Abstract
In Arabidopsis ton2 mutants, abnormalities of the cortical microtubular cytoskeleton, such as disorganization of the interphase microtubule array and lack of the preprophase band before mitosis, markedly affect cell shape and arrangement as well as overall plant morphology. We present the molecular isolation of the TON2 gene, which is highly conserved in higher plants and has a vertebrate homolog of unknown function. It encodes a protein similar in its C-terminal part to B″ regulatory subunits of type 2A protein phosphatases (PP2As). We show that the TON2 protein interacts with an Arabidopsis type A subunit of PP2A in the yeast two-hybrid system and thus likely defines a novel subclass of PP2A subunits that are possibly involved in the control of cytoskeletal structures in plants.
INTRODUCTION
Plants possess unique features in many aspects of development compared with animals. At the cellular level, plants are characterized by specific features such as the presence of a pecto-cellulosic cell wall, continuous cytoplasmic connections through the plasmodesmata, and the lack of cell motility during morphogenesis (Kaplan and Hagemann, 1991). Reflecting these unique characteristics, the highly dynamic cortical arrays of microtubules (MTs) and actin filaments have adopted various specialized arrangements in plants. The organization of cortical arrays is coordinated tightly with other cellular events, and the cortical cytoskeleton, plasmalemma, and cell wall operate as a continuum (Wyatt and Carpita, 1993). The cortical cytoskeleton reorganizes steadily during all stages of the plant cell life and plays a crucial role in governing the orientation of both cell division and expansion (Cyr and Palevitz, 1995). In contrast to animals, fungi, and protists, in land plants the establishment of division planes depends on (1) the positioning of the preprophase band (PPB), a transient cortical ring of MTs that precisely foretells the position of the cell division plane at the G2/prophase transition, and (2) guidance of the phragmoplast to this predetermined cortical site during cytokinesis. During interphase, arrays of parallel MTs encircle the cell at the cortex. The positioning of MTs within these cortical arrays appears to involve both MT dynamicity (nucleation, growth/shrinkage, stabilization, and severing) and translocation, but the relative roles of these events and the proteins involved are unknown.
To understand the molecular mechanisms underlying MT arrangements, mutants impaired in MT functions are essential, and a number of proteins involved in MT organization or dynamics have been identified through a genetic approach (Azimzadeh et al., 2001). These include the Arabidopsis botero1 mutant (which also is allelic to the fra2 mutant) (Zhong et al., 2001), which displays incorrect orientation of interphase cortical MTs and defects in anisotropic growth in root tip cells (Bichet et al., 2001). The FRA2 gene encodes a protein with high similarity to the sea urchin p60 subunit of katanin, which is known to be involved in MT severing (Burk et al., 2001; McClinton et al., 2001). The Arabidopsis temperature-sensitive mor1 mutation causes cortical MT shortening and disorganization concomitant with isotropic cell expansion and left-handed organ twisting (Whittington et al., 2001). The MOR1 gene encodes a protein similar to a family of MT-associated proteins from mammals and fungi (Whittington et al., 2001).
The Arabidopsis zwichel trichome-branching mutation affects the kinesin-like calmodulin binding protein (Oppenheimer et al., 1997). This MT motor protein is proposed to provide the MT stability needed for trichome branch initiation (Mathur and Chua, 2000). The maize tangled1 mutant has an altered cell division pattern associated with defective PPB positioning and phragmoplast guidance (Cleary and Smith, 1998). The TANGLED1 gene was cloned recently and was found to encode a MT-associated protein (Smith et al., 2001). Finally, in Arabidopsis spiral mutants, which exhibit right-handed twisting of organs associated with defects in cell growth anisotropy, cortical MTs form left-handed helices instead of transverse arrays (Furutani et al., 2000). The SPIRAL1 and SPIRAL2 genes define two novel plant-specific gene families involved in cortical MT organization (I. Furutani, H. Tachimoto, and T. Hashimoto, unpublished data).
The Arabidopsis fass/tonneau mutants were isolated originally from visual genetic screens for mutations affecting seedling body organization (Mayer et al., 1991; Torres-Ruiz and Jürgens, 1994; Traas et al., 1995). In these mutants, both the arrangement of interphase cortical MTs and PPB formation are affected (Traas et al., 1995; McClinton and Sung, 1997). The tonneau (ton) mutations fall into two complementation groups, ton1 and ton2 (Traas et al., 1995). Only one mutant allele was isolated for the ton1 locus, segregating in a line obtained from large-scale T-DNA insertional mutagenesis (Nacry et al., 1998). All other mutant alleles belong to the same complementation group (ton2) and are allelic to fass.
Mutations at these loci drastically change plant shape, resulting in thick, dwarf seedlings and plantlets. However, the general body pattern (i.e., the number and relative positions of organs) is not altered, and mutants eventually produce highly compressed inflorescences when grown in vitro (Mayer et al., 1991; Torres-Ruiz and Jürgens, 1994; Traas et al., 1995).
At the cellular level, ton mutants are altered markedly in cell size, shape, and arrangement. This appears to be related to a defect in cell elongation and a random orientation of division planes. Cell differentiation seems mostly unaffected, and most cell types are present, including cell structures such as stomata that result from asymmetrical divisions. Cells of ton mutants have abnormal organization of the interphase cortical cytoskeleton and are unable to form PPBs (Traas et al., 1995; McClinton and Sung, 1997). In interphase cells, cortical MTs most often are arranged randomly instead of forming transverse regular arrays, which are normally associated with cell elongation in plants. Noncortical MTs seem to be unaffected.
Thus, the ton mutations uncouple histogenesis (oriented cell divisions and cell shape changes) and morphogenesis from organogenesis and provide new insights into basic processes of plant development. The elucidation of the molecular bases of these mutations should provide key information on the molecular control of the cytoskeleton and cell division/elongation in plants (Lloyd, 1995). Here, we present the molecular isolation of the TON2 gene. The TON2 locus was isolated by a combination of map-based cloning and insertional mutagenesis. We show that the protein likely functions as a regulatory subunit of type 2A protein phosphatase (PP2A) and may be involved in phosphorylation cascades that control the dynamic organization of the cortical cytoskeleton in plant cells.
RESULTS
Isolation and Genetic Characterization of ton2 Mutants
A total of 16 independent alleles of the ton2 mutation were identified in several visual screens for Arabidopsis morphological mutants affected in cell elongation. Mutant alleles ton2-13 and ton2-14 were isolated from a population of T-DNA insertion lines in ecotype Wassilewskija (Ws). Other ton2 alleles were recovered from ethyl methanesulfonate–mutagenized populations in Columbia (Col), Landsberg erecta, and Ws ecotypes; among those, mutants ton2-5 and ton2-12, both in the Col background, were chosen for further analysis. All ton2 mutations segregate as single recessive Mendelian traits and are allelic to the previously described fass (Mayer et al., 1991) and gordo (Fisher et al., 1996) mutations.
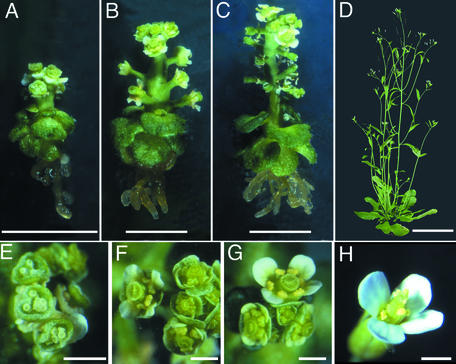
ton2-5 and ton2-13 show extreme phenotypes with a highly compressed apical–basal axis and presumably correspond to a complete loss of function, whereas ton2-12 and ton2-14 have milder phenotypes with less pronounced morphological alterations and better elongation (Figures 1A to 1D). In vitro, plants reach a maximum height of ∼1.5 to 3 cm at maturity for strong and weak alleles, respectively. Some almost morphologically normal, although sterile, flowers are observed in weak alleles (Figures 1E to 1G).
Figure 1.
Phenotypes of Mutant Plants.
Photographs showing strong ([A], ton2-13) and weak ([B], ton2-12; [C], ton2-14) alleles of the ton2 mutation 6.5 weeks after germination, compared with wild-type Arabidopsis grown in the greenhouse (D). Flowers of mutant plants are shown in (E) (ton2-13), (F) (ton2-12), and (G) (ton2-14), and a wild-type flower is shown in (H). Bars = 5 mm in (A) to (C), 5 cm in (D), and 1 mm in (E) to (H).
ton2 Mutations Affect the Organization of the Cortical Microtubular Cytoskeleton
It was demonstrated previously that ton mutant cells show abnormalities in the organization of the cortical microtubular cytoskeleton (Traas et al., 1995; McClinton and Sung, 1997). In this study, we examined the dynamic organization of MTs in ton2 mutants using a newly developed fusion protein, the green fluorescent protein–microtubule binding domain (GFP-MBD) reporter protein (Marc et al., 1998). The GFP-MBD reporter protein enables the labeling of MTs in vivo by a noninvasive method, which allows us to follow MT rearrangements during mitosis and interphase. Ws lines expressing a GFP-MBD marker were obtained by in planta transformation. One morphologically normal line was selected and then crossed to heterozygous ton2-13 (strong allele) and ton2-14 (weak allele) lines. Examples of confocal imaging of wild-type and mutant cells are shown in Figure 2.
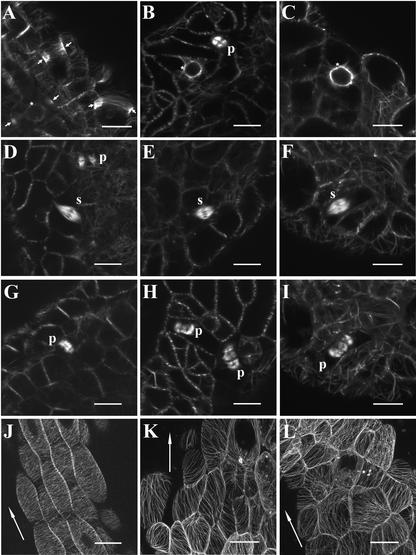
Figure 2.
Confocal Imaging of MT Arrays in Wild-Type, ton2-14 Weak Allele, and ton2-13 Strong Allele Seedlings Expressing a GFP-MBD Fusion (Marc et al., 1998).
(A), (D), (G), and (J) Wild type.
(B), (E), (H), and (K) ton2-14 weak allele.
(C), (F), (I), and (L) ton2-13 strong allele.
(A) to (I) MT arrays in dividing cells from leaf primordia of 7-day-old seedlings. In wild-type cells, MTs are first organized in a PPB encircling the cell at the cortex (arrows in [A]). In ton2 mutant premitotic cells, PPBs are never observed, and only perinuclear MTs are visible (asterisks in [B] and [C]). MTs then organized in the mitotic spindle during metaphase (s in [D] to [F]) and in the phragmoplast during telophase (p in [G] to [I]). Spindles and phragmoplasts of ton2 mitotic cells are similar to those in the wild type. However, guidance of the phragmoplast is largely affected in the mutant background. Note, for example, that (F) and (I) show images of the same cells taken 25 min apart: a spindle with a transverse metaphase plate is followed by a phragmoplast that appears to show erratic growth along a longitudinal plane. (A), (F), and (I) show stacks of five (A), seven (F), and nine (I) optically sectioned images taken 1 μm apart. (B) to (E), (G), and (H) show single optical sections. Bars = 10 μm.
(J) to (L) Interphase MT arrays in epidermal hypocotyl cells of 7-day-old seedlings. Images were taken from the central part of the hypocotyl. In wild-type cells (K), MTs are perpendicular to the hypocotyl and the cell elongation axis (arrows in [J] to [L]). In weak alleles (ton2-14 [K]), cells undergo slight elongation parallel to the hypocotyl axis, which is not observed in strong alleles (ton2-13 [L]). In both mutant alleles, MTs show essentially parallel organization in hypocotyl cells. In the weak allele (K), MTs are in oblique or longitudinal orientation with respect to the hypocotyl axis, whereas the strong allele does not show any preferential MT orientation (L). Arrows in (J) to (L) indicate the axes of hypocotyl growth. (J) to (L) show stacks of 5 (K), 8 (L), and 11 (J) optically sectioned images taken 2 μm apart. Bars = 50 μm.
Division figures were difficult to observe in mutant roots because of their shape and thickness, and they were observed mainly from leaf primordia at the apex. Together, our results confirm the absence of observable PPBs in premitotic cells of the mutants (Figures 2A to 2C). The mitotic arrays of MTs are not affected visibly in ton2 mutants, and division proceeds normally, as judged from the sequence of events observed with the GFP-MBD marker. Spindles have a normal appearance (Figures 2D to 2F), as do the phragmoplasts, which grow centrifugally, as expected (Figures 2G to 2I). However, the guidance of the phragmoplast is clearly affected, as shown by its erratic growth (Figure 2I). Oblique, misshapen phragmoplasts often are observed in mutant cells. Ultimately, they attach to random sites at the cortex, generating the irregularly shaped cells observed in the mutants. We observed no difference in the MT organization of weak and strong alleles in dividing cells. There also was no difference in the duration of the cell cycle phases between wild-type and weak and strong allele cells.
GFP-MBD expression also was observed in hypocotyl cells to examine MT interphase arrays in the mutant background (so-called interphase MTs, even if these cells are not strictly in G1 but are in G0 phase). In wild-type epidermal hypocotyl cells of 7-day-old seedlings, cortical MTs have a parallel and transverse alignment (Figure 2J). In contrast to wild-type seedlings, ton2 mutant epidermal hypocotyl cells are more irregular in shape, radially swollen, and only partially elongated compared with wild-type cells, especially in the strong allele (Figures 2K and 2L). In hypocotyl cells of ton2-13 (strong allele), interphase MTs have mostly parallel arrays whose orientation is not well fixed, with cells in the same area having a transverse, oblique, or longitudinal MT array orientation (Figure 2L). In ton2-14 (weak allele), these defects are less pronounced, and most MTs are oriented obliquely with respect to the growth axis (Figure 2K). This parallel organization is in contrast to the organization of cortical MTs in root epidermal cells observed previously in random arrays (Traas et al., 1995; McClinton and Sung, 1997). Such random MT organization also was observed in cells located in the elongation zone of ton2 mutant roots, next to the region of emerging root hairs (data not shown).
Molecular Characterization of the TON2 Locus
A map-based cloning strategy was devised originally to isolate the TON2 locus. This locus was mapped approximately to the top arm of chromosome 5 (Torres-Ruiz and Jürgens, 1994). An F2 population of 500 individuals was obtained from a cross between ton2-5 (Col ecotype) and wild-type Landsberg erecta. This population was used for fine mapping of ton2 using several markers in the mi438 (33 centimorgan) region of chromosome 5, and a bacterial artificial chromosome (BAC) contig of ∼250 kb covering the ton2 locus was constructed (data not shown; details and markers are available upon request). In the course of this work, the T-DNA allele ton2-13 was isolated. DNA gel blot analysis of this line revealed a single full-length T-DNA insertion, and segregation studies showed that the ton2 phenotype was linked 100% to the T-DNA insert on the basis of the kanamycin resistance of all mutants. Both sides of the T-DNA insertion site were isolated by inverse polymerase chain reaction (PCR) and shown to map precisely in the TON2 BAC contig. Genomic fragments encompassing this locus were isolated from BAC F20K17 (Col ecotype), and 5.8 kb was sequenced. Arabidopsis 5′ and 3′ expressed sequence tag (EST) sequences corresponding to this region were present in dbEST, deriving from a single cDNA clone (YAY132). We sequenced this cDNA clone in its entirety. Our sequences fully agree with the genome sequence that was released subsequently.
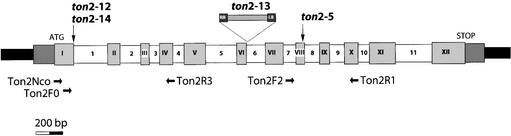
The TON2 cDNA is 1.75 kb long and contains a single open reading frame of 1440 bp, which encodes a predicted polypeptide of 480 residues. The TON2 gene contains 11 introns and 12 exons (Figure 3). In the ton2-13 strong allele, the single T-DNA insertion is located at the very beginning of intron 6. The right border of the T-DNA is associated with a 37-bp deletion of the genomic DNA and a 16-bp insertion of unknown origin. This mutation presumably gives rise to a fusion translation product of 222 amino acids composed of the first 209 residues of the TON2 protein and 23 residues of T-DNA origin.
Figure 3.
Structure of the TON2 Gene.
The TON2 gene comprises 12 exons (gray boxes I to XII) and 11 introns (white boxes 1 to 11). Mutations affecting the four ton2 alleles are indicated. ton2-13 (strong allele) is a T-DNA insertion after the first base of intron 6. ton2-5 (strong allele) is a nonsense mutation in exon VIII. ton2-12 and ton2-14 (weak alleles) are a point mutation and a 23-bp deletion affecting the donor site of intron 1, respectively. Arrows indicate the locations and orientations of the primers used in RT-PCR experiments (see Figures 4 and 5). LB, left border; RB, right border.
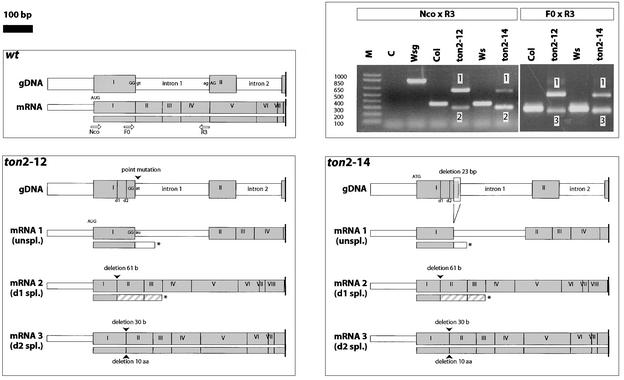
The molecular defects in the ton2-5, ton2-14, and ton2-12 alleles were characterized by sequencing of reverse transcriptase–mediated (RT)–PCR-amplified cDNA and genomic DNA fragments. Compared with wild-type parental sequences, all alleles carry mutations in the same gene (Figure 3). In the ton2-5 strong allele, a nonsense mutation (C960T in cDNA clone YAY132) causes the premature termination of translation after Leu-267 (Gln-268 to Stop) (Figure 3). Therefore, the N-terminal part of the protein (up to residue 267) presumably is not enough for the correct translation and/or activity of the protein. Interestingly, mutations in both weak alleles (ton2-12 and ton2-14) affect the same splice site, the donor site of intron 1 (Figure 4). In both mutants, two major transcripts were revealed by RT-PCR experiments that are expected to give nonfunctional truncated polypeptides comprising (part of) exon I with small extensions (Figure 4). However, in both weak mutants, an alternative misspliced transcript of lower abundance can be detected using specific RT-PCR primers (Figure 4). This second misspliced transcript results in a 10–amino acid truncation of the wild-type predicted polypeptide (Gly-38 to Met-47). This transcript most likely is responsible for the leaky phenotype of the ton2-12 and ton2-14 alleles. Whether the weak mutant phenotype is caused by a low abundance of transcript and protein, a partial functionality of the deleted polypeptide, or both is not known at present.
Figure 4.
Molecular Defects in ton2 Weak Alleles ton2-12 and ton2-14.
The structures of genomic DNA (gDNA) and transcripts (mRNA) are represented for wild-type, ton2-12, and ton2-14 alleles. For clarity, only the 5′ part of the ton2 locus is indicated. Gray boxes represent exons (numbered I to VIII), and white boxes represent introns. The predicted structure of the encoded polypeptide is indicated under each transcript (gray indicates an in-frame translation product of exons; hatched gray indicates out-of-frame translation of exons; white indicates translation of introns; and the asterisk indicates the stop codon). The positions of RT-PCR primers are indicated by white arrows. The positions of alternative donor sites d1 and d2 are indicated. Sequencing of genomic DNA revealed that both weak alleles have a mutation affecting the donor site of intron 1. ton2-12 has a single-base change that converts this site from TGGgt to TGGat, and ton2-14 has a 23-bp deletion that removes the same splice site. RT-PCR analyses of wild-type and weak alleles are shown at top right. All transcripts were characterized by RT-PCR and sequencing of RT-PCR products. As a result of the mutations affecting the donor site of intron 1, splicing of this intron is perturbed and three transcripts are detected in each mutant (bands 1, 2, and 3). In both mutants, two major transcripts were revealed by RT-PCR using primers TON2Nco and TON2R3. Transcript 1 is unspliced and contains intron 1. Transcript 2 is misspliced (d1 donor site) and has the last 61 nucleotides of exon I deleted. These two abnormal transcripts are expected to give nonfunctional truncated polypeptides comprising (part of) exon I with small extensions. However, in both weak mutants, an alternative misspliced transcript (mRNA 3, d2 donor site) of lower abundance can be detected using other RT-PCR primers (Ton2F0 and Ton2R3) that are not able to amplify transcript 2. This second misspliced transcript contains a 30-nucleotide in-frame deletion at the end of exon I, resulting in a 10–amino acid truncation (Gly-38 to Met-47) in the predicted translation product. aa, amino acids; C, control (reverse transcriptase omitted); M, molecular mass markers; spl., spliced; unspl., unspliced; Wsg, genomic DNA; wt, wild type.
The TON2 Gene Is Expressed Ubiquitously
RNA gel blot analysis (data not shown) and RT-PCR experiments (Figure 5) revealed that the TON2 transcript is produced ubiquitously in Arabidopsis. The TON2 transcript was detected at a low level in seedlings and in all organs of the adult plants tested (Figure 5), even those in which cell elongation and division rates are the lowest (i.e., rosette and cauline leaves). It was detected at the same level in dividing suspension-cultured cells and in highly elongated dark-grown seedlings. TON2 expression level also was not modified in ton1 mutants, which share the same phenotype with ton2 mutants.
Figure 5.
Analysis of TON2 Expression by RT-PCR.
A 248-bp RT-PCR product (bottom band) is obtained using Ton2F2 and Ton2R1 primers (see Figure 3) for PCR. The constitutively expressed APRT1 gene (Moffatt et al., 1994) is used as a quantitative control (564-bp RT-PCR product; top band). (−), negative RT-PCR control (reverse transcriptase omitted); Gn, Ws genomic DNA amplified with TON2-specific primers; Et, etiolated Ws seedlings; R, roots; RL, rosette leaves; S, stems; CL, cauline leaves; F, flowers from adult plants; ¢, Arabidopsis suspension cultured cells. The ton2-13 mutant produces a fusion transcript containing the Basta resistance gene of the T-DNA used for mutagenesis, the left border, and the 3′ end of the TON2 mRNA (see Figure 3).
The TON2 Protein Has Strong Similarities to a Regulatory Subunit of PP2A
The TON2 gene encodes a 55-kD protein with a predicted pI of 4.7. Database searches revealed that the gene is highly conserved in higher plants, and nearly identical potential orthologs are available as EST sequences for tomato, Medicago truncatula, poplar, sorghum, and maize (data not shown).
The C-terminal part of TON2 shows significant similarity to the C terminus of a 72-kD human protein (PR72) (Figure 6B). PR72 was purified originally from rabbit skeletal muscle as a component of the trimeric form of Ser/Thr PP2A (Hendrix et al., 1993) and defines the B″ family of PP2A regulatory subunits. PP2As are either heterodimeric enzymes composed of a catalytic subunit (PP2Ac) and a regulatory subunit A or heterotrimeric complexes composed of PP2Ac, subunit A, and various type B regulatory subunits of different masses. B-type subunits associate with the regulatory A subunits and provide targeting and substrate specificity to the enzymatic complex (Luan, 2000). At least three distinct families of B subunits (B, B′, and B″) exist that do not share sequence similarity. The plant TON2 proteins can be assigned to the B″ regulatory subunit family based on the high similarity of their C-terminal regions. The N terminus shows no significant similarity to previously known members of this family.
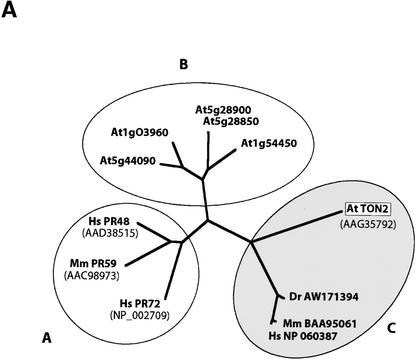
Figure 6.
Sequence Analysis of the TON2 Protein.
(A) Unrooted ClustalX (version 1.8.1) tree of protein sequences similar to TON2. Full (deduced) protein sequences were used for alignment. Branch lengths are proportional to calculated sequence divergence. GenBank accession numbers are indicated for each protein. At, Arabidopsis thaliana; Dr, Danio rerio; Hs, Homo sapiens; Mm, Mus musculus. Group A contains mammalian B′′ subunits, including the founding member human PR72 (Hendrix et al. 1993). Group B contains the five putative B′′ subunits identified from the complete genome sequence of Arabidopsis. Group C includes Arabidopsis TON2 and proteins of unknown function identified by sequencing in human, mouse, and zebrafish. Many highly similar TON2 homologs are found in plant ESTs as partial sequences and are not represented here.
(B) Amino acid alignment of the TON2 protein, its human homolog , and human PR72, the founding member of the B′′ class of PP2A subunit. The mouse and zebrafish TON2-like sequences are essentially identical to the human sequence and are not shown here. Identical residues are indicated by black boxes, and similar residues are indicated by gray boxes (Clustal groups). Note the N-terminal extension in the TON2 sequence that is found in all plant sequences but not in animal sequences.
In addition, the TON2 protein is similar over its entire length to vertebrate proteins of unknown function (Figure 6A), showing 57% similarity (36% identity) to human (Figure 6B) and mouse homologs. Partial EST sequences indicate that an ortholog exists in Danio rerio (Figure 6A). With respect to the mammalian polypeptide, all plant TON2 proteins possess an N-terminal extension of variable length (22 to 31 amino acids; 24 for Arabidopsis) and sequence (Figure 6B). Software available for the prediction of organellar or secretion targeting signals failed to attribute a clear functional significance to this N-terminal extension. All of these results indicate that the plant TON2 proteins and their vertebrate homologs define a novel subclass of PP2A B″ subunits (Figure 6A).
The TON2 Protein Interacts with an Arabidopsis Type A Subunit of PP2A in the Yeast Two-Hybrid System
The yeast two-hybrid system provides a fast and efficient way of testing physical interactions between components of protein phosphatase complexes (Haynes et al., 1999). We used this system to assay the interaction of TON2 with known Arabidopsis type A PP2A regulatory subunits (Table 1).
Table 1.
Interaction between TON2 and Arabidopsis PP2A Subunits Assayed Using the Yeast Two-Hybrid System
| pGBT9 insert | ||||||
|---|---|---|---|---|---|---|
| pGAD424 Insert |
No Insert |
PP2Ac-1 | B′β | TON2 | TON2Nterm | TON2Cterm |
| No insert | − | − | − | − | − | + |
| Aα | − | + | + | + | − | + |
| AαΔ | − | − | − | − | − | + |
Yeast HF7c cells were cotransformed with the plasmids pGBT9 and pGAD424 (Clontech) encoding the GAL4 DNA binding domain and activation domain, respectively. Plasmids pGBT9 or pGAD424 with in-frame cDNAs encoding the PP2Ac-1, B′β, Aα, and AαΔ subunits of Arabidopsis PP2A have been described by Haynes et al. (1999). The AαΔ cDNA encodes a truncated version of the Aα subunit lacking the last 193 amino acids and unable to interact with PP2Ac-1. Interaction between TON2 and A subunit was tested with the TON2 full-length protein, the N-terminal part (TON2Nterm, up to Glu-268), and the C-terminal part (TON2Cterm, from Glu-269). Positive controls consisted of the Aα regulatory subunit and the PP2Ac-1 or B′β subunit. Protein interaction in yeast cells was tested by growth on medium lacking Trp, Leu, and His. Positive results are indicated by +, and negative results are indicted by −. No insert indicates the introduction of the pGAD424 or the pGBT9 plasmid without insert. The interactions with TON2Cterm were meaningless because of the strong background generated by the TON2 C-terminal construct itself.
The full-length TON2 protein clearly is able to interact with the entire A-type subunit AtAα, as is the genuine B-type regulatory subunit AtB′β. This interaction is abolished either when the C-terminal part of TON2 (from Glu-269) is deleted or when a truncated form of the A subunit lacking the last 193 amino acids (AtAαΔ) is used (Table 1). These results suggest that TON2 could act as a B-type regulatory subunit in PP2A enzymatic complexes. As expected from sequence data, the C-terminal part of the protein is necessary for interaction with other subunits.
DISCUSSION
For more than a decade, Arabidopsis mutants have been used to characterize the molecular bases of development in flowering plants, and several mutants impaired in cytoskeleton organization have been isolated. To address the question of the control of MT organization in plants, we have undertaken the isolation of the TON2 gene. The ton2 mutation affects the cortical MT cytoskeleton and has been shown to perturb a basic mechanism(s) involved in both cell elongation and cell division. Here, we report the identification of the TON2 gene. The TON2 protein is similar to a type B″ PP2A regulatory subunit and interacts in the two-hybrid system with known PP2A subunits. We also characterized in greater detail the MT organization of this mutant.
The use of the GFP-MBD MT reporter protein allowed us to visualize accurately dynamic changes in the organization of the MT cytoskeleton in living cells of ton2 and wild-type plants. Time-lapse observations of cell division in GFP-MBD–expressing plants confirm unambiguously the absence of the MT PPB in both strong and weak mutant alleles, in agreement with tubulin-immunolabeling studies from McClinton and Sung (1997). Lack of PPB in the ton2 mutant most likely is responsible for the mispositioning of new cell walls between daughter cells after mitosis. Indeed, the position of the PPB has been correlated in a number of previous reports to the position of the division site in vegetative cells. PPB is proposed to leave signals at the cell cortex that allow the centrifugally forming cell plate and phragmoplast to be guided toward the division site (Mineyuki and Palevitz, 1990). Monitoring GFP-MBD fluorescence in ton mutants revealed that the phragmoplast initially grows normally and becomes misoriented later, before anchoring to the wall. This might be explained by a deficiency in phragmoplast guidance attributable to an absence of appropriate cortical signals at the division site. Comparable defects in the establishment of division planes are described in discordia mutants, which are impaired in an actin-dependent mechanism involved in phragmoplast guidance (Gallagher and Smith, 1999).
Thus, the ton2 phenotype reinforces hypotheses linking the PPB and the division site. It also confirms that PPBs are not required for mitosis per se, the progression of which is normal in mutant plants (McClinton and Sung, 1997). The PPB, rather, is viewed as a component of the specific cytokinetic apparatus that evolved in land plants, although cell cycle regulators may control its assembly and disassembly (Katsuta and Shibaoka, 1992; Colasanti et al., 1993).
To determine whether PPB formation is the main target of the ton2 mutation or if the mutation also primarily affects interphase MTs, we observed MT organization in nondividing cells. In hypocotyls of both ton2 strong and weak mutant alleles, most epidermal cells were found to form parallel arrays of MTs. This ability to align parallel arrays of MTs favors the idea that the loss of growth anisotropy in the ton2 mutant results only from the mispositioning of division planes caused by the absence of the PPB. However, two other observations prove that interphase cortical MT organization is a primary defect of the ton2 mutation. (1) In agreement with previous studies (Traas et al., 1995; McClinton and Sung, 1997), we observed that interphase MTs are oriented randomly in root cells (data not shown). This finding indicates that the loss of cortical MT alignment likely is responsible for the reduced anisotropy of ton2 cells, as proposed by Traas et al. (1995) and McClinton and Sung (1997). Indeed, transversely oriented cortical MTs in young cells undergoing rapid elongation appear to be essential to determine the main growth axis in plant cells (Baskin et al., 1999; Bichet et al., 2001). (2) The phenotype of ton2 weak mutant alleles also supports this hypothesis. Indeed, hypocotyl cells of weak mutant alleles, in which the TON2 function is not impaired completely, also lack a PPB but exhibit a higher rate of cell elongation, combined with a better alignment of cortical MTs. This finding demonstrates that the lack of PPBs cannot explain all of the cell expansion defects observed in the ton2 strong alleles and that TON2 function likely is required for the control of directional expansion by cortical interphase arrays.
It is surprising that in hypocotyl epidermal cells of the ton2 mutant, MTs are able to form parallel arrays even in strong allele cells in which TON2 most likely is nonfunctional. This phenomenon was not observed in root epidermal cells. We propose that the alignment of cortical MTs in hypocotyl cells may be a consequence of cell shape and/or wall architecture during the late stages of the elongation process. For example, cellulose microfibrils, which are the strongest component of the primary wall, could be involved in providing spatial cues for cortical MT organization during the late elongation phase. For instance, MT organization is inhibited after treatment of tobacco cells with the cellulose synthase inhibitor isoxaben (Fisher and Cyr, 1998). In addition, in Arabidopsis root epidermal cells, MTs acquire a transverse orientation after microfibril transverse alignment (Sugimoto et al., 2000). Similarly, the structure of the wall itself could influence the organization of cortical MT arrays in ton2 cells.
These observations favor a role for TON2 in organizing the cortical microtubular network. How this is achieved remains to be determined. TON2 may act directly on the MT arrays or indirectly by influencing the actin cytoskeleton. Indeed, actin microfilaments are aligned in the cell cortex parallel to MTs (Palevitz, 1987; Traas et al., 1987) and colocalize with the PPB, forming a broad band at the end of interphase or during preprophase that narrows as prophase progresses (Palevitz, 1987; Traas et al., 1987; Cleary et al., 1992). Cytochalasin treatments do not affect PPB formation, despite their inhibitory effect on PPB narrowing (Mineyuki and Gunning, 1990; Eleftheriou and Palevitz, 1992). In this regard, the absence of PPBs in ton2 mutants is unlikely to result from a perturbation of the actin cytoskeleton. Future work will make use of GFP markers available (Kost et al., 1998) for the examination of the F-actin network to determine whether the organization of actin filaments is perturbed in the mutant.
Cortical MT arrays are of major importance for plant morphogenesis, but their dynamics and interrelation remain poorly understood. To our knowledge, TON2 is the only plant protein identified that is essential for the organization of both arrays, and its molecular identification provides the first clue to the regulatory processes of such cellular structures. The C-terminal part of the TON2 protein is highly similar to members of the B″ regulatory subunit family of type 2A Ser/Thr protein phosphatases. Two major structurally distinct families of Ser/Thr phosphatases, PP1/PP2A and PP2C, are present in plants as in animals. Additional studies also suggest the presence of other types of phosphatases in Arabidopsis (Andreeva et al., 1998). Most PP2A is in the form of heterotrimeric enzyme comprising a catalytic (C) subunit and two distinct regulatory subunits, A and B. The sequence of the C subunit is highly conserved in evolution, and its function has been shown to be essential in yeast, Drosophila, and mouse. The C subunit forms distinct complexes with a wide array of regulatory A and B subunits. The A subunits are hook-shaped proteins that act as scaffolding subunits (Groves et al., 1999). The C and B subunits are recruited by the A subunit to form specific heterotrimers.
The B-type subunits have been shown to modulate the activity, substrate specificity, and subcellular localization of the PP2A complex (Goldberg, 1999). A remarkable diversity of B-type subunits is present in eukaryotes, and basic types of such subunits include B (PR55), B′ (PR56), B″ (PR72/PR130): each subfamily generally is encoded by several homologous genes and splice variants. The B″ family comprises three related mammalian proteins: human PR72/PR130 (splice variants) and PR48, and mouse PR59. Proteins from Caenorhabditis elegans and Arabidopsis also may be included in this family on the basis of their sequence similarity (Yan et al., 2000). The recent release of the complete genomic sequence of Arabidopsis reveals at least three type A, five type C, two type B, nine type B′, and five type B″ PP2A subunits, not including the TON2 protein. Such a variety of subunits likely reflects a large combinatorial diversity of functions for PP2A complexes.
As expected for a bona fide PP2A B-type subunit, we observed a clear interaction between TON2 and an A-type subunit from Arabidopsis in the yeast two-hybrid system. Together with sequence similarity, these results support TON2's involvement in the formation of a PP2A complex, which presumably controls the phosphorylation status of proteins important for the structure of the plant cortical cytoskeleton.
In yeast and animals, PP2A activity regulates a wide range of cell processes (Virshup, 2000). In particular, PP2A controls phosphorylation cascades involved in the control of cell growth and division (Millward et al., 1999). Although most PP2A activity is soluble, distinct pools of enzyme are associated with subcellular structures such as membranes or MTs. In animal cells, antibodies against A, B, and C PP2A subunits label interphase and mitotic MTs as well as centrosomes. It was confirmed recently (Hiraga and Tamura, 2000) that MT–PP2A interaction requires the B subunit. Such a MT-associated PP2A fraction has not been observed in plants to date. Nevertheless, the regulation of plant MT cytoskeleton dynamics clearly involves protein phosphorylation/dephosphorylation: the alignment of cortical MTs is lost in Arabidopsis roots, and the cortical MT cytoskeleton is disrupted in BY-2 tobacco cells treated with PP1 and PP2A inhibitors (Hasezawa and Nagata, 1992; Baskin and Wilson, 1997); PPB formation also is perturbed by the inhibition of PP1 and PP2A activities (Hasezawa and Nagata, 1992) or PP2A activity only (Ayaydin et al., 2000). Reciprocally, kinase inhibitor treatments delay the formation and disappearance of PPB (Katsuta and Shibaoka, 1992), induce cortical MT cold stabilization (Mizuno, 1992), and lead to the inhibition of gibberellin-induced cortical MT reorientation (Mizuno, 1994).
The finding that TON2 likely encodes a PP2A regulatory subunit provides molecular evidence of a key role for the phosphorylation/dephosphorylation of proteins in the control of cortical MT organization. The target of such phosphatase activity is not known at present, although a possible candidate could be the protein(s) encoded by the TON1 locus (Nacry et al., 1998), because a mutation at this locus gives a phenotype identical to the ton2 phenotype in every respect (Traas et al., 1995). In addition, ton1 ton2 double mutants are indistinguishable from both single mutants (data not shown), giving a strong indication that these mutations affect the same process.
Most basic processes that determine the architecture of cortical MT arrays remain unknown. As suggested by Baskin and Wilson (1997), three steps leading to MT cytoskeleton reorganization should be distinguished: emission of a positional signal, either from cell wall or intracellular factors (such as cell cycle regulators); perception and transmission of the signal to the MTs by putative transmembrane or cytoplasmic proteins; and MT reorientation. Whether this last step involves intact MT repositioning or MT depolymerization followed by selective stabilization of MTs polymerized in the correct orientation, or both, remains to be determined. One or more of these processes are potential targets for regulation by phosphorylation/dephosphorylation. The identification of proteins involved in these processes, including putative TON2 targets, and their link to recently identified plant cytoskeletal components (Smertenko et al., 2000; Smith et al., 2001; Whittington et al., 2001), should allow a better understanding of the manner in which the cortical cytoskeleton is organized in plant cells.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown in vitro as described previously (Nacry et al., 1998). T-DNA insertion lines were obtained by vacuum infiltration transformation (Bechtold et al., 1993) using Agrobacterium tumefaciens strain MP5-1 (Bouchez et al., 1993). Ethyl methanesulfonate mutants were obtained from a screen of individual M2 families derived from ethyl methanesulfonate–mutagenized seed of Columbia ecotype (Santoni et al., 1994).
Green Fluorescent Protein–Microtubule Binding Domain Marker Lines and Confocal Imaging
Wassilewskija plants expressing the green fluorescent protein–microtubule binding domain (GFP-MBD) marker were constructed as follows. A HindIII-EcoRI fragment containing the GFP-MBD reporter cassette was isolated from the original construct (Marc et al., 1998) and inserted into pCAMBIA1300. This vector was introduced into Agrobacterium strain C58C1(MP90). Transgenic Arabidopsis plants were obtained using the infiltration procedure (Bechtold et al., 1993; Clough and Bent, 1998), followed by in vitro selection on hygromycin. Several transformants showed slight morphological defects, presumably as a result of the cytotoxicity of the reporter cassette. One line was chosen for further use on the basis of its lack of morphological alterations and good GFP expression, and it was crossed with ton2-13 and ton2-14 heterozygous lines.
Roots, hypocotyls, and flower primordia of plants expressing the GFP-MBD fusion protein were mounted in low-melting-point agarose (0.4% in water) and viewed directly using a Leica TCS-NT confocal laser scanning microscope (Leica, Heidelberg, Germany) with an argon/krypton laser (Omnichrome, Chino, CA) and an acousto-optical tunable filter (AOTF) for excitation. GFP fluorescence was excited with the 488-nm line and collected through a bandpass filter (BP 530/30). Slow-scan (220 lines/sec) images (1024 × 1024 pixels) were generated using a ×40/1.00-0.50 PL FLUO-TAR objective.
Molecular Cloning Techniques and Sequence Analysis
Cloning and sequencing procedures were performed essentially as described (Sambrook et al., 1989; Nacry et al., 1998). Sequence comparisons and alignments were performed using the BLAST and Clustal programs. Subcellular localization predictions were made using PSORT (http://psort.nibb.ac.jp/), ChloroP (www.cbs.dtu.dk/services/), and Predotar (www.inra.fr/Internet/Produits/Predotar/).
Reverse Transcriptase–Mediated Polymerase Chain Reaction
Total RNA was prepared by the guanidinium isothiocyanate/cesium chloride method, as described previously (Sambrook et al., 1989), using modified homogenization buffer (5 M guanidinium isothiocyanate in 0.5% sarkosyl and 25 mM Na citrate) and cushion solution (5.7 M cesium chloride in 1 mM EDTA and 25 mM Na citrate). Single-stranded cDNAs were synthesized from 5 μg of total RNA using 100 pmol of oligo(dT)18 and 200 units of Moloney murine leukemia virus reverse transcriptase (Invitrogen, Cergy-Pontoise, France) in a 30-μL reaction mixture containing 1 mM DTT, 40 units of RNase inhibitor (Boehringer Mannheim), and 250 μM each of dATP, dCTP, dGTP, and dTTP in 1 × first-strand buffer (Invitrogen). Reverse transcription was performed for 2 hr at 39°C, and the reaction mixture was diluted to a final volume of 800 μL.
Polymerase chain reaction (PCR) was performed using 5 μL of the diluted cDNA sample with 250 pmol of the primers in a 25-μL reaction. PCR conditions were as follows: 94°C for 2 min; 28 cycles of 94°C for 15 sec, 58°C for 20 sec, and 72°C for 45 sec; and a final elongation step of 5 min at 72°C. APRT1- and TON2-specific primers were used in separate reactions. Five microliters of each reaction were electrophoresed on an agarose gel. Primers used for PCR were as follows: Ton2Nco (5′-CCGAATTCCCCATGGCTAGCGGATCTA- GCGATGGTGAGA-3′), Ton2F0 (5′-GCTTTGGGTTAGAAATCTACG- AC-3′), Ton2F2 (5′-AATCAAATGTATTGCCGCATAGCTT-3′), Ton2R1 (5′-GCACAGCGATCCACTCATATCTT-3′), and Ton2R3 (5′-ATAGCA- GCCAAATCATCAGCGTTTA-3′). For APRT1, primers 5′-TCCCAG- AATCGCTAAGATTGCC-3′ and 5′-CCTTTCCCTTAAGCTCTG-3′ were used.
Yeast Two-Hybrid Assays
The TON2 coding sequence was amplified by PCR from clone YAY132 using primers Ton2Nco (see above) and Ton2Stop (5′-CCC- GAATTCGTCGACTCACTGAGACTCTTCCTCAGGTGG-3′) using Pfu polymerase as indicated by the supplier (Stratagene). The resulting amplification product was introduced into the EcoRI site of pGEX5X-1 (Pharmacia Biotech) and sequenced fully. The full-length TON2 coding sequence was isolated as an EcoRI fragment and introduced into the yeast vector pGBT9 (Clontech, Palo Alto, CA) to give pGBT9-TON2, which contained an in-frame fusion of the GAL4 DNA binding domain with TON2. The N-terminal (up to Gln-268) and C-terminal (from Glu-269) parts of TON2 also were introduced in frame in the same vector. The C-terminal PstI fragment was removed from pGBT9-TON2 to give pGBT9-TON2Nterm, which contains the GAL4 domain fused to the first 268 residues of the TON2 protein; the same PstI fragment was isolated from pGBT9-TON2, blunt ended using T4 Polymerase (Invitrogen), and introduced in the SmaI site of pGBT9 to give pGBT9-TON2Cterm, a fusion of the GAL4 DNA binding domain with the C-terminal part of TON2 from Glu-269.
Plasmids pGBT9 and pGAD424 (Clontech) containing cDNAs encoding Arabidopsis type 2A protein phosphatase subunits Aα, AαΔ, B′β, and c-1, which were generously provided by S. Rundle (Western Carolina University, Cullowhee, NC), were described by Haynes et al. (1999). Constructs were cotransformed into yeast strain HF7c (Clontech) using the lithium acetate procedure and plated onto yeast minimal medium lacking either Leu and Trp or Leu, Trp, and His (Bartel et al., 1993).
Accession Numbers
The accession numbers for the sequences described in this article are AF290025 (BAC F20K17), Z35033 and Z35032 (Arabidopsis 5′ and 3′ ESTs, respectively), AF280057 (cDNA clone YAY132), and NP_060387 (HsTON2).
Acknowledgments
We are grateful to Gerd Jürgens and Ulrike Mayer (University of Tübingen, Germany), Jérôme Giraudat (CNRS, Gif-sur-Yvette, France), Richard Cyr (Pennsylvania State University), and Sabine Rundle (Western Carolina University) for kindly providing materials used in this work. We thank Josette Rousse and Anna Christodoulidou for their help and Herman Höfte for critical reading of the manuscript. J.A. was the recipient of a fellowship from the French Ministry for Research. This work was supported by a grant from the French Ministry for Research (Action Concertée Incitative Biologie du Développement Grant 47).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010402.
References
- Andreeva, A.V., Evans, D.E., Hawes, C.R., Bennett, N., and Kutuzov, M.A. (1998). PP7, a plant phosphatase representing a novel evolutionary branch of eukaryotic protein Ser/Thr phosphatases. Biochem. Mol. Biol. Int. 44, 703–715. [DOI] [PubMed] [Google Scholar]
- Ayaydin, F., Vissi, E., Meszaros, T., Miskolczi, P., Kovacs, I., Feher, A., Dombradi, V., Erdodi, F., Gergely, P., and Dudits, D. (2000). Inhibition of serine/threonine-specific protein phosphatases causes premature activation of cdc2MsF kinase at G2/M transition and early mitotic microtubule organisation in alfalfa. Plant J. 23, 85–96. [DOI] [PubMed] [Google Scholar]
- Azimzadeh, J., Traas, J., and Pastuglia, M. (2001). Molecular aspects of microtubule dynamics in plants. Curr. Opin. Plant Biol. 4, 513–519. [DOI] [PubMed] [Google Scholar]
- Bartel, P.L., Chien, C., Strenglanz, R., and Fields, S. (1993). Using the two-hybrid system to detect protein-protein interactions. In Cellular Interactions in Development: A Practical Approach, D.A. Harley, ed (Oxford, UK: Oxford University Press), pp. 153–179.
- Baskin, T., and Wilson, J. (1997). Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 113, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, T.I., Meekes, H.T.H.M., Liang, B.M., and Sharp, R.E. (1999). Regulation of growth anisotropy in well-watered and water-stressed maize roots. II. Role of cortical microtubules and cellulose microfibrils. Plant Physiol. 119, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. 316, 1188–1193. [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Cleary, A.L., and Smith, L.G. (1998). The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 10, 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, A.L., Gunning, B.E.S., Wasteneys, G.O., and Hepler, P.K. (1992). Microtubule and F-actin dynamics at the division site in living Tradescantia stamen hair cells. J. Cell Sci. 103, 977–988. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., Cho, S.-H., Wick, S., and Sundaresan, V. (1993). Localization of the functional p34cdc2 homolog of maize in root tip and stomatal complex cells: Association with predicted division sites. Plant Cell 5, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr, R., and Palevitz, B. (1995). Organization of cortical microtubules in plant cells. Curr. Opin. Cell Biol. 7, 65–71. [DOI] [PubMed] [Google Scholar]
- Eleftheriou, E.P., and Palevitz, B.A. (1992). The effect of cytochalasin D on preprophase band organization in root tip cells of Allium. J. Cell Sci. 103, 989–998. [Google Scholar]
- Fisher, D., and Cyr, R. (1998). Extending the microtubule/microfibril paradigm. Plant Physiol. 116, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R.H., Barton, M.K., Cohen, J.D., and Cooke, T.J. (1996). Hormonal studies of fass, an Arabidopsis mutant that is altered in organ elongation. Plant Physiol. 110, 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., Thitamadee, S., Shikanai, T., and Hashimoto, T. (2000). The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Gallagher, K., and Smith, L.G. (1999). discordia mutations specifically misorient asymmetric cell divisions during development of the maize leaf epidermis. Development 126, 4623–4633. [DOI] [PubMed] [Google Scholar]
- Goldberg, Y. (1999). Protein phosphatase 2A: Who shall regulate the regulator? Biochem. Pharmacol. 57, 321–328. [DOI] [PubMed] [Google Scholar]
- Groves, M., Hanlon, N., Turowski, P., Hemming, B., and Barford, D. (1999). The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110. [DOI] [PubMed] [Google Scholar]
- Hasezawa, S., and Nagata, T. (1992). Okadaic acid as a probe to analyse the cell cycle progression in plant cells. Bot. Acta 105, 63–69. [Google Scholar]
- Haynes, J.G., Hartung, A.J., Hendershot, J.D., Passingham, R.S., and Rundle, S.J. (1999). Molecular characterization of the B′ regulatory subunit gene family of Arabidopsis protein phosphatase 2A. Eur. J. Biochem. 260, 127–136. [DOI] [PubMed] [Google Scholar]
- Hendrix, P., Mayer-Jackel, R., Cron, P., Goris, J., Hofsteenge, J., Merlevede, W., and Hemmings, B. (1993). Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A: Evidence for different size forms produced by alternative splicing. J. Biol. Chem. 268, 15267–15276. [PubMed] [Google Scholar]
- Hiraga, A., and Tamura, S. (2000). Protein phosphatase 2A is associated in an inactive state with microtubules through 2A1-specific interaction with tubulin. Biochem. J. 346, 433–439. [PMC free article] [PubMed] [Google Scholar]
- Kaplan, D., and Hagemann, W. (1991). The relationship of cell and organism in vascular plants. Bioscience 41, 693–703. [Google Scholar]
- Katsuta, J., and Shibaoka, H. (1992). Inhibition by kinase inhibitors of the development and the disappearance of the preprophase band of microtubules in tobacco BY-2 cells. J. Cell Sci. 103, 397–405. [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Lloyd, C. (1995). Life on a different plane. Curr. Biol. 5, 1085–1087. [DOI] [PubMed] [Google Scholar]
- Luan, S. (2000). Protein phosphatases: Structure, regulation, and function. Adv. Bot. Res. 32, 67–107. [Google Scholar]
- Marc, J., Granger, C.L., Brincat, J., Fisher, D.D., Kao, T., McCubbin, A.G., Cyr, R.J., and Kao, T.H. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N.H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, Y., Torres Ruiz, R.A., Berleth, T., Misera, S., and Jurgens, G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407. [Google Scholar]
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201, 252–260. [DOI] [PubMed] [Google Scholar]
- McClinton, R.S., Chandler, J.S., and Callis, J. (2001). Isolation, characterization, and intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma 216, 181–190. [DOI] [PubMed] [Google Scholar]
- Millward, T.A., Zolnierowicz, S., and Hemmings, B.A. (1999). Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 24, 186–191. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y., and Gunning, B.E.S. (1990). A role for preprophase bands of microtubules in maturation of new cell walls, and a general proposal on the function of preprophase band sites in cell division in higher plants. J. Cell Sci. 97, 527–537. [Google Scholar]
- Mineyuki, Y., and Palevitz, B.A. (1990). Relationship between preprophase band organization, F-actin and the division site in Allium: Fluorescence and morphometric studies on cytochalasin-treated cells. J. Cell Sci. 97, 283–295. [Google Scholar]
- Mizuno, K. (1992). Induction of cold stability of microtubules in cultured tobacco cells. Plant Physiol. 100, 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K. (1994). Inhibition of gibberellin-induced elongation, reorientation of cortical microtubules and change of isoform of tubulin in epicotyl segments of azuki beans by protein kinase inhibitors. Plant Cell Physiol. 35, 1149–1157. [Google Scholar]
- Moffatt, B.A., McWhinnie, E.A., Agarwal, S.K., and Schaff, D.A. (1994). The adenine phosphorisosyltransferase-encoding gene from Arabidopsis thaliana. Gene 143, 211–216. [DOI] [PubMed] [Google Scholar]
- Nacry, P., Camilleri, C., Courtial, B., Caboche, M., and Bouchez, D. (1998). Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, M.D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevitz, B.A. (1987). Actin in the preprophase band of Allium cepa. J. Cell Biol. 104, 1515–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Santoni, V., Bellini, C., and Caboche, M. (1994). Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta 192, 557–566. [Google Scholar]
- Smertenko, A., Saleh, N., Igarashi, H., Mori, H., Hauser-Hahn, I., Jiang, C.J., Sonobe, S., Lloyd, C.W., and Hussey, P.J. (2000). A new class of microtubule-associated proteins in plants. Nat. Cell Biol. 2, 750–753. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., Gerttula, S.M., Han, S., and Levy, J. (2001). TANGLED1: A microtubule binding protein required for the spatial control of cytokinesis in maize. J. Cell Biol. 152, 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz, R.A., and Jürgens, G. (1994). Mutations in the Fass gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967–2978. [DOI] [PubMed] [Google Scholar]
- Traas, J.A., Doonan, J.D., Rawlins, D.J., Shaw, P.J., Watts, J., and Lloyd, C.W. (1987). An actin network is present in the cytoplasm throughout the cell cycle of carrot cells and associates with the dividing nucleus. J. Cell Biol. 105, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676–677. [Google Scholar]
- Virshup, D.M. (2000). Protein phosphatase 2A: A panoply of enzymes. Curr. Opin. Cell Biol. 12, 180–185. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Wyatt, S., and Carpita, N. (1993). The plant cytoskeleton-cell-wall continuum. Trends Cell Biol. 3, 413–417. [DOI] [PubMed] [Google Scholar]
- Yan, Z., Fedorov, S.A., Mumby, M.C., and Sanders Williams, R. (2000). PR48, a novel regulatory subunit of protein phosphatase 2A, interacts with cdc6 and modulates DNA replication in human cells. Mol. Cell. Biol. 20, 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D., and Ye, Z. (2001). Fibers: A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]