Abstract
To investigate how roots respond to directional cues, we characterized a T-DNA–tagged Arabidopsis mutant named sku5 in which the roots skewed and looped away from the normal downward direction of growth on inclined agar surfaces. sku5 roots and etiolated hypocotyls were slightly shorter than normal and exhibited a counterclockwise (left-handed) axial rotation bias. The surface-dependent skewing phenotype disappeared when the roots penetrated the agar surface, but the axial rotation defect persisted, revealing that these two directional growth processes are separable. The SKU5 gene belongs to a 19-member gene family designated SKS (SKU5 Similar) that is related structurally to the multiple-copper oxidases ascorbate oxidase and laccase. However, the SKS proteins lack several of the conserved copper binding motifs characteristic of copper oxidases, and no enzymatic function could be assigned to the SKU5 protein. Analysis of plants expressing SKU5 reporter constructs and protein gel blot analysis showed that SKU5 was expressed most strongly in expanding tissues. SKU5 was glycosylated and modified by glycosyl phosphatidylinositol and localized to both the plasma membrane and the cell wall. Our observations suggest that SKU5 affects two directional growth processes, possibly by participating in cell wall expansion.
INTRODUCTION
Roots grow by coordinating cell division and cell expansion at their tips. In Arabidopsis, this occurs within a 1-mm region of the tip composed of a meristem along with two zones of cell expansion termed the distal and main elongation zones (Mullen et al., 1998a). Within the meristem, cell division and differentiation begin with four sets of progenitor cells that surround the nondividing quiescent center cells (Dolan et al., 1993; Benfey and Scheres, 2000). Cells of each lineage divide to form concentric rings around the developing vasculature, first expanding quasiisodiametrically and then anisotropically as they pass through the elongation zones. Differentiating cells divide and expand anisotropically along the longitudinal axis of the root, forming distinct files (Dolan et al., 1993).
The cell files are in large part linear, although a slight twisting bias or handedness occurs naturally in roots of some Arabidopsis ecotypes, including Wassilewskija and Landsberg erecta (Mirza, 1987; Rutherford and Masson, 1996). This twisting, or cell file rotation, develops within the elongation zones and also occurs in both clockwise and counterclockwise directions in conjunction with root tip bending during responses to certain environmental stimuli (Okada and Shimura, 1990).
As a root grows within a constant environment, cells of a given type divide and expand approximately the same amount around the periphery of the root, resulting in a straight root. Various stimuli induce a hormone-mediated differential growth process to occur, thereby altering the orientation of the root tip. For instance, a change in the root tip's alignment with the gravity vector induces an increase in cell expansion on the upper flank of the tip along with a decrease in expansion on the lower flank, causing the root tip to bend downward along a two-dimensional plane (Mullen et al., 1998a; Chen et al., 1999).
More complex directional growth responses occur when roots are exposed to multiple stimuli. An example of this is the wavy root growth pattern. Okada and Shimura (1990) reported that Arabidopsis roots growing on a tilted impenetrable agar surface formed a consistent and reproducible sinusoidal wave pattern. Upon close examination, it was found that the back-and-forth movement made by the root tip as it waved down the agar surface occurred in three dimensions and was accompanied by an alternating root tip rotation about its axis (Okada and Shimura, 1990; Rutherford and Masson, 1996; J.C. Sedbrook and C.R. Somerville, unpublished data).
Okada and Shimura (1990) found that the wavy root growth pattern was surface dependent and probably attributable to a combination of root tip responses to gravity and touch. Others have noted that circumnutation may regulate the wave frequency (Mullen et al., 1998b). The biomechanical properties of the root result in its buckling during wave formation (J.C. Sedbrook and C.R. Somerville, unpublished data), and varied media compositions and conditions greatly influence the root waving pattern (Buer et al., 2000; J.C. Sedbrook and C.R. Somerville, unpublished data).
To investigate the processes underlying regulated cell growth, we used root waving as a phenotype to facilitate the identification of mutants with alterations in growth processes. Here, we report the characterization of a novel mutant, sku5, which develops roots that skew away from the typical downward direction of growth on agar surfaces. sku5 roots and etiolated hypocotyls also twist more than normal in a counterclockwise direction about their longitudinal axes as they grow.
Analyses of the growth response of the mutant shows that directional growth on agar surfaces can be uncoupled from axial root tip rotation, two processes that normally occur together. The SKU5 gene encodes a glycoprotein that is related structurally to the multiple-copper oxidases. SKU5 becomes glycosyl phosphatidylinositol (GPI) modified and localized to the plasma membrane and cell wall. Our results suggest that SKU5 plays an important role in regulating directional root growth.
RESULTS
Isolation and Characterization of sku5
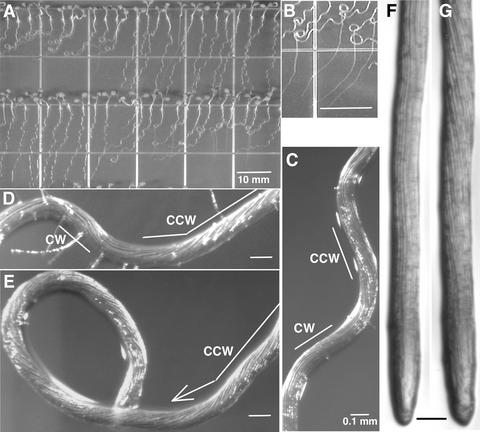
The sku5 mutant was identified by screening a population of T-DNA–mutagenized seedlings for altered root growth on the surface of 1.5% agar-solidified germination medium (GM) that was tilted 30° from the vertical. Roots of wild-type seedlings grew in a waving pattern that was skewed slightly to the left when viewed from above the agar surface (Figure 1A, Table 1). The sku5 mutant was indistinguishable from the wild type except that the growth of mutant roots skewed strongly to the left and, in most cases, the roots looped to the left to form coils (Figure 1A).
Figure 1.
Root Growth Phenotypes of sku5 and Wild-Type Seedlings.
(A) Seedlings grown for 7 days on 1.5% agar-solidified GM tilted at 30° from the vertical. sku5 seedlings are at left on the top row and at right on the bottom row.
(B) sku5 roots grow straight down after penetrating the agar until they touch the bottom of the plate, after which they slant again.
(C) to (E) Close-up of wild-type (C) and sku5 ([D] and [E]) roots that had gone through the root-waving assay. Note the cell file rotation. CCW, counterclockwise; CW, clockwise.
(F) and (G) Wild-type (F) and sku5 (G) roots that had been growing for 7 days in 0.8% agar-solidified GM. Note the greater amount of cell file rotation of the sku5 root.
Bars in (A) and (B) = 10 mm; bars in (C) to (G) = 0.1 mm.
Table 1.
Growth Measurements of Wild-Type and sku5 Organs under Various Conditions
| Roots on Tilted 1.5% Agar |
Roots on Vertical 0.8% Agar |
Roots in 0.8% Agar |
Roots in Liquid GM |
Etiolated Hypocotyls in Air |
|
|---|---|---|---|---|---|
| WTa lengthb | 23.9 ± 2.5 | 25.1 ± 2.8 | 24.0 ± 3.2 | NDc | 14.8 ± 1.1 |
| sku5 lengthb | 21.5 ± 2.4 | 21.0 ± 2.5 | 20.7 ± 2.8 | ND | 13.3 ± 1.0 |
| P value | 6.3 × 10−7 | 1.9 × 10−14 | 2.7 × 10−48 | NAd | 5.3 × 10−29 |
| WT anglee | 14.2° ± 4.8 | 2.8° ± 8.0 | 0.14° ± 4.9 | NA | NA |
| sku5 anglee | 59.4° ± 7.0 | 31.7° ± 11.3 | 0.55° ± 5.6 | NA | NA |
| P value | 5.9 × 10−97 | 1.5 × 10−38 | 0.47 | NA | NA |
| WT CFRf | ND | 2.8 ± 4.1 | 2.9 ± 3.1 | 2.6 ± 2.6 | NA |
| sku5 CFRf | ND | 17.1 ± 4.9 | 16.7 ± 5.2 | 13.2 ± 5.2 | NA |
| P value | NA | 3.3 × 10−30 | 1.2 × 10−31 | 2.5 × 10−24 | NA |
Measurements are averages ± sd; n = sample size; P value = Student's t test probability, where <0.05 is statistically significant. The P values correspond to a comparison between wild-type and sku5 samples of a given treatment. 47<n<93 for all samples except “Roots in 0.8% Agar” wild type and sku5 length (418<n<422) and “Etiolated Hypocotyls in Air” wild type and sku5 length (145<n<164).
WT, Wild type.
Average organ length in millimeters.
ND, Not determined.
NA, Not applicable.
Average angle that roots grew, where 0° is vertical and positive numbers were to the left of vertical, as viewed from above the agar surface.
The number of epidermal cell files that crossed a 1-mm-long line drawn down the longitudinal axis of the root, from ∼1.5–2.5 mm from the tip. CFR, Cell file rotation.
When seedlings were grown on vertically oriented 0.8% agar-solidified GM, which did not impede vertical root growth, sku5 roots still slanted much more in a leftward direction than did the wild type (Table 1). On the other hand, when sku5 roots growing in a skewed direction on inclined agar medium penetrated the agar, they assumed a normal vertical trajectory of growth until they touched the bottom of the plate, after which they skewed again (Figure 1B). The growth directions of sku5 and wild-type roots that were grown for 7 days embedded in 0.8% agar-solidified GM also did not deviate significantly from the vertical (Table 1). These observations indicate that the sku5 skewed root growth phenotype was surface dependent and was not caused by a difference in the gravitropic set point angle.
Root waving has been associated with the formation of twisted epidermal cell files in places along the root where it has changed directions (Okada and Shimura, 1990; Rutherford and Masson, 1996). We observed that sku5 roots grown on tilted 1.5% agar-solidified GM had many more cell files twisted in a counterclockwise direction than the wild type, especially in regions where the roots had looped (Figures 1C to 1E). In general, the direction of twisting correlated with the direction of root growth for both mutant and wild-type seedlings (i.e., counterclockwise twisting indicates leftward growth direction). However, along stretches of the sku5 roots where the cell file rotation was constant, the rate of change in growth direction was variable (Figure 1E), indicating that the two processes were not strictly coupled.
To determine if the sku5 counterclockwise cell file rotation bias also occurred in roots that were not subject to the directional stimuli found at a surface, we grew seedlings in 0.8% agar-solidified GM and in liquid GM, which presented the roots with minimal touch stimulation. Under these two conditions, sku5 roots twisted in a counterclockwise direction much more than wild-type roots, albeit slightly less than when grown on agar surfaces (Figures 1F and 1G, Table 1). The maximum angles at which the cell files crossed the longitudinal axes of the roots were approximately the same for both wild-type and sku5 roots.
Under all of these conditions, the rate of epidermal cell file rotation varied along the lengths of the roots, with no apparent regularity. In some cases, both wild-type and sku5 roots rotated briefly in a clockwise direction about their axes. Cell file rotation was not seen in the first 400 μm of the root tip but became visible within the elongation zones (Figure 1G). This finding suggests that the axial twisting was caused by differential cell expansion that began in the elongation zones and was not the result of a skewed pattern of cell division in the meristem.
Confocal microscopic imaging of propidium iodide–stained sku5 and wild-type roots revealed that in regions in which no cell file rotation occurred, epidermal and the underlying cortical cell files were parallel to each other (data not shown). In regions in which epidermal cell files were twisted, sku5 and wild-type cortical cell files also were twisted, albeit generally slightly less than the overlying epidermal cell files (Figures 2A and 2B).
Figure 2.
Confocal Microscopic Sections through Epidermal and Cortical Root Cells Revealing the Relative Alignments of the Cell Files.
Images of wild-type (A) and sku5 (B) root mature zones that were stained with propidium iodide. For each image, a cross-section through epidermal cells was colored green and then overlaid on a cross-section through cortical cells (colored red). The arrows point to the cell walls of individual cell files to highlight the fact that in regions in which cell file rotation occurs in both wild-type and sku5 roots, the epidermal cell files twist slightly more than the underlying cortical cell files.
We also examined the hypocotyl cell files of sku5 and wild-type seedlings grown in the dark. We found that sku5 hypocotyls twisted more in a counterclockwise direction compared with the wild type (viewed along the axis toward the apex), indicating that the SKU5 gene also affects directional growth in this organ (average angles of cortical cells in relation to longitudinal axes: wild type, 3.8° ± 4.5°; sku5, 6.4° ± 5.1° [P = 0.001, 76 < n < 81]). We observed no obvious twisting or morphological defects in light-grown sku5 hypocotyls, petioles, and inflorescences (data not shown).
In addition to the surface-dependent directional growth defect of sku5 roots and the axial rotation defects of sku5 roots and etiolated hypocotyls, we found that sku5 roots and etiolated hypocotyls were ∼15 and 10% shorter, respectively, than those of the wild type (Table 1). The sku5 root length differences occurred when seedlings were grown both in and on agar, indicating that this phenotype was not surface dependent. These length differences were statistically significant and were not attributable to differences in germination times (Table 1).
To determine if sku5 roots were shorter than wild-type roots because the cells did not expand longitudinally to the same length as wild-type cells, we measured the lengths of cortical cells in the mature zones of sku5 and wild-type roots. Seedlings were grown for 7 days in 0.8% agar-solidified GM, and the roots were stained with propidium iodide and imaged with a confocal microscope. We found that sku5 cortical cells were the same length as wild-type cortical cells (sku5 average cell length was 203.5 ± 46.1 μm [n = 212], and wild-type average cell length was 200.4 ± 45.3 μm [n = 285]; P = 0.48 by Student's t test). We also observed no differences in the widths of cortical cells or other cell types within the root or the hypocotyl (data not shown). Cell sizes and shapes within the root tip, including the meristem and the elongation zones of sku5 roots, also appeared normal. We were unable to measure the lengths of sku5 epidermal cells because they twisted out of view.
Given that gravitropism plays an important role in root waving (Rutherford and Masson, 1996; J.C. Sedbrook and C.R. Somerville, unpublished data), we tested whether sku5 roots had altered bending kinetics upon gravistimulation. Three-day-old agar-embedded seedlings were rotated 90°, and root tip angles were measured over time. At time points up to 12 h, there were no differences between the mutant and the wild type (see supplemental data online). At 12 to 15 h, sku5 roots were slightly less oriented to the gravity vector than wild-type roots, possibly as a result of the slightly reduced growth rate or of the fact that the axial rotation continually changes the orientation of the gravity-sensing columella cells in the tip.
We also germinated and grew sku5 and wild-type seedlings on 1.5% agar-solidified GM plates that slowly rotated in a clockwise direction on a clinostat. This treatment essentially randomized the seedling's exposure to the gravity vector, allowing observation of root growth character when this stimulus was minimized. After 5 days of growth, nearly all of the sku5 seedling roots had formed tight clockwise coils, whereas wild-type seedling roots had grown randomly or had taken on a slight clockwise growth direction (Figure 3).
Figure 3.
Root Growth Phenotypes.
Phenotypes of wild-type (A) and sku5 (B) seedlings after 5 days of growth on 1.5% agar-solidified GM rotated at 2 rpm on a clinostat in a clockwise direction about a line drawn through the center of the plate (viewed from above the agar surface).
Although we did observe a “clinostat effect” in these experiments associated with the direction of seedling rotation (i.e., counterclockwise clinostat rotation tended to make roots grow in a counterclockwise direction [Mirza, 1988; Rutherford and Masson, 1996]), the results always showed that sku5 roots coiled much more in a clockwise direction than did wild-type roots (data not shown). Therefore, because wild-type roots did not coil like sku5 roots under randomized gravity, we conclude that an altered gravity response of sku5 roots is not the cause of the surface-dependent skewing phenotype. Rather, the gravitropic response counteracts the sku5 skewing phenotype.
We also examined how the microtubule-interacting drug propyzamide affected sku5 root growth. Furutani et al. (2000) found that 1 μM propyzamide suppressed the clockwise epidermal axial rotation phenotype as well as the anisotropic growth defects found in spiral1 (spr1) and spiral2 (spr2) mutants. Like sku5, spr1 mutant roots exhibit a surface-dependent directional growth defect, except that spr1 roots slant to the right instead of to the left on tilted agar surfaces. Growth on 3 μM propyzamide induced wild-type Landsberg erecta, spr1, and spr2 root epidermal cell files to twist in a counterclockwise direction and caused their roots to slant similarly to the left on tilted agar surfaces.
This treatment had the same effects on sku5 and wild-type Wassilewskija roots, except that the sku5 growth parameters were much more exaggerated than those seen in the wild type (data not shown). Additionally, propyzamide equally affected sku5 and wild-type root morphologies and lengths. Therefore, it appears that the sku5 mutant is not affected directly in the same processes as the spr mutants, which have defects in the orientations of their cortical microtubule arrays (Furutani et al., 2000).
Cloning of the SKU5 Gene
sku5 was isolated from a collection of T-DNA insertion mutants derived from a plasmid carrying neomycin phosphotransferase as the selectable marker (Bouchez et al., 1993). Analysis of F1 plants and F2 segregants from a cross between the mutant and the wild type indicated that the sku5 mutation was recessive and segregated as a single nuclear mutation. Analysis of the sku5 and kanamycin resistance phenotypes of 50 seedlings from each of 149 F3 families showed that the sku5 mutation cosegregated with the kanamycin resistance marker on the T-DNA. DNA gel blot analysis of genomic DNA derived from homozygous sku5 plants, using T-DNA fragments as probes, revealed that sku5 plants contained only one T-DNA insertion (data not shown). Together, these data indicate that a T-DNA element may have inserted within the SKU5 gene.
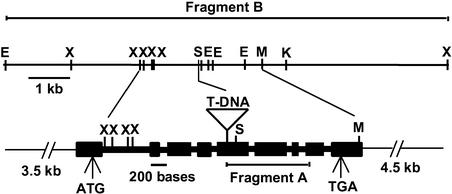
An adaptor PCR method (Siebert et al., 1995) was used to isolate a 1.1-kb region of genomic DNA (fragment A) flanking the T-DNA insertion in sku5 plants (Figure 4). The sequence of fragment A had regions of identity to two Arabidopsis EST clone sequences. At the time, no matches with Arabidopsis genomic sequences were identified because that portion of the genome had not been sequenced. Fragment A was used to probe a BAC library filter set, identifying two BAC clones (1M5 and 9F23). The candidate gene was cloned on a 10.7-kb EcoRI-XbaI fragment (fragment B; Figure 4).
Figure 4.
Restriction Map of the 10.7-kb Genomic Fragment B (top) and Structure of the SKU5 Gene (bottom).
For the SKU5 gene, black boxes represent exons and thick connecting lines represent introns. ATG and TGA represent translational start and stop sites, respectively. The location of the T-DNA insertion is shown within the fifth exon. The location of fragment A is delineated below the SKU5 gene. E, EcoRI; K, KpnI; M, SmaI; S, SacI; X, XbaI.
Fragment A also was used as a probe to screen a cDNA library (Kieber et al., 1993), resulting in the identification of four full-length or nearly full-length cDNA clones (pJS1 to pJS4). Sequence analysis of these clones, along with that of fragment B, revealed a 3.3-kb gene with nine exons encoding a putative 1764-bp open reading frame predicted to encode a 66-kD protein (Figure 4). The T-DNA had inserted within the fifth exon of that gene (Figure 4). To determine if the putative SKU5 gene could rescue the sku5 mutant phenotype, fragment B was cloned into the pBIB binary vector (Becker, 1990) to produce pJS5, which was introduced into sku5 plants via Agrobacterium tumefaciens–mediated transformation.
T1 transformants carrying this construct were isolated on hygromycin-containing Murashige and Skoog (1962) agar and allowed to self-fertilize, and the resultant T2 seedlings were scored for root waving and for hygromycin resistance (hygromycin resistance originated from the pBIB T-DNA). The analysis of six independent T2 families (50 seedlings each) revealed a 3:1 segregation of the wild type to skewed root waving coupled with a 3:1 segregation of hygromycin resistance to sensitivity (data not shown). Thus, fragment B complemented the sku5 mutation.
As a control, a 1.4-kb SacI-SmaI fragment within the putative SKU5 gene on pJS5 was removed, and the modified construct (pJS6) was transformed into sku5 plants. Phenotypic analysis of 15 T2 families from plants transformed with pJS6 revealed that all seedlings exhibited the skewed root-waving phenotype. Thus, we conclude that the gene carrying the T-DNA insertion was SKU5.
SKU5 Belongs to a Multigene Family That Is Related Structurally to the Multiple-Copper Oxidases
The Arabidopsis genome contains 18 genes that are predicted to encode proteins sharing 42 to 65% amino acid sequence identity with SKU5. We named these genes SKS (SKU5 Similar). GenBank contains EST sequences corresponding to 10 of these genes. ClustalX and TreeView analyses placed the predicted proteins from these genes in an unrooted bootstrap tree consisting of four branches or clades (see supplemental data online). Hydropathy profiles of these proteins along with SKU5 revealed that all of them contained hydrophobic domains at their N termini (data not shown). The computer program SignalP version 2.0 (http://www.cbs.dtu.dk/services/SignalP-2.0/; Nielsen et al., 1997) predicted that all of these hydrophobic sequences were cleavable signal sequences, whereas PSORT (http://psort.nibb.ac.jp/form.html; Nakai and Horton, 1999) predicted that all but two of these sequences (SKS10 and SKS15) could be cleaved proteolytically.
Two SKU5-like proteins from tobacco and tomato (NTP303, which has 45% amino acid sequence identity with SKU5 [Wittink et al., 2000], and PE3, which has 50% identity [Tucker and Zhang, 1996]) have been purified previously and their N-terminal peptides sequenced. Both proteins lacked the predicted N-terminal hydrophobic domain, presumably because they were removed proteolytically. Furthermore, a search of the Arabidopsis Plant Plasma Membrane Database (http://sphinx.rug.ac.be:8080/ppmdb/) identified a 15–amino acid sequence obtained from the N terminus of an anonymous plasma membrane protein (sample HD022_1_0 ADPY.FYNFEV.YFT) that corresponds to a region of the SKU5 protein sequence (5′-ADPYSFYNFEVSYIT-3′). This peptide sequence begins at residue 21 of SKU5, the position adjacent to the predicted cleavage site of the SKU5 leader peptide. Therefore, it appears likely that SKU5, along with many, if not all, of the SKS proteins, has an N-terminal cleavable signal sequence.
Database searches also revealed that the predicted SKU5 protein shares 23 to 27% amino acid sequence identity with ascorbate oxidases and laccases. These two families of multiple-copper oxidases coordinate four copper ions within three spectroscopically distinct centers (types 1, 2, and 3), allowing for the one-electron oxidation of a reducing substrate coupled to the four-electron reduction of oxygen to water (Messerschmidt and Huber, 1990). The amino acid residues that act as ligands to coordinate the copper ions within the different copper centers are conserved completely in both of these families. Interestingly, the ligands for the type 1 and 3 copper centers are absent in SKU5 (residues Asn-82, Phe-124, Ser-126, Gln-449, Ser-454, Arg-510, and Glu-512 should be His), whereas those for the type 2 copper centers are present (His-80 and His-452). None of the three types of copper center motifs is intact within the predicted SKS proteins (data not shown).
Searches of EST databases identified SKU5-related genes in at least 20 different monocotyledonous and dicotyledonous plant species (data not shown). SKU5 amino acid sequence alignments with the predicted protein portions encoded by these ESTs exhibited 51 to 86% amino acid sequence identity. This level of identity, along with the fact that they also do not contain all of the copper binding motifs, suggest that the genes corresponding to these ESTs are in the same family as SKU5 and the SKS genes.
SKU5 Is Expressed Ubiquitously
To verify that SKU5 is expressed in all tissues, we raised a polyclonal antibody against a peptide with a sequence unique to SKU5 and used this antibody to probe gel blots of proteins from various tissue extracts. This antibody specifically recognized a band corresponding to a 90-kD protein in wild-type plant extracts that was absent in sku5 plant extracts (Figures 5A and 5B). This antibody recognized SKU5-related bands in extracts from wild-type roots, hypocotyls, cotyledons, leaves, inflorescence stems, and flowers (Figures 5A and 5B).
Figure 5.
SKU5 Is Expressed in All Tissues.
(A) Protein gel blot of extracts from various seedling and adult plant organs probed with anti-SKU5 antibody. Equal amounts of protein were loaded in all lanes. Note the absence of a band corresponding to a 90-kD protein in the sku5 seedling sample.
(B) The same blot stained with Ponceau S.
SKU5 Is Glycosylated
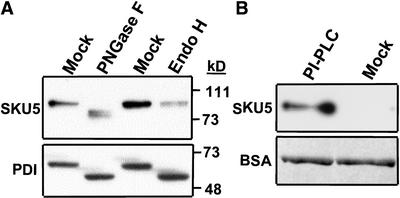
SKU5 migrated more slowly than expected on SDS-PAGE gels (apparent mass of 90 kD instead of the predicted 66 kD; Figure 5A). To determine whether SKU5 was glycosylated, crude seedling extracts were treated with peptide N-glycosidase F (PNGase F) or endoglycosidase H (Endo H), and the mobility of SKU5 was measured on protein gel blots of SDS-PAGE gels (Figure 6A).
Figure 6.
SKU5 Can Be Deglycosylated by PNGase F and Released from Membranes by PI-PLC.
(A) Protein gel blot probed first with the anti-SKU5 antibody (top) and then with an anti–protein disulfide isomerase (PDI) antibody (positive control; bottom). Wild-type seedling extracts were treated with PNGase F, Endo H, or buffer solution (mock). Note that both enzymes removed N-glycans from PDI, whereas only PNGase F removed them from SKU5.
(B) Gel blot analysis of proteins released from wild-type microsomal membranes by treatment with PI-PLC (top). Microsomal membranes were treated with either PI-PLC or a buffer solution (mock) and then partitioned with Triton X-114 to separate the soluble protein (shown here) from the membrane-bound protein. BSA was added to the samples as a check for equal recovery during the manipulations, as described in Methods. Proteins were resolved by SDS-PAGE, transferred to a filter that was probed with the anti-SKU5 antibody (top), and then stained with Ponceau S to reveal the BSA protein loading control (bottom).
SKU5 within the PNGase F–treated sample migrated faster than that in the mock-treated sample, whereas SKU5 within the Endo H–treated sample migrated at the same rate as that in the mock-treated sample. We inferred that SKU5 was glycosylated with N-linked glycans that could be cleaved by PNGase F but not by Endo H. This pattern of differential glycanase susceptibility is symptomatic of proteins that pass through the Golgi, where the N-linked glycans become modified by a mannosidase, which makes them resistant to Endo H cleavage (Kornfeld and Kornfeld, 1985).
Another line of evidence showing that SKU5 becomes modified post-translationally was obtained by in vitro translation of SKU5 in a reticulocyte lysate. This produced a protein that migrated on a SDS-PAGE gel at the predicted size of 66 kD (data not shown). When canine microsomal membranes capable of glycosylation were added to the system, the apparent mass of SKU5 increased to 90 kD (data not shown).
SKU5 Is Anchored by GPI
Except for hydrophobic N and C termini, SKU5 was hydrophilic (data not shown) and possessed a 20–amino acid region just upstream of the hydrophobic C terminus containing features consistent with GPI modification (Eisenhaber et al., 2000). GPI modification of proteins occurs in the endoplasmic reticulum, where the C-terminal hydrophobic signal sequence is cleaved proteolytically and replaced by a GPI moiety. The GPI-modified protein then is transported through the trans-Golgi network to the plasma membrane, where the lipid portion of the GPI moiety becomes inserted in the outer leaf of the plasma membrane bilayer, anchoring the protein in an extracellular orientation (Ramalingam et al., 1996; Yan et al., 1998).
Three computer programs predicted that the SKU5 protein has the essential characteristics to become GPI modified: PSORT (http://psort.nibb.ac.jp/form.html; Nakai and Horton, 1999); big-PI Predictor (http://mendel.imp.univie.ac.at/gpi/gpi_server.html; Eisenhaber et al., 1998); and DGPI (http://129.194.186.123/GPI-anchor/index_en.html; D. Buloz and J. Kronegg, unpublished data).
We tested the possibility that SKU5 was anchored by GPI by determining whether the protein was released from membrane fractions by phosphatidylinositol-specific phospholipase C (PI-PLC). PI-PLC cleaves the lipid component of the GPI anchor, solubilizing the protein. Microsomal membranes were fractionated by Triton X-114 phase partitioning to remove proteins that were associated peripherally with the membranes and then incubated with PI-PLC. The samples then were reextracted with Triton X-114, which resulted in the partitioning of the GPI anchor–released proteins into the aqueous phase. SKU5 was present in the PI-PLC–treated aqueous phase sample but not in the mock-treated control, confirming that SKU5 had been GPI anchored to membranes (Figure 6B).
SKU5 Localized to the Plasma Membrane and the Cell Wall
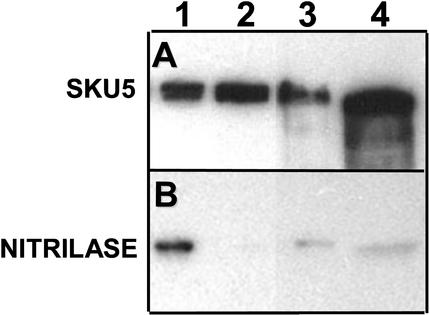
To identify the subcellular location of SKU5, we separated membrane-associated and soluble proteins by centrifugation. We also separately isolated cell wall fluid by first vacuum infiltrating seedlings with a cold buffer and then removing the fluids by centrifugation at low speeds. These fractions were electrophoresed on SDS-PAGE gels, and protein gel blots were probed with the anti-SKU5 antibody (Figure 7A).
Figure 7.
SKU5 Localizes to Microsomal Membranes, Supernatant, and Cell Wall Fluid Fractions.
Protein gel blot probed first with anti-SKU5 antibody (A) and then with an anti-nitrilase antibody (B) (cytoplasmic control; 38 kD). Lane 1, wild-type supernatant from the 100,000g spin; lane 2, wild-type pellet from the 100,000g spin; lane 3, wild-type cell wall fluid obtained from a low-speed spin of intact seedlings; lane 4, same as lane 3 except that the extraction buffer contained 1 M NaCl.
SKU5 was present in similar amounts in the 100,000g supernatant of wild-type extracts (Figure 7A, lane 1) and in the 100,000g pellet (lane 2). The amount of cytoplasmic contamination of the 100,000g pellet was low, as indicated by reprobing of the protein gel blot with an antibody against the cytoplasmic protein nitrilase (Figure 7B, lane 2). SKU5 also was detected in intracellular wash fluids extracted in a buffer with or without 1 M NaCl (Figure 7B, lanes 3 and 4). Thus, approximately half of the SKU5 protein is located in microsomal membranes, and the remainder is soluble and appears to be located in the cell wall.
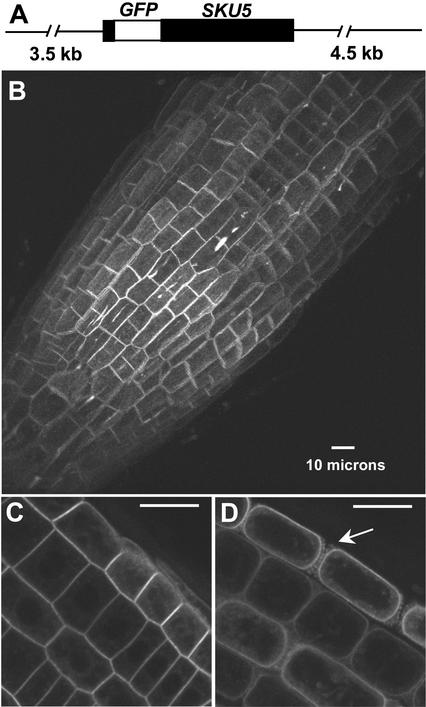
We also generated transgenic plants expressing a green fluorescent protein (GFP)::SKU5 fusion protein by placing the GFP gene in frame within the SKU5 gene of fragment B in a position just after the N-terminal leader peptide–encoding sequence (Figure 8A). This construct was transformed into sku5 seedlings, in which it rescued the sku5 mutant phenotype (data not shown). Confocal microscopic analysis identified GFP fluorescence throughout the seedlings, with the brightest fluorescence occurring within the root meristem and the distal elongation zone (Figure 8B). This fluorescence could be seen completely surrounding cells of all tissue types, yet it appeared brighter along some parts of cells compared with others. Bright streaks also were seen along the longitudinal axis of the root, localized to three-cell junctions (Figure 8B).
Figure 8.
Fluorescence from SKU5::GFP Fusion Protein Localizes Predominantly to the Plasma Membrane and the Cell Wall of All Cells throughout the Root.
(A) The SKU5::GFP construct, which is fragment B with the GFP gene (open box) inserted in frame downstream of the SKU5 signal peptide sequence.
(B) to (D) GFP fluorescence in the root tip (B) (pointing toward the lower left; three-dimensional reconstruction) and in root tip cells (cross-section view) before (C) and after (D) treatment with 100 mM mannitol. Arrow in (D) denotes apparent Hechtian strands. Bars = 10 μm.
These distinctive GFP-related fluorescence features did not occur with any consistent pattern. Faint GFP fluorescence also was visible within the cells; the illuminated structures appeared similar to those seen with endoplasmic reticulum– and Golgi-localized proteins (Figure 8C). With the filter set and gain settings used for these experiments, there was no detectable autofluorescence from nontransgenic plants (data not shown).
To determine if any of the peripherally localized GFP fluorescence was associated with the plasma membrane, we treated roots with 100 mM mannitol to plasmolyze the cells. Confocal images of these roots showed that a considerable amount of fluorescence localized to the plasma membranes as they pulled away from the cell wall during plasmolysis (Figure 8D). Fluorescence also was associated with strands emanating from these membranes, which appeared to be Hechtian strands.
Protein gel blot analysis of extracts from seedlings expressing the SKU5::GFP fusion protein, using either anti-SKU5 or anti-GFP antibody, identified a band of the predicted size of the fusion protein and bands of the sizes of SKU5 (detected by anti-SKU5 antibody) and the GFP protein (detected by anti-GFP antibody) (data not shown). Therefore, it appears that some of the fusion protein had been cleaved proteolytically, and consequently, some of the GFP-related fluorescence possibly did not reflect SKU5 localization accurately (data not shown).
To clarify this issue, we isolated cellular fractions from SKU5::GFP-expressing transgenic seedlings and performed protein gel blot analysis. Our results showed that the anti-SKU5 antibody clearly detected the SKU5::GFP fusion protein in the microsomal membrane fraction from these preparations, whereas the anti-GFP antibody detected very little free GFP (data not shown). This finding indicates that the GFP-related fluorescence observed at the plasma membrane emanated predominantly (and perhaps exclusively) from the SKU5::GFP fusion protein.
However, the anti-GFP antibody detected a large amount of free GFP in the cell wall fluid and soluble protein fractions, whereas the anti-SKU5 antibody detected relatively little SKU5::GFP fusion protein (data not shown). This finding suggests that the bright fluorescent streaks seen within the cell wall at the three-cell junctions probably came mostly from free GFP; hence, SKU5 may not localize normally to these spots at such high concentrations. We do not know why the three-cell junction fluorescence occurred only in distinct locations instead of uniformly.
DISCUSSION
Plants regulate root growth by controlling cell division and cellular expansion in the meristematic region and the elongation zones of the root tip. In doing so, they are able to respond to the numerous stimuli within their environment, avoiding those that are harmful and growing toward those that increase their likelihood of survival. Little is known about how plant roots sense multiple environmental stimuli and integrate the ensuing physiological signals into an optimal growth response.
To dissect one aspect of root growth, we characterized a T-DNA–tagged Arabidopsis mutant named sku5 with roots that responded abnormally to directional stimuli. The sku5 mutation was caused by a T-DNA insertion within the middle of the SKU5 coding sequence. RNA and protein gel blot analyses revealed that no mRNA or SKU5 protein was detectable in sku5 plants, so the mutation appears to be null. Phenotypic and molecular analyses of this mutant have provided novel insights into a previously uncharacterized process that appears to be required for normal root growth.
When sku5 roots were grown in a homogeneous agar medium, they aligned normally with respect to the gravity vector. However, when grown on a vertical or tilted agar surface, sku5 roots consistently skewed to the left, away from the gravity vector at a characteristic angle. sku5 roots also skewed to the left when they were grown at the bottom of an agar plate against the plastic. This environment provided a differential touch stimulus but did not have the same directional moisture and ion gradients found at the surface. Thus, we infer that the surface-induced growth response is derived from a touch stimulus. sku5 roots grown on agar in the absence of a single gravity vector formed clockwise coils.
Therefore, it is likely that the growth angle of sku5 roots on the surface of inclined agar is maintained by an equilibrium between gravity-induced and surface-induced growth responses, not unlike that seen with the sku1 and sku2 mutants (Rutherford and Masson, 1996). sku5 roots were able to change their direction of growth as well as their direction of axial rotation to form nearly normal-looking sinusoidal waves but had a tendency to loop instead of wave as they grew, which was associated with prolonged counterclockwise root tip axial rotation. Thus, another factor appears to be involved in mediating the back-and-forth oscillations in directional growth.
The sku5 root tip axial rotation defect appeared constitutive, occurring while the roots were growing on an agar surface as well as in agar or liquid medium. This is in contrast to the directional growth phenotype, which appeared only when roots grew on an agar surface. The average rate of axial rotation, however, was slightly greater when the roots were grown on the agar surface, suggesting that a touch stimulus enhances the twisting. Most if not all mutants reported to have surface-dependent directional growth defects produce roots with an axial rotation defect.
In fact, mutants with roots that skew to the left exhibit counterclockwise axial rotations, whereas roots that skew to the right exhibit clockwise axial rotations (Mirza, 1987; Simmons et al., 1995; Rutherford and Masson, 1996; Marinelli et al., 1997; Rutherford et al., 1998; Furutani et al., 2000; J.C. Sedbrook and C.R. Somerville, unpublished data). Therefore, the direction of the axial rotation bias coincides with the direction of surface growth. Even though the growth process underlying root axial rotation is not sufficient to cause root skewing, these two processes probably are linked by a common physical process or signaling mechanism.
sku5 roots were ∼15% shorter than wild-type roots when grown in or on agar. Etiolated sku5 hypocotyls also were shorter than normal and twisted more in a counterclockwise direction about their axes than those of the wild type. The axial rotation defect was not observed in light-grown hypocotyls, petioles, or inflorescences, yet it occurred in dark-grown roots. Dark-grown petioles and inflorescences were not analyzed. Therefore, either the twisting phenotype is too subtle to measure in light-grown hypocotyl cells, which expand much less than in the dark, or the SKU5-related growth process is light regulated in hypocotyls but not in roots.
Only a few percent of the reduced root and hypocotyl length was attributable to the fact that roots composed of twisted cells are shorter than straight roots with the same number of cells. sku5 and wild-type root cortical cells are the same size. Therefore, it appears that sku5 roots were mostly shorter because of slower rates of cell expansion and cell division. Apparently, both expansion and division rates decreased proportionally, because we observed no differences in the sizes of sku5 meristems and elongation zones compared with those of the wild type (data not shown). It is unclear if the sku5 mutation directly affects cell division rates or if cell division is linked to a cell expansion defect. A sku5 effect on the rate of cell expansion seems likely because of the axial rotation defect, which is manifested in the elongation zones.
If cortical cells grew proportionally less than the overlying epidermal cells, then the epidermal cell files would have to twist to release the tension. Indeed, we observed that sku5 epidermal cell files twisted more than the underlying cortical cell files. This greater amount of epidermal cell file twisting also was observed in wild-type roots, suggesting that it was a naturally occurring phenomenon. Because the twisting almost always occurred in the same direction in sku5 roots, there must be an underlying directional growth bias. Furutani et al. (2000) postulated that the direction of root axial rotation was dictated by the orientation of cortical microtubules.
The SKU5 protein exhibits sequence similarity with the multiple-copper oxidases ascorbate oxidase and laccase. Ascorbate oxidases are cell wall–localized glycoproteins that catalyze the oxidation of ascorbic acid. This reaction appears to affect cell expansion, but the mechanism has not been determined (DeTullio et al., 1999; Kato and Esaka, 2000; Smirnoff, 2000). Ascorbate oxidase activity also may play a role in regulating cell division (Kerk and Feldman, 1995; Kerk et al., 2000; Potters et al., 2000). Laccases are cell wall–localized enzymes that have been shown to oxidize a number of aromatic (normally phenolic) and inorganic substances, in some cases producing radicals that polymerize spontaneously (Messerschmidt and Huber, 1990).
There has been much debate regarding whether laccases are involved in lignin polymerization (Ranocha et al., 1999). Even though SKU5 is related structurally to these two families of proteins, it lacks most of the highly conserved His residues necessary to coordinate type 1 and type 3 copper ions. Therefore, SKU5 likely has a different enzymatic function. The two His residues present in ascorbate oxidases and laccases that coordinate the type 2 copper ion are present in SKU5. A number of proteins exist that coordinate a single type 2 copper ion (McGuirl and Dooley, 1999). Because these proteins lack the other copper ions necessary to perform electron transfer during substrate oxidation, they use a topaquinone redox cofactor (Whittaker, 1999). This cofactor forms autocatalytically by the oxidation of a Tyr residue to form the second redox active center. Thus, our working hypothesis is that SKU5 is a monocopper oxidase of unknown specificity.
The Arabidopsis genome contains 18 other genes, provisionally designated SKS1 to SKS18, that are predicted to encode proteins with 44 to 65% amino acid sequence identity with SKU5. The predicted SKS proteins lack intact types 1, 2, and 3 copper centers. Therefore, these proteins probably do not possess oxidase activity unless they have evolved to bind copper in sites distinct from those seen in ascorbate oxidase and laccase. All of the predicted SKS proteins appear to have N-terminal leader sequences, like SKU5, while lacking any endoplasmic reticulum retention signals. This fact suggests that, like SKU5, they are secreted.
Orthologs of a few SKS proteins have been described previously as pollen-specific genes, including NTP303 from tobacco (Albani et al., 1992) and BP10 from Brassica napus (Weterings et al., 1992). The functions of these proteins are not known. NTP303, which shares 45% amino acid sequence identity with SKU5, was found to localize to the vegetative plasma membranes of pollen and to the cell walls and callose plugs of pollen tubes (Wittink et al., 2000). Tucker and Zhang (1996) reported that a tomato protein (PE3) that shares 50% amino acid sequence identity with SKU5 possessed pectin esterase activity, but subsequently, they were unable to confirm these results (G. Tucker and J. Zhang, personal communication). We also have been unable to identify esterase activity associated with heterologously expressed SKU5 using a variety of activity assays and esterase substrates (data not shown).
Several lines of evidence indicate that SKU5 is GPI modified, N-glycosylated, and anchored to the plasma membrane. (1) An N-terminal peptide sequence in the Arabidopsis Plant Plasma Membrane Database corresponds to the site at which the SKU5 leader peptide is predicted to be cleaved. (2) PNGase F treatment showed that SKU5 is glycosylated. For glycosylation to occur, SKU5 must have been targeted to the endoplasmic reticulum. (3) Endo H treatment was unable to remove N-linked glycans, so the protein likely passed through the Golgi, where mannosidase made the glycans resistant to Endo H cleavage.
(4) Detection of SKU5 in crude membrane preparations showed that it was membrane bound. (5) Plasma membrane localization was confirmed by the analysis of GFP-related fluorescence in seedlings expressing a SKU5::GFP fusion protein. (6) PI-PLC treatment released SKU5 from membranes, indicating that SKU5 had been anchored by GPI. (7) SKU5 was recovered in fluids extracted from the cell wall, suggesting that it was released from the GPI anchor in vivo, perhaps in a regulated manner.
The GPI modification of proteins has been found to occur in a variety of organisms, including animals, plants, fungi, and protozoa (McConville and Ferguson, 1993; Kinoshita et al., 1997; Takos et al., 1997). In Arabidopsis, as many as 200 proteins with a wide variety of functions are predicted to be GPI modified (Sherrier et al., 1999; Borner et al., 2002). The primary role of the GPI anchor is to attach proteins to the outer leaflet of the plasma membrane. These anchored proteins can be organized into microdomains, perhaps forming functional complexes (Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998).
Some GPI-anchored proteins become localized within the lumen of intracellular vesicles until they are secreted in a regulated manner (Lisanti et al., 1990). Additionally, the GPI anchor can act as an intracellular sorting signal, directing the attached protein to polar locations (Hooper, 1997; Schindelman et al., 2001). Work is in progress to determine what role the GPI modification of SKU5 plays in organ growth.
The analysis of transgenic seedlings expressing the SKU5::GFP fusion protein under the control of the SKU5 promoter showed that GFP-related fluorescence occurred all around cells in all cell layers of the root. When root cells were plasmolyzed with mannitol, some of the fluorescence localized to the plasma membrane. Although protein gel blot analysis of whole plant extracts showed that some of the SKU5::GFP fusion protein was cleaved to form free GFP and SKU5, protein gel blot analysis of microsomal membranes purified from the transgenic seedlings identified predominantly intact SKU5::GFP, with very little free GFP. Hence, it seems likely that the plasma membrane fluorescence originated from the fusion protein. The fluorescence appeared fairly uniform around the root cells, with only some random brighter patches of fluorescence occurring along cells in parts of the tip. Because these patches exhibited no obvious patterns, we conclude that the localization of SKU5 is not polarized.
We envision a number of different models to explain SKU5 function. For instance, SKU5 might modify linkages between cell wall structural components, allowing for turgor-driven cell extension to occur in such a way to provide a directionality to cell expansion without affecting the cell's ability to achieve normal size. Because sku5 mutant roots twist, there must be an underlying growth imbalance that SKU5 activity normally counteracts. SKU5 activity also must counteract the surface-induced growth response for roots to grow in a normal downward direction. Alternatively, SKU5 may affect a property of the middle lamella that would allow cells to slide past each other as they grow.
Because epidermal cells normally grow to be longer than the underlying cortical cells, a torsional strain will develop if the primary cell walls of these two cell types do not grow smoothly past each other. The strain could be released by the kind of twisting of the organ that is observed in the sku5 mutant. It also is conceivable that instead of modifying cell wall structural properties directly, SKU5 might act on a component in the cell wall that is in a signaling pathway, affecting directed cell expansion.
Additional studies will be required to discern exactly what role SKU5 plays in controlling growth. Perhaps detailed structural studies will reveal an alteration in a component of the sku5 cell wall. This would facilitate the identification of potential substrates and enzymatic activity of the SKU5 protein. Along those lines, Fourier transform infrared spectroscopic analysis of plant tissues as well as quantitative analysis of neutral sugars derived from cell walls revealed no differences between sku5 and the wild type (data not shown).
To examine the possibility of a regulatory function, comparison of gene expression in the mutant and the wild type using DNA microarrays may be useful. It also is possible that analysis of mutations in some of the SKS genes may result in a phenotype that is more suggestive of enzymatic function than is the sku5 mutation. The large collections of insertion mutants available in Arabidopsis should greatly facilitate the implementation of this approach.
METHODS
Plant Stocks and Manipulation
Wild-type Arabidopsis thaliana seeds of the ecotype Wassilewskija were provided by Tim Caspar (DuPont, Wilmington, DE). The sku5 mutant, ecotype Wassilewskija, was isolated from the Versailles collection of T-DNA insertion mutants (pGKB5 binary vector) (Bechtold et al., 1993; Bouchez et al., 1993). All techniques and conditions used to grow and manipulate Arabidopsis seeds, seedlings, and plants were as described previously (Rutherford and Masson, 1996).
Analysis and Quantification of Organ Growth
The root-waving assay and quantification of root slanting were performed as described in Rutherford and Masson (1996), except that an Olympus D-600L digital camera (Tokyo, Japan) was used for photography. Root length measurements were performed using NIH Image software (version 1.62b7) to analyze digital camera images. Close-up images were obtained with a Leica Wild M8 microscope (Wetzlar, Germany) attached to a Diagnostic Instruments SPOT digital camera (Sterling Heights, MI). The root gravitropism assay was performed and analyzed as described in Sedbrook et al. (1999).
Confocal imaging of organs was performed with a Nikon Diaphot 200 inverted microscope equipped with a Nikon ×40 1.15 or a Nikon ×60 1.2 numerical aperture water immersion objective (Tokyo, Japan) and a Bio-Rad MRC 1024 confocal head with a krypton-argon laser. Enhanced green fluorescent protein (EGFP) was excited at 488 nm, and emitted fluorescence was collected through a 525/30 bandpass filter. Three-dimensional reconstructions of image stacks were generated with Lasersharp software (Bio-Rad) or NIH Image software. For confocal imaging, seedlings were immersed in half-strength Murashige and Skoog (1962) medium, 1.5% Suc, and 10 μg/mL propidium iodide (Sigma) between two cover slips.
Exogenous propyzamide (Chem Service, Inc.; www.chemservice.com/) treatments were performed as follows. Seeds were sown onto autoclaved 3MM paper strips that were positioned on 0.8% agar (type E; Sigma)-solidified germination medium (half-strength Murashige and Skoog [1962] salts and 1.5% Suc), incubated at 4°C in the dark for 3 days, and then grown on vertically oriented plates for 4 days in a growth chamber (22°C with 16 h of light). The strips containing the seedlings were transferred onto 1.5% agar-solidified germination medium and allowed to grow tilted 30° from the vertical for another 5 days, after which the plates were imaged and analyzed as described above.
Cloning and Analysis of the SKU5 Gene and cDNAs
Procedures used to manipulate plant and bacterial DNA were as described by Sedbrook et al. (1996)(1999). PCR amplification of a genomic DNA fragment flanking the T-DNA inserted in the sku5 gene was performed as described in Siebert et al. (1995), with the sku5 genomic DNA being digested with EcoRV before adaptor ligation. Texas A&M University BAC filters and clones, as well as the SKU5 cDNA clone 164E21T7, were obtained from the ABRC (Columbus, OH). SKU5 cDNA clones were isolated from a cDNA library in a λ vector (Kieber et al., 1993), and the DNA was sequenced and analyzed as described in Sedbrook et al. (1999). Complementation of the sku5 mutation also was performed as described previously (Sedbrook et al., 1999), except the pBIB binary vector was used (Becker, 1990) and no vacuum was used for Agrobacterium tumefaciens transformation.
Generation and Analysis of Transgenic Plants Carrying a SKU5::Green Fluorescent Protein Reporter Construct
To make the SKU5::green fluorescent protein (GFP) fusion, the EGFP gene was amplified by PCR from the pEGAD vector (Cutler et al., 2000) without a stop codon and placed in frame within PCR-engineered sites in the SKU5 gene just downstream of the nucleotide sequences predicted to encode the SKU5 leader peptide, within the 10.7-kb fragment B. Confocal imaging of T2 and T3 generation seedlings was performed as described above.
Generation of Anti-SKU5 Antibodies and Testing for SKU5 Glycosylation
Anti-SKU5 antibodies were generated by Quality Controlled Biochemicals (Hopkinton, MA) by injecting the synthetic peptide CSKGKAKGQLPPGPQDEF conjugated to limpit hemocyanin into two rabbits and then affinity purifying the serum after two boosts. Various wild-type and sku5 tissues were ground in urea extraction buffer (9.5 M urea, 100 mM Tris, pH 6.8, 2% sodium lauryl sulfate, and 2% β-mercaptoethanol), electrophoresed on SDS-PAGE gels, transferred onto an Immobilon-P polyvinylidene difluoride membrane (Millipore, Bedford, MA), and analyzed using standard procedures.
The anti-SKU5 antibody was used at a dilution of 1:1000, followed by 1:7500 dilution of a goat anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody (Pierce). The Supersignal West Pico chemiluminescent substrate (Pierce) was used for horseradish peroxidase detection. To determine if SKU5 was glycosylated, 1-week-old seedlings were ground in the denaturing buffer supplied with the peptide N-glycosidase F and endoglycosidase H enzymes (New England Biolabs, Beverly, MA) and treated according to the manufacturer's instructions. The anti–protein disulfide isomerase antibody was purchased from Rose Biotechnology (www.rosebiotech.com/).
Fractionation of SKU5 and Glycosyl Phosphatidylinositol Anchor Determination
Microsomal membranes and membrane-free supernatant were isolated as follows. Seven-day-old wild-type seedlings were ground in ice-cold extraction buffer (50 mM potassium phosphate, pH 7.5, 2 mM MgCl2, 50 mM NaCl, and 1 × protease inhibitor cocktail [Calbiochem] with or without 400 mM Suc), and the insoluble debris were removed by centrifugation at 10,000g for 10 min. Microsomal membranes then were separated from the supernatant by centrifugation at 100,000g for 1 h.
Cell wall fluid was isolated by vacuum infiltrating 7-day-old intact seedlings for 15 min with ice-cold buffer (100 mM Tris-HCl, pH 7.3, 5 mM EDTA, and 0.05% β-mercaptoethanol with or without 1 M NaCl) and spinning at 500g for 15 min to collect the fluid (modified from Sieber et al., 2000). The protein was precipitated with 10% trichloroacetic acid and resuspended in an SDS-PAGE loading buffer (see below for details).
The glycosyl phosphatidylinositol (GPI) modification of SKU5 was demonstrated by its removal from membrane fractions with phosphatidylinositol-specific phospholipase C, as described in Sherrier et al. (1999), except that microsomal membranes were used. In summary, microsomal membranes that were resuspended in 100 mM potassium phosphate buffer, pH 7.4 were first partitioned three times in 2.4% precondensed Triton X-114 (Ausubel et al., 1994) to remove proteins peripherally associated with the membranes. This was done by incubating the Triton X-114/membrane mixture for 15 min on ice, removing insoluble material by centrifugation at 10,000g at 4°C for 10 min, heating the mixture at 37°C for 5 min, and then separating the phases by spinning at 1000g at room temperature for 10 min.
The lower Triton X-114/membrane–containing phase was diluted 10-fold with 100 mM potassium phosphate buffer, pH 7.4, and divided into two equal parts; then, 1 unit/100 μL of phosphatidylinositol-specific phospholipase C (Sigma) was added to one part while an equal volume of 100 mM potassium phosphate buffer was added to the other part as a control. These samples were incubated for 1.5 h at 30°C with periodic mixing and then centrifuged at 1000g for 10 min.
The aqueous upper phase was collected, 1 μg of BSA was added as an internal standard for protein recovery, and the samples were precipitated with 10% trichloroacetic acid (incubated on ice for 2 h and then centrifuged at full speed for 10 min in a microcentrifuge). The pellets were resuspended in SDS-PAGE loading buffer, the pH was neutralized with ammonium hydroxide fumes (an ammonium hydroxide–saturated Q-tip was held in the tube until the bromphenol blue turned from yellow to blue), and the pellets were electrophoresed on an SDS-PAGE gel and protein gel blotted as described above.
Accession Numbers
The accession numbers for the sequences mentioned in this article are R30492 and Z37702 (two Arabidopsis EST clones of sku5) and AF439406 (sku5 complete coding sequence).
Supplementary Material
Acknowledgments
We thank Herman Höfte for providing the Versailles collection of T-DNA insertion mutants, the ABRC for the Texas A&M University BAC filters and clones as well as a SKU5 cDNA clone, Sean Cutler for the anti-nitrilase antibody, Seth Davis for the anti-GFP antibody, David Ehrhardt for assistance with confocal microscopy and for fruitful discussions, Paul Dupree for technical advice on demonstrating GPI anchoring, John Christie for technical advice, and Todd Richmond, Gert de Boer, Sean Cutler, Dario Bonetta, and other members of the Carnegie Institution for technical advice and fruitful discussions. This work was supported in part by grants from the Department of Energy to C.R.S. (Grant DOE-FG02-00ER20133) and from the National Aeronautics and Space Administration (Grants NAG2-1189 and NAG2-1492), the National Science Foundation (Grant MCB-9905675), and Wisconsin Hatch funds (WIS04310) to P.H.M. J.C.S. was supported in part by a National Institutes of Health postdoctoral fellowship (Grant 1 F32 GM20181).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002360.
Footnotes
Online version contains Web-only data.
References
- Albani, D., Sardana, R., Robert, L.S., Altosaar, I., Arnison, P.G., and Fabijanski, S.F. (1992). A Brassica napus gene family which shows sequence similarity to ascorbate oxidase is expressed in developing pollen: Molecular characterization and analysis of promoter activity in transgenic tobacco plants. Plant J. 2, 331–342. [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III 316, 1194–1199. [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, P.N., and Scheres, B. (2000). Root development. Curr. Biol. 10, R813–R815. [DOI] [PubMed] [Google Scholar]
- Borner, G.H.H., Sherrier, D.J., Stevens, T.J., Arkin, I.T., and Dupree, P. (2002). Prediction of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis: A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Ser. III 316, 1188–1193. [Google Scholar]
- Buer, C.S., Masle, J., and Wasteneys, G.O. (2000). Growth conditions modulate root-wave phenotypes in Arabidopsis. Plant Cell Physiol. 41, 1164–1170. [DOI] [PubMed] [Google Scholar]
- Chen, R.J., Rosen, E., and Masson, P.H. (1999). Gravitropism in higher plants. Plant Physiol. 120, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTullio, M.C., Paciolla, C., Dalla-Veechia, F., Rascio, N., D'Emerico, S., DeGara, L., Liso, R., and Arrigoni, O. (1999). Changes in onion root development induced by the inhibition of peptidyl-prolyl hydroxylase and influence of the ascorbate system on cell division and elongation. Planta 209, 424–434. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organization of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Eisenhaber, B., Bork, P., and Eisenhaber, F. (1998). Sequence properties of GPI-anchored proteins near the omega-site: Constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Eisenhaber, B., Bork, P., Yuan, Y.P., Loffler, G., and Eisenhaber, F. (2000). Automated annotation of GPI anchor sites: Case study C. elegans. Trends Biochem. Sci. 25, 340–341. [DOI] [PubMed] [Google Scholar]
- Friedrichson, T., and Kurzchalia, T.V. (1998). Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394, 802–805. [DOI] [PubMed] [Google Scholar]
- Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., Thitamadee, S., Shikanai, T., and Hashimoto, T. (2000). The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Hooper, N.M. (1997). Glycosyl-phosphatidylinositol anchored membrane enzymes. Clin. Chim. Acta 266, 3–12. [DOI] [PubMed] [Google Scholar]
- Kato, N., and Esaka, M. (2000). Expansion of transgenic tobacco protoplasts expressing pumpkin ascorbate oxidase is more rapid than that of wild-type protoplasts. Planta 210, 1018–1022. [DOI] [PubMed] [Google Scholar]
- Kerk, N.M., and Feldman, L.J. (1995). A biochemical model for the initiation and maintenance of the quiescent center: Implications for organization of root meristems. Development 121, 2825–2833. [Google Scholar]
- Kerk, N.M., Jiang, K.N., and Feldman, L.J. (2000). Auxin metabolism in the root apical meristem. Plant Physiol. 122, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). Ctr1: A negative regulator of the ethylene response pathway in Arabidopsis encodes a member of the Raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Ohishi, K., and Takeda, J. (1997). GPI-anchor synthesis in mammalian cells: Genes, their products, and a deficiency. J. Biochem. 122, 251–257. [DOI] [PubMed] [Google Scholar]
- Kornfeld, R., and Kornfeld, S. (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664. [DOI] [PubMed] [Google Scholar]
- Lisanti, M.P., Rodriguez-Boulan, E., and Saltiel, A.R. (1990). Emerging functional roles for the glycosyl phosphatidylinositol membrane protein anchor. J. Membr. Biol. 117, 1–10. [DOI] [PubMed] [Google Scholar]
- Marinelli, B., Gomarasca, S., and Soave, C. (1997). A pleiotropic Arabidopsis thaliana mutant with inverted root chirality. Planta 202, 196–205. [DOI] [PubMed] [Google Scholar]
- McConville, M.J., and Ferguson, M.A.J. (1993). The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 294, 305.–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuirl, M.A., and Dooley, D.M. (1999). Copper-containing oxidases. Curr. Opin. Chem. Biol. 3, 138–144. [DOI] [PubMed] [Google Scholar]
- Messerschmidt, A., and Huber, R. (1990). The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin: Modeling and structural relationships. Eur. J. Biochem. 187, 341–352. [DOI] [PubMed] [Google Scholar]
- Mirza, J.I. (1987). The effects of light and gravity on the horizontal curvature of roots of gravitropic and agravitropic Arabidopsis thaliana L. Plant Physiol. 83, 118–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza, J.I. (1988). The clinostat effect on the direction of root growth of wild type and aux-1 seedlings of Arabidopsis thaliana. Arabidopsis Inf. Serv. 26, 35–38. [Google Scholar]
- Mullen, J.L., Ishikawa, H., and Evans, M.L. (1998. a). Analysis of changes in relative elemental growth rate patterns in the elongation zone of Arabidopsis roots upon gravistimulation. Planta 206, 598–603. [DOI] [PubMed] [Google Scholar]
- Mullen, J.L., Turk, E., Johnson, K., Wolverton, C., Ishikawa, H., Simmons, C., Soll, D., and Evans, M.L. (1998. b). Root-growth behavior of the Arabidopsis mutant rgr1: Roles of gravitropism and circumnutation in the waving/coiling phenomenon. Plant Physiol. 118, 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–35. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Okada, K., and Shimura, Y. (1990). Reversible root-tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250, 274–276. [DOI] [PubMed] [Google Scholar]
- Potters, G., Horemans, N., Caubergs, R.J., and Asard, H. (2000). Ascorbate and dehydroascorbate influence cell cycle progression in a tobacco cell suspension. Plant Physiol. 124, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam, S., Maxwell, S.E., Medof, M.E., Chen, R., Gerber, L.D., and Udenfriend, S. (1996). COOH-terminal processing of nascent polypeptides by the glycosylphosphatidylinositol transamidase in the presence of hydrazine is governed by the same parameters as glycosylphosphatidylinositol addition. Proc. Natl. Acad. Sci. USA 93, 7528–7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha, P., McDougall, G., Hawkins, S., Sterjiades, R., Borderies, G., Stewart, D., Cabanes-Macheteau, M., Boudet, A.M., and Goffner, D. (1999). Biochemical characterization, molecular cloning and expression of laccases—a divergent gene family—in poplar. Eur. J. Biochem. 259, 485–495. [DOI] [PubMed] [Google Scholar]
- Rutherford, R., Gallois, P., and Masson, P.H. (1998). Mutations in Arabidopsis thaliana genes involved in the tryptophan biosynthesis pathway affect root waving on tilted agar surfaces. Plant J. 16, 145–154. [DOI] [PubMed] [Google Scholar]
- Rutherford, R., and Masson, P.H. (1996). Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiol. 111, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., Chen, R.J., and Masson, P.H. (1999). ARG1 (Altered Response to Gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc. Natl. Acad. Sci. USA 96, 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., Kronebusch, P.J., Borisy, C.G., Trewavas, A.J., and Masson, P.H. (1996). Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiol. 111, 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Metraux, J.P., and Nawrath, C. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12, 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, C., Migliaccio, F., Masson, P., Caspar, T., and Soll, D. (1995). A novel root gravitropism mutant of Arabidopsis thaliana exhibiting altered auxin physiology. Physiol. Plant. 93, 790–798. [PubMed] [Google Scholar]
- Smirnoff, N. (2000). Ascorbic acid: Metabolism and functions of a multi-faceted molecule. Curr. Opin. Plant Biol. 3, 229–235. [PubMed] [Google Scholar]
- Takos, A.M., Dry, I.B., and Soole, K.L. (1997). Detection of glycosyl-phosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett. 405, 1–4. [DOI] [PubMed] [Google Scholar]
- Tucker, G., and Zhang, J. (1996). Expression of polygalacturonase and pectinesterase in normal and transgenic tomatoes. In Pectins and Pectinases, Progress in Biotechnology, J. Visser and A.G.J. Voragen, eds (Amsterdam: Elsevier Science).
- Varma, R., and Mayor, S. (1998). GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801. [DOI] [PubMed] [Google Scholar]
- Weterings, K., Wim, R.J., Vanaarssen, R., Kortstee, A., Spijkers, J., Vanherpen, M., Schrauwen, J., and Wullems, G. (1992). Characterization of a pollen-specific cDNA clone from Nicotiana tabacum expressed during microgametogenesis and germination. Plant Mol. Biol. 18, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Whittaker, J.W. (1999). Oxygen reactions of the copper oxidases. Essays Biochem. 34, 155–172. [DOI] [PubMed] [Google Scholar]
- Wittink, F.R.A., Knuiman, B., Derksen, J., Capkova, V., Twell, D., Schrauwen, J.A.M., and Wullems, G.J. (2000). The pollen-specific gene Ntp303 encodes a 69-kDa glycoprotein associated with the vegetative membranes and the cell wall. Sex. Plant Reprod. 12, 276–284. [Google Scholar]
- Yan, W., Shen, F., Dillon, B., and Ratnam, M. (1998). The hydrophobic domains in the carboxyl-terminal signal for GPI modification and in the amino-terminal leader peptide have similar structural requirements. J. Mol. Biol. 275, 25–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.