Abstract
Biotic and abiotic stresses stimulate the synthesis of jasmonates and ethylene, which, in turn, induce the expression of genes involved in stress response and enhance defense responses. The cev1 mutant has constitutive expression of stress response genes and has enhanced resistance to fungal pathogens. Here, we show that cev1 plants have increased production of jasmonate and ethylene and that its phenotype is suppressed by mutations that interrupt jasmonate and ethylene signaling. Genetic mapping, complementation analysis, and sequence analysis revealed that CEV1 is the cellulose synthase CeSA3. CEV1 was expressed predominantly in root tissues, and cev1 roots contained less cellulose than wild-type roots. Significantly, the cev1 mutant phenotype could be reproduced by treating wild-type plants with cellulose biosynthesis inhibitors, and the cellulose synthase mutant rsw1 also had constitutive expression of VSP. We propose that the cell wall can signal stress responses in plants.
INTRODUCTION
Jasmonates (JAs) are a family of cyclopentanone compounds synthesized from linolenic acid via the octadecanoic pathway. They inhibit plant growth generally but also promote diverse processes, including fruit ripening, senescence, tuber formation, tendril coiling, pollen formation, and defense responses against insect pests and pathogens (for review, see Creelman and Mullet, 1997). In plants undergoing normal development, JAs are produced predominantly in germinating seeds and floral tissues (Creelman and Mullet, 1995). The fundamental role of JAs in Arabidopsis fertility and defense was revealed by mutants unable to synthesize or respond to these compounds, which were male sterile and had enhanced susceptibility to pests and pathogens (Staswick et al., 1992; Feys et al., 1994; McConn and Browse, 1996).
JA synthesis is induced by a number of biotic and abiotic stresses, including wounding, water deficit, and pathogen attack (Creelman et al., 1992; Creelman and Mullet, 1995; Penninckx et al., 1998). Elicitors of JA responses include chitin, oligogalacturonides, and cell wall–degrading enzymes (Rojo et al., 1999; Norman-Setterblad et al., 2000). These treatments stimulate the expression of genes such as those encoding vegetative storage proteins (VSPs; Benedetti et al., 1995), thionin (THI2.1; Epple et al., 1995), and plant defensin (PDF1.2; Penninckx et al., 1998), which are expressed characteristically in plants during response to stress and pathogens.
To identify genes involved in JA signaling, we joined the upstream region of the JA-responsive gene VSP1 to the open reading frame of luciferase (LUC) as a reporter and isolated mutants that constitutively expressed LUC, one of which was designated cev1 (Ellis and Turner, 2001). cev1 plants have stunted root growth, constitutively express the JA- and ethylene-induced genes THI2.1 and PDF1.2 and the basic chitinase gene CHI-B, and have enhanced resistance to pests and pathogens (Ellis and Turner, 2001). Here, we further characterize cev1 and show that the mutation lies within the cellulose synthase gene CeSA3, also named Ath-B (Arioli et al., 1998) and IXR1 (Scheible et al., 2001). We present evidence that the inhibition of cellulose synthesis activates JA- and ethylene-dependent stress responses.
RESULTS
cev1 Constitutively Produces JA and Ethylene
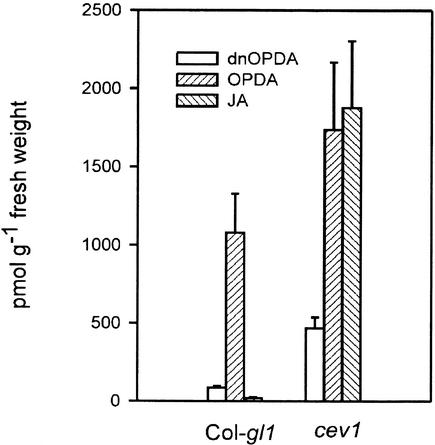
cev1 plants have constitutive expression of the JA-responsive genes VSP and THI2.1 and the JA- and ethylene-responsive genes PDF1.2 and CHI-B (Ellis and Turner, 2001). There-fore, we analyzed JA and ethylene production in cev1 plants. cev1 plants contained >1500 pmol/g fresh weight, and wild-type plants contained <25 pmol/g fresh weight. The JA biosynthetic intermediate 12-oxo-phytodienoic acid and the 16:3 fatty acid derivative dinor-12-oxo-phytodienoic acid also were present in higher amounts in cev1 plants than in wild-type plants (Figure 1). Dark-grown and light-grown cev1 seedlings produced approximately twice as much ethylene as wild-type seedlings (Table 1).
Figure 1.
The cev1 Mutant Constitutively Produces JA.
Four-week-old plants were assayed for JA, its intermediate 12-oxo-phytodienoic acid (OPDA), and the 16:3 fatty acid derivative dinor-12-oxo-phytodienoic acid (dnOPDA).
Table 1.
cev1 Plants Overproduce Ethylene
| Col-gl1 | cev1 | |
|---|---|---|
| Dark-grown seedlings | 2.1 ± 0.6 nL | 4.2 ± 0.3 nL |
| Light-grown seedlings | 22 ± 9 nL g−1 | 43 ± 6 nL g−1 |
Ethylene produced by ∼500 4-day-old dark-grown seedlings during 3 days of growth or by 200 mg of 10-day-old light-grown seedlings over 24 hours was determined (see also Methods).
The Mutant Phenotype of cev1 Plants Is Partially Suppressed by etr1 and coi1
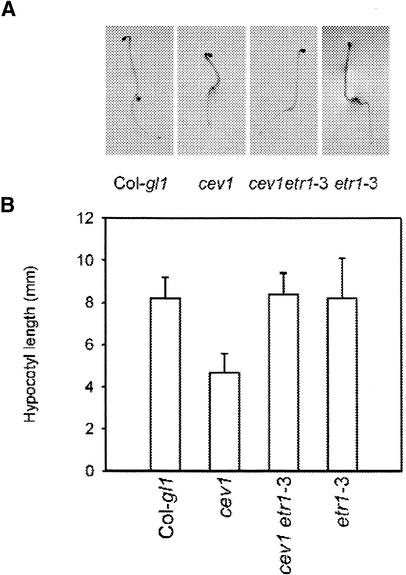
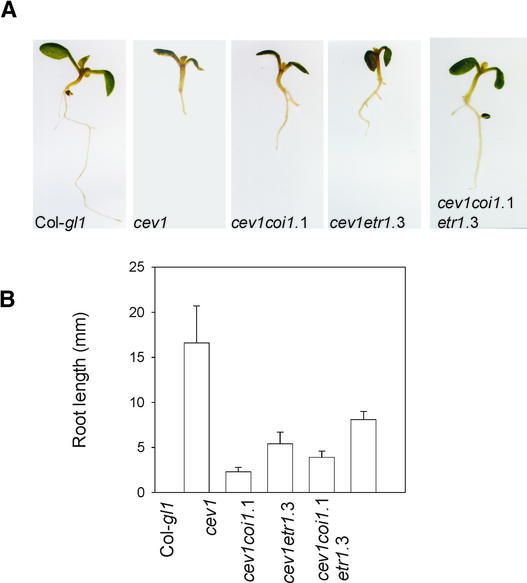
Previously, we had shown that VSP expression and anthocyanin accumulation in cev1 plants requires COI1 and that prolific root hair formation requires ETR1 (Ellis and Turner, 2001). Two other phenotypes that characterize cev1 plants are shortened hypocotyls in dark-grown seedlings (Figure 2) and shortened roots in light-grown seedlings (Figure 3). In Arabidopsis, ethylene inhibits hypocotyl elongation in dark-grown seedlings, and this response requires ETR1, and JA inhibits root growth, which requires COI1. Therefore, we examined the contribution of ethylene and JA to these cev1 mutant phenotypes in the double mutants cev1 etr1-3 and cev1 coi1-1 and in the triple mutant cev1 etr1-3 coi1-1.
Figure 2.
The Ethylene Receptor ETR1 Is Required for Shortened Hypocotyls in the cev1 Mutant.
(A) Seedlings grown in the dark for 4 days on Murashige and Skoog (1962) (MS) agar.
(B) Lengths of hypocotyls of dark-grown seedlings. T-bars indicate sd values for 10 seedlings.
Figure 3.
ETR1 and COI1 Are Partially Required for Root Growth Inhibition in the cev1 Mutant.
(A) Eight-day-old seedlings grown on MS agar.
(B) Root lengths of 12-day-old seedlings grown on MS agar. T-bars indicate sd values for 10 roots.
Hypocotyls of dark-grown double mutant cev1 etr1-3 seedlings were longer than those of cev1 and not significantly different from those of wild-type and etr1-3 seedlings (Figure 2). This finding indicates that the shortened hypocotyls of dark-grown cev1 seedlings can be accounted for by the action of ethylene. Roots of light-grown seedlings of the double mutants cev1 etr1-3 and cev1 coi1-1 were significantly longer than those of the cev1 mutant, and roots of the triple mutant cev1 etr1-3 coi1-1 were significantly longer than those of the double mutants but only half the length of those of wild-type seedlings (Figure 3). This finding indicates that the shortened roots of cev1 plants can be accounted for in part by the action of JA and ethylene.
Positional Cloning of CEV1
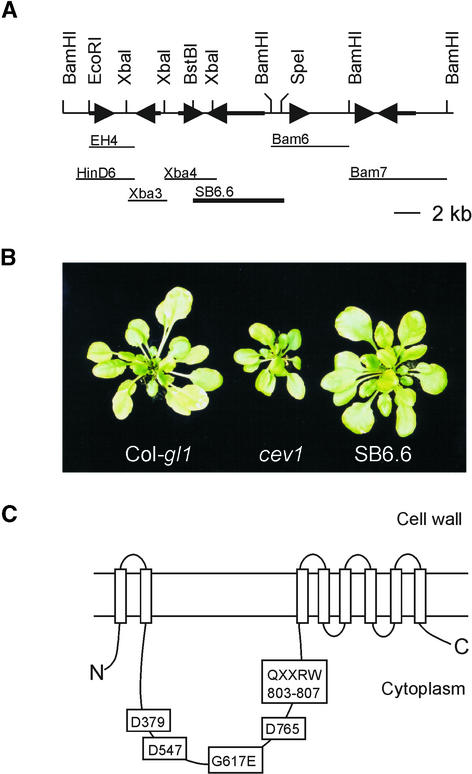
Cleaved amplified polymorphic sequence and simple sequence length polymorphism markers were used to map the cev1 mutation to an ∼20-kb region on top of chromosome 5 near nga225 and located on the transformation-competent artificial chromosome (TAC) clone K2A11 (Figure 4A). The K2A11 insert was introduced into the cev1 mutant by Agrobacterium tumefaciens–mediated transformation. Plants con-taining the transgene were selected, and expression of the LUC reporter for VSP and root length was measured. Transgenic plants had wild-type activity of the LUC reporter, and root length was not different from that of wild-type seedlings (Table 2). Significantly, the progeny of most self-pollinated primary transformants segregated to give progeny with the cev1 mutant phenotype and wild-type plants, indicating that the cev1 mutation had been complemented by sequences in K2A11.
Figure 4.
Complementation Analysis of cev1.
(A) Partial restriction endonuclease map of the K2A11 TAC clone and DNA fragments used in complementation analysis of cev1. Arrows indicate putative open reading frames. The DNA fragment SB6.6 complemented the cev1 phenotype.
(B) Photographs of 4-week-old wild-type, cev1, and SB6.6 plants.
(C) Scheme of the predicted CEV1 protein showing the cellulose synthase signature motif D,D,D,QXXRW and the position of the cev1 mutation.
Table 2.
Complementation of the cev1 Phenotype
|
VSP1::luciferase Activity (mV mg−1 min−1) |
Root Length (mm) | |
|---|---|---|
| Col-gl1 | 6.4 ± 2.8 | 18.5 ± 2.5 |
| cev1 | 98.1 ± 6.8 | 5.4 ± 0.8 |
| K2A11;cev1 | 9.3 ± 2.7 | 18.5 ± 2.9 |
| SB6.6;cev1 | 4.6 ± 2.7 | 17.1 ± 2.2 |
All plants contained the VSP1::LUC transgene. Seedlings were grown for 12 days on Murashige and Skoog agar. Luciferase activity and the length of the primary roots were determined for at least 10 seedlings.
Subclones of K2A11 in the SLJ75515 binary vector were introduced into cev1 plants by Agrobacterium-mediated transformation. Of these, only clone SB6.6 complemented the cev1 mutation (Figure 4B). SB6.6 is a 6.6-kb SpeI-BstBI fragment containing one full open reading frame for the putative cellulose synthase, also designated CeSA3 (Delmer, 1999), and a partial open reading frame from a gene bearing homology with receptor kinase genes. However, clone Xba4 (Figure 4A) containing the entire receptor kinase-like gene failed to complement the cev1 phenotype. Furthermore, sequencing of cev1 DNA revealed no mutations in the receptor kinase-like open reading frame.
cev1 plants containing the SB6.6 transgene had wild-type levels of expression of the VSP1::LUC reporter gene, their roots were of similar length to those of wild-type seedlings (Table 2), and they produced rosettes similar to those of wild-type plants (Figure 4B). The sequence of the CeSA3 gene from cev1 plants revealed a single C-to-T transition in the coding region of the gene that alters the predicted amino acid sequence G617E. Characterization of the cDNA from this gene indicated that the transcript extended from 16,008 to 10,782 of the TAC clone K2A11 and contained 14 introns, including one in the predicted 5′ untranslated region, as noted previously (Scheible et al., 2001). The cev1 mutation G617E is in a cytoplasmic loop (Figure 4C) in a region conserved within plant cellulose synthases (Delmer, 1999). These results indicate that the G617E mutation in CEV1, the predicted cellulose synthase CeSA3, is responsible for the cev1 mutant phenotype.
cev1 Roots Have Reduced Amounts of Cellulose
Analysis of the cellulose content of the cev1 mutant revealed that its roots contained ∼45% of the cellulose of wild-type roots (12.0% ± 2.4% versus 26.8% ± 7.8% of the ethanol-insoluble cell wall material). However, the cellulose content of leaf tissues from cev1 plants was not significantly different from that of wild-type plants.
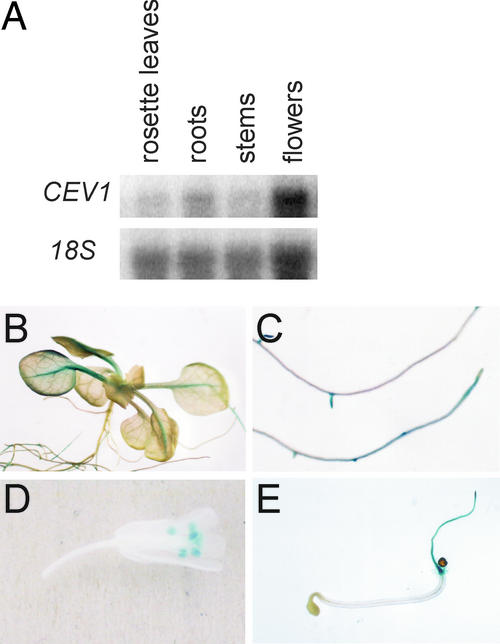
CEV1 Is Expressed in Anthers, Roots, and Midvein Tissue
Expression of the CEV1 gene was examined by RNA gel blot analysis (Figure 5A) and by histochemical staining of CEV1::β-glucuronidase (GUS) reporter plants (Figures 5B to 5E). CEV1 mRNA was detected in flower and root tissues and to a lesser extent in leaves and stems (Figure 5A), consistent with previous results (Scheible et al., 2001). Histochemical staining for GUS activity in transgenic plants revealed that expression in roots was confined to the apex of the primary and secondary roots and to root hairs (Figures 5C and 5E). Expression also was observed in the midveins of leaves and in the anthers (Figures 5B and 5D). GUS expression generally was absent from older root tissues and from older leaves and was not observed in inflorescences (data not shown). Wounding or treatment with methyl jasmonate (MeJA) did not increase CEV1 mRNA by RNA gel blot analysis, nor did it induce GUS expression (data not shown).
Figure 5.
Expression Pattern of the CEV1 Gene.
(A) Total RNA was extracted, and 1 μg was electrophoresed, blotted, and probed for CEV1 or 18S mRNA.
(B) to (E) Histochemical assays of GUS activity in CEV1::GUS transgenic plants.
(B) Rosette of a 2-week-old seedling.
(C) Roots of 2-week-old seedlings.
(D) Flower.
(E) Three-day-old dark-grown seedling.
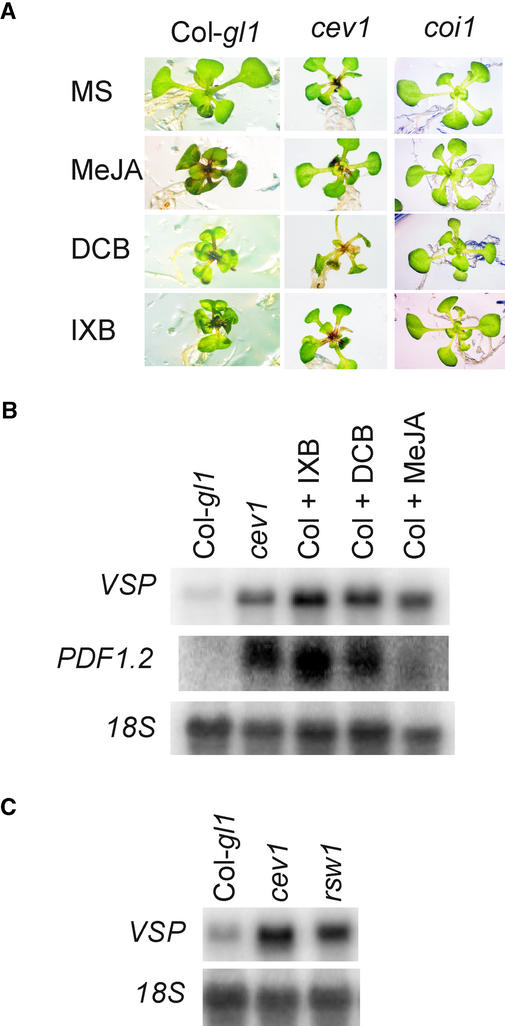
Cellulose Biosynthesis Inhibitors Induce a cev1 Phenocopy
The cellulose biosynthesis inhibitors 2,6-dichlorobenzonitrile (DCB) and isoxaben (IXB) were found to reproduce the cev1 phenotype. Twelve-day-old wild-type Arabidopsis seedlings placed on 5 μM DCB or 5 μM IXB for 3 days became stunted, leaves displayed epinasty, and there was visible accumulation of anthocyanin in the midveins (Figure 6). These effects also were caused by MeJA and are characteristic of the cev1 phenotype. Significantly, growth of the aerial parts of the coi1 mutant was not affected visibly by any of these treatments (Figure 6A). However, root growth of coi1 plants was inhibited by DCB and IXB but not by MeJA. Treatment with DCB or IXB induced the production of both PDF1.2 and VSP mRNA in wild-type plants, whereas treatment with 20 μM MeJA induced the production of VSP mRNA but little PDF1.2 mRNA (Figure 6B).
Figure 6.
Cellulose Biosynthesis Inhibitors Induce COI1-Dependent JA Responses.
(A) and (B) Seedlings were grown on MS agar for 12 days, transplanted onto MS agar supplemented with 20 μM MeJA, 5 μM IXB, or 5 μM DCB, and incubated for another 3 days.
(A) Seedlings of wild-type, cev1, and coi1-1 plants.
(B) Total RNA was extracted, and 1 μg was electrophoresed, blotted, and probed for VSP or PDF1.2 mRNA.
(C) Seedlings were germinated for 5 days on MS agar at 22°C and transferred to 30°C for another 7 days. Total RNA was extracted, and 1 μg was electrophoresed, blotted, and probed for VSP mRNA.
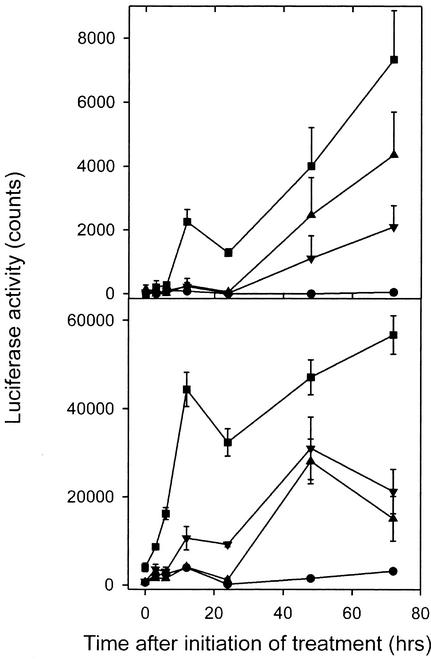
We compared the time course for induction of the LUC reporter for VSP after exposure to MeJA and the cellulose biosynthesis inhibitors IXB and DCB (Figure 7). MeJA induced LUC activity after 3 h, but IXB and DCB increased LUC activity only after 12 to 24 h.
Figure 7.
Time Course of the Induction of VSP Expression by Cellulose Biosynthesis Inhibitors.
(A) VSP1::LUC activity.
(B) VSP2::LUC activity.
Seedlings were grown on MS agar for 12 days and transplanted onto MS agar supplemented with 20 μM MeJA, 5 μM IXB, or 5 μM DCB. LUC activity was measured at the times indicated using low-light images of plants taken with a liquid nitrogen–cooled charge-coupled device imaging system. T-bars indicate sd values for eight seedlings. Circles, control; squares, MeJA; triangles, IXB; inverted triangles, DCB.
rsw1 Expresses Increased Levels of a JA-Responsive Gene
The cellulose synthase mutants rsw1 and prc1 were examined for the expression of JA-responsive genes. The rsw1-1 mutant appears normal at 22°C, but when it is incubated at 30°C, it has defective cellulose synthesis and its growth is stunted (Arioli et al., 1998). Dark-grown prc1 plants are stunted, although light-grown plants appear normal (Fagard et al., 2000). When grown at 30°C, rsw1 seedlings had considerably higher levels of VSP mRNA than wild-type plants (Figure 6C), and at 22°C, no differences in VSP expression were observed (data not shown). At 30°C, neither rsw1 nor cev1 plants expressed PDF1.2 mRNA (data not shown), indicating that the higher temperature may adversely affect the expression of this gene. Dark-grown prc1, cev1, and wild-type plants were examined for the expression of VSP and PDF1.2 mRNA, but these transcripts were not detected in any of the seedlings. However, light is required for VSP expression (Berger et al., 1995) and apparently for PDF1.2 as well.
DISCUSSION
The cev1 mutant was isolated in a screen for mutants with constitutive expression of the LUC reporter for the VSP1 promoter, and it was found subsequently to have constitutive expression of other JA- and ethylene-responsive genes, including VSP2, PDF1.2, CHI-B, and THI2.1 (Ellis and Turner, 2001), and enhanced resistance to pests and pathogens (C. Ellis, I. Karafyllidis, and J.G. Turner, unpublished results). Here, we show that cev1 plants overproduce JA and ethylene, and this is largely responsible for the altered morphology and pattern of gene expression in this mutant. Plants that are wounded or treated with the fungal pathogen Alternaria brassisicola also have increased synthesis of both JA and ethylene and increased expression of genes regulated by JA and ethylene (Creelman et al., 1992; Penninckx et al., 1998; Rojo et al., 1999). Therefore, cev1 plants reproduce many of the physiological alterations that characterize wounded and infected plants.
Surprisingly, the constitutive defenses of the cev1 mutant result from a recessive mutation in the cellulose synthase gene CeSA3. This raises the significant question of whether JA and ethylene signaling in the cev1 mutant results from an effect of the altered enzymatic activity of the mutant protein CEV1/G617E, possibly induced by reduced wall synthesis or by increased substrate level, or from a signaling property of the CEV1/G617E protein.
To address this question, we examined other cellulose synthase mutants and the effects of inhibitors of cellulose biosynthesis. Plants treated with the cellulose biosynthesis inhibitors IXB and DCB were stunted and epinastic, accumulated anthocyanin, and had enhanced expression of VSP and the JA- and ethylene-responsive gene PDF1.2. These phenotypes, except for the stunted roots, were suppressed in coi1 plants, indicating that JA signaling was required for many of the effects of IXB and DCB.
We also found that plants containing the rsw1 mutation in the cellulose synthase gene CeSA1 constitutively expressed the JA-responsive gene for VSP. In addition, dark-grown cev1 plants, such as prc1 and rsw1 plants (Fagard et al., 2000), have stunted hypocotyls. We have demonstrated that in cev1 plants, these are effects of increased JA and ethylene signaling.
However, not all cellulose synthase mutants induce stress signaling. For example, ixr1 and ixr2 in CeSA3 and CeSA6, respectively, are gain-of-function mutations that confer resistance to the cellulose biosynthesis inhibitor IXB (Scheible et al., 2001). These alleles do not cause stunted growth, anthocyanin accumulation, or epinasty, unlike the loss-of-function cev1 and prc1 alleles. Also, with the exception of irregular xylem and a slight decrease in growth, mutations in the IRX1 and IRX3 genes, which are required for cellulose synthesis in secondary cell walls, do not have morphological abnormalities (Turner and Somerville, 1997).
The time course for induction of the JA-responsive VSP::LUC transgenes by IXB and DCB indicated that LUC increased only 24 h after treatment, whereas LUC activity increased 3 h after treatment with MeJA, which is approximately the same time for the wound induction of VSP expression (Creelman et al., 1992; I. Karafyllidis and J.G. Turner, unpublished data). This finding indicates that IXB- and DCB-induced JA signaling is likely a secondary effect of these inhibitors. IXB and DCB inhibit the incorporation of radiolabeled Glc into cellulose within 1 h of treatment (Heim et al., 1990).
Hence, alterations in metabolite levels or in turgor pressure resulting from less Glc being used for cell wall synthesis should occur much more rapidly than the observed induction of stress responses. Therefore, we suggest that the delay is attributable to the time required for the growth and production of cells with altered cell walls and that JA and ethylene signaling in cev1, rsw1, and in plants treated with IXB and DCB is a consequence of altered wall composition or structure.
Relatively little is known about the function of the plant cell wall in signaling. However, in yeast, signaling in response to several stimuli, including temperature, osmotic stress, and pheromones, may involve the cell wall (Davenport et al., 1995; Kamada et al., 1995; Buehrer and Errede, 1997). These signals are mediated by the RHO1 GTPase, which is a regulatory subunit of the β-1,3-glucan synthase complex that synthesizes one of the major cell wall components in yeast (Qadota et al., 1996). RHO1 normally is activated by TOR2, a protein that has homology with phosphatidylinositol kinases (Schmidt et al., 1997).
However, mutations in genes for cell wall biosynthesis, and chemically induced cell wall defects, activate RHO1 GTPase-dependent signaling directly and independently of TOR2 (Bickle et al., 1998). This finding agrees with our finding that in Arabidopsis, mutations in genes for cell wall biosynthesis activate JA and ethylene signaling. In addition, mutations in an Arabidopsis chitinase-like gene, which is presumed to be involved in cell wall modification, also activate the production of ethylene (Zhong et al., 2002). It appears that cell wall integrity in Arabidopsis, like that in yeast, can mediate signaling events.
The cell wall of plants has some functional similarities to the extracellular matrix (ECM) of mammalian cells, which plays a key role in cell signaling (Reuzeau and Pont-Lezica, 1995). In mammalian cells, integrin receptors interact with the ECM and, in response to stimuli, including mechanical stress, activate RhoA, a homolog of RHO1 (Qadota et al., 1994; Clark and Brugge, 1995). In mammals, peptides containing the amino acid sequence RGD disrupt interactions between integrin and protein ligands within the ECM (Ruoslahti, 1996).
RGD peptides also disrupt the formation of Hechtian strands in plasmolyzed Arabidopsis cells, indicating that integrin-like proteins occur in Arabidopsis even though integrin genes have not been identified in its genome (Canut et al., 1998). RGD peptides also decrease plant defense responses at sites of fungal infection, including the production of hydrogen peroxide and callose (Mellersh and Heath, 2001). Evidently, adhesion between the cell wall and the plasmalemma occurs through integrin-like linkages and can signal plant defense responses.
Oligosaccharides and cell wall material have long been known to have signaling properties in plants (Darvill and Albersheim, 1984). Oligosaccharides and fungus-derived cell wall fragments have been demonstrated to elicit JA synthesis (Gundlach et al., 1992; Doares et al., 1995). However, in Arabidopsis, oligosaccharides promote ethylene production and inhibit the expression of JA-regulated genes (Rojo et al., 1999), and in tomato, JA induces polygalacturonase expression, suggesting that the oligogalacturonides produced in planta likely act downstream of JA synthesis (Ryan, 2000). Although clarification of their roles is required, the evidence indicates a connection between oligosaccharides and wound response.
Inhibitors of cellulose synthesis, and mutations in cellulose synthase genes, may cause the release of oligosaccharides and cell wall–derived compounds that subsequently activate JA synthesis and stress signaling. Mutations in cellulose synthase genes cause alterations in wall composition. For example, rsw1 root cell walls contain approximately half the amount of cellulose and twice the amount of starch of wild-type plants, along with a modest increase in pectin (Peng et al., 2000). prc1 cell walls contain 50% more hemicellulose and slightly less pectin than wild-type plants (Fagard et al., 2000), and cev1 cell walls have reduced cellulose. These altered walls may contain high levels of elicitors of JA and ethylene signaling, or the altered wall architecture may allow the untimely release of such molecules. We were not able to “wash out” elicitors from cev1 plants, although this may be the result of the limited solubility or mobility of potential elicitors.
We show here that the cev1 phenotype is caused by a mutation in the putative cellulose synthase gene CeSA3, that cev1 roots have reduced cellulose content, and that the most striking phenotypes of this mutant are caused by its overproduction of JA and ethylene. Furthermore, other genetic or chemical disruptions of cellulose synthesis result in the activation of these signaling pathways. Therefore, we propose that the cell wall can mediate JA- and ethylene-dependent stress and defense responses.
METHODS
Nomenclature
Analysis of the Arabidopsis thaliana genome has identified at least 10 cellulose synthase genes, which have been designated CeSA1 to CeSA10 (Delmer, 1999; Richmond and Somerville, 2000). The cellulose synthase gene responsible for the cev1 phenotype, CeSA3, also has been designated Ath-B (Arioli et al., 1998) and IXR1 (Scheible et al., 2001). Other mutants discussed in this article include rsw1, also designated CeSA1 (Arioli et al., 1998), and prc1, also designated ixr2 and CeSA6 (Fagard et al., 2000).
Growth Conditions
prc1-1 and rsw1-1 seeds were a gift from M. McCann (John Innes Centre, Norwich, UK) and R.E. Williamson (Australian National University, Canberra, Australia). Unless stated otherwise, plants were grown as described previously (Ellis and Turner, 2001) at 22°C in a 16-h photoperiod on 0.5 × Murashige and Skoog (1962) (MS) agar (seedlings) or soil (mature plants). Seedlings for hypocotyl measurements were grown on MS medium for 4 days in the dark. Seedlings for root measurements were grown for 12 days under a 16-h photoperiod. Construction and selection of the cev1 coi1-1 and cev1 etr1-3 double mutant lines were as described previously (Ellis and Turner, 2001). The cev1 coi1-1 etr1-3 triple mutant line was constructed from a cross between the cev1 coi1-1 and cev1 etr1-3 double mutant lines. The triple mutants were selected on the basis of their lack of anthocyanin accumulation and their lack of prolific root hairs.
For treatment with cellulose biosynthesis inhibitors, 12-day-old seedlings grown on MS agar were transferred to MS medium supplemented with 5 μM 2,6-dichlorobenzonitrile (Aldrich) or isoxaben (RdH Laborchemikalian, Seelze, Germany) and grown for another 1 to 3 days. To test VSP levels in the rsw1-1 mutant, plants were germinated on MS medium for 5 days and transferred to 30°C for 7 days before harvest and extraction of RNA.
Cloning of the CEV1 Gene
The cev1 mutant, in a Columbia (Col)-gl background, was crossed with Landsberg erecta plants, and 416 F2 individuals exhibiting the short-root phenotype of cev1 were selected for analysis. DNA from these plants was used to map the CEV1 gene to an ∼20-kb region on chromosome 5 using a combination of cleaved amplified polymorphic sequence and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) with data obtained from TAIR (http://www.arabidopsis.org). This region was located on the transformation-competent artificial chromosome clone K2A11, which was obtained from the ABRC (Columbus, OH).
The clone was introduced into cev1 plants by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998), and it complemented the mutation. Restriction endonuclease fragments of the K2A11 clone were ligated into the binary vector pSLJ75515, introduced into cev1 plants by Agrobacterium-mediated transformation, and tested for complementation of the mutant phenotype. Clone SB6.6 was found to complement the mutant phenotype.
Overlapping fragments of the CEV1 gene and the cev1 mutant were amplified by PCR using the following pairs of primers: 5′-CCATCCCAAGATTCTCCTCTTCGTCTTC-3′ and 5′-AAGTGAGGG-TGACTAGATGAC-3′, 5′-GTACAGGAATGACGAAGCGAG-3′ and 5′-CCCTCTTCTATGTCATCGAG-3′, 5′-CATTTGTCAAAGATCGTAGAG-C-3′ and 5′-AGAGGAACAATAGAACCAGCTG-3′, and 5′-CTGGAGAAGCGATTTGGACAG-3′ and 5′-GAACGAACAGACAGATAC-ACAC-3′. These fragments were sequenced with the aid of BigDye dideoxy terminator mix (Applied Biosystems, Foster City, CA).
Reporter Gene Construction and Assays
To construct the CEV1::β-glucuronidase (GUS) reporter gene, the region corresponding to −3 to −970 upstream of the CEV1 gene was amplified using primers 5′-GTCATCTAGATGCTTCCAACACCG-3′ and 5′-CACATCTAGACATGCACCACCAG-3′. This fragment was confirmed by DNA sequencing, cloned into the XbaI site of pBI101 (Clontech, Palo Alto, CA), and introduced into Col-gl1 plants by Agrobacterium-mediated transformation (Clough and Bent, 1998). The VSP1::luciferase (LUC) transgenic line has been described previously (Ellis and Turner, 2001). To create the VSP2::LUC transgenic line, the LUC gene from pGEMluc (Promega) was inserted into a pBI101 vector (Clontech) in place of the GUS gene using the BamHI and SacI restriction endonuclease sites.
A 1.5-kb DNA fragment containing the 5′ untranslated region of VSP2 was isolated via PCR of genomic DNA using primers 5′-CTTCTTAATTAAGCTTATATCTTC-3′ and 5′-GAGGATTTTCATGGATCCTAATGG-3′. This fragment was cloned into the HindIII and BamHI sites of the pBI101/LUC vector and introduced into Col-gl1 plants by Agrobacterium-mediated transformation (Clough and Bent, 1998). Histochemical staining for GUS activity and in vitro and in vivo LUC activities were assayed as described previously (Ellis and Turner, 2001).
Jasmonate and Ethylene Determinations
Jasmonate was determined as described previously (Hause et al., 2000). Ethylene was determined according to Guzman and Ecker (1990) with slight modifications. To determine ethylene in etiolated seedlings, ∼500 seeds were placed in a vial containing MS medium that was closed with a SUBA-seal, leaving a headspace volume of ∼5 mL. Seeds were kept at 4°C for 3 days and then incubated in the dark for 3 days at 22°C before samples were removed for analysis. To determine ethylene produced by light-grown seedlings, ∼200 mg fresh weight of 10-day-old light-grown seedlings was placed into vials containing MS medium. The vials were sealed and incubated in the light for 24 h before samples were removed for analysis. Headspace samples (10 μL) were analyzed using a 610 series gas chromatograph (ATI Unicam, Thermo DNIX, Houston, TX). Ethylene standards were from Aldrich. All experiments were performed in triplicate.
Cellulose Measurements
Seedlings were grown on vertical surfaces of MS agar for 3 weeks, at which time samples were harvested, frozen in liquid nitrogen, ground with a mortar and pestle, and homogenized in 50 mM Tris, pH 7.2, and 1% SDS. Samples were filtered through a fine mesh (5 μm), washed with ethanol and acetone, and resuspended in water. An aliquot was reserved for determination of total cell wall sugar content. The cell wall material was digested with 2 M trifluoroacetic acid for 90 min at 120°C, cooled, and centrifuged (4000g for 5 min). The resulting cellulose pellet was washed with water and quantified colorimetrically using the phenol/sulfuric acid method (Dubois et al., 1956).
Root and Hypocotyl Measurements
Roots and hypocotyls were measured with a caliper. At least 10 plants of each genotype were measured for each data set.
RNA Gel Blot Analysis
RNA gel blot analysis was performed as described previously (Ellis and Turner, 2001).
Accession Number
The accession number for CeSA3 is AF027174.1. The accession number for clone K2A11 is AB018111.
Acknowledgments
We thank Otto Miersch for JA and 12-oxo-phytodienoic acid measurements, Maureen McCann and Nick Carpita for helpful discussions and advice regarding cellulose analysis, and the editor and reviewers for constructive comments. We acknowledge funding from Syngenta and from the Biotechnology and Biological Sciences Research Council (UK).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002022.
References
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Benedetti, C.E., Xie, D., and Turner, J.G. (1995). COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Bell, E., Sadka, A., and Mullet, J.E. (1995). Arabidopsis thaliana AtVSP is homologous to soybean VSPA and VSPB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light, and phosphate. Plant Mol. Biol. 27, 933–942. [DOI] [PubMed] [Google Scholar]
- Bickle, M., Delley, P.-A., Schmidt, A., and Hall, M.N. (1998). Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 8, 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer, B.M., and Errede, B. (1997). Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canut, H., Carrasco, A., Galaud, J., Cassan, C., Bouyssou, H., Vita, N., Ferrara, P., and Pont-Lezica, R. (1998). High affinity RGD-binding sites at the plasma membrane of Arabidopsis thaliana link the cell wall. Plant J. 16, 63–71. [DOI] [PubMed] [Google Scholar]
- Clark, E.A., and Brugge, J.S. (1995). Integrins and signal transduction pathways: The road taken. Science 268, 233–238. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1995). Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 92, 4114–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., Tierney, M.L., and Mullet, J.E. (1992). Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc. Natl. Acad. Sci. USA 89, 4938–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvill, A.G., and Albersheim, P. (1984). Phytoalexins and their elicitors: A defense against microbial infection in plants. Annu. Rev. Plant Physiol. 35, 243–275. [Google Scholar]
- Davenport, K.R., Sohaskey, M., Kamada, Y., Levin, D.E., and Gustin, M.C. (1995). A second osmosensing signal transduction pathway in yeast. J. Biol. Chem. 270, 30157–30161. [DOI] [PubMed] [Google Scholar]
- Delmer, D.P. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. [DOI] [PubMed] [Google Scholar]
- Doares, S.H., Syrovets, T., Weiler, E.W., and Ryan, C.A. (1995). Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoic pathway. Proc. Natl. Acad. Sci. USA 92, 4095–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, M., Gilles, K.A., Hamilton, J.K., Rebers, P.A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 38, 350–361. [Google Scholar]
- Ellis, C., and Turner, J.G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1995). An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 109, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach, H., Muller, M.J., Kutchan, T.M., and Zenk, M.H. (1992). Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. USA 89, 2389–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: Allene oxide cyclase is highly ex-pressed in distinct flower organs and vascular bundles. Plant J. 24, 113–126. [DOI] [PubMed] [Google Scholar]
- Heim, D.R., Skomp, J.R., Tschabold, E.E., and Larrinua, I.M. (1990). Isoxaben inhibits the synthesis of acid insoluble cell wall materials in Arabidopsis thaliana. Plant Physiol. 93, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, Y., Jung, U.S., Piotrowski, J., and Levin, D.E. (1995). The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9, 1559–1571. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh, D.G., and Heath, M.C. (2001). Plasma membrane–cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Norman-Setterblad, C., Vidal, S., and Palva, E.T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant-Microbe Interact. 4, 430–438. [DOI] [PubMed] [Google Scholar]
- Peng, L., Hocart, C.H., Redmond, J.W., and Williamson, R.E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., Anraku, Y., Botstein, D., and Ohya, Y. (1994). Conditional lethality of a yeast strain expressing human RHOA in place of RHO1. Proc. Natl. Acad. Sci. USA 91, 9317–9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., Python, C.P., Inoue, S.B., Arisawa, M., Anraku, Y., Zheng, Y., Watanabe, T., Levin, D.E., and Ohya, Y. (1996). Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272, 165–170. [DOI] [PubMed] [Google Scholar]
- Reuzeau, C., and Pont-Lezica, R.F. (1995). Comparing plant and animal extracellular matrix-cytoskeleton connections: Are they alike? Protoplasma 186, 113–121. [Google Scholar]
- Richmond, T.A., and Somerville, C.R. (2000). The cellulose synthase superfamily. Plant Physiol. 124, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Leon, J., and Sanchez-Serrano, J.J. (1999). Cross-talk between wound signaling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20, 135–142. [DOI] [PubMed] [Google Scholar]
- Ruoslahti, E. (1996). RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12, 697–715. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (2000). The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477, 112–121. [DOI] [PubMed] [Google Scholar]
- Scheible, W.-R., Eshed, R., Richmond, T., Delmer, D., and Somerville, C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. USA 98, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., Bickle, M., Beck, T., and Hall, M.N. (1997). The phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531–542. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Kays, S.J., Schroeder, B.P., and Ye, Z.-H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14, 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]