Abstract
p63 is a recently identified homolog of p53 that is found in the basal layer of several stratified epithelial tissues such as the epidermis, oral mucosa, prostate, and urogenital tract. Studies with p63−/− mice and analysis of several human autosomal-dominant disorders with germ line p63 mutations suggest p63 involvement in maintaining epidermal stem cell populations. The p63 gene encodes six splice variants with reported transactivating or dominant-negative activities. The goals of the current study were to determine the splice variants that are expressed in primary human epidermal keratinocytes (HEKs) and the biochemical activity p63 has in these epithelial cell populations. We found that the predominant splice variant expressed in HEKs was ΔNp63α, and it was present as a phosphorylated protein. During HEK differentiation, ΔNp63α and p53 levels decreased, while expression of p53 target genes p21 and 14-3-3σ increased. ΔNp63α had transcriptional repressor activity in vitro, and this activity was reduced in ΔNp63α proteins containing point mutations, corresponding to those found in patients with Hay-Wells syndrome. Further, we show that ΔNp63α and p53 can bind the p21 and 14-3-3σ promoters in vitro and in vivo, with decreased binding of p63 to these promoters during HEK differentiation. These data suggest that ΔNp63α acts as a transcriptional repressor at select growth regulatory gene promoters in HEKs, and this repression likely plays an important role in the proliferative capacity of basal keratinocytes.
Recently, p63 and p73, two p53 homologues, were identified (2, 27, 39, 54, 57). These proteins exhibit a high sequence and structural homology to the p53 protein. Each gene encodes an amino-transactivating domain, a core DNA-binding domain, and a carboxy-oligomerization domain. However, there are significant differences between these homologues and p53. Both p63 and p73 genes contain two transcriptional start sites that are used to generate transcripts that encode proteins with or without an amino-transactivating domain. Proteins with the transactivating domain are termed TAp63 or TAp73, and proteins lacking the transactivation domain are termed ΔNp63 or ΔNp73. In addition, both genes can be alternatively spliced to generate proteins with different carboxy termini. For example, six splice variants can be generated from the two promoters of the p63 gene with three different C termini termed α, β, and γ (7, 57). The p63α and p73α proteins also contain an additional region not found in p53 known as a sterile alpha motif (SAM) domain. This domain is found in the α form of p63 and p73 (27, 57) and is a protein-protein interaction domain implicated in developmental processes (46, 53).
In addition to the structural differences within the p53 gene family, differing functional properties were discovered. These differences became apparent after analysis of p63−/− and p73−/− mice. Whereas p53−/− mice are developmentally normal but prone to neoplastic disease (14), the p63−/− and p73−/− mice have severe developmental abnormalities. The p63−/− mice are born but die shortly after birth and are deficient in the development of limbs and several epithelial tissues such as skin, prostate, mammary gland, and urothelia (36, 58). The p73−/− mice exhibit neurological, pheromonal, and inflammatory defects (59).
The p63 protein is localized to the nucleus of basal cells of stratified epithelia such as skin, oral mucosa, cervix, vaginal epithelium, urothelium, prostate, breast, and other tissues (12, 13, 57). The ΔNp63α splice variant is the predominant, if not the only, form expressed in these basal epithelial cells (13, 41, 57). Ectopic expression of the ΔN splice variants can decrease p53 target gene promoter activity, suggesting a role for ΔNp63α in maintaining the proliferative capacity of cells by repressing p53 target genes involved in growth arrest (27, 57). This hypothesis is supported by the data of Parsa et al. and Pellegrini et al. showing that a decrease in p63 was associated with a reduced proliferative potential and subsequent terminal differentiation of skin keratinocytes (41, 42).
We analyzed here the role of p63 in primary human epidermal keratinocyte (HEK) differentiation. Our results indicate that ΔNp63α is the predominant form of p63 protein expressed in primary cultures of HEKs and is downregulated during differentiation. We also show that ΔNp63α is a phosphoprotein that can function as a transcriptional repressor and bind consensus p53-binding sites in the p21waf1 (p21) and 14-3-3σ promoters in vivo.
MATERIALS AND METHODS
Cell culture and treatment.
Second-passage primary HEKs were obtained from the Vanderbilt Skin Disease Research Core. HEKs were isolated as previously described (17) and were cultured in EpiLife M-EPI-500 keratinocyte growth medium (Cascade Biologics, Portland, Oreg.) supplemented with human keratinocyte growth supplement S-001-5 (Cascade Biologics) and 0.06 mM CaCl2. The human colorectal carcinoma cell lines HCT116 and RKO were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. The human embryonic kidney cell line 293 was kindly provided by S. Hiebert (Vanderbilt University Department of Biochemistry, Nashville, Tenn.) and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. All cells were cultured at 37°C with 5% CO2.
p63 cloning.
ΔNp63 splice variants, α, β, and γ were cloned from primary human oral mucosa. Tissue was obtained from patient specimens collected at Vanderbilt University School of Medicine. The Access RT-PCR system (Promega, Madison, Wis.) was used with mRNA isolated from the primary human oral keratinocytes. Primers used to generate the splice variants were as follows: ΔNp63 N-terminal 5′-CCCAAGCTTAATACGACTCACTATAGGGAGACCATGGAACAAAAACTCATCTCAGAAGAGGATCTGATGTTGTACCTGGAAAACAATG-3′, ΔNp63α C-terminal 5′-CGGGATCCTCACTCCCCCTCCTCTTTG-3′, ΔNp63β C-terminal 5′-CGGGATCCTCAGACTTGCCAGATCCTG-3′, and ΔNp63γ C-terminal 5′-CGGGATCCCTATGGGTACACTGATCGG-3′. The N-terminal primer also encodes for an in frame myc-epitope tag. p63 splice variants were subsequently cloned into the pCEP4 vector (Invitrogen, Carlsbad, Calif.) for transient transfections.
HEK differentiation assays.
HEKs were grown to ∼90% confluency, washed twice with phosphate-buffered saline (PBS), and induced to differentiate by the addition of 1 mM CaCl2 in growth factor-deficient medium. Cells were harvested at days 0, 2, 4, 6, and 8 after the induction of differentiation. Culture medium was changed every 2 days. Cells were harvested as described for Western and Northern analyses.
Protein lysate preparation and Western analysis.
Dishes (100 mm) of primary HEKs were washed twice with ice-cold PBS, and harvested by scraping into 750 μl of kinase lysis buffer (KLB; 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Nonidet P-40, 0.1% Triton X-100, 4 mM EDTA, 1 mM dithiothreitol [DTT]) containing the phosphatase inhibitors 50 mM NaF, 0.2 mM sodium vanadate, 10 mM p-nitrophenyl phosphate, and 10 mM β-glycerophosphate and the protease inhibitors antipain (10 μg/ml), leupeptin (10 μg/ml), pepstatin A (10 μg/ml), chymostatin (10 μg/ml; Sigma), and 4-(2-aminoethyl)-benzenesulfonylfluoride (200 μg/ml; Calbiochem, San Diego, Calif.). Cells were incubated on ice 30 to 45 min, and the protein supernatant was clarified by centrifugation at 13,000 × g for 10 min at 4°C. Protein concentration was determined by the Bio-Rad protein quantification kit (Bio-Rad Laboratories, Hercules, Calif.). Western analysis was performed as previously described (17) with the following primary antibodies: α-p63 monoclonal antibody Ab-1 (Oncogene Research Products, Calbiochem), α-p53 monoclonal antibody Ab-2 (Oncogene Research Products, Calbiochem), α-p21Waf1 antibody Ab-1 (Oncogene Research Products, Calbiochem), α-14-3-3σ polyclonal antibody N-14 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), α-β-actin polyclonal antibody I-19 (Santa Cruz), and α-Gal4 monoclonal antibody DBD-RK5C1 (Santa Cruz). Uniformity of protein loading was assessed by β-actin analyses, as well as by fast green staining of the membranes.
Northern analysis.
Dishes (150 mm) of HEK cells were harvested for mRNA isolation as previously described (18). mRNA (1 to 3 μg) was lyophilized, resuspended in sample buffer (1× morpholinepropanesulfonic acid [MOPS]; 0.1 M MOPS [pH 7.0], 40 mM sodium acetate, 5 mM EDTA [pH 8.0], 50% formamide, 6.5% formaldehyde), and heated at 55°C for 15 min. A 10× loading buffer (50% glycerol, 1 mM EDTA, 0.25% bromophenol blue, 0.25% xylene cyanol, 0.3 mg of ethidium bromide per ml) was added to the sample at a 1× concentration, and mRNA was resolved by gel electrophoresis on a 1% agarose gel containing 2% formaldehyde and 1× MOPS. The gel was washed two times for 40 min each time in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer, and mRNA was transferred to a supported nitrocellulose membrane (Gibco-BRL). ΔNp63, p53, 14-3-3σ, p21, and cyclophilin cDNAs were labeled with [α-32P]dCTP by using Prime-It II (Stratagene, La Jolla, Calif.). After a 2-h prehybridization in Express Hyb (Clontech Laboratories, Inc., Palo Alto, Calif.), membranes were incubated 1 h with 106 cpm of labeled cDNA per ml in Express Hyb. Membranes were washed two times for 1 min each time at room temperature in 0.1× SSC-0.1% sodium dodecyl sulfate (SDS), followed by 1 h in 0.3 × SSC-0.1% SDS. Blots were subsequently exposed for autoradiography.
Phosphatase assay.
p63 was immunoprecipitated from HEK protein lysates (75 μg of control or 150 μg of differentiation day 4) by rocking at 4°C for 1 h in KLB with a p63 antibody (H-129; Santa Cruz) and 15-μl bed volume of protein A-Sepharose (PAS; Amersham Biosciences Corp., Piscataway, N.J.). Immunoprecipitates were washed once with KLB and twice with phosphatase buffer (50 mM Tris [pH 8.0], 10% glycerol). Samples were resuspended in 20 μl of phosphatase buffer with or without phosphatase inhibitors. A total of 40 U (2 μl of 20 U/μl) of calf intestinal alkaline phosphatase (CIAP; Roche, Indianapolis, Ind.) was added, and samples were incubated 37°C for 3 h. The phosphatase reaction was stopped by the addition of Laemmli SDS sample buffer. The control and phosphatase-treated lysates were analyzed by Western blotting.
Luciferase assays.
HCT116 cells were transiently transfected with the p21-luciferase reporter constructs and expression vectors encoding p53, ΔNp63α, or Gal4-ΔNp63α. p21-luciferase was kindly provided by B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, Md.) (15), and p21-lucΔRE1 was kindly provided by S. Hiebert. The p21-lucΔRE1/RE2 was generated by PCR amplification with the full-length p21-luciferase reporter as a template and the following primers: 5′-CGGGATCCGAGATTTCCAGACTCTGAGC-3′ and 5′-CAGCCGGTCCCGGAACC-3′. The PCR product was cloned into the BamHI-PstI-digested luciferase reporter vector. 293 cells were transiently transfected with a Gal4 DNA-binding domain-TK-luciferase reporter construct (kindly provided by S. Hiebert) previously described (47) and Gal4-p63 fusion constructs. Gal4-p63 fusion cDNAs were cloned into the pM2/pY2 vector at the SalI-HindIII sites. The following primers were used to generate the Gal4-p63 cDNAs: Gal4-ΔNp63 N-terminal 5′-ACGCGTCGACTTGTACCTGGAAAACAATG-3′, ΔNp63α C-terminal 5′-CCCAAGCTTTCACTCCCCCTCCTCTTTG-3′, ΔNp63β C-terminal 5′-CCCAAGCTTTCAGACTTGCCAGATCCTG-3′, ΔNp63γ C-terminal 5′-CCCAAGCTTCTATGGGTACACTGATCGG-3′, Gal4-SAM N-terminal 5′-ACGCGTCGACCCTCCGTATCCCACAGAT-3′, and Gal4-SAM C-terminal 5′-CCCAAGCTTTCAGAATTCGTGGAGCTGCCG-3′. Gal4-ΔNp63α SAM domain point mutants (35) were kindly provided by H. van Bokhoven (Department of Human Genetics, University Hospital, Nijmegen, The Netherlands) and served as templates to generate additional Gal4 fusion cDNAs with the above primers. All transfections were performed with Lipofectamine (Gibco-BRL), and cells were harvested 24 to 48 h after transfection. Luciferase activity measurements were performed by using the dual-luciferase assay kit (Promega).
DNA-binding assay.
Radiolabeled oligonucleotide duplexes containing the two p53 DNA-binding sites of p21 and 14-3-3σ were generated by using the following oligonucleotides: p21 site 1, 5′-TGGCCATC AGGAACATGTCCCAACATGTTGAGCTCTGGCA-3′; p21 site 2, 5′-TAGAGGAAGAAGACTGGGCATGTCTGGGCAGAGATTTCCA-3′; 14-3-3σ site 1, 5′-CTGGGACTACAGGCATGTGCCACCATGCCCGGCTAATTTT-3′; and 14-3-3σ site 2, 5′-TGGAAACCCTGTAGCATTAGCCCAGACATGTCCCTACTCCTCC-3′. For end labeling, 500 ng of each oligonucleotide were incubated with 167 μCi of [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) in kinase buffer (70 mM Tris [pH 7.6], 10 mM MgCl2, 5 mM DTT) for 2 h at 37°C. DNA was ethanol precipitated two times, allowed to air dry, and resuspended in 100 μl of annealing buffer (20 mM Tris [pH 7.5], 2 mM MgCl2, 50 mM NaCl). The complementary oligonucleotides for each binding site were mixed, boiled for 5 min, and allowed to anneal.
The human large cell lung carcinoma cell line H1299, which does not express p53 or p63 protein, was transfected with pCEP4 expression vectors encoding either myc-tagged p53 or myc-tagged p63 proteins or with the empty pCEP4 expression vector (Invitrogen). Western analyses of the transfected cell lysates with α-myc antibody (clone 9E-10) were performed, and p53 and p63 were quantified in triplicate by using a Fluor-S Max MultiImager (Bio-Rad Laboratories). Equivalent amounts of p53 and p63 protein were immunoprecipitated by rocking at 4°C for 1 h in DNA binding buffer (DBB; 20 mM Tris [pH 7.2], 100 mM NaCl, 10% glycerol, 1% Nonidet P-40) with the myc antibody and a 15-μl bed volume of PAS (Amersham Biosciences). Immunoprecipitated proteins were washed twice with DBB and then once in DBB containing 1 mM DTT. The immunopurified protein was rocked for 1 h at 4°C in 100 μl of DBB with 2 × 106 cpm of α-32P-labeled DNA fragments prepared as described above. The protein-DNA complexes were rocked for 1 h at 4°C with 10 μg of poly(dI-dC) (Roche). After three washes with DBB, the proteins were digested with SDS-proteinase K (VWR Scientific Products, West Chester, Pa.) in TE8 (20 mM Tris [pH 8.0], 10 mM EDTA) for 30 min at 55°C prior to electrophoresis on 10% polyacrylamide gels (acrylamide-bisacrylamide [19:1]) at 40 V. Radiolabeled DNA was quantified by using an Instant Imager (Packard Instrument Company, Downers Grove, Ill.).
Formaldehyde cross-linking.
Growth medium was aspirated from ∼5 × 106 cells, and cell cultures were washed with PBS and incubated with a 1.6% formaldehyde (EM Sciences) solution in PBS for 13 min at room temperature. The cross-linking was terminated by the addition of glycine to a final concentration of 0.144 M for 5 min. Monolayers were washed twice with PBS. Extracts were prepared by scraping cells in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0], 5 mM EDTA) containing the protease inhibitors antipain (10 μg/ml), leupeptin (10 μg/ml), pepstatin A (10 μg/ml), chymostatin (10 μg/ml), and 4-(2-aminoethyl) benzenesulfonylfluoride (200 μg/ml) and the phosphatase inhibitors 50 mM NaF and 0.2 mM sodium vanadate. Cell lysates were sonicated to yield chromatin fragments of ∼600 bp as assessed by agarose gel electrophoresis. Debris was pelleted by centrifugation for 15 min at 13,000 × g. The lysate was divided into aliquots, and 0.8 mg of protein extract was precleared with 10 μg of mouse immunoglobulin G bound to PAS for p53 immunoprecipitation or with 20 μg of rabbit immunoglobulin G bound to PAS for p63 immunoprecipitation. Protein lysates were precleared for 1 h at 4°C. After centrifugation for 2 min at 13,000 × g, supernatants were transferred to a new tube. A 15-μl bed volume of PAS and 2 μg of α-p53 antibody (Ab-2; Oncogene Research Products) or α-p63 antibody (H129; Santa Cruz) was added to extracts precleared with nonspecific antibodies, and immunoprecipitation was performed by rocking the extracts overnight at 4°C. To control for nonspecific binding during immunoprecipitation, cross-linked lysates were also immunoprecipitated with mouse monoclonal α-cyclin B1 antibody (GNS1; Santa Cruz) or rabbit polyclonal α-Bax antibody (N20; Santa Cruz) that did not cross-react with p53 or p63, respectively.
Immunocomplexes were washed twice with RIPA buffer, four times with immunoprecipitation wash buffer (100 mM Tris [pH 8.5], 500 mM LiCl, 1% Nonidet P-40, 1% deoxycholic acid), and twice more with RIPA buffer. Between washes, samples were rocked for 5 min at 4°C; 200 μl of cross-linking reversal buffer (125 mM Tris [pH 6.8], 10% β-mercaptoethanol, 4% SDS) was added to the washed PAS pellet. Samples were boiled for 30 min to reverse the formaldehyde cross-links. DNA was phenol-chloroform extracted, and the phenol-chloroform phase was back extracted with 10 mM Tris (pH 8.3), ethanol precipitated, allowed to air dry, and dissolved in sterile H2O.
PCR amplification.
p21 site 1 and 14-3-3σ site 1 PCR amplifications were performed in 16.6 mM (NH4)2SO4, 0.67 mM Tris (pH 8.8), 6.7 mM MgCl2, 10 mM β-mercaptoethanol, 10% dimethyl sulfoxide, 1.5 mM nucleotides, and 1.25 U of Taq polymerase (Promega). A total of 175 ng of each primer was used per 25-μl reaction. Forty-five PCR cycles were performed for p21 site 1, with each cycle consisting of 20 s at 94°C, 45 s at 61°C, and 25 s at 72°C. 14-3-3σ site 1 was amplified by using 45 PCR cycles each consisting of 20 s at 94°C, 45 s at 57.5°C, and 25 s at 72°C. p21 site 2 and 14-3-3σ site 2 PCR amplifications were performed by using Ready-To-Go PCR beads (Amersham Biosciences) according to the manufacturer's directions with a final primer concentration of 0.4 μM. Thirty PCR cycles were performed for p21 site 2. Each cycle consisted of 30 s at 95°C, 45 s at 66°C, and 25 s at 72°C. Thirty-eight PCR cycles were performed for 14-3-3σ site 2. Each cycle consisted of 20 s at 94°C, 45 s at 61°C, and 25 s at 72°C. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) PCR amplification was performed in 10 mM Tris (pH 9.0), 50 mM KCl, 0.1% Triton-X, 0.5 mM MgCl2, 0.25 mM concentrations of nucleotides, and 1.25 U of Taq polymerase (Promega). Each primer was used at 0.2 μM per 25-μl reaction. Thirty-five cycles of PCR were performed for GAPDH amplification, each cycle consisted of 20 s at 94°C, 45 s at 62°C, and 25 s at 72°C. The primers used for PCR amplifications were as follows: p21 site 1, 5′-GCTTGGGCAGCAGGCTG-3′ and 5′-AGCCCTGTCGCAAGGATCC-3′; p21 site 2, 5′-GCAGTGGGGCTTAGAGTGGGG-3′ and 5′-CAGGCTTGGAGCAGCTACAATTAC-3′; 14-3-3σ site 1, 5′-CATTTAGGCAGTCTGATTCC-3′ and 5′-GCTCACGCCTGTCATCTC-3′; and 14-3-3σ site 2, 5′-CTCACTACCTCAAGATACCC-3′ and 5′-CACAGGCCTGTGTCTCCC-3′. GAPDH was amplified with 5′-CACCAGCCATCCTGTCCTCC-3′ and 5′-GTTCCTTCCCAGCCCCCACT-3′ primers. PCR DNA products were resolved by using 8% polyacrylamide gels (acrylamide-bisacrylamide [19:1]) in 1× Tris acetate-EDTA buffer. Gels were stained with ethidium bromide. Relative levels of DNA were determined by using Quantity One software (Bio-Rad Laboratories).
RESULTS
ΔNp63α is the predominant splice variant expressed at the protein level in primary HEKs.
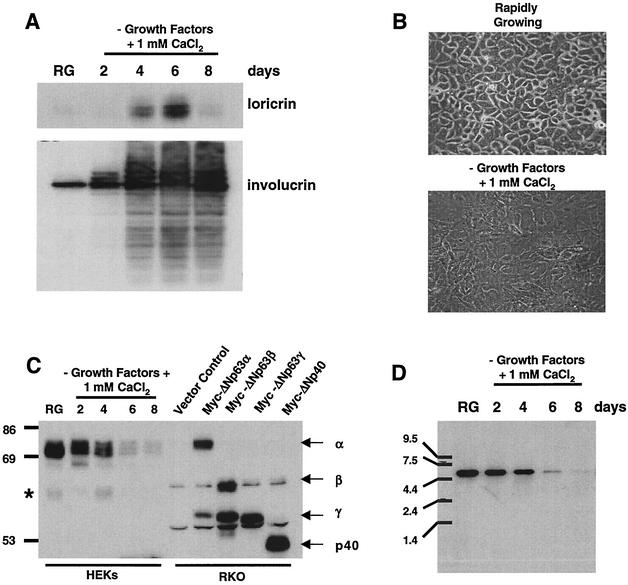
To examine the role p63 plays in rapidly growing and differentiating epithelial cells, we analyzed the specific splice variant(s) expressed in the primary cultures of HEKs. Using well-characterized conditions for in vitro keratinocyte growth (31), we generated protein lysates from HEKs that were rapidly growing or induced to differentiate by the removal of growth factors and the addition of 1 mM CaCl2. This differentiation resulted in increased protein levels of keratinocyte differentiation markers loricrin and involucrin (20), as well as the appearance of a differentiated morphology compared to proliferating cells (Fig. 1A and B). In contrast to the increase in differentiation markers, p63 protein levels decreased during differentiation (Fig. 1C). Since p63 expression has been shown to decrease during keratinocyte terminal differentiation in vivo (57), our in vitro culture system replicated this process.
FIG. 1.
Differentiation-induced modulation of ΔNp63α protein levels in primary HEKs. HEKs were induced to differentiate as described in Materials and Methods. (A) Western analysis of HEK lysates from rapidly growing and differentiating cells for expression of the terminal differentiation markers loricrin and involucrin. (B) Micrograph of rapidly growing HEKs and HEKs at day 4 after induction of differentiation. (C) Western analysis with antibody Ab-1 for p63 expression in rapidly growing and differentiating HEKs. Myc epitope-tagged ΔNp63 splice variants were ectopically expressed in RKO cells, and lysates were analyzed to serve as molecular weight markers for comparison to HEK p63. The asterisk to the left of the blot aligns with a cross-reactive band that may represent ΔNp63β expression. (D) Northern analysis of p63 transcript expression in HEKs differentiated as in panel A. The results shown are representative of three independent experiments with separate primary cultures of HEKs.
Previous studies have shown p63 expression in the basal layer of numerous epithelial tissues (57) with the predominant p63 splice variant expressed at the transcript level being ΔNp63α (12, 13, 38, 41, 42). Using reverse transcription-PCR, we generated cDNAs representative of all ΔNp63 splice variants from primary human oral keratinocytes (data not shown). However, when we performed Western analysis to determine the p63 levels in protein extracts harvested from rapidly growing and differentiating epidermal keratinocytes, the ΔNp63α splice variant was the predominant form expressed (Fig. 1C).
Expression vectors encoding myc-tagged ΔNp63α, ΔNp63β, ΔNp63γ, and ΔNp40AIS were generated and transfected into the colon epithelial cell line, RKO, that does not express p63. Protein lysates were generated from transfected RKO cells to serve as controls for the Western analysis. The predominant p63 protein expressed in the HEKs aligned with ectopically expressed myc-tagged ΔNp63α from RKO cells and decreased during differentiation (Fig. 1C). The faster-migrating cross-reactive band on the immunoblot in Fig. 1C, designated with an asterisk, may represent low-level ΔNp63β expression. We also examined the p63 transcript levels by Northern (Fig. 1D). We only detected one 4.5-kb transcript in mRNA isolated in HEKs and, consistent with our Western analysis results, the level of transcript decreased during differentiation. Due to small differences in cDNA lengths of p63 splice variants, it is possible that the p63 transcript in Fig. 1D represents several splice variants not distinguishable by Northern analysis. However, the Western blot results show that ΔNp63α is the predominant protein expressed in primary cultures of HEKs that are either rapidly growing or induced to differentiate.
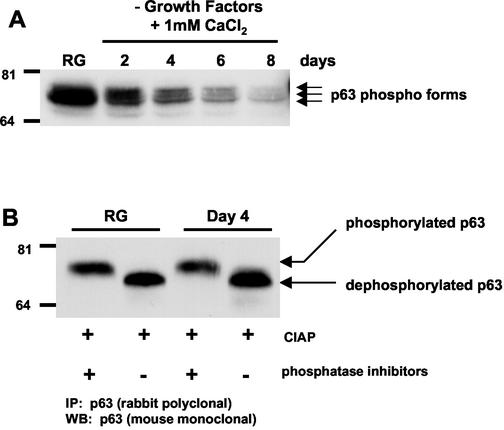
Western analysis revealed multiple, slower-migrating bands that cross-reacted with the p63-specific antibody, suggesting that p63 may also be a phosphoprotein (Fig. 1C and 2A). To determine whether the differential migration of ΔNp63α was due to phosphorylation, p63 was immunoprecipitated from HEK protein lysates prepared from rapidly growing and differentiated cells (day 4) and then treated with CIAP in the presence or absence of phosphatase inhibitors (Fig. 2B). Twice the amount of protein was used in the immunoprecipitations of the lysates prepared from day 4 differentiated cultures compared to rapidly growing cultures to allow comparison of equal p63 levels. Western analysis revealed a collapse of the slower-migrating ΔNp63α protein form to a faster-migrating form after CIAP treatment, a finding consistent with the conclusion that p63 is a phosphorylated protein.
FIG. 2.
ΔNp63α is a phosphoprotein. (A) Protein lysates were prepared from rapidly growing and differentiating HEKs and analyzed by Western blotting with mouse monoclonal antibody Ab-1. Arrows denote three differentially migrating ΔNp63α forms. (B) p63 was immunoprecipitated with rabbit polyclonal antibody H129, and immoprecipitates were treated with CIAP in the presence or absence of phosphatase inhibitors. Immunoprecipitated protein was analyzed by Western blotting for p63 by using mouse monoclonal antibody Ab-1. Resolution of the multiple bands by SDS-PAGE in Fig. 2B was not possible due to the percentage of the acrylamide used. The results shown are representative of four independent experiments.
Changes in expression of cell cycle regulatory proteins and corresponding transcripts in differentiating keratinocytes.
As epidermal cells migrate from the basal layer, they begin the process of terminal differentiation characterized by loss of DNA replication and cell cycle arrest, loss of colony-forming ability, and an increase in cell size (3, 4, 51). Upregulation of the p53 target genes p21 and 14-3-3σ is associated with this phenomenon (11, 34, 37, 48, 60). p21 is a cyclin-dependent kinase inhibitor originally identified as a p53 target gene and a cyclin/cyclin-dependent kinase-associated protein (15, 22, 23, 56). 14-3-3σ has been linked to p53-dependent regulation of cell cycle progression at the G2/M transition (6, 24).
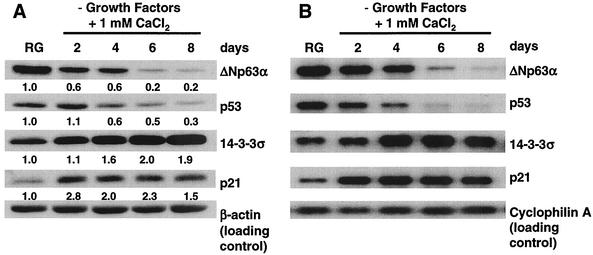
We evaluated p21 and 14-3-3σ protein and mRNA levels relative to ΔNp63α and p53 levels during HEK differentiation. Both ΔNp63α and p53 proteins decreased five- and twofold, respectively (Fig. 3A, compare RG and day 8). In contrast, 14-3-3σ and p21 protein levels increased by 2- and 2.8-fold, respectively (Fig. 3A). Similar to the changes in protein levels, ΔNp63α and p53 transcript levels decreased during differentiation (Fig. 3B, compare RG and day 8), and p21 and 14-3-3σ increased as cells differentiated (Fig. 3B). These data suggest that increases in 14-3-3σ and p21 expression during keratinocyte differentiation are due to either increased activity of the remaining p53 protein or loss of ΔNp63α-mediated transcriptional repression resulting from decreased ΔNp63α protein levels.
FIG. 3.
Differentiation-induced changes in select protein and mRNA levels in primary HEKs. HEKs were induced to differentiate as described in Materials and Methods. (A) Western analysis of p63 (Ab-1), p53 (Ab-2), 14-3-3σ (N-14), and p21 (Ab-1) in rapidly growing and differentiating HEKs with the antibodies listed in Materials and Methods. Note that p63 migrates as a single band due to the use of electrophoresis conditions that allow for analysis of the molecular weight range of proteins shown in panel A. The numbers below the Western panels represent the fold change relative to rapidly growing HEKs. (B) Northern analysis of transcripts for proteins shown in panel A. The results are representative of three independent experiments with independent primary cultures of HEKs.
ΔNp63α represses transcription.
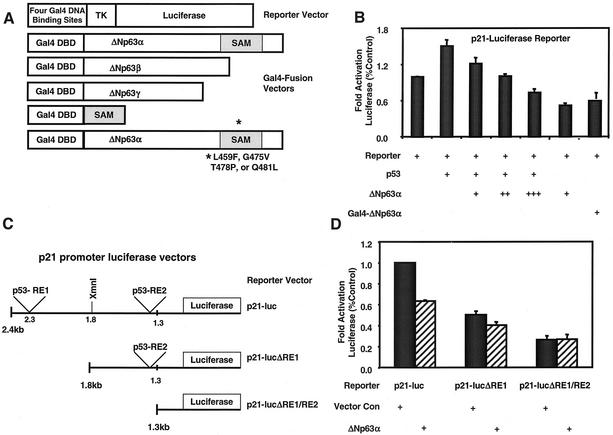
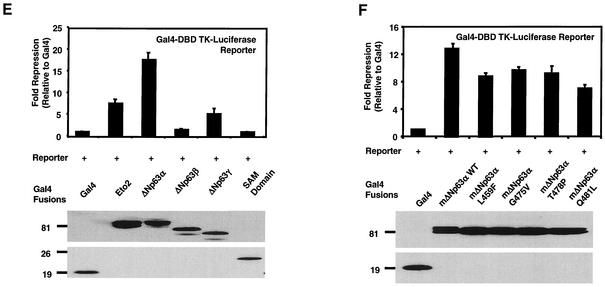
Previous studies have shown that ectopic expression of p63 can repress transcription from reporter vectors containing tandem repeats of the p53 consensus DNA-binding site upstream of a minimal promoter (7, 57). However, the mechanism by which this occurs has not been determined. To explore this mechanism, we analyzed the ability of ΔNp63 splice variants to repress transcription as fusion proteins. We generated expression vectors that encode Gal4-ΔNp63α, -ΔNp63β, and -ΔNp63γ fusion proteins (Fig. 4A). All ΔNp63 splice variants used to generate the fusion proteins were full length. To verify the Gal4 fusion did not impair ΔNp63 activity, we tested the ability of the Gal4-ΔNp63α to repress transcription from a p21-luciferase vector (Fig. 4B). We transiently transfected HCT116 cells with expression vectors encoding ΔNp63α or Gal4-ΔNp63α, as well as the p21-luciferase vector, and found that Gal4-ΔNp63α functions similarly to wild-type ΔNp63α in its ability to inhibit transcription from the p21 promoter (Fig. 4B). Transfection of p53 served as a positive control for activation of the p21 promoter in this assay.
FIG. 4.
ΔNp63α represses transcription. (A) Schematic representing vectors used in panels B, D, E, and F. Abbreviations: DBD, DNA-binding domain; TK, thymidine kinase. The SAM mutants were L459F, G475V, T478P, and Q481L. (B) Analysis of ΔNp63α and Gal4-ΔNp63α activity in a p21 promoter-luciferase reporter assay. HCT116 cells were transfected with a p21 promoter-luciferase reporter construct (containing 2,400 bp of p21 promoter sequence) and expression vector encoding either p53, ΔNp63α, or Gal4-ΔNp63α. +, ++, and +++, indicate 1:1, 1:3, and 1:10 ratios of p53 to ΔNp63α expression vectors, respectively. (C) Schematic representing reporter vectors used in panel D. Abbreviations: p53-RE, p53-response element; luc, luciferase. (D) HCT116 cells were transfected as in panel B with the indicated reporter vectors and an empty expression vector (Vector Con) or one expressing ΔNp63α. All values were normalized to those generated with lysates prepared from cells cotransfected with the full-length p21-luciferase reporter vector and the empty expression vector, pCEP4. (E) 293 cells were transfected with the luciferase reporter vector in panel A and the indicated Gal4 fusion vectors. Gal4 alone served as the negative control, and Gal4-ETO2 served as the positive control. (F) 293 cells were transfected with expression vectors encoding murine Gal4-ΔNp63α and Gal4-ΔNp63α proteins containing the indicated SAM domain point mutations. All luciferase assays were normalized for transfection efficiency with a renilla reporter vector. Western analyses with a Gal4-specific antibody were performed to verify protein expression, and results are shown in the lower portions of panels E and F. The results shown are representative of five independent experiments performed each time in triplicate, and error bars indicate the standard deviation.
We also determined whether ΔNp63α could function as a dominant-negative inhibitor of p53 transactivation. Cotransfection of expression vectors encoding p53 and ΔNp63α with the p21-luciferase reporter vector resulted in a significant decrease in p53-mediated luciferase expression (Fig. 4B). Further, the ability of ΔNp63α to repress transactivation of the p21 promoter-luciferase vector was dose dependent and was consistent with a dominant-negative activity of ΔNp63α and the previous findings of Yang et al. (57). To determine the necessity of the p53 response elements for the repressive activity of ΔNp63α, we used the assay described above and analyzed p21 promoter deletions that lack either one (p21-lucΔRE1) or both (p21-lucΔRE1/RE2) of the reported p53 response elements in the p21 promoter (15, 16) (Fig. 4C). We transiently transfected the HCT116 cells with an expression vector encoding ΔNp63α and the full-length or deletion mutant p21-luciferase luciferase reporter vectors. Similar to our results in Fig. 4B, ΔNp63α reduced transcriptional activity of the full-length p21-reporter vector by ∼45% compared to vector control (Fig. 4D). Loss of one p53 binding site reduced this repression, and loss of both p53 binding sites abrogated the ability of ΔNp63α to repress transcriptional activity from the p21 promoter (Fig. 4D). However, the basal activity of the p21-lucΔRE1/RE2 was significantly lower than that of the full-length p21 promoter reporter.
The results in Fig. 4B established that the Gal4 DNA-binding domain did not impair ΔNp63α function; thus, we analyzed the Gal4-ΔNp63α, -ΔNp63β, and -ΔNp63γ splice variants for their ability to repress transcription. Each Gal4 fusion expression vector was transfected into 293 cells, and the encoded protein was analyzed for its ability to repress luciferase reporter gene transcription regulated by four Gal4 DNA-binding sites upstream of a thymidine kinase promoter (Fig. 4A). The ΔNp63α splice variant repressed the TK-luciferase reporter construct by ∼20-fold, whereas the ΔNp63β did not repress transcription and the ΔNp63γ repressed transcription only by ∼5-fold (Fig. 4E). Western analysis of the luciferase assay extracts demonstrated that all ΔNp63 proteins were expressed at similar levels (Fig. 4E, lower panel). These results suggest that the transcriptional repression activity of ΔNp63α is contained within the carboxy terminus of ΔNp63α that contains the SAM domain. All three splice variants bind DNA in in vitro assays (data not shown), indicating that the loss of repression may be due to loss of protein-protein interactions within the carboxy terminus of the ΔNp63α splice variant. Transfection of Gal4-ETO2, a previously described transcriptional corepressor protein (1), served as a positive control for repression of the luciferase reporter vector. These data demonstrate that ΔNp63α can repress transcription directly or through recruitment of transcriptional corepressor molecules.
Analysis of SAM domain mutations.
Recent genetic studies link p63 to the proper development of limbs, ectodermal appendages, and the lip and palate in humans (5, 35). Specifically, the autosomal-dominant disorder, ankyloblepharon-ectodermal dysplasia-clefting syndrome or Hay-Wells syndrome, is characterized by point mutations within the SAM domain of p63α splice variants (35). To further explore a potential role of the SAM domain in transcriptional repression, we generated expression vectors encoding Gal4-ΔNp63α fusion proteins containing SAM domain point mutations corresponding to mutations found in the p63 gene of individuals with Hay-Wells syndrome. The four point mutations—L459F, G475V, T478P, and Q481L—and a corresponding wild-type fusion expression vector were generated by using murine p63 (Fig. 4A). Murine p63 was previously used for analysis of SAM domain point mutations by McGrath et al. (35). As before, each vector was transiently transfected into 293 cells and analyzed for its ability to repress transcription from the reporter vector shown in Fig. 4A. Wild-type murine ΔNp63α repressed transcription ∼13-fold (Fig. 4F). This level of repression activity is similar to that observed with human ΔNp63α protein (Fig. 4E). In the context of full-length ΔNp63α, the SAM domain point mutants repressed transcription ∼7- to 10-fold, suggesting that these mutations led to a reduction of p63 activity rather than complete loss of function. Western analysis of the protein lysates used in this assay demonstrated that all mutant proteins were expressed at relatively equal, if not greater, levels than wild-type ΔNp63α. These results suggest that the difference in activity between wild-type and SAM domain point mutant proteins was not due to differences in protein expression (Fig. 4F, lower panel). Because of the reduced activity of the SAM domain mutant proteins, we generated and analyzed a human Gal4-SAM fusion protein for analysis in the repression assay. The results presented in Fig. 4E indicate that the SAM domain alone is not sufficient to repress transcription but is likely required for repressor activity in the context of the C-terminal domain of ΔNp63α.
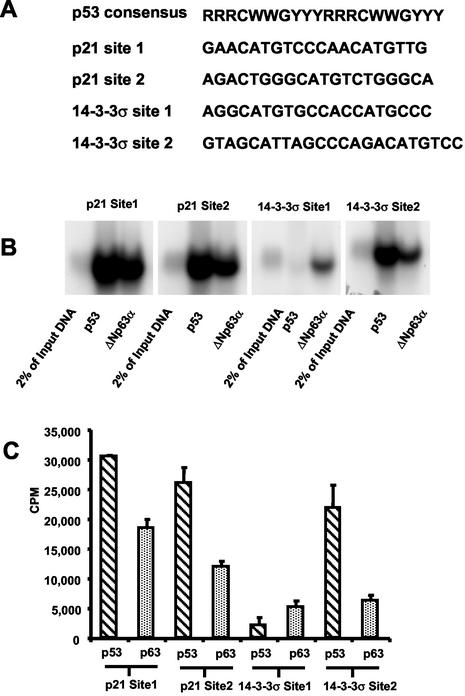
Relative binding affinities of ΔNp63α and p53 to p53 consensus DNA-binding sites.
The p53 protein binds DNA in a sequence-specific manner and regulates transcription of gene products involved in processes such as growth arrest, DNA repair, and apoptosis (49). To compare the relative affinities of ΔNp63α and p53 binding to known p53-binding sites, in vitro DNA-binding assays were performed with radiolabeled duplex oligonucleotides representing p53-binding sites in the p21 and 14-3-3σ promoters as previously described (15, 24) (Fig. 5A). H1299 cells were transfected with myc-tagged p53 or myc-tagged ΔNp63α, and equal amounts of p53 or ΔNp63α were immunoprecipitated with α-myc epitope antibody. The immunoprecipitates were assayed for their ability to bind the radiolabeled oligonucleotide duplexes. p53 and ΔNp63α bound to both p53 consensus sites present in the p21 promoter (Fig. 5B). Similarly, ΔNp63α bound to both sites present in the 14-3-3σ promoter, whereas p53 only displayed significant binding to site 2. Quantification of bound radiolabeled DNA illustrated that p53 has ∼1.5- and ∼2-fold-greater relative binding affinities than ΔNp63α for p21 sites 1 and 2, respectively (Fig. 5C). Similarly, p53 had a ∼4-fold-higher relative binding affinity for 14-3-3σ binding site 2. In contrast, ΔNp63α bound to the 14-3-3σ site 1 oligonucleotide with a relative binding affinity that was ca. four- to fivefold greater than that of p53. Similar assays were performed with the myc epitope-tagged SAM mutants analyzed in Fig. 4. However, comparison of SAM mutants to wild-type ΔNp63α showed no significant difference in the relative DNA-binding affinity (data not shown). Thus, the reduction in the transcriptional repression activity of the SAM mutations observed in Fig. 4D was not due to differences in DNA-binding affinity.
FIG. 5.
Relative binding affinities of p53 and ΔNp63α for p53 consensus sites in the p21 and 14-3-3σ promoters. (A) The p53 consensus binding sequence and p53 binding sites in the p21 and 14-3-3σ promoters. Abbreviations: R, purine; Y, pyrimidine; W, adenine or thymine. (B) H1299 cells were transfected with myc-tagged p53 or ΔNp63α, and protein lysates were quantified by using the Fluor-S Max MultiImager. Based on Fluor-S Max quantification, equal amounts of myc-tagged p53 and myc-tagged ΔNp63α were immunoprecipitated with a myc epitope antibody. Immunoprecipitated p53 and ΔNp63α were assayed for their ability to bind radiolabeled oligonucleotides representing p53-binding sites in the p21 and 14-3-3σ promoters as described in Materials and Methods. (C) Bound oligonucleotides were separated on acrylamide gels, exposed for autoradiography, and quantified. Each autoradiograph shows one representative result of at least three independent experiments that are quantified and displayed with the standard deviation.
p53 and/or p63 occupancy at the p21 and 14-3-3σ and promoters in vivo during HEK differentiation.
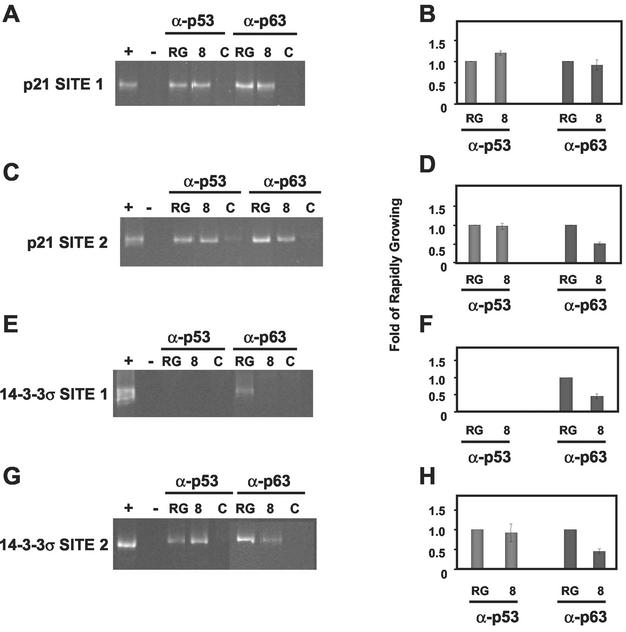
Since ΔNp63α exhibited significant binding to p53 consensus sites in the p21 and 14-3-3σ promoters, we studied the ability of p63 to bind these consensus sites in vivo by using a chromatin immunoprecipitation (ChIP) methodology previously described in an earlier study from our laboratory (52). Either rapidly growing or differentiated cultures (day 8) of HEKs were cross-linked by exposure to 1.6% formaldehyde as described in Materials and Methods. After cross-linking, p53 and p63 were immunoprecipitated, and the DNA to which the proteins bound was purified. The DNA was PCR amplified with primers specific for sequences that flank the p53 response elements in the promoters studied. To assure that the amplified DNA was the correct size, control PCRs were performed with or without genomic HEK DNA (Fig. 6A, C, E, and G, lanes + or −, respectively). To control for nonspecific binding during immunoprecipitation, cross-linked lysates were immunoprecipitated with mouse monoclonal α-cyclin B1 or rabbit polyclonal α-Bax antibodies that did not cross-react with p53 or p63, respectively (Fig. 6A, C, E, and G, lanes C).
FIG. 6.
ChIP analyses for p53 and p63 binding at the 14-3-3σ and p21 promoters during keratinocyte differentiation. Abbreviations: RG, rapidly growing; 8, day 8 after induction of differentiation; C, control. HEKs were induced to differentiate as described in Materials and Methods. At the time of harvest, HEKs were treated with formaldehyde (X-Link) and processed as described in Materials and Methods. The DNA immunoprecipitated with p53- or p63-specific antibodies was PCR amplified by using primers flanking the p53 binding sites in the p21 (A and C) and 14-3-3σ (E and G) promoters. DNA fragments generated by PCR were resolved by PAGE, the gels were stained with ethidium bromide, and the PCR products were quantified by densitometry. Lanes marked “+” indicate PCR products that were generated by using DNA template derived from total genomic DNA harvested from rapidly growing HEKs. Lanes marked “−” indicate the absence of DNA input for the PCR. The lanes marked “C” indicate PCRs performed with templates immunoprecipitated with antibodies specific to cyclin B1 and Bax for p53 and p63 analyses, respectively. Each ethidium bromide-stained gel shows one representative result of at least three independent experiments that are quantified and displayed with standard deviations in the corresponding panels B, D, F, and H.
The ChIP experiments revealed that p53 and p63 bind both p21 promoter site 1 and site 2 in rapidly growing (RG) and differentiating (day 8) HEKs (Fig. 6A and C). p53 occupancy at p21 site 1 increased modestly from rapidly growing cells to differentiation day 8 (<1.5-fold), whereas p63 occupancy did not change (Fig. 6B). In contrast to site 1 binding, p53 binding to p21 site 2 did not change and p63 decreased twofold (Fig. 6D). Analysis of the 14-3-3σ promoter showed that p53 binding to site 2 remained unchanged in rapidly growing and differentiated keratinocytes (Fig. 6G and H); however, there was no appreciable binding of p53 to 14-3-3σ site 1 (Fig. 6E and F), a finding consistent with the in vitro DNA-binding results shown in Fig. 5. Like the p21 promoter, p63 bound to both 14-3-3σ site 1 and site 2 in vivo, and by day 8 of differentiation this binding decreased twofold at both site 1 and site 2 (Fig. 6F and H). These data suggest that increased p21 and 14-3-3σ expression are due to the loss of p63 binding and subsequent decreased transcriptional repression of these promoters by p63.
DISCUSSION
Since the identification of the p53 homologue p63, several studies have investigated its function in epithelial cell growth and development. Using HEKs as a model system, we sought to further analyze the biochemical role p63 plays in keratinocyte growth and differentiation. We demonstrated that the primary splice variant of p63 expressed in HEKs is ΔNp63α, and its expression decreases as cells differentiate. Further, the ΔNp63α protein was present in differentiating HEKs as several phosphoforms. In addition to the reduction in p63 transcript and protein levels during differentiation, we also observed a decrease in p53 transcript and protein. The reduction in p53 and p63 expression correlated with an increase in expression of the cell cycle regulatory proteins p21 and 14-3-3σ. Using Gal4 fusion proteins, we determined that the p63 protein represses transcription and that the ΔNp63α splice variant has the highest activity. In addition, in vitro and in vivo DNA-binding assays showed that ΔNp63α binds to both p53 response elements in the p21 and 14-3-3σ promoters with p63 occupancy at p21 site 2 and 14-3-3σ sites 1 and 2 decreasing as cells differentiated.
Consistent with previously published reports (41, 42), we observed a decrease in p63 transcript and protein during differentiation. Analysis of the limited number of keratinocytes in the p63−/− mouse showed expression of epithelial terminal differentiation markers (58), suggesting that epithelial defects were due to the lack of cell survival and/or proliferation and not to impaired terminal differentiation. In support of this, a recent study in zebrafish using antisense oligonucleotides demonstrated that the ΔNp63 splice variant(s) were required for epithelial proliferation (30). These model systems suggest a role for p63 in maintaining the survival or proliferation of basal keratinocytes and, in conjunction with our HEK data, indicate that the loss of ΔNp63α facilitates the growth arrest associated with differentiation.
We determined that p63 migrated as multiple phospho-forms by SDS-polyacrylamide gel electrophoresis (PAGE), suggesting that phosphorylation is a mechanism by which the p63 protein is regulated. This hypothesis is supported by the findings that phosphorylation is a key posttranslational modification for regulation of p53 (49). However, ΔNp63α lacks a transactivation domain where many of the regulatory phosphorylation sites are found in p53. Future studies are required to identify the phosphoresidues in ΔNp63α, upstream kinases, and phosphorylation-dependent associated proteins.
It has been suggested that ΔNp63α-mediated repression can occur through direct protein-protein interaction, and several groups have examined the association of p63 proteins encoded by the various splice variants with other p53 family members. Davison et al. and Irwin et al. determined that p63 and p73 can form homodimers or have weak heterotypic interactions through their oligomerization domain but do not interact with the p53 oligomerization domain (10, 26). Kojima et al. found similar results by using a yeast two-hybrid system (28). Further, several studies have shown that p63 and p53 can interact through the core/DNA-binding domain (21, 44, 50). One consequence of p53 association with ΔNp63α may be caspase-dependent degradation of select ΔNp63 proteins (p40 and ΔNp63α) (44). The significance of these findings remains to be determined in the context of proliferating and differentiating epithelial cells.
Through the use of Gal4 fusion proteins, we determined that the C-terminal domain of ΔNp63α is involved in transcriptional repression. Further, single amino acid substitutions within the SAM domain of ΔNp63α resulted in reduced transcriptional repression. Similar results were obtained for the ΔNp63α proteins containing SAM domain mutations by using a p53-reporter assay and cotransfections of the mutant proteins with p53 or TA-p63γ (35). Chi et al. (8) have shown that the SAM domain of p73 contains a folded, globular α-helical structure and suggest that this domain interacts with additional, as-yet-uncharacterized signaling proteins. However, the SAM domains of p73 and p63 are monomeric and do not interact with one another, leaving the possibility that the p63 SAM domain may play a role in recruiting transcriptional corepressors to select target genes, and these protein-protein interactions are disrupted in individuals with Hay-Wells syndrome (35). Consistent with this hypothesis is our finding that ΔNp63α proteins containing the Hay-Wells mutations bind DNA with the same relative affinity as the wild-type protein. Studies have also demonstrated that a frameshift mutation found in ectrodactyl, ectodermal dysplasia, and cleft lip patients, causing loss of the SAM domain and carboxy-terminal sequence, results in the total loss of transcriptional repressive ability (5). Taken together, the data suggest that the carboxy-terminal region of ΔNp63α containing the SAM domain plays an integral role in ΔNp63α-mediated transcriptional repression.
Since ΔNp63α has been identified as the primary splice variant expressed at the protein level in epithelial cells, several questions remain to be addressed. In particular, what target genes does ΔNp63α regulate and which of these genes are coordinately regulated by p53? Our in vitro DNA-binding assays and ChIP analyses support the hypothesis that p53 and p63 can coordinately bind target genes such as p21 and 14-3-3σ. These results are in agreement with those of Flores et al. showing that increased association of both p53 and p63 with p21, mdm2, PERP, and NOXA promoters in mouse embryo fibroblasts expressing E1A after DNA damage (19).
If ΔNp63α functions as a transcriptional repressor in vivo, as our Gal4 fusion experiments support, then protein levels, promoter binding affinity, and coassociated proteins are likely factors involved in this coordinate regulation of downstream target genes. Similar to our results, Weinberg et al. reported that p53 transcript and protein decreased during differentiation whereas p21 promoter activity increased (55). Does this increase reflect an elevation of p53 activity, an elevation of the activity of other transcriptional activators, or the loss of ΔNp63α repressor activity at the promoter? In support of a role for other transcriptional activators, several studies show that Sp1 and Sp3 can transcriptionally activate the p21 promoter (29, 40, 43, 45) and transcriptional regulators such as these may activate the p21 promoter when p53 levels are decreased during keratinocyte differentiation. In support of the theory that loss of ΔNp63α repressor activity at the p21 promoter allows p53 or other transactivators to act unopposed, Liefer et al. showed a decrease in p63 in mouse keratinocytes in vitro and mouse epidermis in vivo after UV-B exposure (32), a treatment which leads to elevated p53 transcriptional activity (33). Further, ectopic expression of ΔNp63α in the mouse epidermis resulted in decreased UV-B-induced apoptosis (32), a phenotype thought to be primarily dependent on p53 activity (61). Our findings that p63 and p53 can bind the same promoter elements in vivo support the role of ΔNp63α acting coordinately with p53 to regulate select target genes during keratinocyte proliferation and differentiation. Our observations also favor the possibility that p53 and ΔNp63α compete for consensus DNA-binding sites with p53 having a relatively higher binding affinity than ΔNp63α for select promoters, such as p21.
Previous studies and findings reported here support the following model. When rapidly proliferating basal epithelial cells are exposed to cell stress, increased p53 protein combined with the higher binding affinity of p53 for select promoter sites displaces ΔNp63α. Further, as is the case after exposure of keratinocytes to UV radiation, the p63 protein levels decrease. These events lead to subsequent transactivation of genes whose products are involved in growth arrest and apoptosis. In the absence of cell stress, constitutively expressed ΔNp63α protein levels exceed those of p53, and thus select target gene promoters are repressed, allowing for continued proliferation of keratinocytes in the basal layer where ΔNp63α is localized in stratified epithelium. During differentiation, both p53 and ΔNp63α levels decrease; however, it is the loss of ΔNp63α-mediated repression of select target genes that plays a role in differentiation. This model is consistent with the the proposed oncogenic role of p63 overexpression in squamous cell carcinomas of the head and neck (9, 25) and the observations that ectopic expression of the p40AIS splice variant in Rat 1a cells results in increased growth of these cells in soft agar and athymic, nude mice (25). However, as suggested above, it is likely that ΔNp63α also regulates gene expression independently of p53, since mutation of p53 and amplification of p63 both occur during genesis of squamous cell carcinomas (25) (J. Sniezek and J. Pietenpol, unpublished results). Clearly, additional experimentation is required to further link p63 biochemistry to biology and to determine the interplay of p63 and p53 signaling pathways. New technologies, including in vivo DNA-binding assays and mass spectrometry, will aid in the identification of key posttranslational modifications, associated proteins, and novel target genes that are regulated by p63.
Acknowledgments
This work was supported by the National Institutes of Health grant CA70856 and the Burroughs-Wellcome Fund (J.A.P.), U.S. Army grant DAMD17-01-1-0439 (M.D.W.), and National Institutes of Health grants ES00267 and CA68485 (Core Services).
We thank members of the Pietenpol laboratory for critical reading of the manuscript.
REFERENCES
- 1.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augustin, M., C. Bamberger, D. Paul, and H. Schmale. 1998. Cloning and chromosomal mapping of the human p53-related KET gene to chromosome 3q27 and its murine homolog Ket to mouse chromosome 16. Mamm. Genome 9:899-902. [DOI] [PubMed] [Google Scholar]
- 3.Barrandon, Y., and H. Green. 1985. Cell size as a determinant of the clone-forming ability of human keratinocytes. Proc. Natl. Acad Sci. USA 82:5390-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrandon, Y., and H. Green. 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 84:2302-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celli, J., P. Duijf, B. Hamel, M. Bamshad, B. Kramer, A. Smits, R. Newbury-Ecob, R. Hennekam, G. Van Buggenhout, A. van Haeringen, C. Woods, A. van Essen, R. de Waal, G. Vriend, D. Haber, A. Yang, F. McKeon, H. Brunner, and H. van Bokhoven. 1999. Heterozygous germline muations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99:143-153. [DOI] [PubMed] [Google Scholar]
- 6.Chan, T. A., H. Hermeking, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature 401:616-620. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. 1999. The p53 family: same response, different signals? Mol. Med. Today 5:387-392. [DOI] [PubMed] [Google Scholar]
- 8.Chi, S. W., A. Ayed, and C. H. Arrowsmith. 1999. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 18:4438-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, H. R., J. G. Batsakis, F. Zhan, E. Sturgis, M. A. Luna, and A. K. El-Naggar. 2002. Differential expression of p53 gene family members p63 and p73 in head and neck squamous tumorigenesis. Hum. Pathol. 33:158-164. [DOI] [PubMed] [Google Scholar]
- 10.Davison, T. S., C. Vagner, M. Kaghad, A. Ayed, D. Caput, and C. H. Arrowsmith. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274:18709-18714. [DOI] [PubMed] [Google Scholar]
- 11.Dellambra, E., O. Golisano, S. Siviero Bondanza, E. P. Lacal, M. Molinari, S. D'Atri, and M. DeLuca. 2000. Downregulation of 14-3-3σ prevents clonal evolution and leads to immortalization of primary human keratinocytes. J. Cell Biol. 149:1117-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dellavalle, R. P., T. B. Egbert, A. Marchbank, L. J. Su, L. A. Lee, and P. Walsh. 2001. CUSP/p63 expression in rat and human tissues. J. Dermatol. Sci. 27:82-87. [DOI] [PubMed] [Google Scholar]
- 13.Di Como, C. J., M. J. Urist, I. Babayan, M. Drobnjak, C. V. Hedvat, J. Teruya-Feldstein, K. Pohar, A. Hoos, and C. Cordon-Cardo. 2002. p63 expression profiles in human normal and tumor tissues. Clin. Cancer. Res. 8:494-501. [PubMed] [Google Scholar]
- 14.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 15.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry, W. S., T. Tokino, T. Waldman, J. D. Oliner, V. E. Velculescu, M. Burrell, D. E. Hill, E. Healy, J. L. Rees, S. R. Hamilton, K. W. Kinzler, and B. Vogelstein. 1995. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 55:2910-2919. [PubMed] [Google Scholar]
- 17.Flatt, P. M., J. O. Price, A. Shaw, and J. A. Pietenpol. 1998. Differential cell cycle checkpoint response in normal human keratinocytes and fibroblasts. Cell Growth Differ. 9:535-543. [PubMed] [Google Scholar]
- 18.Flatt, P. M., L. J. Tang, C. D. Scatena, S. T. Szak, and J. A. Pietenpol. 2000. p53 Regulation of G2 checkpoint is retinoblastoma protein dependent. Mol. Cell. Biol. 20:4210-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560-564. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs, E., and C. Byrne. 1994. The epidermis: rising to the surface. Curr. Opin. Genet. Dev. 4:725-736. [DOI] [PubMed] [Google Scholar]
- 21.Gaiddon, C., M. Lokshin, J. Ahn, T. Zhang, and C. Prives. 2001. A subset of tumor-derived mutant forms of p53 downregulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, Y., C. W. Turck, and D. O. Morgan. 1993. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature 366:707-710. [DOI] [PubMed] [Google Scholar]
- 23.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 24.Hermeking, H., C. Lengauer, K. Polyak, T.-C. He, L. Zhang, S. Thiagalingam, K. W. Kinzler, and B. Vogelstein. 1997. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1:3-11. [DOI] [PubMed] [Google Scholar]
- 25.Hibi, K., B. Trink, M. Patturajan, W. Westra, O. Caballero, D. Hill, E. Ratovitski, J. Jen, and D. Sidransky. 2000. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 97:5462-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelan, D. I. Smith, L. Wanguo, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin. 2002. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407:642-645. [DOI] [PubMed] [Google Scholar]
- 27.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J. Biscan, A. Valent, A. Minty, P. Chalon, J. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:808-819. [DOI] [PubMed] [Google Scholar]
- 28.Kojima, T., Y. Ikawa, and I. Katoh. 2001. Analysis of molecular interactions of the p53-family p51(p63) gene products in a yeast two-hybrid system: homotypic and heterotypic interactions and association with p53-regulatory factors. Biochem. Biophys. Res. Commun. 281:1170-1175. [DOI] [PubMed] [Google Scholar]
- 29.Koutsodontis, G., I. Tentes, P. Papakosta, A. Moustakas, and D. Kardassis. 2001. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21WAF1/Cip1 gene by the p53 tumor suppressor protein. J. Biol. Chem. 276:29116-29125. [DOI] [PubMed] [Google Scholar]
- 30.Lee, H., and D. Kimelman. 2002. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell 2:607-616. [DOI] [PubMed] [Google Scholar]
- 31.Leigh, I., E. Lane, and F. Watt. 1994. The keratinocyte handbook. Cambridge University Press, Cambridge, United Kingdom.
- 32.Liefer, K., M. Koster, X. Wang, A. Yang, F. McKeon, and D. Roop. 2000. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 60:4016-4020. [PubMed] [Google Scholar]
- 33.Liu, M., and J. C. Pelling. 1995. UV-B/A irradiation of mouse keratinocytes results in p53-mediated WAF1/CIP1 expression. Oncogene 10:1955-1960. [PubMed] [Google Scholar]
- 34.Macleod, K. F., N. Sherry, G. Hannon, D. Beach, T. Tokino, K. Kinzler, B. Vogelstein, and T. Jacks. 1995. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 9:935-944. [DOI] [PubMed] [Google Scholar]
- 35.McGrath, J. A., P. H. G. Duijf, V. Doetsch, A. D. Irvine, R. de Waal, K. R. J. Vanmolkot, V. Wessagowit, A. Kelly, D. J. Atherton, W. A. D. Griffiths, S. J. Orlow, A. van Haeringen, M. G. E. M. Ausems, A. Yang, F. McKeon, M. A. Bamshad, H. G. Brunner, B. C. J. Hamel, and H. van Bokhoven. 2001. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum. Mol. Genet. 10:221-229. [DOI] [PubMed] [Google Scholar]
- 36.Mills, A. A., B. H. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 37.Missero, C., E. Calautti, R. Eckner, J. Chin, L. H. Tsai, D. M. Livingston, and G. P. Dotto. 1995. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc. Natl. Acad. Sci. USA 92:5451-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nylander, K., P. J. Coates, and P. A. Hall. 2000. Characterization of the expression pattern of p63α and ΔNp63α in benign and malignant oral epithelial lesions. Int. J. Cancer 87:368-372. [PubMed] [Google Scholar]
- 39.Osada, M., M. Ohba, C. Kawahara, C. Ishioka, R. Kanamaru, I. Katoh, Y. Ikawa, Y. Nimura, A. Nakagawara, M. Obinata, and S. Ikawa. 1998. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 40.Pagliuca, A., G. Pasquale, and L. Lania. 2000. Differential role for Sp1/Sp3 transcription factors in the regulation of the promoter activity of multiple cyclin-dependent kinase inhibitor genes. J. Cell Biochem. 76:360-367. [DOI] [PubMed] [Google Scholar]
- 41.Parsa, R., A. Yang, F. McKeon, and H. Green. 1999. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J. Investig. Dermatol. 113:1099-1105. [DOI] [PubMed] [Google Scholar]
- 42.Pellegrini, G., E. Dellambra, O. Golisano, E. Martinelli, I. Fantozzi, S. Bondanza, D. Ponzin, F. McKeon, and M. De Luca. 2001. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA 98:3156-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prowse, D. M., L. Bolgan, A. Molnar, and G. P. Dotto. 1997. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 272:1308-1314. [DOI] [PubMed] [Google Scholar]
- 44.Ratovitski, E. A., M. Patturajan, K. Hibi, B. Trink, K. Yamaguchi, and D. Sidransky. 2001. p53 associates with and targets ΔNp63 into a protein degradation pathway. Proc. Natl. Acad. Sci. USA 98:1817-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santini, M. P., C. Talora, T. Seki, L. Bolgan, and G. P. Dotto. 2001. Cross talk among calcineurin, Sp1/Sp3, and NFAT in control of p21WAF1/CIP1 expression in keratinocyte differentiation. Proc. Natl. Acad. Sci. USA 98:9575-9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz, J., C. P. Ponting, K. Hofmann, and P. Bork. 1997. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6:249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibata, H., Z. Nawaz, S. Y. Tsai, B. W. O'Malley, and M. J. Tsai. 1997. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT). Mol. Endocrinol 11:714-724. [DOI] [PubMed] [Google Scholar]
- 48.Steinman, R. A., B. Hoffman, A. Iro, C. Guillouf, D. A. Liebermann, and M. E. el-Houseine. 1994. Induction of p21WAF-1/CIP1 during differentiation. Oncogene 9:389-396. [PubMed] [Google Scholar]
- 49.Stewart, Z. A., and J. A. Pietenpol. 2001. p53 signaling and cell cycle checkpoints. Chem. Res. Toxicol. 14:243-263. [DOI] [PubMed] [Google Scholar]
- 50.Strano, S., G. Fontemaggi, A. Costanzo, M. G. Rizzo, O. Monti, A. Baccarini, G. Del Sal, M. Levrero, A. Sacchi, M. Oren, and G. Blandino. 2002. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem. 277:18817-18826. [DOI] [PubMed] [Google Scholar]
- 51.Sun, T. T., and H. Green. 1976. Differentiation of the epidermal keratinocyte in cell culture: formation of the cornified envelope. Cell 9:511-521. [DOI] [PubMed] [Google Scholar]
- 52.Szak, S. T., D. Mays, and J. A. Pietenpol. 2001. Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 21:3375-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thanos, C. D., and J. U. Bowie. 1999. p53 Family members p63 and p73 are SAM domain-containing proteins. Protein Sci. 8:1708-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trink, B., K. Okami, L. Wu, V. Sriuranpong, J. Jen, and D. Sidransky. 1998. A new human p53 homologue. Nat. Med. 4:747-748. [DOI] [PubMed] [Google Scholar]
- 55.Weinberg, W. C., C. G. Azzoli, K. Chapman, A. J. Levine, and S. H. Yuspa. 1995. p53-mediated transcriptional activity increases in differentiating epidermal keratinocytes in association with decreased p53 protein. Oncogene 10:2271-2279. [PubMed] [Google Scholar]
- 56.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 57.Yang, A., M. Kaghad, Y. Wang, E. Gillet, M. Fleming, V. Dotsch, N. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 58.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 59.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, W., L. Grasso, C. D. McClain, A. M. Gambel, Y. Cha, S. Travali, A. B. Deisseroth, and W. E. Mercer. 1995. p53-independent induction of WAF1/CIP1 in human leukemia cells is correlated with growth arrest accompanying monocyte/macrophage differentiation. Cancer Res. 55:668-674. [PubMed] [Google Scholar]
- 61.Ziegler, A., A. S. Jonason, D. J. Leffell, J. A. Simon, H. W. Sharma, J. Kimmelman, L. Remington, T. Jacks, and D. E. Brash. 1994. Sunburn and p53 in the onset of skin cancer. Nature 372:773-776. [DOI] [PubMed] [Google Scholar]