Abstract
Phytochrome A signaling shows two photobiologically discrete outputs: so-called very-low-fluence responses (VLFR) and high-irradiance responses (HIR). By modifying previous screening protocols, we isolated two Arabidopsis mutants retaining VLFR and lacking HIR. Phytochrome A negatively or positively regulates phytochrome B signaling, depending on light conditions. These mutants retained the negative but lacked the positive regulation. Both mutants carry the novel phyA-302 allele, in which Glu-777 (a residue conserved in angiosperm phytochromes) changed to Lys in the PAS2 motif of the C-terminal domain. The phyA-302 mutants showed a 50% reduction in phytochrome A levels in darkness, but this difference was compensated for by greater stability under continuous far-red light. phyA-302:green fluorescent protein fusion proteins showed normal translocation from the cytosol to the nucleus under continuous far-red light but failed to produce nuclear spots, suggesting that nuclear speckles could be involved in HIR signaling and phytochrome A degradation. We propose that the PAS2 domain of phytochrome A is necessary to initiate signaling in HIR but not in VLFR, likely via interaction with a specific partner.
INTRODUCTION
Phytochrome is a small family of red light (R) and far-red light (FR) photoreceptors comprising five isoforms, phytochrome A through phytochrome E (phyA through phyE), in Arabidopsis (Neff et al., 2000). phyA and phyB are dimeric cytoplasmic proteins in dark-grown seedlings that migrate to the nucleus upon light activation (Kircher et al., 1999; Yamaguchi et al., 1999). The N-terminal domain bears an open chain tetrapyrrol chromophore. The C-terminal domain contains motifs that are important for interaction with the nuclear transcription factor PIF3 (Ni et al., 1998, 1999; Martinez-García et al., 2000), the phytochrome kinase substrate PKS1 (Fankhauser et al., 1999), nucleoside diphosphate kinase 2 (Choi et al., 1999), and cryptochrome (Ahmad et al., 1998).
The pattern of a response—graded, multiphasic, with a threshold, with biorhythmic oscillations, et cetera—depends on the architecture of the signaling network (Becskei et al., 2001). Seedling deetiolation under continuous FR is mediated by phyA (Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993). Even if a single end point process is considered (e.g. inhibition of hypocotyl growth), two components of phyA activity can be distinguished, demonstrating the bistable or binary nature of the response (Casal et al., 2000): the very-low-fluence response (VLFR) component, which saturates with infrequent (e.g., single or hourly pulses) activation of phyA by very low fluences of R or FR, and the high-irradiance response (HIR) component, which requires sustained activation of phyA with higher fluences of FR.
The architectural features of the phyA signaling network that give rise to the biphasic response have not been elucidated. Nevertheless, VLFR and HIR can be dissected genetically by the vlf1, vlf2, fhy3, and dim/dwarf1/eve1 loci that operate downstream of phyA (Yanovsky et al., 1997, 2000; Luccioni et al., 2001). These two components also can be separated molecularly because the HIR requires a region of target gene promoters that is not necessary for VLFR (Cerdán et al., 2000). Furthermore, activation of phyA in the VLFR mode (by a pulse of R) negatively regulates phyB signaling, whereas activation of phyA in the HIR mode (by several hours of FR) positively regulates phyB signaling initiated by a subsequent R pulse (Cerdán et al., 1999).
Together, these observations are consistent with a model in which signaling downstream of phyA branches out into two cascades, depending on the mode in which the photoreceptor is activated. The point at which divergence is initiated has not been established. The aim of this work was to identify mutants selectively impaired in the HIR and not in the VLFR component to gain insight into the molecular differences between these two response modes.
RESULTS
Novel Alleles of phyA That Show Deficient Hypocotyl Growth Inhibition but Normal Seed Germination
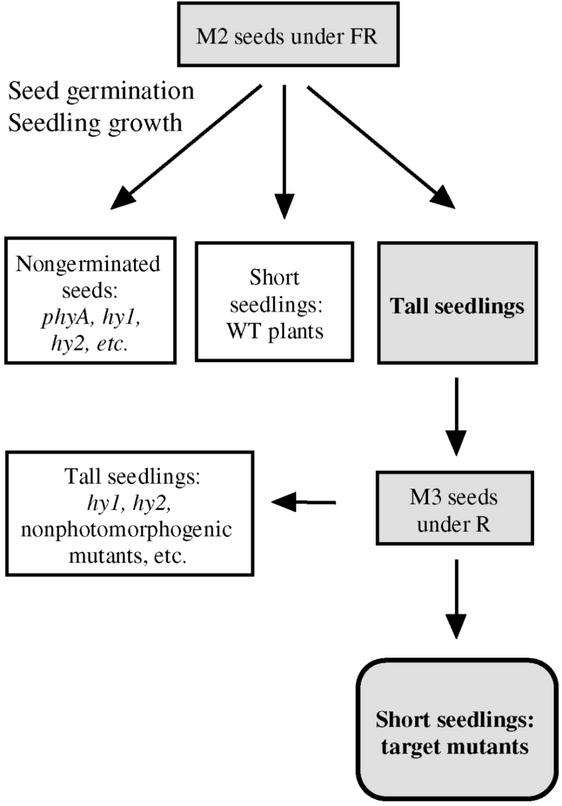
To obtain Arabidopsis mutants with impaired HIR but retaining VLFR, both germination of mutagenized seed and subsequent seedling growth were conducted under continuous FR (Figure 1). phyA-mediated promotion of seed germination is a VLFR (Botto et al., 1996; Shinomura et al., 1996) because the response is saturated with hourly pulses of FR and continuous FR has no additional effect (Figure 2A). In contrast, phyA-mediated inhibition of hypocotyl growth is predominantly a HIR because continuous FR is significantly more effective than hourly pulses of FR.
Figure 1.
Screening Protocol Used to Identify Mutants Affected More Strongly in HIR Than in VLFR of phyA.
WT, wild type.
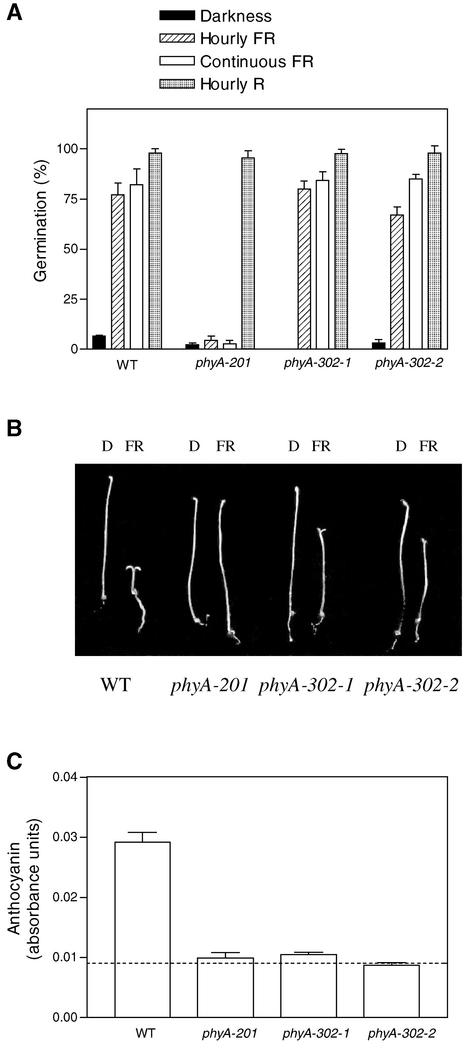
Figure 2.
phyA-302 Mutants Show Normal Induction of Seed Germination (A), Weak Hypocotyl Growth Inhibition and Cotyledon Unfolding (B), and No Promotion of Anthocyanin Synthesis (C) under Continuous FR.
Data are means and se of six (A) or five (C) replicate boxes. In (C), the dashed line shows absorbance levels in dark-grown seedlings. D, darkness; WT, wild type.
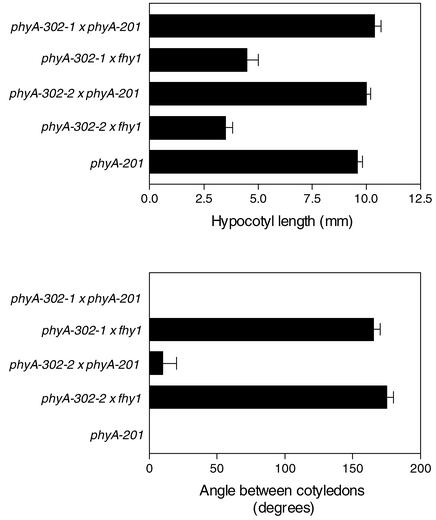
Tall seedlings under continuous FR were selected from M2 seeds (Figure 1). Normal seedlings under pulsed R were selected from M3 seeds of these mutants to avoid phytochrome chromophore and nonphotomorphogenic hypocotyl growth mutants (Figure 1). Two independent lines (designated phyA-302-1 and phyA-302-2 based on the evidence described below), showing seed germination under FR (Figure 2A), normal inhibition of hypocotyl growth under R, and long hypocotyls under continuous FR (Figure 2B), were chosen for further studies. The two mutants failed to complement the null phyA-201 mutant (Figure 3).
Figure 3.
Complementation Tests of the Mutants That Germinate but Fail to Deetiolate Normally under FR.
The novel mutants were used as mother plants, and phyA-201 and fhy1 were used as pollen donors. Data are means and se of 8 to 12 replicate plants.
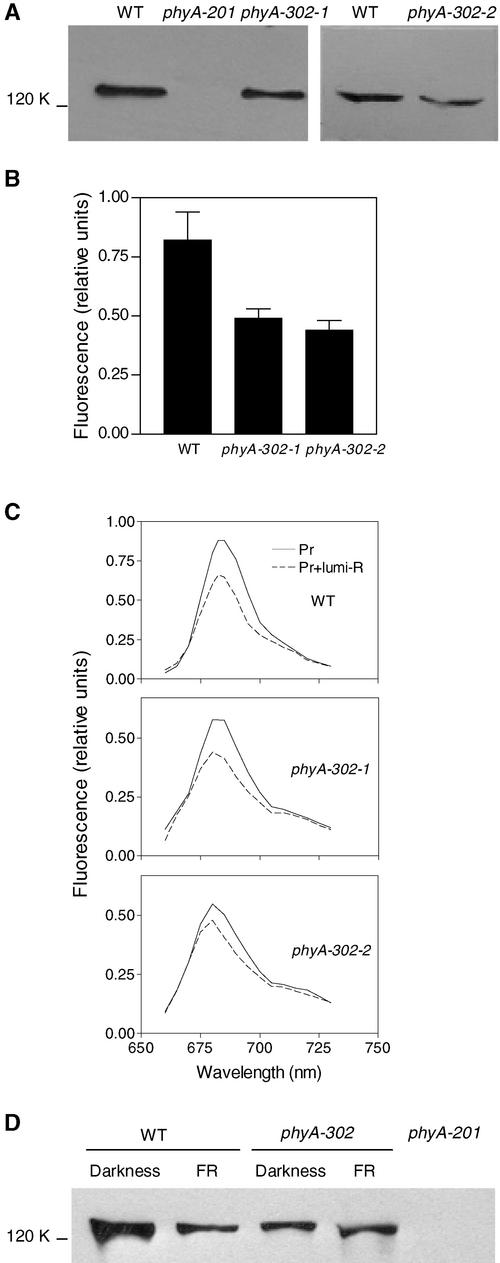
In etiolated seedlings, the levels of phyA detectable immunologically and by fluorescence signals were reduced by ∼50% in both mutant lines (Figures 4A and 4B). The fluorescence spectra (Figure 4C), the extent of Pr-to-lumi–R (the first photoproduct stable at low temperatures) conversion at 85K upon illumination with 639 nm (0.24 to 0.3), and the extent of Pr-to-Pfr conversion at 273K upon illumination with 680 nm (0.55 to 0.65) were not affected significantly by the phyA-302 mutations. Because the abundance of phyA normally is reduced by continuous FR compared with darkness or hourly FR pulses (Casal et al., 2000), immunoblot analysis also was conducted with dark-grown 3-day-old seedlings transferred to continuous FR for 8 h. Although phyA-302 mutants had less phyA in dark-grown control seedlings, the residual phyA was more stable than in the wild type (Figure 4D).
Figure 4.
Etiolated Seedlings of the phyA-302 Mutants Contain Less phyA, but This phyA Is More Stable under Continuous FR.
(A) Immunoblots of 4-day-old etiolated seedlings.
(B) Low-temperature fluorescence of phytochrome in etiolated hypocotyls.
(C) Low-temperature fluorescence spectra of phytochrome in etiolated hypocotyls.
(D) Immunoblot of 3-day-old seedlings transferred to FR for 8 h.
Protein gel blots are representative of three independent determinations. Fluorescence data are means and se of seven independent sets of measurements. WT, wild type.
The Two Missense Mutations Occur in Amino Acid 777 of phyA
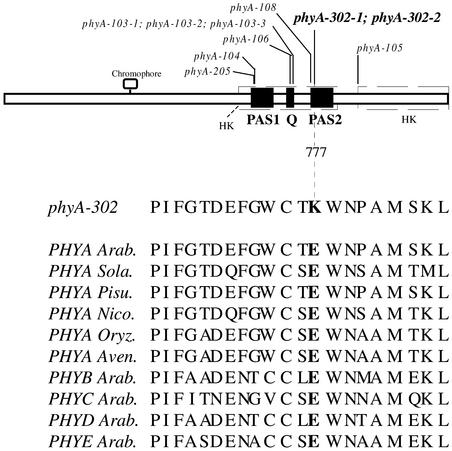
The phyA genes of the two mutants and of the wild type were cloned and sequenced completely. The two independent mutants contain the same Guanidine-to-Adenine mutation that results in the change of amino acid 777 from Glu to Lys (Figure 5; amino acid numbering based on the chromophore attachment site at Cys-323).
Figure 5.
Location of the Missense Mutation in phyA-302-1 and phyA-302-2.
The positions of other C-terminal missense phyA mutations are indicated for comparative purposes. The amino acid sequence in the region of the phyA-302 mutation is compared with sequences of wild-type PHYA from Arabidopsis and other angiosperms as well as other Arabidopsis phytochromes. Arab., Arabidopsis; Aven., Avena sativa; HS, His kinase–related domains; Nico., Nicotiana tabacum; Oryz., Oryza sativa; Pisu., Pisum sativum; Q, Quail box; Sola., Solanum tuberosum.
phyA-302:Green Fluorescent Protein Fusion Proteins Fail to Produce Nuclear Speckles
Null phyA mutant seedlings of Arabidopsis were transformed with the Arabidopsis PHYA:green fluorescent protein (GFP) or phyA-302:GFP genes under the control of a 35S promoter. The PHYA:GFP transgene fully rescues the phyA null mutant phenotype (Kim et al., 2000). The phyA-302:GFP transgene rescued the phyA null mutant phenotype under hourly pulses of FR (hypocotyl length [mm] in the wild type was 11.8 ± 0.2 in darkness and 10.8 ± 0.2 in pulsed FR [P < 0.05]; hypocotyl length in the null phyA mutant was 11.3 ± 0.4 in darkness and 11.2 ± 0.2 in pulsed FR [P > 0.1]; hypocotyl length in the null phyA mutant expressing the phyA-302:GFP transgene was 13.3 ± 0.3 in darkness and 12.2 ± 0.3 in pulsed FR [P < 0.05]).
Continuous FR reduced hypocotyl length in the wild type dramatically (2.0 ± 0.1 mm), but it had no more effect than pulsed FR in the phyA-302:GFP transgenic plants (12.1 ± 0.3 mm). This pattern of response agrees with the behavior of the phyA-302 mutants (see below). The localization of phyA:GFP and phyA-302:GFP fusion proteins was investigated in transgenic seedlings grown in darkness and subsequently transferred to continuous FR.
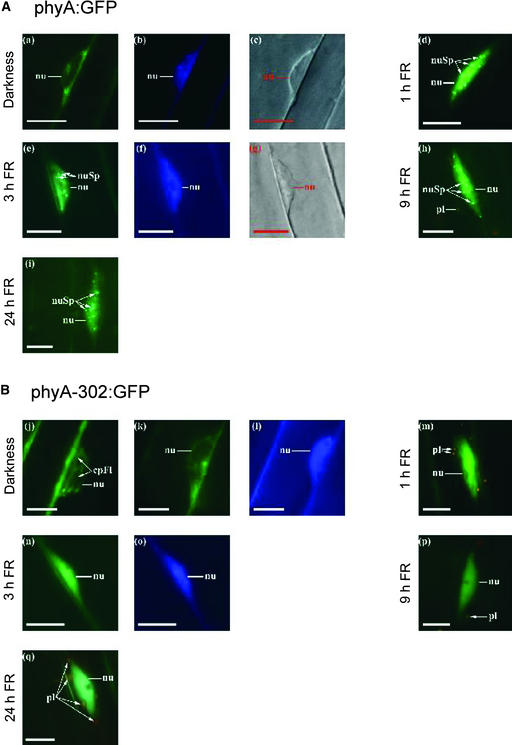
In the seedlings that remained as dark controls, the nuclei showed no fluorescence associated with the presence of phyA (Figure 6). phyA:GFP or phyA-302:GFP was present in the nuclei of plants exposed to 1, 3, 9, or 24 h of continuous FR, indicating that the phyA-302 mutation did not affect nuclear translocation. However, although phyA:GFP was able to form speckles in >90% of the nuclei (Kim et al., 1999; Kircher et al., 1999; Hisada et al., 2000), plants bearing the phyA-302:GFP fusion protein failed to form these spots (Figure 6), which were observed in only 10% of the nuclei after 24 h of FR and never in nuclei corresponding to the hook region of the hypocotyl.
Figure 6.
Arabidopsis phyA:GFP and phyA-302:GFP Fusion Proteins Translocate to the Nucleus, but phyA-302:GFP Fails to Produce Nuclear Speckles under FR.
(A) Representative nuclei of transgenic seedlings expressing phyA:GFP.
(B) Representative nuclei of transgenic seedlings expressing phyA-302:GFP.
The seedlings were kept in darkness ([a], [b], [c], [j], [k], and [l]) or transferred to continuous FR for 1 h ([d] and [m]), 3 h ([e], [f], [g], [n], and [o]), 9 h ([h] and [p]), or 24 h ([i] and [q]). The subcellular localization of the GFP fusion proteins was investigated by fluorescence microscopy ([a], [d], [e], [h], [i], [j], [k], [m], [n], [p], and [q]). 4′,6-Diamidino-2-phenylindole staining ([b], [f], [l], and [o]) and differential interference contrast images ([c] and [g]) are included ([a] to [c], [e] to [g], [k] to [l], and [n] to [o]; each show identical nuclei). Nuclei (nu), nuclear spots (nuSp), plastids (pl), and cytoplasmic fluorescence (cpFl) are indicated. Bars = 10 μm.
Sustained and Transient Activation with FR Are Equally Effective in phyA-302
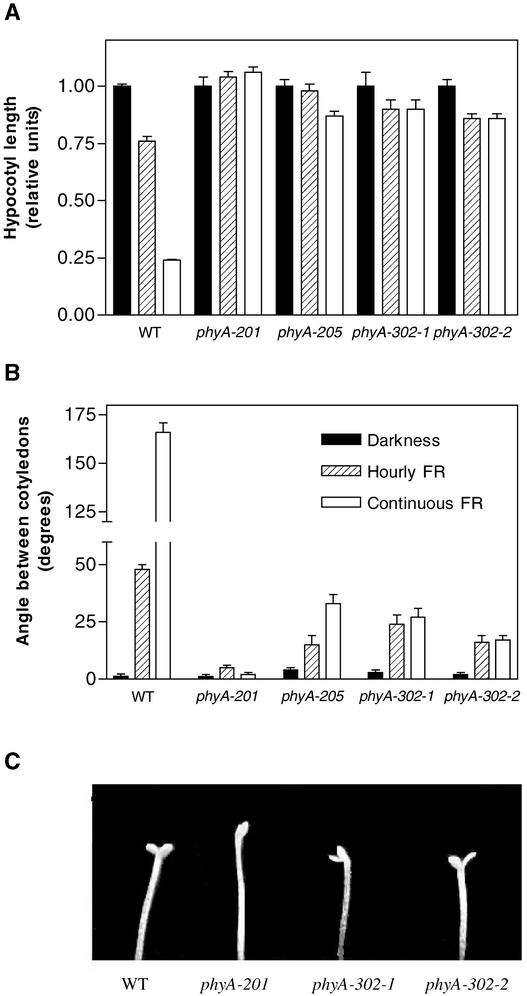
In the phyA-302 mutants, hypocotyl growth inhibition, which is dominated by the HIR component, was more affected than seed germination, which is dominated by the VLFR component. To compare VLFR and HIR of the same end point responses (hypocotyl growth and cotyledon unfolding), etiolated seedlings were allowed to germinate in darkness for 24 h after a saturating R pulse and transferred to either hourly pulses of FR, which saturate the VLFR but are insufficient to cause a HIR, or continuous FR at a fluence rate that is sufficient to initiate a HIR (Casal et al., 2000).
The wild type showed hypocotyl growth inhibition and cotyledon unfolding under hourly FR (i.e., VLFR), but continuous FR was significantly more efficient, suggesting a strong contribution of a HIR (Figure 7). In the phyA-302-1 and phyA-302-2 mutants, pulsed and continuous FR produced similar effects, indicating the absence of a HIR. The weak phyA-205 allele (Nagatani et al., 1993; Reed et al., 1994), included for comparative purposes, showed reduced responses to both pulsed and continuous FR, but the latter was significantly more effective than hourly FR. Thus, in contrast to the phyA-302 mutants, the phyA-205 allele showed a reduced VLFR but retained some HIR (Figure 7).
Figure 7.
At Equal Total Fluence, Hourly and Continuous FR Are Equally Effective at Inhibiting Hypocotyl Growth and Promoting Cotyledon Unfolding in the phyA-302 Mutants, whereas Continuous FR Is More Effective Than Pulsed FR in Wild-Type and phyA-205 Seedlings.
The seedlings were grown under continuous FR of 10 μmol·m−2·s−1, hourly FR (3 min/h) of 200 μmol·m−2·s−1, or in darkness. Data are means and se of 12 replicate boxes. WT, wild type.
(A) Hypocotyl length relative to dark controls (symbols as in [B]).
(B) Angle between the cotyledons.
(C) Detail of the cotyledons under hourly pulses of FR.
We also investigated the effects of pulsed and continuous FR in the hy1 and hy2 mutants, which have severely reduced levels of phytochrome (Koornneef et al., 1980). These mutations affect chromophore synthesis genes and not apoprotein genes (Davis et al., 1999; Muramoto et al., 1999; Kohchi et al., 2001). Although photomorphogenesis under FR is very weak in these mutants, both showed stronger effects under continuous FR than under pulsed FR (angle between the cotyledons [°] in the wild type was 4 ± 1 in darkness, 108 ± 3 in pulsed FR, and 180 ± 0 in continuous FR; angle between the cotyledons in hy1 was 1 ± 1 in darkness, 3 ± 1 in pulsed FR, and 11 ± 2 in continuous FR; angle between the cotyledons in hy2 was 1 ± 1 in darkness, 5 ± 1 in pulsed FR, and 17 ± 5 in continuous FR). These results indicate that a reduction of photoactive phyA levels does not necessarily eliminate the HIR in Arabidopsis.
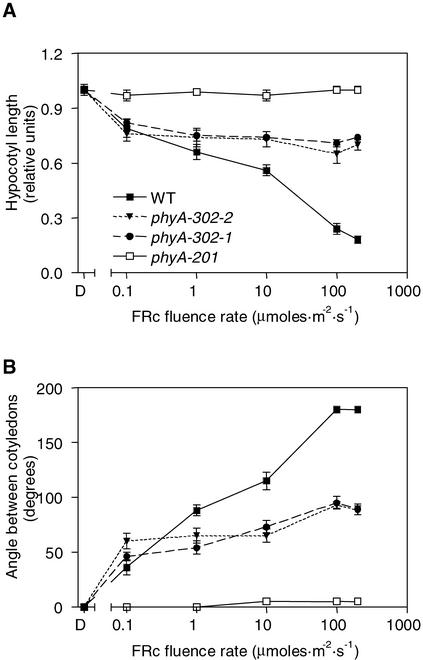
Weak Fluence Rate Dependence of the Effects of Continuous FR in phyA-302
One of the distinctive features of phyA activity under continuous FR is its strong fluence rate dependence. In the wild type, both hypocotyl growth and cotyledon unfolding increased in response to the fluence rate of continuous FR between 0 and 100 μmol·m−2·s−1 (Figure 8). In the phyA-302 mutants, hypocotyl growth inhibition saturated at the lowest fluence rate of continuous FR tested (0.1 μmol· m−2·s−1), and cotyledon unfolding showed weak fluence rate dependence at >0.1 μmol·m−2·s−1. At this fluence rate, hypocotyl growth inhibition and cotyledon unfolding showed wild-type levels (Figure 8).
Figure 8.
Weak Fluence Rate Dependence of Hypocotyl Growth Inhibition (A) and Cotyledon Unfolding (B) by Continuous FR in the phyA-302 Mutants.
Symbols in (B) are as indicated in (A). Data are means and se of six replicate boxes. WT, wild type.
Continuous FR Fails to Promote Anthocyanin Synthesis in phyA-302
In Arabidopsis, continuous FR promotes anthocyanin synthesis, but hourly pulses of FR have negligible effects on this process compared with dark controls (Yanovsky et al., 2000). This finding indicates that anthocyanin synthesis is dominated by the HIR component, whereas the VLFR component makes at most a weak contribution. If the phyA-302 mutations cause a general failure of HIR, the promotion of anthocyanin synthesis by continuous FR should be impaired severely in phyA-302-1 and phyA-302-2. This expectation was met by the data (Figure 2C). Thus, the phyA-302 mutants showed wild-type promotion of seed germination, intermediate inhibition of hypocotyl growth and cotyledon unfolding, and no promotion of anthocyanin by continuous FR (Figure 2). Across different processes, the effect of the mutations was inversely related to the effect of pulsed FR compared with continuous FR.
Inductive Responses in phyA-302
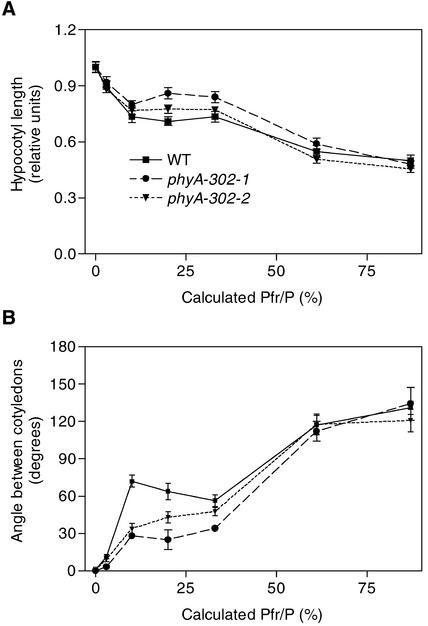
To investigate inductive responses (i.e., the effects of light pulses) in more detail, the seedlings were exposed to hourly pulses predicted to establish a series of calculated proportions of phytochromes in the Pfr form (Pfr/P). The wild type showed biphasic responses of hypocotyl growth and cotyledon unfolding (Figure 9). The first phase (which saturates at Pfr/P of 10%) is the VLFR mediated by phyA, and the second phase (observed between Pfr/P of 33 and 87%) is the R/FR reversible response mediated by phyB (Yanovsky et al., 1997). In the phyA-302 mutants, the VLFR was affected only partially (Figure 9). Protein levels are similar under hourly FR pulses compared with those in darkness (Casal et al., 2000), suggesting that on a protein basis (Figure 4), the mutated phyA photoreceptor retained normal efficiency for VLFR.
Figure 9.
VLFR of phyA- and phyB-Mediated Responses of Hypocotyl Growth (A) and Cotyledon Unfolding (B) in Wild-Type and phyA-302 Seedlings.
Seedlings were grown under hourly R/FR pulses providing the indicated calculated proportion of phytochrome in the Pfr form. The first phase is the VLFR and the second phase (Pfr/P > 30%) is the phyB-mediated effect. Symbols in (B) are as indicated in (A). Data are means and se of 16 replicate boxes. WT, wild type.
Analysis of the Interaction between phyA-302 and phyB Signaling
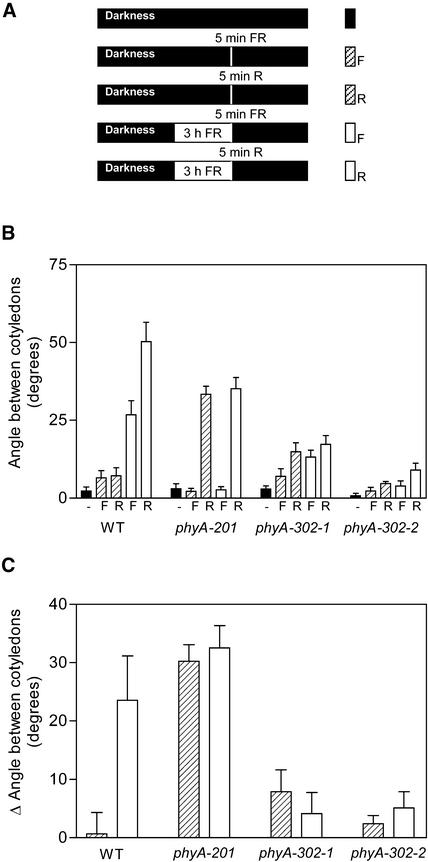
The interaction between phyA and phyB signaling is synergistic when phyA operates in the HIR mode and antagonistic when phyA operates in the VLFR mode (Cerdán et al., 1999). To investigate whether the phyA-302 mutants retained these interactions, the seedlings were exposed daily to a brief pulse of R or FR in factorial combination with or without 3 h of FR (Figure 10A). The effect of R pulses compared with FR pulses is mediated by phyB (Cerdán et al., 1999). In seedlings exposed only to brief pulses, phyA operated in the VLFR mode, whereas in those seedlings exposed to 3 h of FR, phyA operated predominantly in the HIR mode. In the absence of 3-h FR exposures, wild-type seedlings failed to unfold the cotyledons in response to a daily pulse of R compared with FR (Figure 10B; the difference is shown in Figure 10C).
Figure 10.
The phyA-302 Mutants Retain the phyA-Mediated Negative Modulation of phyB Signaling but Fail to Amplify phyB Signaling in Response to Prolonged Exposures to FR.
(A) Protocol and symbols. Seedlings were grown in complete darkness or exposed daily to a 5-min pulse of FR (F) or R, or to 3 h of FR followed immediately by a 5-min pulse of either FR or R.
(B) Angle between the cotyledons. Data are means and se of seven or eight replicate boxes. WT, wild type.
(C) The difference in angle between cotyledons caused by a R compared with a FR 5-min daily pulse was calculated for the seedlings exposed or not exposed to 3 h of FR.
Because daily exposure to a pulse of R induces cotyledon unfolding in phyA null mutants but not in the phyA phyB double mutant, phyA signaling in the VLFR mode is predicted to antagonize phyB signaling (Cerdán et al., 1999). In wild-type seedlings exposed daily to 3 h of FR, a subsequent R pulse caused cotyledon unfolding. Both phyA and phyB were required for this amplification effect, indicating a synergistic action of both photoreceptors when phyA operates in the HIR mode (Cerdán et al., 1999). In the phyA-302 mutants, a pulse of R given alone was not effective (Figure 10). This finding indicates that in terms of the antagonistic effect, the mutated phyA molecule behaved like wild-type phyA.
However, in contrast to what was seen in the wild type, daily exposure to 3 h of FR completely failed to amplify the response to a R versus FR test pulse (Figure 10). This finding indicates that in terms of the synergistic effect, the mutated phyA molecule lacks functional activity. In other words, in seedlings exposed to 3 h of FR followed by a R pulse, cotyledon unfolding was observed in the wild type (as a result of the amplification of the response by 3 h of FR) and in the phyA null mutant (in which the response to R is constitutive) but was very poor in the phyA-302 mutants (in which the constitutive response was minimal and amplification was null).
DISCUSSION
Novel phyA Alleles Affected in the PAS2 Domain
Screening for mutants able to germinate but not to deetiolate normally under continuous FR resulted in the isolation of two independent phyA-302 plants carrying a missense mutation (Glu to Lys) at amino acid 777 (Figure 5). The traditional protocol for the screening of phyA mutants, which involves the induction of germination of chilled seeds by white light before transfer to FR (Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993), has yielded a number of null and leaky phyA alleles that do not coincide with the mutation reported here (Dehesh et al., 1993; Whitelam et al., 1993; Reed et al., 1994; Xu et al., 1995).
Several phyA and phyB mutations that do not alter phyA or phyB protein levels, spectral activity, or dimer formation are located between amino acids 632 and 768 (amino acid numbering based on the chromophore attachment site at Cys-323), particularly amino acids 715, 716, and 727 (Figure 5) (Quail et al., 1995; Wagner and Quail, 1995; Xu et al., 1995). This region, often designated the Quail box (amino acids 714 to 731) (Song, 1999), is important for phytochrome interaction with partners such as PIF3 (Ni et al., 1998, 1999) and nucleoside diphosphate kinase 2 (Choi et al., 1999). The phyA-302 mutations are not in the Quail box (Figure 5). They do not affect the His kinase–related domain present between amino acids 889 and 1117 (Schneider-Poetsch and Braun, 1991; Yeh and Lagarias, 1998) that is involved in phytochrome interaction with PKS1 (Fankhauser et al., 1999) and that participates in phyB function (Krall and Reed, 2000).
There is a second region within the C terminus, between amino acids 593 and 825, related to the His kinase domain of the cyanobacterial phytochrome Cph1 (Yeh and Lagarias, 1998), that contains the Quail box, two individual PAS domains (amino acids 621 to 684 and 767 to 817) (Taylor and Zhulin, 1999), and the phyA-302 mutations. Actually, these mutations fall within the PAS2 domain (Figure 5) in an amino acid conserved in PAS2 of angiosperm phytochromes but not in the PAS1 domain of phyA or the PAS domain of photoreactive yellow protein (Taylor and Zhulin, 1999).
phyA-302 Mutants Retain VLFR but Lack HIR
phyA-mediated responses have two components whose relative contributions to the output depends on the developmental context: the VLFR, which saturates with infrequent pulses of very low fluences of R or FR, and the HIR, which requires sustained excitation with FR and shows strong fluence rate dependence. The phyA-302 mutants retained VLFR but lacked HIR (Figures 3, 7, and 8).
The failure of HIR in phyA-302 is not a general (nonspecific) consequence of reductions in phyA signaling. The phyA-205 allele, which carries a missense mutation (Val to Met) at amino acid 631 (Reed et al., 1994) (Figure 5) and retains normal protein levels and photochemical activity (Quail et al., 1995), shows weak phyA-mediated effects but retains a significant HIR (the difference between hourly and continuous FR in Figure 7). Similarly, phyA-103, phyA-104, phyA-106, and phyA-108, which carry missense mutations within the Quail box, and phyA-105, which carries a Ala-to-Val mutation at position 893 (Figure 5), all affect VLFR (Parks et al., 1996), but they retain the strong fluence rate dependence typical of HIR (Xu et al., 1995).
The loss of HIR in phyA-302 is not the consequence of reduced phyA levels. Although the phyA-302 mutation caused a 50% reduction in protein levels in dark-grown seedlings, this difference was compensated for by the greater stability of phyA under FR in phyA-302 than in the wild type (Figure 4D). Furthermore, reduced phyA levels do not eliminate HIR per se. The phyA-109 mutant has low protein levels but retains fluence rate–dependent inhibition of hypocotyl growth (Xu et al., 1995), and hy1 and hy2 chromophore mutants have very little active phyA (Koornneef et al., 1980), yet they show a stronger residual response to continuous FR than pulsed FR, which is typical of HIR.
The phyB-101 mutation involves a Glu-to-Lys change at the amino acid position equivalent to phyA-302, and the recombinant mutant protein produced by the phyB-101 allele shows enhanced Pfr-to-Pr dark reversion (Elich and Chory, 1997). Increased dark reversion accounts for the phyB-101 phenotype, which is weak under continuous R (which counteracts dark reversion) and resembles phyB-null alleles in its failure to respond to the Pfr level established before darkness (Elich and Chory, 1997). We have not analyzed dark reversion in phyA-302, but it is obvious that if this mutation causes Pfr dark reversion of phyA (which is absent in Landsberg erecta; Eichenberg et al., 2000), this should have a greater effect on the response to pulsed FR than to continuous FR (i.e., the phenotype should be opposite to the one observed here).
The region between amino acids 730 and 821 serves as a homodimerization interface bearing two surface anti-parallel β-strands (amino acids 735 to 750 and 780 to 800) (Song, 1999). However, the phyA-302 mutation did not affect dimerization (data not shown), which is consistent with observations in mutated phyB (Wagner and Quail, 1995). A response to continuous FR typical of phyA is mediated by chimeric phytochrome molecules bearing the N-terminal domain of phyA and the C-terminal domain of phyB (Wagner et al., 1996). This finding implies that any determinant of HIR present in the C terminus also should be found in phyB. This is the case for the Glu residue present at amino acid 777 in Arabidopsis phyA and conserved in phyB (Figure 5).
The Glu-to-Lys mutation observed in phyA-302 mutants severely affected the fluence rate dependence of phyA action under FR (Figure 8), but the same mutation in the equivalent position of phyB did not impair fluence rate dependence under continuous R (Wagner and Quail, 1995). Thus, different processes would underlie the fluence rate dependence of phyA- and phyB-mediated responses. In accordance with this interpretation, the fluence rate dependence of phyB-mediated responses to continuous R appears to be attributable to Pfr-to-Pr dark reversion (Kretsch et al., 2000), a kinetic component that is absent for phyA in Landsberg erecta (Eichenberg et al., 2000).
The wild type of Arabidopsis shows little morphological response to a daily pulse of R. This treatment, however, is effective in both phyA null mutants and wild-type seedlings exposed daily to a few hours of continuous FR before the R pulse. These observations have led to a model in which phyA in the VLFR mode (i.e., under R) negatively regulates phyB signaling, whereas phyA in the HIR mode (i.e., under continuous FR) positively regulates phyB signaling (Cerdán et al., 1999). The phyA-302 mutants behaved neither like the wild type nor like a phyA null mutant because they failed to respond to daily R pulses given alone or after a daily FR pretreatment (Figure 10). This observation is consistent with the ability of phyA-302 to retain VLFR and its failure to exhibit HIR.
phyA-302:GFP Fusion Proteins Fail to Produce Nuclear Speckles
Both phyA and phyB are present in the cytosol in etiolated seedlings and migrate to the nucleus upon illumination, where they localize to discrete subnuclear sites, forming spots that are smaller and more numerous in the case of phyA (Kim et al., 1999; Kircher et al., 1999; Yamaguchi et al., 1999; Hisada et al., 2000). The phyA-302:GFP fusion protein showed no obvious abnormalities with regard to translocation to the nucleus but failed to produce nuclear speckles (Figure 6). This observation suggests that Glu-777 in the PAS2 domain of phyA could be part of a speckle retention signal (Kim et al., 1999).
In animal cells, many nuclear factors localize either partially or completely in distinct “bodies” or subnuclear compartments, some of which contain factors involved in the processing and transcription of RNA (Lamond and Earnshaw, 1998). In addition to phytochromes, few plant proteins are known to form nuclear spots: COP1, a nuclear repressor of photomorphogenesis (Stacey and von Arnim, 1999); cryptochromes, blue light photoreceptors (Mas et al., 2000); LAF1, a transcription factor involved in photomorphogenesis (Ballesteros et al., 2001); and Rpn6, a proteasome non-ATPase regulatory subunit that is part of complexes apparently involved in responses to environmental stresses (Peng et al., 2001).
Mutations that occur in the WD-40 repeat domain of COP1 (Stacey and von Arnim, 1999) or in Glu-777 of the PAS2 domain of phyA affect both subnuclear localization and biological activity, suggesting a functional role for spot formation. The phenotype of phyA-302 mutants could result from either a failure of the mutated protein to colocalize with partners or impaired protein–protein interactions. Nuclear spots that contain phyA might represent foci for transcriptional complexes, a role that would be consistent with the action of phytochrome as a direct regulator of transcription factor activity (Ni et al., 1999). Failure to form such foci could impair HIR-specific transcriptional changes. Nuclear foci also might be involved in protein degradation, as speculated by Mas et al. (2000). This effect could account for the enhanced stability of the phyA-302 protein under continuous FR.
Implications for phyA Signaling
The change from Glu (acidic side chain) to Lys (basic side chain) caused by the phyA-302 mutations is predicted to alter the electrostatic environment of the peptide region, which could impair phyA interaction with other proteins. Actually, PAS domains often are involved in protein–protein interactions in signaling (Taylor and Zhulin, 1999). Furthermore, the mutation affects a peptide region containing Trp residues 774 and 778 (773 and 777 in oat phyA) that undergoes detectable conformational changes during phototransformation and becomes exposed preferentially in the Pfr form (Park et al., 2000). At least in the context of the full-length C-terminal domain of phyB, the Glu-to-Lys mutation at position 838 (which is equivalent to the phyA-302 mutation) impairs the binding of PIF3 (Ni et al., 1998).
Binding of nucleoside diphosphate kinase 2 to the C-terminal domain of phyA is disrupted by the phyA-103 mutation (Choi et al., 1999), but to the best of our knowledge, more C-terminal mutations have not been tested. The region close to the junction between the Quail box and the kinase motif (Figure 5) also could affect phyA kinase activity, as proposed recently by Park et al. (2000). However, the putatively impaired binding or activity of PIF3, nucleoside diphosphate kinase 2, or PKS1 does not offer a direct explanation for the lack of HIR in phyA-302. Antisense or knockout seedlings with reduced or no PIF3 (Ni et al., 1998), nucleoside diphosphate kinase 2 (Choi et al., 1999), or PKS1 (Fankhauser et al., 1999) retain full or weakly affected HIR.
Taking into account the fact that VLFR and HIR can be dissected genetically (Yanovsky et al., 1997, 2000; Luccioni et al., 2001) and at the level of target genes (Cerdán et al., 2000), it is tempting to speculate that the Glu-777 residue in the PAS2 domain is involved in HIR signaling via interaction with a partner not necessary for VLFR. Nuclear speckles could be a site for such interaction, and eventually a site for phyA destruction after signaling. There are several examples in nonplant systems in which signaling bifurcates at the receptor protein itself via specific docking sites for different signaling proteins (Pawson, 1995), creating a network of interconnected pathways that help coordinate cellular responses to the environment.
METHODS
Plant Material
Mutagenized seeds of Arabidopsis thaliana (ecotype Landsberg erecta) were purchased from Lehle Seeds (Round Rock, TX). For the mutant screening, the seeds were incubated in boxes (175 × 225 mm2 × 45 mm in height) containing 0.8% agar for 3 days at 7°C before transfer to the specific protocol conditions (Figure 1). The wild type was Landsberg erecta. phyA-201 (Nagatani et al., 1993), phyA-205 (Reed et al., 1994), and hy1 and hy2 (Koornneef et al., 1980; Davis et al., 1999; Muramoto et al., 1999; Kohchi et al., 2001) were included in some experiments for comparative purposes.
The construction of the 35S:PHYA:green fluorescent protein (GFP) chimeric gene containing the full-length PHYA cDNA from Arabidopsis in the binary vector pPCV 812 and its introduction in the phyA null background of Arabidopsis have been described (Kim et al., 2000). To produce the 35S:PHYA-302:GFP chimeric gene, the phyA-302 point mutation was introduced in the full-length PHYA cDNA by PCR and transferred to the 35S-GFP/nopaline synthase binary expression plasmid described previously (Kircher et al., 1999; Kim et al., 2000). Transgenic plants harboring GFP constructions were produced in the phyA-211 mutant Columbia background according to the method of Koncz et al. (1994). Several positive lines with varying levels of PHYA-302:GFP were analyzed in preliminary experiments, and the wild-type and mutant lines chosen exhibited similar intensities of GFP fluorescence.
Experimental Setting
In germination experiments, 25 seeds were sown in clear plastic boxes (40 × 33 mm2 × 15 mm in height) on two layers of filter paper saturated with distilled water, incubated in darkness at 6°C for 3 days, and further incubated at 25°C either in darkness or exposed to different light treatments. Germinated seeds (radicle protrusion) were counted 4 days later. In hypocotyl growth and cotyledon unfolding experiments, 15 seeds of each genotype were sown as described for germination experiments. After chilling, the boxes were exposed to a red light (R) pulse to induce homogeneous germination, incubated in darkness at 25°C for 24 h, and transferred to the light or dark treatments for 3 days before measurements.
Hypocotyl length was measured to the nearest 0.5 mm with a ruler, and the largest 10 seedlings from each box (i.e., one replicate) were averaged. The angle between the cotyledons was measured with a protractor in the same seedlings used for length measurements, and the 10 values obtained per box also were averaged before statistical analysis. For anthocyanin experiments, 50 seeds were sown per box according to the procedure used for hypocotyl growth experiments. After 3 days of light treatment, the seedlings were harvested for extraction with 1 mL of 1% (w/v) HCl methanol. Absorbance at 530 nm was measured and corrected for chlorophyll absorption by subtracting 0.25 absorbance at 657 nm.
Hourly pulses of far-red light (FR; 3 min/h at 200 μmol·m−2·s−1) and continuous FR (10 μmol·m−2·s−1) were provided by incandescent lamps in combination with a water filter, a red acetate filter, and six 2-mm-thick blue acrylic filters (Paolini 2031; La Casa del Acetato, Buenos Aires, Argentina). The calculated Pfr/P was 10%. In the experiments in which responses were plotted against calculated Pfr/P, the seedlings were exposed to 3-min hourly light pulses (10 to 50 μmol·m−2·s−1). Incandescent lamps were combined with a water filter and an RG9 filter (calculated Pfr/P of 3%; Schott, Mainz, Germany), one red plus one green acetate filter (calculated Pfr/P of 33%), or one red acetate filter (calculated Pfr/P of 61%). R (calculated Pfr/P of 87%) was provided by light-emitting diodes.
Protein Extraction and Immunoblot Analysis
Extracts were prepared from samples harvested on ice according to Martinez-García et al. (1999) and subjected to SDS-PAGE in 1.5-mm-thick, 4.5/7.5% stacking/resolving gels (Mini Protean II; Bio-Rad, Richmond, CA). Proteins were electroblotted to nitrocellulose (0.45-μm pore size; Sigma, St. Louis, MO) for 45 min in a Milliblot-SDE system (Millipore, Bedford, MA) according to the manufacturer's instructions. The remaining protein binding capacity was blocked with 1% skim milk, 50 mM Tris-Cl, and 200 mM NaCl, pH 7.4, for 30 min at 37°C. The monoclonal antibody 073D raised in mouse against Avena phyA (Somers et al., 1991) was kindly provided by Peter Quail (University of California, Berkeley, and U.S. Department of Agriculture Plant Gene Expression Center, Albany, CA).
The blot was incubated overnight at 7°C with 10 mL of this primary antibody at a dilution of 1:1000. After washing, the membrane was incubated with a 1:500 dilution of affinity-isolated alkaline phosphatase–conjugated antibody to mouse IgG developed in goat (Sigma). The bands were visualized by incubating the blots in 0.1 M Tris, pH 9.5, 100 mM NaCl, and 5 mM MgCl2 containing 0.165 mg/mL 5-bromo-4-chloro-3-indolyl phosphate (p-toluidine salt) and 0.33 mg/mL nitroblue tetrazolium (both Sigma).
Phytochrome Fluorescence
Fluorescence data were obtained as described in Sineshchekov et al. (1998) from 3- to 4-day-old seedlings grown in complete darkness at 26°C on filter paper moistened with tap water. The cotyledons were cut off to eliminate protochlorophyllide interference, and 10 hypocotyls plus roots were glued with a 50:50 water:glycerol mixture to a Plexiglas plate of the sample holder. Fluorescence emission spectra at low temperature (85K) were recorded with the use of a laboratory-designed spectrofluorimeter based on two double-grating monochromators. The source of exciting light (reduced by ∼100-fold with the use of neutral filters to avoid photochemical effects) and actinic light at 639 nm was a 5-mW laser diode (RLD-635; Taiwan).
For each experimental sample, three emission spectra usually were measured: (1) in the etiolated state (state 0); (2) after phototransformation induced by the saturating actinic light of Pr into the first photoproduct stable at low temperatures (lumi–R) (state 1); and (3) after Pr photoconversion into Pfr at 273K by monochromatic light at 680 nm (state 2). To obtain emission spectra of phytochrome, the spectrum of the phyA-201 phyB-1 Arabidopsis double mutant was subtracted from the experimental spectra of the samples after normalization at 660 nm (at which phytochrome emission is negligible), and the phytochrome difference spectrum was divided by the intensity of background emission at 660 nm to correct for sample mass. The noise level during the registration of the emission spectra was 5 to 7%, and the smoothing of the spectra was performed by eye.
DNA Sequencing
Genomic DNA was isolated from plants of the wild type and of the phyA alleles identified here according to the procedure described by Rogers and Bendich (1988). The complete PHYA gene (including exons and introns) was amplified by PCR using a pair of primers designed on the basis of a previously published sequence of the gene (Dehesh et al., 1993). The PCR mixture was 20 mM Tris-HCl, pH 8.4, 50 mM KCl, 2 mM MgCl2, 200 μM each deoxynucleotide triphosphate, 25 pmol of each primer, 2 μL of the genomic DNA preparation, and 2.5 units of Taq polymerase (Gibco BRL) in a final volume of 25 μL.
The mixture was heated to 94°C for 4 min and subjected to 35 cycles of 30 s at 94°C, 30 s at 55°C, and 210 s at 72°C. The reaction products were analyzed on 0.8% agarose gels. The size of the products was ∼4.8 kb. The corresponding fragments were extracted from the gels and subcloned in the pUC19 plasmid (Sambrook et al., 1989) according to standard protocols. Both strands of the cloned phyA genes were sequenced using the recombinant plasmids as templates with an automated DNA sequencer (Alf Express II; Amersham Pharmacia) and 10 pairs of primers internal to the PCR product designed on the basis of a published sequence (Dehesh et al., 1993).
Epifluorescence and Light Microscopy
Dark-grown seedlings (3 days old) were either kept in the dark or exposed to 1, 3, 9, or 24 h of continuous FR (21 μmol·m−2·s−1). All subsequent manipulations were performed under dim green light. The upper halves of the hypocotyls were collected, transferred to glass slides, and analyzed with an Axiovert microscope (Zeiss, Oberkochem, Germany). Excitation of GFP was performed with standard fluorescein isothiocyanate filters. Representative nuclei were photographed with an Axiocam camera and Axiovision software (Zeiss). Only epidermal cells of the upper quarter of the hypocotyl were analyzed.
To minimize the nuclear import of fusion proteins induced by microscopic light, photographs of the GFP fluorescence were taken during the first minute of microscopic analysis. 4′,6-Diamidino-2-phenylindole staining of nuclei was performed by incubating the seedlings in an aqueous solution containing 10% DMSO (v/v) and 10 μg/mL 4′,6-diamidino-2-phenylindole. For optimal presentation, the images were processed using Adobe Photoshop 5.0 (Adobe Systems Europe, Edinburgh, UK).
Acknowledgments
We thank Peter Quail (University of California, Berkeley) for his kind provision of the phyA monoclonal antibody and the ABRC (Ohio State University, Columbus) for seed batches. This work was supported by grants from Fondo Nacional de Ciencia y Técnica (BID 1201/OC-AR-PICT 06739), the University of Buenos Aires (TG59), and the Fundación Antorchas (A-13622/1-40) to J.J.C., Consejo Nacional de Investigaciones Científicas y Técnicas Grant 4527/96 to R.J.S., a Howard Hughes Medical Institute International Scholar Fellowship, a Hungarian Foundation for Basic Science Grant (T-02584), and a Wolfgang Paul Award to F.N., Deutsche Forschungsgemeinschaft Grant SFB 592 to E.S., and Russian Foundation for Fundamental Research Grant 99-04-48730 to V.A.S.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.000521.
References
- Ahmad, M., Jarrillo, J.A., Smirnova, O., and Cashmore, A. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1, 939–948. [DOI] [PubMed] [Google Scholar]
- Ballesteros, M.L., Bolle, C., Lois, L.M., Moore, J.M., Vielle-Calzada, J.-P., Grossniklaus, U., and Chua, N.-M. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15, 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei, A., Séraphin, B., and Serrano, L. (2001). Positive feedback in eukaryotic gene networks: Cell differentiation by graded to binary response conversion. EMBO J. 20, 2528–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, J.F., Sánchez, R.A., Whitelam, G.C., and Casal, J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal, J.J., Yanovsky, M.J., and Luppi, J.P. (2000). Two photobiological pathways of phytochrome A activity, only one of which shows dominant negative suppression by phytochrome B. Photochem. Photobiol. 71, 481–486. [DOI] [PubMed] [Google Scholar]
- Cerdán, P.D., Staneloni, R.J., Ortega, J., Bunge, M.M., Rodriguez-Batiller, J., Sánchez, R.A., and Casal, J.J. (2000). Sustained but not transient phytochrome A signaling targets a region of a Lhcb1*2 promoter that is not necessary for phytochrome B action. Plant Cell 12, 1203–1211. [PMC free article] [PubMed] [Google Scholar]
- Cerdán, P.D., Yanovsky, M.J., Reymundo, F.C., Nagatani, A., Staneloni, R.J., Whitelam, G.C., and Casal, J.J. (1999). Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J. 18, 499–507. [DOI] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.-K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.-R., and Song, P.-S. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., Kurepa, J., and Vierstra, R.D. (1999). The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenase. Proc. Natl. Acad. Sci. USA 96, 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh, K., Franci, C., Parks, B.M., Seeley, K.A., Short, T.W., Tepperman, J.M., and Quail, P.H. (1993). Arabidopsis hy8 locus encodes phytochrome A. Plant Cell 5, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberg, K., Hennig, L., Martin, A., and Schäfer, E. (2000). Variation in dynamics of phytochrome A in Arabidopsis ecotypes and mutants. Plant Cell Environ. 23, 311–319. [Google Scholar]
- Elich, T.D., and Chory, J. (1997). Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell 9, 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Hisada, A., Hanzawa, H., Weller, J.L., Nagatani, A., Reid, J.B., and Furuya, M. (2000). Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunochemical procedures. Plant Cell 12, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, L., Kircher, S., Toth, R., Adam, E., Schäfer, E., and Nagy, F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22, 125–133. [DOI] [PubMed] [Google Scholar]
- Kim, Y.H., Choi, C.Y., and Kim, Y. (1999). Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci. USA 96, 12350–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochromes A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A., and Lagarias, J.C. (2001). The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C.S., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, B.S. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Press), pp 1–22.
- Koornneef, M., Rolf, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Krall, L., and Reed, J.W. (2000). The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl. Acad. Sci. USA 97, 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsch, T., Poppe, C., and Schäfer, E. (2000). A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J. 22, 177–186. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and Earnshaw, W.C. (1998). Structure and function in the nucleus. Science 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Luccioni, L.G., Oliverio, K.A., Yanovsky, M.J., Boccalandro, H., and Casal, J.J. (2001). Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 178, 173–181. [PMC free article] [PubMed] [Google Scholar]
- Martinez-García, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Martinez-García, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257. [DOI] [PubMed] [Google Scholar]
- Mas, P., Devlin, P.F., Panda, S., and Kay, S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211. [DOI] [PubMed] [Google Scholar]
- Muramoto, T., Kohchi, T., Yokota, A., Hwang, I., and Goodman, H.M. (1999). The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plasmid heme oxygenase. Plant Cell 11, 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Park, C.M., Bhoo, S.-H., and Song, P.S. (2000). Inter-domain crosstalk in the phytochrome molecules. Semin. Cell Dev. Biol. 11, 449–456. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1993). hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, B.M., Quail, P.H., and Hangarter, R.P. (1996). Phytochrome A regulates red-light induction of phototropic enhancement in Arabidopsis. Plant Physiol. 110, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson, T. (1995). Protein modules and signalling networks. Nature 373, 573–580. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Staub, J.M., Serino, G., Kwok, S.F., Kurepa, J., Bruce, B.D., Vierstra, R.D., Wei, N., and Deng, X.-W. (2001). The cellular level of PR500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol. Biol. Cell 12, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.O., and Bendich, A.J. (1988). Extraction of DNA from plant tissue. In Plant Molecular Biology Manual, S. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–10.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schneider-Poetsch, H., and Braun, B. (1991). Proposal on the nature of phytochrome action based on the C-terminal sequences of phytochrome. J. Plant Physiol. 137, 576–580. [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and phytochrome B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov, V.A., Ogorodnikova, O.B., Devlin, P.F., and Whitelam, G.C. (1998). Fluorescence spectroscopy and photochemistry of phytochromes A and B in wild-type, mutant and transgenic strains of Arabidopsis thaliana. J. Photochem. Photobiol. B Biol. 42, 133–142. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Sharrock, R.A., Tepperman, J.M., and Quail, P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3, 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, P.-S. (1999). Inter-domain signal transmission within phytochromes. J. Biochem. Mol. Biol. 32, 215–225. [Google Scholar]
- Stacey, M.G., and von Arnim, A.G. (1999). A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J. Biol. Chem. 274, 27231–27236. [DOI] [PubMed] [Google Scholar]
- Taylor, B.L., and Zhulin, I.B. (1999). PAS domains: Internal sensors of oxygen, redox potential, and light. Microb. Mol. Biol. Rev. 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Fairchild, C.D., Kuhn, R.M., and Quail, P.H. (1996). Chromophore-bearing NH2-terminal domains of phytochromes A and B determine their photosensory specificity and differential light lability. Proc. Natl. Acad. Sci. USA 93, 4011–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., and Quail, P.H. (1995). Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proc. Natl. Acad. Sci. USA 92, 8596–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Parks, B.M., Short, T.W., and Quail, P.H. (1995). Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell 7, 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 3, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Casal, J.J., and Luppi, J.P. (1997). The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signalling pathways that correspond to the very-low fluence and high-irradiance responses of phytochrome. Plant J. 12, 659–667. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Whitelam, G.C., and Casal, J.J. (2000). fhy3-1 retains inductive responses of phytochrome A. Plant Physiol. 123, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.C., and Lagarias, C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95, 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]