclear pore-dense regions of the mammalian spermatocyte (Fawcett and Chemes 1979) are largely void of telomeres, and that the round-up of NPCs during meiotic prophase most likely relates to the kinetic activity of NE-bound meiotic telomeres and changes in nuclear envelope architecture (see below).
DISCUSSION
During the onset of meiotic prophase, mammalian telomeres attach to the nuclear envelope, where they develop motility, which leads to their transient clustering during the leptotene/zygotene transition (e.g., Rasmussen and Holm 1980, Scherthan et al., 1996). This perinuclear localization and motility of meiotic telomeres contrasts with somatic telomeres, which are largely confined to the nuclear interior (Vourc'h et al., 1993; Luderus et al., 1996; Scherthan et al., 1996). Here we determined whether the protein composition of the somatic telomere complex differs from that of meiotic telomeres. The data of our IF experiments show that the duplex telomere repeat binding proteins TRF1 and TRF2 (Chong et al., 1995; Bilaud et al., 1997; Broccoli et al., 1997a) locate in abundance to premeiotic as well as meiotic telomeres during all stages of prophase I of humans and rodents. Similarly, mammalian Rap1, a TRF2-interacting protein that shows considerable homology to scRap1 (Li et al., 2000) is a component of the mammalian telomeric complex. The observations in mammalian meiosis reveal an analogy to the situation in S. cerevisiae, where scRap1, the major telomere repeat-binding protein of yeast (Shore and Nasmyth 1987; Longtine et al., 1989), is also present at vegetative and meiotic telomeres (Klein et al., 1992; Gotta et al., 1996). The abundance of TRF and Rap1 proteins at meiotic telomeres of mammals and yeast (this report, Klein et al., 1992) demonstrate a cross-kingdom conservation of the telomeric complex at meiosis. However, in contrast to the yeast protein, human Rap1 locates to the telomere through interaction with TRF2 (Li et al., 2000).
Recently, it has been noted that Taz1 of the fission yeast S. pombe (Cooper et al., 1997) represents an orthologue of TRF1 and TRF2. Like the human proteins, Taz1 binds to duplex telomere repeats, is a negative regulator of telomere length (see Li and de Lange 2000), and localizes to mitotic and meiotic telomeres (Cooper et al., 1998; Nimmo et al., 1998). Disruption of Taz1 function leads to abrogation of telomere-mediated transcriptional silencing (Cooper et al., 1997) and induces failure of meiotic telomere clustering at the spindle pole body and defective nuclear motility during the horsetail stage of the asynaptic fission yeast meiosis (for review see de Lange, 1998b; Cooper 2000). By analogy, we propose that, during mammalian meiosis, TRF1 and/or TRF2 are required for tethering telomere repeats to the attachment plaque, which connects the ends of the axial/lateral elements with the nuclear envelope (see von Wettstein et al., 1984).
In spread spermatocytes, we observed fibrous TRF1 connections between nonhomologous chromosome ends. These associations could represent persisting contacts of meiotic telomeres, which are frequent during the bouquet stage. Such interactions could involve TRF1 and TRF2, which bind to telomeres as homodimers or higher order oligomers (Bianchi et al., 1997, 1999; Broccoli et al., 1997a). Furthermore, TRF1 has been shown to form synthetic synaptic complexes between parallel aligned TTAGGG arrays in vitro (Griffith et al., 1998). Telomeric sequences are bound by the two myb domains of TRF1 with great flexibility, and sequences between TRF1-bound half sites created looped structures in vitro (Bianchi et al., 1999). Telomeric DNA loops of spread pachytene chromosomes have been shown to be significantly smaller than DNA loops more distant from the telomere (Heng et al., 1996), which could reflect the special protein composition and TTAGGG repeat binding properties at the meiotic chromosome end. A persisting telomeric complex may explain why telomere attachment plaques remain prominent while the SC is gradually disintegrated during diplotene.
Confluent IF signals between peripheral telomere complexes could involve further telomere proteins like tankyrase (see below) and proteins of the meiotic nuclear envelope, such as meiosis specific lamin C2, which accumulates around attachment plaques of SC at the inner nuclear envelope (Alsheimer et al., 1999). Lamin C2 is capable of forming larger structures with lamin B1 that shows a more continuous perinuclear distribution in the pachytene meiocyte (see Alsheimer et al., 1999). Generation of a more discontinuous meiotic nuclear lamina throughout prophase I may be the underlying mechanism for the patchy distribution of nuclear pore complexes. A more fluid nuclear envelope may facilitate motility of peripheral meiotic telomeres.
Recently, it has been shown that somatic nuclear pore complexes are associated with Tpr protein filaments which extend into the nuclear interior in human, flies, and yeast (Cordes et al. 1993, 1997, Zimowska et al. 1997, Strambio-de Castillia et al. 1999, for review see Paddy 1998). Nuclear pores are the gateways of protein and RNA traffic in and out of the eukaryotic nucleus (for recent reviews see Nakielny and Dreyfuss, 1999; Blobel and Wozniak, 2000). In meiosis of some species, the accumulation of pores near the bouquet basis has been thought to facilitate synapsis initiation near closely spaced chromosome ends (see Loidl 1990). Recently, it has been demonstrated that Mlps, homologs of mammalian Tpr, are involved in tethering vegetative telomeres to the nuclear envelope via their interaction with nuclear pore complexes (Galy et al. 2000). However, vegetative telomere positioning and Mlp/Tpr localization are unaffected in nuclear pore clustering strains of yeast (LaRoche et al. 1998, Strambio-deCastillia et al. 1999), suggesting that telomere/pore connections can be resolved. In human meiosis, we observed that pores and telomeres are rearranged largely independently from each other. Similar observations have been made in mouse prophase I nuclei (M.J. and H.S., unpublished results). Therefore, it will be interesting to determine whether telomere/Tpr interactions exist in mammalian nuclei. Analysis is under way to determine Tpr and telomere distribution in mammalian meiosis.
Another protein potentially involved in telomere/nuclear membrane interactions could be tankyrase, a TRF1 interacting telomere-specific PARP which has been shown to modulate the telomere binding activity of TRF1 through poly-ADP-ribosylation in vitro (Smith et al. 1998). Similar to ankyrins, tankyrase contains 24 ANK repeat motifs in its TRF1 interacting domain (Smith et al., 1998). Ankyrins anchor cytoskeletal components to transmembrane proteins (for review see Bennett 1992). A role for tankyrase during earliest prophase could involve anchorage of the attachment plaque to the nuclear envelope via its ANK repeats. In this respect it will be interesting to determine potential interactions of tankyrase with lamins or integral proteins of the inner nuclear membrane (see Foisner and Gerace, 1993; Dechat et al., 1998). Another role of tankyrase could be to modulate the affinity of TRF1 to telomeric repeats or telomere/envelope interactions through poly-ADP-ribosylation of TRF1 and itself. However, such a function would most likely be involved in the resolution of attachment plaques and telomere associations during diplotene/anaphase I. Most interestingly, in grasshopper anaphase I, the final point of resolution of sister chromatid cohesion during anaphase I is the telomere (Suja et al., 1999). The dearth of anaphases and the relative compacted and clumped mammalian chromosomes in our formaldehyde-fixed preparations precluded a test for the involvement of telomere proteins in such a mechanism.
The only telomere protein of a synaptic organism whose expression is restricted to meiosis is, up to now, the product of Ndj1 of budding yeast (Chua and Roeder, 1997; Conrad et al., 1997). Ndj1 localizes to yeast telomeres throughout prophase, and its absence confers defects in the distribution of telomeric Rap1 at pachytene, and impairs cross-over interference and segregation of nonrecombinant chromosomes (Chua and Roeder, 1997; Conrad et al., 1997). Recently, we observed that Ndj1 is required for bouquet formation in the synaptic meiosis of budding yeast (Trelles-Sticken et al. 2000). Further molecular and biochemical studies are required to identify an orthologous protein in higher eukaryotes and other specific players in the meiotic telomere act.
ACKNOWLEDGMENTS
These studies were supported by a Deutsche Forschungsgemeinschaft grant to H.S. (no.: Sche350/8–3). T.d.L. acknowledges grant support from the NIH. B.L. is a recipient of a fellowship from the Leukemia Society of America. We thank Peter de Boer, Wageningen, NL, for help with the materials, C. Heyting, Wageningen, NL, for providing SCP3 antiserum, and Jan Karlseder, Rockefeller University, New York, NY, for serum #644 to mouse Trf1.
REFERENCES
- Albini SM, Jones GH. Synaptonemal complex-associated centromeres and recombination nodules in plant meiocytes prepared by an improved surface-spreading technique. Exp Cell Res. 1984;155:588–592. doi: 10.1016/0014-4827(84)90219-2. [DOI] [PubMed] [Google Scholar]
- Alsheimer M, von Glasenapp E, Hock R, Benavente R. Architecture of the nuclear periphery of rat pachytene spermatocytes: distribution of nuclear envelope proteins in relation to synaptonemal complex attachment sites. Mol Biol Cell. 1999;10:1235–1245. doi: 10.1091/mbc.10.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris JP, Blobel G. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J Cell Biol. 1989;108:2059–2067. doi: 10.1083/jcb.108.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V. Ankyrins. Adaptors between diverse plasma membrane proteins and the cytoplasm. J Biol Chem. 1992;267:8703–8706. [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H., and Greider, C.W. (1995). Telomeres. Cold Spring Harbor Monograph Series, Cold Spring Harbor, NY.
- Blobel G, Wozniak RW. Proteomics for the pore. Nature. 2000;403:835–836. doi: 10.1038/35002687. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins TRF1 and TRF2. Nat Genet. 1997a;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Chong L, Oelmann S, Fernald AA, Marziliano N, van Steensel B, Kipling D, Le Beau MM, de Lange T. Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression and conserved protein domains. Hum Mol Genet. 1997b;6:69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst O, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- Church K. Arrangement of chromosome ends and axial core formation during early meiotic prophase in the male grasshopper Brachystola magna by 3D, EM reconstruction. Chromosoma. 1976;58:365–376. doi: 10.1007/BF00292844. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Cooper JP. Telomere transitions in yeast: the end of the chromosome as we know it. Curr OpinGenet & Dev. 2000;10:169–177. doi: 10.1016/s0959-437x(00)00070-8. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandjinou AT, Dionne I, Gravel S, LeBel C, Parenteau J, Wellinger RJ. Cytological and functional aspects of telomere maintenance. Histol Histopathol. 1999;14:517–524. doi: 10.14670/HH-14.517. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci U S A. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Telomeres and senescence: ending the debate. Science. 1998a;279:334–335. doi: 10.1126/science.279.5349.334. [DOI] [PubMed] [Google Scholar]
- de Lange T. Ending up with the right partner. Nature. 1998b;392:753–754. doi: 10.1038/33790. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gotzmann J, Stockinger A, Harris CA, Talle MA, Siekierka JJ, Foisner R. Detergent-salt resistance of LAP2alpha in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J. 1998;17:4887–4902. doi: 10.1093/emboj/17.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A.F., Sedat, J.W., Cande, W.Z., and Bass, H.W. (1995). The cytology of telomeres. In: Telomeres. ed. E.H. Blackburn and C.W. Greider. pp. 295-338, Cold Spring Harbor Monograph Series, Cold Spring Harbor, NY.
- Fawcett DW, Chemes HE. Changes in distribution of nuclear pores during differentiation of the male germ cells. Tissue & Cell. 1979;11:147–162. doi: 10.1016/0040-8166(79)90015-6. [DOI] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- Gillies CB. Synaptonemal complex and chromosome structure. Annu Rev Genet. 1975;9:91–109. doi: 10.1146/annurev.ge.09.120175.000515. . . [DOI] [PubMed] [Google Scholar]
- Gilson ET, Laroche T, Gasser SM. Telomeres and the functional architecture of the nucleus. Trends Cell Biol. 1993;3:128–134. doi: 10.1016/0962-8924(93)90175-z. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Bianchi A, de Lange T. TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol. 1998;278:79–88. doi: 10.1006/jmbi.1998.1686. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Heng HHQ, Chamberlain JW, Shi XM, Spyropoulos B, Tsui LC, Moens PB. Regulation of meiotic chromatin loop size by chromosomal position. Proc Natl Acad Sci U S A. 1996;93:2795–2800. doi: 10.1073/pnas.93.7.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm PB. Three-dimensional reconstruction of chromosome pairing during the zygotene stage of meiosis in Lillium longiflorum (Thunb.) Carlsb. Res Comm. 1977;42:103–151. [Google Scholar]
- Ishikawa F, Naito T. Why do we have linear chromosomes? A matter of Adam and Eve. MutatRes. 1999;434:99–107. doi: 10.1016/s0921-8777(99)00017-8. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53-, and ATM-dependent. apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Meiosis: how could it work? Proc Natl Acad Sci U S A. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF-X, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers JHM, Offenberg HH, van Aalderen M, Vink AC, Dietrich AJ, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: Implications for telomere evolution. Cell, 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Loidl J. The initiation of meiotic chromosome pairing: the cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Wilson NM, Petracek ME, Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable fromRAP1. Curr Genet. 1989;16:225–39. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- Luderus MEE, van Steensel B, Chong L, Sibon OCM, Cremers FFM, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca M, Müller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111:1363–1370. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- Moses MJ. Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus). I. Morphology of the autosomal complement in spread preparations. Chromosoma. 1977;60:99–125. doi: 10.1007/BF00288459. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram CS, Dani M, Deaven LL, Jones DJ, Meyne J, Ratcliff RL, Wu JR. A highly conserved DNA sequence, (TTAGGG)n present at the telomeres of human chromosomes. Proc Natl Sci USA. 1988;85:6622–6628. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. The remaking of chromosomes. The Collecting Net (Woods Hole) 1938;13:181–195. [Google Scholar]
- Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–90. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Paddy MR. The Tpr protein: linking structure and function in the nuclear interior? Am J Hum Genet. 1998;63:305–310. doi: 10.1086/301989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandita TK, Westphal CH, Anger M, Sawant SG, Geard CR, Pandita RK, Scherthan H. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol Cell Biol. 1999;19:5096–5105. doi: 10.1128/mcb.19.7.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SW, Holm PB. Mechanics of Meiosis. Hereditas. 1980;93:187–216. doi: 10.1111/j.1601-5223.1980.tb01360.x. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 1998;12:2574–86. doi: 10.1101/gad.12.16.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: it takes two to tango. Genes Develop. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Cremer T. Methodology of non isotopic in situ-hybridization in paraffin embedded tissue sections. In: Adolph KW, editor. Methods in Molecular Genetics. San Diego: Academic Press; 1994. , 5, 223–238. [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Härle M, Heyting C, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T. TRF1, a mammalian telomeric protein. Trends Genet. 1997;13:21–26. doi: 10.1016/s0168-9525(96)10052-4. [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112:3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stick R, Schwarz H. Disappearance and reformation of the nuclear lamina structure during specific stages of meiosis in oocytes. Cell. 1983;33:949–958. doi: 10.1016/0092-8674(83)90038-7. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suja JA, Antonio C, Debec A, Rufas JS. Phosphorylated proteins are involved in sister-chromatid arm cohesion during meiosis I. J Cell Sci. 1999;112:2957–2969. doi: 10.1242/jcs.112.17.2957. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Loidl J, Scherthan H. Bouquet formation in budding yeast: Initiation of recombination is not required for meiotic telomere clustering. J Cell Sci. 1999;112:651–658. doi: 10.1242/jcs.112.5.651. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., Dresser, M.E., and Scherthan, H. (2000). Meiotic telomere protein Ndj1p is required for meiosis specific telomere distribution and bouquet formation in budding yeast. J. Cell Biol., in press. [DOI] [PMC free article] [PubMed]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- Vourc'h C, Taruscio D, Boyle AL, Ward DC. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res. 1993;205:142–151. doi: 10.1006/excr.1993.1068. [DOI] [PubMed] [Google Scholar]
- Wagenaar EB. End-to-end attachments in mitotic interphase and their possible significance to meiotic chromosome pairing. Chromosoma. 1969;26:410–426. [Google Scholar]
- Zalensky AO, Tomilin NV, Zalenskaya IA, Teplitz RL, Bradbury EM. Telomere-telomere interactions and candidate telomere binding protein(s) in mammalian sperm cells. Exp Cell Res. 1997;232:29–41. doi: 10.1006/excr.1997.3482. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]