Abstract
Most 5-methylcytosine in Neurospora crassa occurs in A:T-rich sequences high in TpA dinucleotides, hallmarks of repeat-induced point mutation. To investigate how such sequences induce methylation, we developed a sensitive in vivo system. Tests of various 25- to 100-bp synthetic DNA sequences revealed that both T and A residues were required on a given strand to induce appreciable methylation. Segments composed of (TAAA)n or (TTAA)n were the most potent signals; 25-mers induced robust methylation at the special test site, and a 75-mer induced methylation elsewhere. G:C base pairs inhibited methylation, and cytosines 5′ of ApT dinucleotides were particularly inhibitory. Weak signals could be strengthened by extending their lengths. A:T tracts as short as two were found to cooperate to induce methylation. Distamycin, which, like the AT-hook DNA binding motif found in proteins such as mammalian HMG-I, binds to the minor groove of A:T-rich sequences, suppressed DNA methylation and gene silencing. We also found a correlation between the strength of methylation signals and their binding to an AT-hook protein (HMG-I) and to activities in a Neurospora extract. We propose that de novo DNA methylation in Neurospora cells is triggered by cooperative recognition of the minor groove of multiple short A:T tracts. Similarities between sequences subjected to repeat-induced point mutation in Neurospora crassa and A:T-rich repeated sequences in heterochromatin in other organisms suggest that related mechanisms control silent chromatin in fungi, plants, and animals.
DNA methylation represents the most obvious and most studied epigenetic process. Nevertheless, remarkably little is known about what controls its specificity. In mammals, approximately 75% of CpG dinucleotides are typically methylated (49). Certain sequences, such as the highly repeated Alu, L1, and retroviral elements, seem to “attract” methylation; others, such as most CpG islands seem virtually immune to methylation or show characteristic methylation states that vary depending on the tissue, the developmental stage, or the parent of the origin of the sequence (3, 4, 54, 57, 58). DNA methylation in mammals presumably results from the interplay of numerous factors, including several DNA methyltransferases, specific DNA-binding proteins, such as the transcription factor SP1 (4, 5, 14, 22), and other factors that are poorly characterized (34).
The situation is much simpler in Neurospora crassa, making this organism an attractive model to investigate control of DNA methylation. One DNA methyltransferase, encoded by the dim-2 (defective in methylation) gene (21), is responsible for all known methylation in the organism. Approximately 1.5% of the cytosines in Neurospora DNA are methylated in vegetative cells (11, 36). Methylation is not limited to CpG dinucleotides or other symmetrical sites (43). Most of the Neurospora genome is unmethylated, but DNA methylation is found in ribosomal DNA and in approximately gene-sized sequences that show signs of having been exposed to repeat-induced point mutation (RIP), a genetic process that operates in the stage between fertilization and karyogamy in the sexual phase of the life cycle (39, 41, 44). RIP detects repeated DNA such as transposons in a pairwise manner and riddles them with G:C to A:T transition mutations (7, 12). Sequences mutated by RIP are typically found methylated in vegetative cells (8, 39, 50), and most such methylated sequences function as portable methylation “signals,” i.e., when introduced in an unmethylated form into the genome by transformation, they trigger methylation of themselves and flanking sequences (24, 25, 46, 48). In a study of a series of am alleles generated by RIP, all alleles that had at least an ≈3% change in sequence (51% G+C compared with 54% G+C in the wild-type sequence) served as signals for de novo methylation (50). Although required for initiating RIP, repeats are not required to trigger DNA methylation in vegetative cells (24, 38, 42).
What causes certain sequences, such as products of RIP, to trigger DNA methylation? In principle, methylation of a chromosomal segment may reflect the presence of a positive signal that induces methylation, the absence of a signal that prevents methylation, or both. To investigate these possibilities, Miao and colleagues dissected a 1.6-kb relic of RIP, the zeta-eta (ζ−η) region (15), that directs de novo methylation at a variety of chromosomal locations (24). Different nonoverlapping segments of the region, including fragments as short as 171 bp, triggered methylation of the DNA and some surrounding sequences, suggesting that RIP had created positive-acting signals. Considering that a variety of sequences mutated by RIP trigger DNA methylation, one would not expect to identify a single critical DNA sequence. Indeed, no oligonucleotide consensus sequence could be identified by comparison of methylated and unmethylated alleles of N. crassa sequences (24, 50). Thus, RIP-generated methylation signals are probably degenerate and/or discontinuous.
Because RIP causes exclusively G:C to A:T changes and has a marked preference for 5′CpA/5′TpG dinucleotide pairs, generating 5′TpA/5′TpA dinucleotide pairs (which are underrepresented in all organisms that have been examined [6]), an attractive possibility was that methylation is signaled by A+T-rich and/or TpA-rich sequences. To test this, two similar 226-bp DNA fragments were tested: an A:T-rich fragment with an A:T content equivalent to that of η but with the low TpA density found in sequences not exposed to RIP, such as the pristine similar sequence θ, and a TpA-rich fragment, with a high TpA density, like η, but with a normal A:T content, as in θ. Curiously, both the A:T-rich and TpA-rich fragments induced methylation, albeit not as efficiently as the original η sequence (24). These results suggested that both A:T richness and high densities of TpA dinucleotides promote de novo methylation but that neither is an essential feature of de novo methylation signals.
In order to systematically investigate how certain sequences trigger DNA methylation, we developed a sensitive in vivo assay system to test short sequences, e.g., synthetic oligonucleotides. The system was built on our observation of the additivity of methylation signals in Neurospora cells and on our finding of reproducible de novo methylation of sequences that are targeted, in single copy, to a given chromosomal site (24, 25). Our system involves placing test sequences in a defined site flanked by sequences with a low number of mutations caused by RIP. Using this assay, we conducted systematic tests of various simple sequences to induce de novo methylation. We demonstrated that establishment of cytosine methylation in N. crassa is not sequence specific, is favored by the presence of both A and T residues on the same strand, and is inhibited by G:C base pairs. To explore the possibility that methylation of such sequences may be mediated by an AT-hook type protein, we tested binding of HMG-I to signal sequences and tested the effect of the AT-hook analogue distamycin on DNA methylation. The results support the hypothesis that methylation signals are read by proteins that interact with the minor groove of A:T rich sequences.
MATERIALS AND METHODS
Construction of his-3 targeting vectors carrying ζ−η segments imbedded in am+ or amRIP4 sequences.
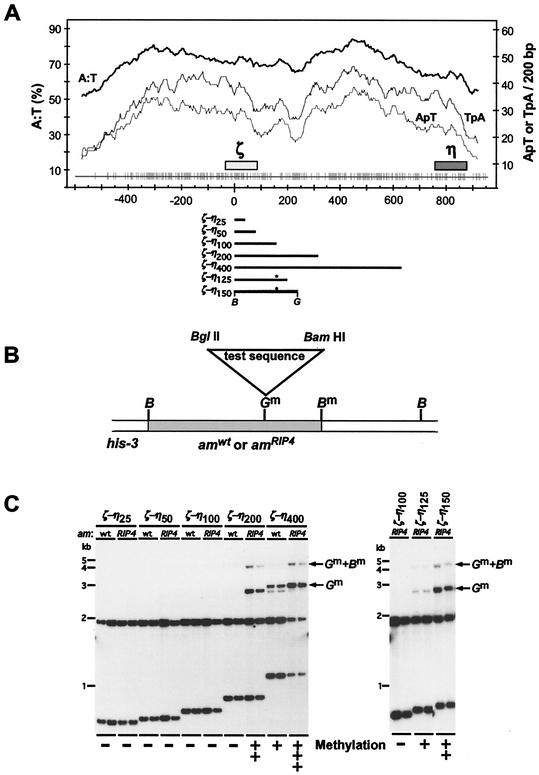
Specialized targeting vectors were constructed by modification of plasmids pBM62 and pBM63 (B. Margolin and E. U. Selker, unpublished data). These plasmids consist of the his-3 targeting vector pBM61 (23) with an inserted 2.6-kb BamHI fragment containing the am (glutamate dehydrogenase) gene from am+ and amRIP4 (50) strains, respectively. Fragments of the ζ−η region containing a BamHI site were amplified by PCR from pMR27 (obtained from M. Rountree and E. U. Selker) with an SP6 primer (5′-CGATTTAGGTGACACTATAG-3′) and ζ−η-specific primer 01 (5′-GAAGATCTCTTATTTATAGGAAAATGGA-3′), 02 (5′-GAAGATCTTAACTACTAGTTAAGTAATT-3′), 03 (5′-GAAGATCTAAGTAGAAAAAAAATATACA-3′), 04 (5′-GAAGATCTTAGGGAGAGACCGGATACTT-3′), or 05 (5′-GAAGATCTTTTCTACTATTATACAATAT-3′), and digested with BamHI and BglII to generate ζ−η25, ζ−η50, ζ−η100, ζ−η200, and ζ−η400, respectively (Fig. 1A).
FIG. 1.
Development of a sensitive assay system to test the capacity of various oligonucleotides to signal de novo methylation. A series of different-size segments of the methylated ζ−η region were tested as signals for DNA methylation when inserted into am+ or amRIP4 sequences and targeted to the his-3 locus. (A) Base composition, dinucleotide frequencies, and distribution of mutations from RIP in the ζ−η region (adapted from reference 24). The percentages of A:T base pairs and total TpA or ApT dinucleotides on one DNA strand were tabulated in 200-nucleotide windows, shifted in increments of 3 bp, along the ζ−η region with the program Window (Genetics Computer Group). The results are plotted above a graph of the distribution of the G:C to A:T mutations in the ζ−η region inferred by comparison of sequences for this region and its unique, unmethylated homologue, θ (15). The ζ and η 5S rRNA pseudogenes (47) and the ζ−η region fragments tested in this study are indicated by the solid bars. Asterisks (∗) indicate the position of an extra BamHI/BglII hybrid site (not in the native ζ−η region) resulting from the mode of construction. (B) System to test the capacity of various short oligonucleotides to trigger de novo methylation. Test segments with BamHI (B) and BglII (G) sites at their termini were made with synthetic oligonucleotides as described elsewhere and cloned into the BglII site of a his-3 targeting vector such as pHT6 (Table 1), which includes a 100-bp ζ−η segment inserted in amRIP4 sequences. The chimeric am/ζ−η sequences were targeted to the his-3 locus by flanking the am/test segment DNA with sequences in and downstream of his-3 (23). The sites that became methylated with some inserts are indicated by m superscripts. (C) Southern analysis. Genomic DNA (0.5 μg) samples from two independently isolated transformants with the test sequences correctly integrated at his-3 were digested with the methylation-sensitive endonucleases BamHI and BglII, separated on a 1% agarose gel, blotted, and hybridized with the 2.6-kb BamHI am fragment. The left panel shows tests of 25-, 50, 100-, 200-, and 400-bp segments of the ζ−η region, and the right panel shows results of adding 25 or 50 bp of ζ−η DNA to the 100-bp construct. The size of each ζ−η segment tested and the nature of the flanking sequences (am+ or amRIP4) are indicated above the autoradiogram. The 1.9-kb am band and 0.7- to 1.1-kb am/ζ−η bands result when both sites are unmethylated. Arrows indicate bands that result from methylation at one or both sites.
The BamHI-BglII ζ−η segments were gel purified and cloned in the BglII sites of pBM62 and pBM63. A set of five hybrid plasmids having 25, 50, 100, 200, or 400 bp of ζ−η sequences in pBM62, in the same orientation, were identified and designated pHTζ−η25wt, pHTζ−η50wt, pHTζ−η100wt, pHTζ−η200wt, and pHTζ−η400wt, respectively. Similarly, a set of derivatives of pBM63 were constructed and designated pHTζ−η25RIP4, pHTζ−η50RIP4, pHTζ−η100RIP4, pHTζ−η200RIP4, and pHTζ−η400RIP4.
Plasmid pHTζ−η125RIP4 was constructed by cloning a BamHI-BglII fragment of annealed oligonucleotides 06 (5′-AGGGATCCTATATATTATATTTTTATATCGTTTAGATCTCT-3′) and 07 (5′-AGAGATCTAAACGATATAAAAATATAATATATAGGATCCCT-3′) into the BglII site of pHTζ−η100RIP4. Plasmid pHTζ−η150-RIP4 was constructed by cloning a BamHI-BglII fragment of the PCR product amplified from pMR27 with primers 08 (5′-GAGGATCCTATATATTATATTTTTATATCGTTTAA-3′) and 09 (5′-GAAGATCTAAAAAAGGGTTAAGTACGCGAAT-3′) into the BglII site of pHTζ−η100RIP4. BamHI and BglII sites in the oligonucleotide sequences are italicized. Escherichia coli transformants carrying the cloned inserts were screened by PCR with am-specific primers 047 (5′-TCCGTCAACCTTTCCATTCT-3′) and 400 (5′-CCAGTAAGGACACCCTCG-3′) to synthesize fragments spanning the inserts.
Construction of his-3 targeting plasmids carrying synthetic test sequences in the amRIP4::ζ−η100 hybrid construct.
All synthetic oligonucleotides used in this study had 20- to 100-bp test sequences plus common terminal sequences with a BamHI site on one end and a BglII site on the other end (Fig. 1B). Each oligonucleotide was incubated in 20 μl of T7 Sequenase buffer U.S. Biochemicals [USB]) containing 100 μM primer 10 (5′-CGCGGAAGATCT-3′) and 10 mM deoxynucleoside triphosphates at 94°C for 1 s, and then the temperature was reduced 1°C/30 s to 27°C. The annealing product was incubated with 1 to 5 U of T7 Sequenase (USB) at 27°C for 1 s, and the temperature was increased 1°C/13 s to 50°C. The resulting duplex DNA was digested with BamHI and BglII and cloned into the BglII site of pHTζ−η100RIP4. Cloned test sequences were checked by sequencing with primer 047. With the exception of the (GATC)n 100-mer, all cloned test sequences were inserted in the same orientation. Plasmids carrying 100-mer test sequences were obtained by screening for plasmids with two head-to-tail copies of the corresponding 50-mer inserts.
Targeting test constructs to his-3 in N. crassa.
All constructs were targeted to the his-3 locus of N. crassa strain N1445 [mata his-3 (1-234-723); inl am132) as previously described (23, 24). Conidia were harvested from cultures incubated for 5 days at 32°C and then 5 days at 25°C on Vogel's agar medium (55) containing 1.5% sucrose and supplemented with alanine, inositol, and histidine. Plasmids for his-3 targeting were linearized with NdeI and introduced into competent conidia by electroporation (23).
DNA preparation and methylation analysis.
Neurospora strains were grown from conidia (≈107 conidia/ml) at 32°C for 2 days with shaking in 5 ml of Vogel's minimal liquid medium supplemented with 1.5% sucrose, inositol, and alanine. Genomic DNA was prepared from mycelia as described previously (24), and samples (0.5 μg) were digested with the cytosine methylation-sensitive restriction endonucleases BamHI and BglII (8 to 10 U of each) and analyzed by Southern hybridization with the 2.6-kb BamHI am gene fragment as the probe (24). Note that hybridization signals can only come from transformed nuclei because the host strain N1445 has a deletion of the entire am gene and the presence of untransformed nuclei in heterokaryons does not affect de novo methylation (24). Blots were stripped (37) and reprobed for the unmethylated Neurospora cpc-1 gene in pMO31 (29) to verify that digestions were complete. Methylation levels were quantified with a Molecular Dynamics Storm 860 Phosphorimager system with ImageQuant software. The methylation level was calculated by comparing the fraction of signal in the bands resulting from methylation with the total signal in all bands. At least two independent his-3-targeted transformants were analyzed for each construct, and each transformant was tested in at least two independent Southern hybridizations. As found in a related previous study (24), no significant variation in methylation was detected. Table 1 summarizes the sequences and methylation status of most of the fragments tested.
TABLE 1.
Induction of DNA methylation in N. crassa by synthetic DNA fragmentsa
| Plasmid designation | N. crassa strain(s) with targeted sequence(s) | Length tested (bp) | Insert sequence | Repeat unit or feature | Methylationb (%)
|
||
|---|---|---|---|---|---|---|---|
| Mean | SD | ||||||
| pHT1 | N2301 | 26 | AATAAATAAATAAATTTTTTAAAATA | Random, 100% A:T | 37.0 | 1.8 | |
| TTATTTATTTATTTAAAAAATTTTAT | |||||||
| pHT2 | N2302 | 25 | TTTTTTAATTTTATTTTAAATTTTT | Random, 100% A:T | 6.6 | 1.2 | |
| AAAAAATTAAAATAAAATTTAAAAA | |||||||
| pHT3 | N2303 | 24 | AATTTTTTATTTATTATATAATTT | Random, 100% A:T | 3.2 | 1.3 | |
| TTAAAAAATAAATAATATATTAAA | |||||||
| pHT4 | N2304 | 23 | TAAAAATTAAAATAATTAAAAAA | Random, 100% A:T | 6.8 | 1.1 | |
| ATTTTTAATTTTATTAATTTTTT | |||||||
| pHT5 | N2305 | 25 | ATTTTAAAAAAAATATATTAATAAA | Random, 100% A:T | 14.5 | 0.1 | |
| TAAAATTTTTTTTATATAATTATTT | |||||||
| pHT6 | N2306 | 24 | ATTATATATTTTATTAATTTTTTT | Random, 100% A:T | 6.3 | 0.9 | |
| TAATATATAAAATAATTAAAAAAA | |||||||
| pHT7 | N2307 | 25 | TTATTATTTAAATTTTAAATAAATT | Random, 100% A:T | 15.2 | 2.8 | |
| AATAATAAATTTAAAATTTATTTAA | |||||||
| pHT8 | N2308 | 25 | TAAATTTAATAAATTTATTTTTAAT | Random, 100% A:T | 27.0 | 8.1 | |
| ATTTAAATTATTTAAATAAAAATTA | |||||||
| pHT9 | N2309 | 23 | TAAAAATTAAAATAATTAAAAAA | Random, 100% A:T | 9.0 | 2.9 | |
| ATTTTTAATTTTATTAATTTTTT | |||||||
| pHT10 | N2310 | 25 | TTTATTTAAATTTATAATAATTATA | Random, 100% A:T | 14.7 | 0.7 | |
| AAATAAATTTAAATATTATTAATAT | |||||||
| pHT11 | N2311 | 25 | TATTATAATATTTATAATTATTTTT | Random, 100% A:T | 8.7 | 1.1 | |
| ATAATATTATAAATATTAATAAAAA | |||||||
| pHT12 | N2312 | 25 | TATTATAATATTTATAATTATTTTT | Random, 100% A:T | 5.7 | 0.2 | |
| ATAATATTATAAATATTAATAAAAA | |||||||
| pHT13 | N2313 | 25 | AAATAATAAAAAAATATTTTTTTAA | Random, 100% A:T | 9.0 | 1.5 | |
| TTTATTATTTTTTTATAAAAAAATT | |||||||
| pHT14 | N2314 | 25 | TAAAATATTATTTATATTAATAAAA | Random, 100% A:T | 7.1 | 0.7 | |
| ATTTTATAATAAATATAATTATTTT | |||||||
| pHT15 | N2315 | 25 | AATAAAAAATAAATAATTATAATAA | Random, 100% A:T | 12.1 | 3.8 | |
| TTATTTTTTATTTATTAATATTATT | |||||||
| pHT16 | N2316 | 25 | AAAATTTTAAATATATAATAATAAT | Random, 100% A:T | 16.2 | 5.7 | |
| TTTTAAAATTTATATATTATTATTA | |||||||
| pHT17 | N2317 | 25 | AATAAAAATATTTTTATTTATAAAA | Random, 100% A:T | 7.4 | 2.8 | |
| TTATTTTTATAAAAATAAATATTTT | |||||||
| pHT18 | N2318 | 25 | TAATATATATAAAAAATATAAATTA | Random, 100% A:T | 6.1 | 2.3 | |
| ATTATATATATTTTTTATATTTAAT | |||||||
| pHT19 | N2319 | 25 | TATAAATTTATTTATATAATTAAAA | Random, 100% A:T | 12.2 | 0.6 | |
| ATATTTAAATAAATATATTAATTTT | |||||||
| pHT20 | N2320 | 25 | GTAGGACTTACACCACCTGCGGGTC | Random, 50% G:C | 0.0 | ||
| CATCCTGAATGTGGTGGACGCCCAG | |||||||
| pHT21 | N2321 | 23 | GGACGACCCGATACCACGCGACA | Random, 50% G:C | 0.0 | ||
| CCTGCTGGGCTATGGTGCGCTGT | |||||||
| pHT22 | N2322 | 25 | CGCAGTTGCCCCAAATAGGAGGGAT | Random, 50% G:C | 0.0 | ||
| GCGTCAACGGGGTTTATCCTCCCTA | |||||||
| pHT23 | N2323 | 25 | CACCAGCATCATGGTGCGCCTCAAG | Random, 50% G:C | 0.0 | ||
| GTGGTCGTAGTACCACGCGGAGTTC | |||||||
| pHT24 | N2324 | 24 | CGGTGACGGTGAACAAGCTCAACA | Random, 50% G:C | 0.0 | ||
| GCCACTGCCACTTGTTCGAGTTGT | |||||||
| pHT25 | N2325 | 25 | GACTCGTGCCAAGTCTTCAAGGTCC | Random, 50% G:C | 0.0 | ||
| CTGAGCACGGTTCAGAAGTTCCAGG | |||||||
| pHT26 | N2326 | 21 | TAGCGGAGATCACCCCCAATC | Random, 50% G:C | 0.0 | ||
| ATCGCCTCTAGTGGGGGTTAG | |||||||
| pHT27 | N2327 | 25 | GGTAGCCACGGGCCTTGCCAAGCTT | Random, 50% G:C | 0.0 | ||
| CCATCGGTGCCCGGAACGGTTCGAA | |||||||
| pHT28 | N2328 | 25 | TATCCAGACGGACACACGGCAACTT | Random, 50% G:C | 0.0 | ||
| ATAGGTCTGCCTGTGTGCCGTTGAA | |||||||
| pHT29 | N2329 | 24 | TTTCGCTCTGCGCGGCTCATAAAT | Random, 50% G:C | 0.0 | ||
| AAAGCGAGACGCGCCGAGTATTTA | |||||||
| pHT30 | N2330 | 25 | TGGGCCGGCGAAGTTGGTAGAGGGT | Random, 50% G:C | 0.0 | ||
| ACCCGGCCGCTTCAACCATCTCCCA | |||||||
| pHT31 | N2331 | 25 | CTGCACACGTCATAATCCTGGGAGG | Random, 50% G:C | 0.0 | ||
| GACGTGTGCAGTATTAGGACCCTCC | |||||||
| pHT32 | N2332 | 25 | AAAAAAAAAAAAAAAAAAAAAAAAA | A | 2.1 | 0.6 | |
| N2333 | TTTTTTTTTTTTTTTTTTTTTTTTT | T | |||||
| pHT33 | N2334 | 25 | AATAAAAAATAAAAAATAAAAAATA | TAAAAAA | 10.2 | 0.7 | |
| N2335 | TTATTTTTTATTTTTTATTTTTTAT | ATTTTTT | |||||
| pHT34 | N2336 | 25 | AATAAAAATAAAAATAAAAATAAAA | TAAAAA | 10.4 | 1.3 | |
| N2337 | TTATTTTTATTTTTATTTTTATTTT | ATTTTT | |||||
| pHT35 | N2338 | 25 | AATAAAATAAAATAAAATAAAATAA | TAAAA | 12.8 | 1.6 | |
| N2339 | TTATTTTATTTTATTTTATTTTATT | ATTTT | |||||
| pHT36 | N2340 | 25 | AATAAATAAATAAATAAATAAATAA | TAAA | 29.7 | 4.3 | |
| N2341 | TTATTTATTTATTTATTTATTTATT | ATTT | /PICK> | ||||
| Plasmid designation | N. crassa strain(s) with targeted sequence(s) | Length tested (bp) | Insert sequence | Repeat unit or feature | Methylationb (%)
|
||
| Mean | SD | ||||||
| pHT37 | N2342 | 25 | AATTAATTAATTAATTAATTAATTA | AATT | 24.9 | 1.8 | |
| N2343 | TTAATTAATTAATTAATTAATTAAT | TTAA | |||||
| pHT38 | N2344 | 25 | AATAATAATAATAATAATAATAATA | TAA | 10.8 | 1.0 | |
| N2345 | TTATTATTATTATTATTATTATTAT | ATT | |||||
| pHT39 | N2346 | 25 | AATATATATATATATATATATATAT | TA | 5.8 | 2.2 | |
| N2347 | TTATATATATATATATATATATATATATA | AT | |||||
| pHT40 | N2348 | 20 | TAAATAAATAAATAAATAAA | TAAA | 11.4 | 5.2 | |
| N2349 | ATTTATTTATTTATTTATTT | ATTT | |||||
| pHT41 | N2350 | 40 | TAAATAAATAAATAAATAAA... | TAAA | 74.4 | 5.9 | |
| N2351 | ATTTATTTATTTATTTATTT... | ATTT | |||||
| pHT42 | N2352 | 80 | TAAATAAATAAATAAATAAATAAAT... | TAAA | 81.4 | 1.3 | |
| N2353 | ATTTATTTATTTATTTATTTATTTA... | ATTT | |||||
| pHT43 | N2354 | 25 | TAAAATAAAATAAAATAAAATAAAA | TAAAA | 11.6 | 0.2 | |
| N2355 | ATTTTATTTTTATTTTATTTTTATTTT | ATTTT | |||||
| pHT44 | N2356 | 25 | CTAAACTAAACTAAACTAAACTAAA | CTAAA | 1.5 | 0.2 | |
| N2357 | GATTTGATTTGATTTGATTTGATTT | GATTT | |||||
| pHT45 | N2358 | 25 | CAAATCAAATCAAATCAAATCAAAT | TCAAA | 1.2 | 0.1 | |
| N2359 | GTTTAGTTTAGTTTAGTTTAGTTTA | AGTTT | |||||
| pHT46 | N2360 | 25 | CAATACAATACAATACAATACAATA | TACAA | 0.5 | 0.1 | |
| N2361 | GTTATGTTATGTTATGTTATGTTAT | ATGTT | |||||
| pHT47 | N2362 | 25 | CATAACATAACATAACATAACATAA | TAACA | 0.3 | 0.2 | |
| N2363 | GTATTGTATTGTATTGTATTGTATT | ATTGT | |||||
| pHT48 | N2364 | 100 | CAAAACAAAACAAAACAAAACAAAA... | CAAAA | 4.2 | 1.3 | |
| N2365 | GTTTTGTTTTGTTTTGTTTTGTTTT... | GTTTT | |||||
| pHT49 | N2366 | 100 | CTAAACTAAACTAAACTAAACTAAA... | CTAAA | 63.7 | 6.6 | |
| N2367 | GATTTGATTTGATTTGATTTGATTT... | GATTT | |||||
| pHT50 | N2368 | 100 | CAAATCAAATCAAATCAAATCAAAT... | TCAAA | 86.2 | 6.2 | |
| N2369 | GTTAGTTTAGTTTAGTTTAGTTTA... | AGTTT | |||||
| pHT51 | N2370 | 100 | GAAATGAAATGAAATGAAATGAAAT... | TGAAA | 8.0 | 1.2 | |
| N2371 | CTTTACTTTACTTTACTTTACTTTA... | ACTTT | |||||
| pHT52 | N2372 | 100 | CAATACAATACAATACAATACAATA... | TACAA | 34.8 | 8.3 | |
| N2373 | GTTATGTTATGTTATGTTATGTTAT... | ATGTT | |||||
| pHT53 | N2374 | 100 | CATAACATAACATAACATAACATAA... | TAACA | 5.2 | 0.3 | |
| N2375 | GTATTGTATTGTATTGTATTGTATT... | ATTGT | |||||
| pHT54 | N2376 | 100 | CTAATCTAATCTAATCTAATCTAAT... | CTAAT | 24.4 | 10.5 | |
| N2377 | GATTAGATTAGATTAGATTAGATTA... | GATTA | |||||
| pHT55 | N2378 | 100 | CTAAAACTAAAACTAAAACTAAAAA... | CTAAAA | 74.7 | 1.3 | |
| N2379 | GATTTTGATTTTGATTTTGATTTTT... | GATTTT | |||||
| pHT56 | N2380 | 100 | TCAAAATCAAAATCAAAATCAAAAT... | TCAAAA | 70.6 | 4.9 | |
| N2381 | AGTTTTAGTTTTAGTTTTAGTTTTA... | AGTTTT | |||||
| pHT57 | N2382 | 100 | CAAACAAACAAACAAACAAACAAAC... | CAAA | 1.33 | 0.7 | |
| N2383 | GTTTGTTTGTTTGTTTGTTTGTTTG... | GTTT | |||||
| pHT58 | N2384 | 100 | CTAACTAACTAACTAACTAACTAAC... | CTAA | 22.2 | 3.4 | |
| N2385 | GATTGATTGATTGATTGATTGATTG... | GATT | |||||
| pHT59 | N2386 | 100 | CAATCAATCAATCAATCAATCAATC... | TCAA | 45.3 | 6.7 | |
| N2387 | GTTAGTTAGTTAGTTAGTTAGTTAG... | AGTT | |||||
| pHT60 | N2388 | 100 | CTTACTTACTTACTTACTTACTTAC... | GTAA | 10.9 | 1.5 | |
| N2389 | GAATGAATGAATGAATGAATGAATG... | CATT | |||||
| pHT61 | N2390 | 100 | CATTCATTCATTCATTCATTCATTC... | TGAA | 0.7 | 0.4 | |
| N2391 | GTAAGTAAGTAAGTAAGTAAGTAAG... | ACTT | |||||
| pHT62 | N2392 | 100 | CTATCTATCTATCTATCTATCTATC... | CTAT | 9.1 | 1.6 | |
| N2393 | GATAGATAGATAGATAGATAGATAG... | GATA | |||||
| pHT63 | N2394 | 100 | CAACAACAACAACAACAACAACAAC... | CAA | 0.0 | ||
| N2395 | GTTGTTGTTGTTGTTGTTGTTGTTG... | GTT | |||||
| pHT64 | N2396 | 100 | CTTCTTCTTCTTCTTCTTCTTCTTC... | GAA | 0.2 | 0.1 | |
| N2397 | GAAGAAGAAGAAGAAGAAGAAGAAG... | CTT | |||||
| pHT65 | N2398 | 100 | CTACTACTACTACTACTACTACTAC... | CTA | 32.3 | 6.0 | |
| N2399 | GATGATGATGATGATGATGATGATG... | GAT | |||||
| pHT66 | N2400 | 100 | CATCATCATCATCATCATCATCATC... | CAT | 0.0 | ||
| N2401 | GTAGTAGTAGTAGTAGTAGTAGTAG... | GTA | |||||
| pHT67 | N2402 | 100 | GATCGATCGATCGATCGATCGATCG... | GATC | 5.7 | 0.8 | |
| N2403 | CTAGCTAGCTAGCTAGCTAGCTAGC... | CTAG | |||||
| pHT68 | N2404 | 100 | CATGCATGCATGCATGCATGCATGC... | CATG | 0.0 | ||
| N2405 | GTACGTACGTACGTACGTACGTACG... | GTAC | |||||
| pHT69 | N2406 | 100 | CTAGCTAGCTAGCTAGCTAGCTAGC... | CTAG | 1.7 | 0.4 | |
| N2407 | GATCGATCGATCGATCGATCGATCG... | GATC | |||||
Sequences tested are presented 5′ to 3′ on the top strand and 3′ to 5′ on the bottom strand. For sequences longer than 25 bp, three dots indicate that part of the test sequence is not shown. Sequences that were synthesized by incorporating a nucleotide at random from a 1:1 mixture of dATP and dTTP or from a 1:1:1:1 mixture of dGTP, dCTP, dATP, and dTTP are designated random 100% A:T and random 50% A:T, respectively.
Means and standard deviations are shown. In the case of plasmids pHT1 to pHT31, these values were based on two or more independent Southern hybridizations from each of two independent cultures of the corresponding targeted transformant. All others were based on two or more independent targeted transformants (two cultures each). 0.0, no methylation detected.
Test for reactivation of silenced hph with distamycin A.
About 2,000 conidia of strain N644 (16) were mixed with 5 ml of molten Vogel's sorbose medium containing 1.5% agar, 0.05% fructose, 0.05% glucose, 1 mg of alanine per ml, and 50 μg of inositol per ml and poured into a 4-cm-diameter petri dish. A 4-mm-diameter paper disk containing 400 μg of distamycin A (applied as a 4-mg/ml solution in ethanol) was then placed on the solidified bottom agar. After incubation at 32°C for 16 h, hygromycin B (2.5 mg) was overlaid in 5 ml of the same but with 0.75% agar and incubated for 5 days. Control plates contained only hygromycin B or distamycin A.
Gel mobility shift assays.
Duplex DNA was generated from the synthetic oligonucleotides as described above except that the Sequenase buffer (20 μl) contained 100 ng of oligonucleotide, 1 μM primer 10 (described above), 250 μM each dATP, dTTP, and dGTP, and 50 μCi of [α-32P]dCTP (3,000 Ci/mmol; Perkin-Elmer). The radiolabeled duplex DNA was separated in an 8% polyacrylamide gel in 2× TBE (22a) buffer. A gel slice containing the DNA was isolated, crushed, and incubated in 1 ml of elution buffer containing 0.1% sodium dodecyl sulfate, 1 mM EDTA, 0.5 M ammonium acetate, and 10 mM magnesium acetate at 37°C for 12 h. The eluted DNA was precipitated with ethanol, suspended in TE buffer, and used as a probe for mobility shift assays. Then 1 to 2 μg of purified human recombinant HMG-I (kindly provided by R. Reeves, Washington State University) or ≈1 μg of a Neurospora protein fraction (kindly provided by G. Kothe) was incubated with ≈0.2 ng of probe (≈103 cpm) on ice for 15 min in 15 μl of binding buffer [10 mM HEPES (pH 7.9), 50 mM KCl, 1 mM EDTA, 5 mM dithiothreitol, 1 μg of poly(dI-dC) (Pharmacia), 6.7% glycerol], and fractionated in a 6% polyacrylamide gel in 0.25× TBE buffer at 150 V. The gel was dried and autoradiographed. Each probe was assayed independently at least twice.
RESULTS
Generation of de novo methylation signal by combination of DNA segments that individually do not trigger methylation.
Variation in copy number, chromosomal position, and nature of the integration event can influence whether or not or to what degree a particular fragment of transforming DNA will be methylated. It is therefore important to test the methylation potential of single copies of sequences integrated precisely and without extraneous sequences at a common chromosomal position (23-25). Prior to this study, the shortest DNA fragment known to trigger DNA methylation was a natural fragment of 177 bp, still large enough to contain much information.
To improve our understanding of how some sequences trigger de novo methylation, we developed a system capable of testing the effect of simple features in short DNA sequences. Our system was built on two observations: the capacity of a sequence to trigger DNA methylation depends in part on chromosomal position (25, 45, 46), and a strong methylation signal can be generated by juxtaposition of two DNA fragments that each act as weak signals (24). These observations suggested to us that it might be possible to construct an assay site that is exquisitely sensitive to sequences that promote methylation. To explore this possibility, we tested whether a sequence that has been subjected to RIP but is incapable of triggering de novo methylation at our normal test sites (presumably because it was lightly mutated) would promote methylation of segments inserted into it. We chose amRIP4, which is an allele of the am (glutamate dehydrogenase) gene with 56 mutations caused by RIP scattered over 2.6 kb (50). The overall base composition of amRIP4 is 52% G+C, compared with 54% G+C for the am+ allele. We built vectors that included amRIP4 or am+ sequences flanked by his-3 and associated sequences that allow precise targeting to the his-3 locus and then constructed a set of derivative plasmids, each containing one of a nested set of 25- to 400-bp fragments of the ζ−η region inserted at a BglII site in the am sequences (Fig. 1A and B).
Each construct was targeted to the his-3 locus of an N. crassa host strain (N1445) that carries a deletion of the am gene, and its methylation status was analyzed by Southern blot hybridization with two 5′-methylcytosine-sensitive restriction endonucleases, BglII and BamHI (Fig. 1C). In every case, at least two independent transformants were tested, and the duplicate isolates gave identical results. Table 1 summarizes the results of our studies. Based on results from previous studies with DNA from the ζ−η region, we expected that the 400-bp ζ−η fragment but not the 25-, 50-, 100-, or 200-bp subfragments would trigger methylation in the am+ sequence targeted to his-3. As expected, no methylation was detected with the constructs containing the 25-, 50-, or 100-bp ζ−η segments embedded in either am+ or amRIP sequences, whereas the 400-bp fragment (ζ−η400) induced methylation when present in either am+ or amRIP sequences.
Methylation was detected principally at the BglII site where the ζ−η segment was inserted and to a lesser extent at the BamHI site 700 bp from the insert (Fig. 1B). Interestingly, the 400-bp fragment caused greater methylation in amRIP4 than in am+ sequences, i.e., a larger fraction of the molecules showed methylation at the sites examined and methylation spread further from the insert. Most importantly, the 200-bp segment (ζ−η200) triggered de novo methylation only in amRIP4. This indicated that a combination of DNA fragments that do not individually trigger DNA methylation can generate a signal for de novo methylation.
Sensitive system to test potential methylation signals in vivo.
To test whether the amRIP4::ζ−η100 chimera could be used to assay short fragments for their potential to promote DNA methylation, we inserted an additional 25 or 50 bp of ζ−η sequences into this construct and determined whether the hybrid sequence would induce methylation at his-3. Both the ζ−η125 and ζ−η150 constructs triggered methylation (Fig. 1C), supporting our idea. Results of previous studies suggested that both high A:T content and high density of TpA dinucleotides can promote de novo methylation. Nevertheless, it remained possible that G:C pairs are required to trigger methylation. We used the his-3::amRIP4::ζ−η100 assay system to ask whether 25-bp oligonucleotides consisting of only A:T pairs could trigger DNA methylation of this mosaic sequence.
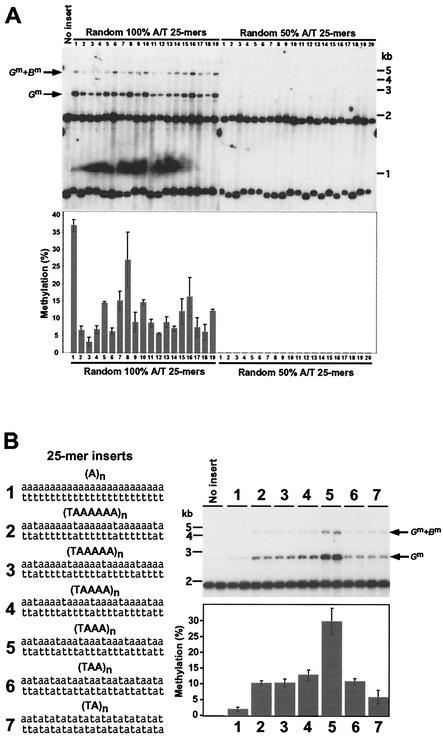
Plasmids with inserts of 25 bp of random sequences synthesized with either 100% A:T or 50% A:T-50% G:C were constructed. Nineteen independently isolated targeted transformants carrying 100% A:T random sequences at his-3 were tested. All showed methylation both at the BglII site where the 25-mers were inserted and at the more distant BamHI site (Fig. 2A). In contrast, none of 20 targeted transformants carrying random sequences synthesized from an even mix of all four deoxynucleotides (≈50% A:T) showed evidence of methylation (Fig. 2A). These observations confirmed that our assay system is sufficiently sensitive to test various synthetic oligonucleotide sequences and supported previous suggestions that high A:T content promotes de novo methylation (24).
FIG. 2.
Induction of DNA methylation by sequences consisting exclusively of A:T base pairs. (A) Capacity of random 100% A+T or 50% A+T 25-mer sequences to induce de novo methylation at the amRIP4::ζ−η100 test site. Nineteen independent his-3-targeted transformants carrying 100% A+T random 25-mers (left panel) and 20 independent his-3-targeted transformants (adjacent lanes) carrying ≈50% A+T random 25-mers (right panel) were analyzed by Southern hybridization as in Fig. 1. Results from an empty (no insert) amRIP4::ζ−η100 control construct are also shown. The bar graph shows quantification of methylation levels as described in Materials and Methods. (B) Role of alternating A and T residues in de novo methylation. The 100% A+T 5-mers with various frequencies of TpA's were tested for the capacity to induce methylation at the amRIP4::ζ−η100 test site. Two independently isolated his-3-targeted transformants were tested for each sequence (1 to 7), and the methylation was quantified as described for panel A. The smallest fragment from the am/ζ−η insert (0.7 kb; Fig. 1A) is not shown in the autoradiogram.
Various DNA sequences consisting exclusively of A:T base pairs can signal de novo cytosine methylation but to different extents.
Strikingly, every 100% A:T 25-mer sequence tested but none of the ≈50% A:T 25-mers tested triggered DNA methylation. Apparently, G:C base pairs are not required for promoting de novo cytosine methylation. The different 100% A:T 25-mers reproducibly induced different levels of methylation, ranging from <5% to >35% (Fig. 2A). It was not obvious what was responsible for the observed variation; no specific oligomeric sequence that might be essential for triggering de novo methylation was evident. It is noteworthy that some sequences that are rich in TpA dinucleotides were less potent than some sequences with fewer TpAs, contrary to expectations based on the fact that RIP preferentially increases the density of this dinucleotide.
As a first step to systematically testing the roles of various sequence elements in the induction of DNA methylation, we constructed and tested a set of 100% A:T 25-mer sequences with TpA (or ApT) dinucleotides separated by 0 to 6 A:T pairs and, as a control, a 25-bp A:T homopolymer (Fig. 2B). The A:T homopolymer induced only a trace of methylation (2.1%), whereas some 25-mers induced substantial methylation, verifying that A:T content is not the only factor determining whether a sequence will trigger methylation. A given strand apparently must contain both T's and A's to induce significant methylation. Surprisingly, (TAAA)n (sequence 5) induced the highest level of methylation (29.7%) in the series. The sequence with the highest number of TpA dinucleotides, (TA)n (sequence 7), induced substantially less methylation (5.8%) than the other five sequences with TpA's (sequences 2 to 6). Fifteen of the 16 possible 4-bp A/T sequences are found in one or more of the constructs described above. We tested the one remaining sequence, TTAA, and found that it too induced methylation; the (TTAA)n 25-mer induced 24.9% ± 1.8% methylation, a level exceeded only by (TAAA)n (Table 1). This firmly established that the density of TpA dinucleotides alone is not the critical determinant of DNA methylation.
G:C base pairs inhibit cytosine methylation.
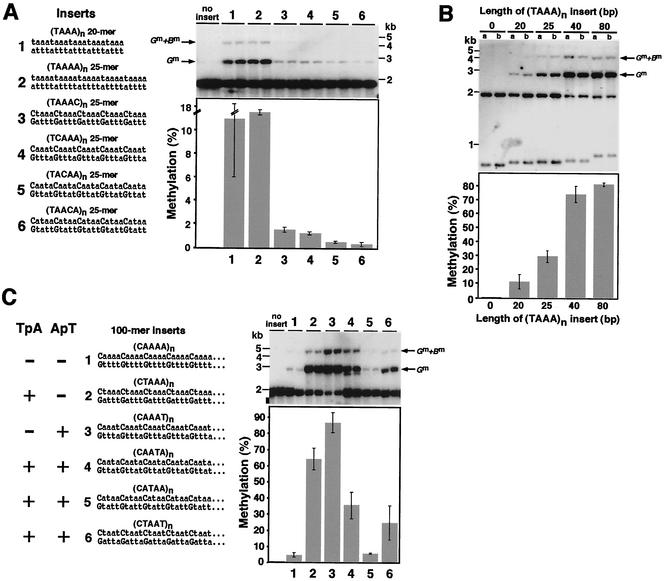
Our observation that various 25-mer 100% A:T sequences but not 50% A:T sequences triggered de novo methylation (Fig. 2A) could be interpreted in two ways: the presence of G:C base pairs directly inhibited signaling or indirectly inhibited de novo methylation by reducing the number of A:T base pairs that signal methylation. To distinguish between these possibilities, we systematically evaluated the effect of G:C base pairs on signaling de novo methylation. G:C pairs were inserted at all possible positions of the repeat sequence that was shown to induce de novo methylation most efficiently [(TAAA)n; Fig. 2B]. Naturally, insertion of G:C pairs into (TAAA)n increases the spacing between interspersed T's and A's from three to four. We knew that a 25-mer (TAAAA)n sequence induced a lower level of methylation than did a 25-mer (TAAA)n sequence (Fig. 2B), but it seemed possible that this was due to the lower number of TAAA repeats in the former sequence or to another factor that could be compensated for with a correspondingly longer sequence.
We therefore compared induction of methylation by a (TAAA)n 20-mer and a (TAAAA)n 25-mer, each of which contained five TpA dinucleotides and four ApT dinucleotides (Fig. 3A, sequences 1 and 2). They induced equivalent levels of methylation (11.4% and 11.6%, respectively), suggesting that increasing the spacing between the TpA's (or ApT's) by the insertion of single base pairs should not, in itself, drastically impair the function of the (TAAA)n methylation signal. Interestingly, all four of the constructs bearing G:C base pair insertions resulted in greatly reduced (7.6- to 38-fold) DNA methylation (Fig. 3A).
FIG.3.
Effect of G:C base pairs, signal length, and TpA and ApT dinucleotides on DNA methylation. (A) Effect of G:C pairs on signals for de novo methylation at the amRIP4::ζ−η100 test site. Two 100% A+T sequences that serve as potent signals for de novo methylation (sequences 1 and 2) were modified by addition or substitution of single G:C pairs at each of the four possible positions in the repeat unit, generating the other sequences tested (sequences 3 to 6). Induction of methylation was tested on two independent targeted transformants for each construct, and the results are presented as in previous figures. (B) De novo methylation induced by different-length (TAAA)n inserts at the amRIP4::ζ−η100 test site. Induction of methylation was tested on two independent targeted transformants for each construct, and the results are presented as in previous figures. (C) Neither TpA nor ApT dinucleotides are essential for induction of methylation. A set of related 100-mer test segments based on the best methylation signal identified, (TAAA)n (see Fig. 2B), were constructed and tested at the amRIP4::ζ−η100 test site. A representative 25-bp segment of each test sequence (sequences 1 to 6) is illustrated, and the results are presented as in previous figures. Note that sequence 2 has TpA's but no ApT's, sequence 3 has ApT's but no TpA's, and sequences 4, 5, and 6 have both.
Increase in signal length promotes de novo cytosine methylation.
Our finding that insertion of G:C base pairs at any of the four positions inhibited induction of methylation encouraged us to use G:C base pairs to separate the influences of individual factors in A:T base pairs upon signaling de novo methylation. The inhibitory effect of G:C base pairs was so strong, however, that we could not confidently measure the relative effects of interrupting the different positions in the (TAAA)n 25-mer. We therefore explored the possibility of amplifying all the methylation signals, including those interrupted by G:C base pairs, by using longer test sequences. To examine the effect of length, we tested 20-, 25-, 40-, and 80-mers of the (TAAA)n sequence targeted to his-3. The results indicated that, indeed, longer fragments induced higher levels of de novo methylation (Fig. 3B). The 25-, 40-, and 80-mer lengths gave 2.6-, 6.5-, and 7.1-fold higher methylation, respectively, than did the 20-mer. The relationship between signal length and level of de novo methylation was nonlinear; the level of methylation induced by the 40-mer (74%) was nearly as high as that induced by the 80-mer (81%). We concluded that use of longer DNA fragments should allow evaluation of methylation induced by weak signals containing G:C base pairs.
Neither TpA nor ApT dinucleotides are essential for signaling de novo cytosine methylation.
To separate the possible roles of TpA and ApT dinucleotides in the induction of DNA methylation, we tested 50- and 100-mer sequences with G:C base pairs inserted at various positions within the (TAAA)n signal sequence. In addition to the sequences tested as 25-mers (Fig. 3A), we also tested two related sequences, one with neither TpA nor ApT [sequence 1 in Fig. 3C; (CAAAA)n] and one with both TpA and ApT dinucleotides [sequence 6 in Fig. 3C; (CTAAT)n]. Sequences 2 to 5 can be regarded as simple variants of sequence 1, derived by inverting a single A:T base pair in the repeat unit. Sequence 2, (CTAAA)n, has TpA dinucleotides but not ApT dinucleotides, while sequence 3, (CAAAT)n, has ApT but not TpA dinucleotides. Sequences 4, (CAATA)n, 5, (CATAA)n, and 6, (CTAAT)n, have both TpA and ApT dinucleotides. Sequence 6 can be thought of as a “recombinant” between sequences 2 and 3.
As expected, greatly increased levels of methylation were induced by the 50-mer (data not shown) and 100-mer (sequences 2 to 5 in Fig. 3C) inserts compared to those induced by the 25-mers with the same sequences (sequences 3 to 6 in Fig. 3A). Nevertheless, in contrast to the situation with the original purely A:T sequence (Fig. 3B), 10 repeats (50 bp) of the G:C-containing sequences did not trigger a near-saturation level of methylation (data not shown). We therefore focused on the 100-mer transformants to assess the effect of G:C base pairs on de novo methylation and used the (TAAA)n 80-mer sequence as the reference standard. The inhibitory effect of G:C base pairs varied with their site of insertion in the original (TAAA)n sequence. Interestingly, the greatest inhibition was found with sequence 5 (CATAA)n, which retains both TpA and ApT dinucleotides. It gave only 5.2% methylation, which is comparable to that induced by sequence 1, (CAAAA)n, which has neither TpA nor ApT dinucleotides (4.2%). In contrast, sequences 2, (CTAAA)n, and 3, (CAAAT)n, which have only ApT and TpA dinucleotides, respectively, induced high levels of methylation (64% and 86%, respectively). These levels are also higher than those induced by two other sequences (sequences 4 and 6) that have both dinucleotides.
Sequences 4, (CAATA)n, and 6, (CTAAT)n, induced methylation of 34.8% and 24.4% of the sites measured, respectively. Finally, no significant inhibitory effect of the G:C base pairs was observed in sequence 3, (CAAAT)n, whose level of methylation (86%) was equivalent to that induced by the 80-mer (TAAA)n sequence (81%; Fig. 3C). This contrasts with the situation with the (CAAAT)n 25-mer, which was a nearly 10-fold less potent methylation signal than the 20-mer (TAAA)n sequence (Fig. 3A). Perhaps the increased length of the 100-mer (CAAAT)n fragment compensated for the weaker signal and led to a saturating level of methylation. It is clear from these results that neither TpA nor ApT dinucleotides are essential to trigger methylation and that the effects of these dinucleotides are not simply additive. This finding is consistent with our observation that the frequency of TpA (or ApT) in sequences consisting exclusively of A:T base pairs is not strictly correlated with the level of methylation induced (Fig. 2 and 3).
Long pure-A:T tracts are not required for signaling cytosine methylation.
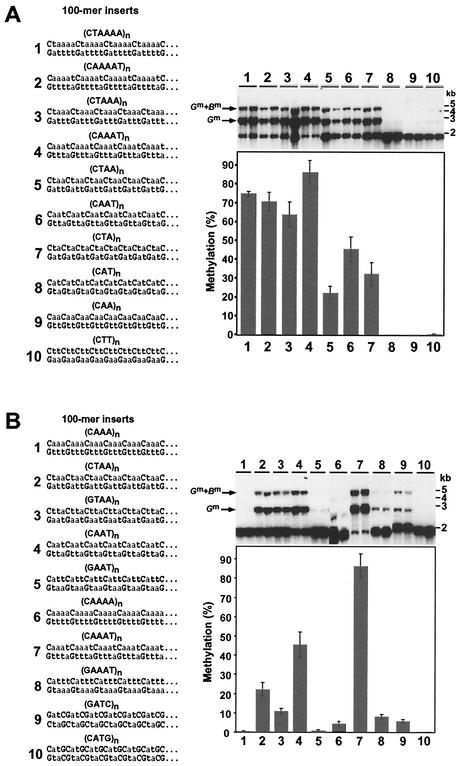
While sequences must have both A and T residues on a given strand to induce appreciable methylation, the results presented above suggest that this requirement does not reflect a simple effect of TpA or ApT dinucleotides. The implication is that longer structures are recognized. Results of previous work suggested that no single 5-bp or larger sequence is responsible for inducing DNA methylation in Neurospora cells (50). We therefore considered all possible A:T sequences up to 4 bp long. In addition, we tested whether long tracts of A:T base pairs uninterrupted by G:C base pairs are required for induction of methylation. To do this, we constructed, targeted and assayed 100-mer sequences consisting of simple repeats with two, three, four, or five A:T base pairs of a signal sequence, [T(A)n]n, separated by G:C base pairs.
We tested constructs that interrupted the ApT step (sequences 1, 3, 5, and 7; Fig. 4A) as well as constructs that interrupted the TpA step (sequences 2, 4, 6, and 8; Fig. 4A). The test sequences with four or five A:T base pair tracts [(CTAAAA)n, (CTAAA)n, (CAAAAT)n, and (CAAAT)n] all yielded similar high levels of methylation (74.7% ± 1.3%, 63.7% ± 6.6%, 70.6% ± 4.9%, and 86.2% ± 6.2%, respectively), indicating that a four-A:T tract is sufficient for strong induction of methylation. Both sequences with 3-bp A/T tracts [(CTAA)n and (CAAT)n] also triggered methylation (22.2% ± 3.4% and 45.3% ± 6.7%, respectively), although less efficiently than those with longer A/T tracts. Surprisingly, one of the sequences with just two contiguous A:T base pairs [sequence 7 in Fig. 4A; (CTA)n] also induced substantial methylation (32.3% ± 6.0%), indicating that long pure-A/T tracts are not required for signaling cytosine methylation in Neurospora. This sequence induced somewhat more methylation than one sequence (sequence 5 in Fig. 4A) that has 3-bp A/T tracts and a higher A+T content overall. One interpretation of this finding is that the higher overall number of TpA dinucleotides in sequence 7 relative to sequence 5 more than offset any negative effects of the shorter A/T tracts and lower A+T content.
FIG. 4.
Effect of A:T tract length and orientation of G:C base pairs on DNA methylation. (A) Long A:T tracts are not required to trigger methylation. The 100-mer sequences related to the best methylation signal identified, (TAAA)n (see Fig. 2B), were constructed and tested as methylation signals at the amRIP4::ζ−η100 test site. Note that sequences 1 and 2 have 5-bp A:T tracts, sequences 3 and 4 have 4-bp A:T tracts, sequences 5 and 6 have 3-bp A:T tracts, and sequences 7 to 10 have 2-bp A:T tracts. (B) Effect of orientation of G:C pairs on de novo methylation at the amRIP4::ζ−η100 test site. Pairs of similar sequences (2 and 3, 4 and 5, 7 and 8, and 9 and 10) that differ only in the orientation of the single G:C in the repeat unit were tested as described for Fig. 2. Inserts 1 and 6 are control test sequences that have neither TpA nor ApT dinucleotides.
Interestingly, sequence 8 [(CAT)n], which has the same base composition and length of contiguous A:T pairs as sequence 7 but differs in that it has ApT dinucleotides rather than TpA dinucleotides, did not induce methylation. As expected, 100-mers of (CAA)n and (CTT)n, which have neither TpA nor ApT dinucleotides, also failed to induce appreciable methylation (sequences 9 and 10 in Fig. 4A). It is noteworthy that while increasing the number of A:T base pairs between the G:C pairs increased methylation up to a point, A/T tracts longer than 4 bp did not appear to further promote methylation. Perhaps the “machine” that recognizes methylation signals reads a 4-bp A/T segment most efficiently. This idea is supported by our findings that a 25-bp (TAAA)n sequence induced greater methylation than did a (TAAAA)n sequence of the same length (29.7% versus 11.6%; Fig. 2B) and that the 20-bp (TAAA)n sequence induced methylation as effectively (11.4%) as the 25-bp (TAAAA)n sequence (Fig. 3A).
Alternative orientations of a G:C base pair differentially inhibit methylation signals.
To characterize the effect of orientation of G:C base pairs relative to TpA and ApT dinucleotides, we constructed and tested pairs of 100-mer sequences with G or C residues 5′ and 3′ of TpA or ApT dinucleotides (Fig. 4B). Insertion of a C residue 5′ of TAA induced about twice the level of methylation induced by the comparable sequence with a G residue 5′ of TAA [compare sequences 2, (CTAA)n, and 3, (GTAA)n, in Fig. 4B, which induced 22.2% and 10.9% methylation, respectively]. An even more dramatic difference was observed with similar sequences with G or C residues before and after ApT dinucleotides (consider both strands in sequences 4 and 5 in Fig. 4B). The sequence (CAAT)n (sequence 4 in Fig. 4B) induced >50-fold greater methylation than did the sequence (GAAT)n (5 in Fig. 4B), 45.3% and 0.7% methylation, respectively. The insignificant induction of methylation by (GAAT)n is comparable to that observed with (CAAA)n (sequence 1 in Fig. 4B), which has neither TpA nor ApT dinucleotides. The distinct effect of G and C residues adjacent to ApT dinucleotides was reconfirmed by comparing sequences (CAAAT)n (sequence 7 in Fig. 4B) with (GAAAT)n (sequence 8 in Fig. 4B), which induced 86.2% and 8.0% methylation, respectively (Table 1).
To explore the possibility that repeats with just 2-bp A/T tracts (only TpA or ApT) can induce detectable methylation if C residues preceding ApT dinucleotides are avoided, we tested 100-mer sequences (GATC)n and (CATG)n (9 and 10 in Fig. 4B). As expected, (CATG)n did not induce detectable methylation; in contrast, (GATC)n induced significant (5.7%) methylation. Note that this sequence is 50% G+C. The finding that (CTA)n and (GATC)n were capable of inducing methylation demonstrates that 2-bp A/T tracts of either TpA or ApT dinucleotides can promote methylation. All of our results are consistent with the idea that C residues 5′ of ApT's (or G residues after ApT's) most drastically inhibit methylation. Interestingly, of all the possible trinucleotides, 5′-CAT-3′ (or 3′-GTA-5′ on the opposite strand) is that most frequently mutated by RIP (15).
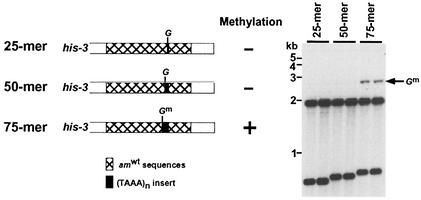
A 75-bp (TAAA)n DNA segment can induce methylation without sequences mutated by RIP.
The shortest natural DNA fragment known to serve as a portable de novo methylation signal is 177 bp (24). As described above, we found that 20 bp of (TAAA)n would trigger methylation in the amRIP4::ζ−η100 artificial test site at the his-3 locus. We wished to determine whether this sequence is more potent than the natural methylation signals from the ζ−η region. To address this question, we inserted one, two, or three copies of a 25-bp (TAAA)n fragment into the BglII site of the wild-type am sequence, targeted the constructs to his-3, and tested for induction of methylation. The 75-mer (TAAA)n but not 25 and 50-mers induced methylation in the am+ sequence background (Fig. 5). The level of methylation established by the 75-mer was similar to that triggered by the 20-mer at the amRIP4::ζ−η100 test site (7.0% and 11.4%, respectively). This finding confirms the high potency of (TAAA)n as a methylation signal, supports our initial observation that mutations caused by RIP in amRIP4 promote de novo methylation (Fig. 1), and provides additional evidence of roles for both the sequence and the length of methylation signals.
FIG. 5.
A 75-nucleotide (TAAA)n DNA fragment can trigger de novo methylation without sequences mutated by RIP. Two independent transformants carrying 25-, 50-, or 75-bp inserts embedded in am+ sequences at the his-3 locus (see diagram) were tested as described for Fig. 2.
Distamycin A, an analogue of the AT-hook DNA binding motif, suppresses DNA methylation and gene silencing in Neurospora.
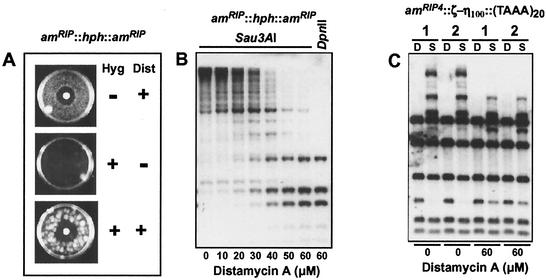
A conserved DNA binding peptide motif called the AT-hook binds preferentially to the minor groove of stretches of A:T-rich sequences, recognizing the structure rather than the particular sequence of such DNA (31-33). Our finding that various A:T-rich sequences, especially those with A:T tracts of at least 4 bp, efficiently signal methylation encouraged us to explore the possibility that a protein harboring the AT-hook or a related motif may be involved in the recognition of de novo DNA methylation signals. As a first step to addressing this question, we tested the effect in Neurospora cells of a minor-groove binder, distamycin, which is an analogue of the AT-hook that directly competes with AT-hook proteins such as HMG-I for binding to A:T tracts in vitro and in vivo (30, 56). We reasoned that distamycin may compete with a protein recognizing the methylation signals and thereby result in loss of DNA methylation. In particular, we tested whether distamycin would activate the methylated, silenced transgene (hph) of strain N644 (16, 40).
About 2,000 asexual spores (conidia) of N644 were challenged with hygromycin B, distamycin, or both (Fig. 6A). Distamycin caused a clear zone of growth inhibition and also induced resistance to hygromycin (Fig. 6A), supporting our hypothesis. To test directly for loss of DNA methylation, we isolated DNA from N644 treated with different concentrations of distamycin and assayed methylation of the amRIP region flanking the hph transgene by Southern hybridization with the isoschizomers Sau3AI and DpnII (Fig. 6B). The Sau3AI digests revealed substantial hypomethylation with 50 μM and 60 μM distamycin A, which caused no appreciable growth inhibition (not shown). Stripping and reprobing the blot with DNA fragments from other methylated regions (psi63 and ribosomal DNA) also revealed hypomethylation, although less than that observed in the amRIP::hph::amRIP region (not shown). We next investigated whether distamycin could also suppress DNA methylation triggered by the (TAAA)20 80-mer, which was found to be the most powerful synthetic de novo methylation signal in this study. Inspection of methylation at the am region flanking this sequence revealed substantial hypomethylation with 60 μM distamycin treatment (Fig. 6C).
FIG. 6.
Distamycin A suppresses DNA methylation and gene silencing in N. crassa. (A) Reactivation of hph induced with distamycin A in strain N644, which carries a methylated silent allele of hph, encoding resistance to hygromycin, surrounded by RIP-altered am sequences (16). Distamycin A (Dist, 772 nmol) was applied to a paper disk on solidified medium containing ≈2,000 conidia of N. crassa N644 in 5 ml of medium without drugs. Then 2.5 mg of hygromycin B (Hyg) was overlaid in 5 ml of medium 16 h later and incubated for 5 days. Control plates contained only distamycin A (top) or hygromycin B (middle). (B) Loss of DNA methylation at amRIP::hph::amRIP induced with distamycin A. Genomic DNA (≈500 ng) of strain N644 treated with the indicated concentrations of distamycin A was digested with 5′-methylcytosine-sensitive and -insensitive isoschizomers (Sau3AI and DpnII, respectively) and analyzed by Southern hybridization with a probe for am. (C) Loss of DNA methylation in amRIP4::ζ−η100::(TAAA)20 induced with distamycin A. Genomic DNA from a strain (N2352) carrying the (TAAA)20 synthetic signal at the amRIP4::ζ−η100 test site (see Fig. 3B) untreated (0 μM) or treated with distamycin A (60 μM) was analyzed by Southern hybridization with Sau3AI (S) or DpnII (D) and an am probe. Results from duplicate cultures (1 and 2) for each treatment are shown.
HMG-I and a Neurospora factor bind efficiently to the best methylation signals.
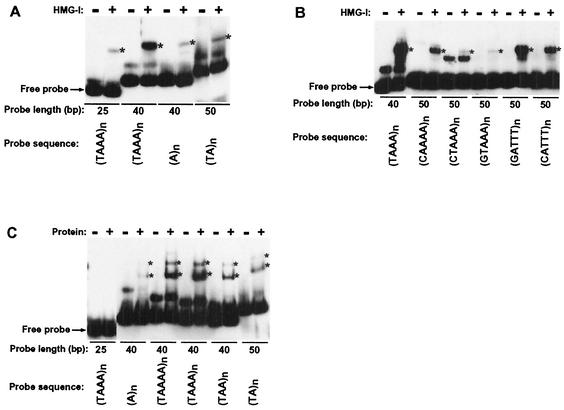
We wished to further explore the idea that a protein with an AT-hook or a functionally similar motif recognizes de novo methylation signals. We therefore performed gel mobility shift assays to test whether human recombinant HMG-I, which carries three AT-hooks, would bind preferentially to sequences that triggered methylation (Fig. 7A and B). We tested the effects of probe length and alternating A and T residues with a set of 100% A:T sequences, 25- and 40-mers of (TAAA)n, an (A)n 40-mer, and a (TA)n 50-mer (Fig. 7A). Robust binding to the best methylation signals was detected even in the presence of poly(dI-dC) competitor. The (TAAA)n 40-mer showed greater binding than the 25-mer of the same sequence, the same length of (A)n or a (TA)n 50-mer, consistent with the capacity of these segments to induce methylation (Fig. 2B and 3B).
FIG. 7.
Correlation of methylation signal strength and binding efficiency of oligonucleotides with N. crassa and mammalian factors. (A) Gel mobility shift assay of HMG-I. Asterisks (∗) indicate bound probe. Each probe was tested with (+) and without (−) protein (1 μg). The position of free probe is indicated. Other lower-mobility bands reflect altered forms of the probes rather than protein-DNA complexes. (B) Gel mobility shift assay of HMG-I was performed as presented in panel A except that 2 μg of protein was used. (C) Gel mobility shift assay of signal oligonucleotides with an N. crassa protein fraction from a carboxymethyl-Sepharose Fast Flow column (courtesy of G. Kothe, University of Oregon). The result is presented as in panel A.
To characterize the effects on HMG-I binding of TpA, ApT dinucleotides, and G:C base pairs in alternate orientations, we assayed 50-mers of (CTAAA)n, (GTAAA)n, (GATTT)n, and (CATTT)n and, as controls, a (TAAA)n 40-mer and a (CAAAA)n 50-mer (Fig. 7B). The presence of G:C base pairs strongly inhibited HMG-I binding, as in the in vivo methylation assays (Fig. 3A), i.e., the 50-mers of (CTAAA)n, (GTAAA)n, (GATTT)n, and (CATTT)n did not bind as well as the (TAAA)n 40-mer. It is noteworthy that (GATTT)n, which has G residues preceding the ApT dinucleotides, bound HMG-I better than did (CATTT)n, which has C residues preceding the ApT dinucleotides. Indeed, binding of the latter sequence was nearly as weak as with (CAAAA)n, again paralleling the behavior of these sequences in our in vivo de novo methylation assays (Fig. 4B). Curiously, (CTAAA)n and (GTAAA)n did not bind HMG-I as well as (CAAAA)n, indicating that TpA dinucleotides inhibit binding, in contrast to the effect of TpA dinucleotides on DNA methylation (Fig, 3C).
Finally, we used crude Neurospora extracts in band shift assays to look for evidence of a factor recognizing the de novo DNA methylation signals that we had identified. Two major binding activities were detected with the pure A:T 40- or 50-mer sequences containing both A and T on a given strand, while no binding was detected with the 25-mer (TAAA)n (Fig. 7C). The relative binding to the probes is strikingly similar to that observed with HMG-I (Fig. 7A) and to the relative strengths of the methylation signals (Fig. 2B). We also found that distamycin A competes with the binding activities (data not shown). The results suggest that the two binding activities in the Neurospora cell extract are due to proteins with AT-hooks or functionally similar motifs.
DISCUSSION
DNA methylation is an obvious and relatively stable modification of the genetic material. Information from a variety of eukaryotes implicates DNA methylation in gene silencing, but remarkably little is known about how DNA methylation is controlled. Control of methylation is typically divided into de novo methylation, i.e., establishment of DNA methylation, and maintenance of methylation. We use N. crassa as a model system to investigate control of DNA methylation largely because we have unparalleled systems to reproducibly test the potential of sequences to trigger de novo methylation (24, 25) and because this organism has a relatively simple methylation machine. Whereas other eukaryotes that have been examined have multiple active DNA methyltransferases, which can vary in their abilities to carry out de novo and maintenance methylation, all DNA methylation in vegetative Neurospora cells appears to result from the action of one DNA methyltransferase, DIM-2 (21), operating under the direction of the histone H3 methyltransferase DIM-5 (53).
To better define signals for de novo DNA methylation in N. crassa, we developed a new system to test short synthetic DNA fragments for their potential to promote methylation. The system was built on our observation that a sequence (amRIP4) with a density of mutations caused by RIP that is insufficient to trigger methylation alone can nevertheless promote methylation of sequences inserted into it. Indeed, combination of a segment of ζ−η DNA that is itself ineffectual as a methylation signal and the lightly mutated amRIP4 triggered robust methylation, providing evidence that sequences can cooperate to induce DNA methylation. We took advantage of this observation to build a construct to test short DNA fragments at a site “poised” for methylation.
We made and tested numerous synthetic oligonucleotide sequences, including some with random sequences of different base compositions and others with specified sequences. None of 20 random 25-mers made with ≈50% A:T triggered methylation, whereas all of 19 random 25-mers with 100% A:T triggered some methylation of flanking sequences, confirming that our assay system is sensitive and that a high A+T content promotes de novo methylation in N. crassa. Tests on pure A:T 25-mers with various sequences showed that both T's and A's are required on a given strand to induce significant methylation and, surprisingly, that sequences with intermediate levels of TpA and ApT dinucleotides induce greater methylation than sequences with a maximal number of these dinucleotides; e.g., a 25-mer consisting of (TAAA)n induced five times higher methylation than did a 25-mer consisting of (TATA)n (Table 1 and Fig. 2B). Indeed, it is not yet certain that TpA or ApT dinucleotides are critical to induce methylation.
Interestingly, the (TAAA)n sequence was strong enough to induce methylation when inserted as a 75-mer in a sequence environment with no mutations caused by RIP (Fig. 5). Results of previous work suggested that no single sequence of 5 bp or larger is responsible for inducing DNA methylation in N. crassa (50). In the present study, we tested all 16 possible A:T sequences up to 4 bp long. The sequences (TAAA)n and (TTAA)n were found to be significantly more effective methylation signals than any of the other sequences. Thus, two general conclusions are that a variety of simple A:T-rich sequences can induce methylation in N. crassa, although to different extents, and that methylation signaling is additive; a feeble signal can induce robust methylation if two or more copies of it are fused or if it is combined with another feeble signal.
Introduction of G:C base pairs at any position in the (TAAA)n repeat strongly inhibited methylation (Fig. 3A), consistent with our analyses of random sequences. This supports the possibility that the high G:C content of CpG islands in mammals contributes to their largely unmethylated status (3). We found that we could increase the sensitivity of the assay by increasing the length of the test sequences (Fig. 3B) and took advantage of this to compare induction of methylation by variants of the optimal sequence with G:C pairs inserted at various positions. In this way, we showed that neither the TpA step nor the ApT step is essential; indeed, the least effective signal in the series, (CATAA)n, retained both TpA and ApT dinucleotides. It may be significant that this sequence includes CpApT, the trinucleotide most frequently mutated by RIP (15, 39).
The observed “fuzzy” nature of signals for DNA methylation in N. crassa is not without precedent. For example, a sequence element required for replication origins in Saccharomyces cerevisiae and Schizosaccharomyces pombe has no strict sequence requirement but must be A:T rich and relatively easy to unwind (27, 28). Most of our best signal sequences are predicted to have very low thermal stabilities (data not shown), which might facilitate flipping out of the substrate cytosine by the DNA methyltransferase (9). It is worth noting, however, that this parameter does not correlate perfectly with signaling capacities [e.g., (TAAA)n, (TAA)n, and (TA)n are predicted to have progressively decreasing stabilities contrary to their order of effectiveness in inducing methylation].
There are other cases in which the function of degenerate A:T-rich sequences appears to depend on additive or cooperative interactions of multiple simple DNA-binding domains. Examples include the DNA elements identified as scaffold attachment regions or matrix attachment regions in chromatin (52, 59) and certain satellite repeats found in heterochromatin (2). Several proteins, including HMG-I (HMG-I/Y) family proteins (31, 32), topoisomerase II (1), histone H1 (18), SAF-A (19), and D1 (2), have been shown to bind to such A:T-rich sequences. In general, these interactions involve special protein motifs that bind in the deep and narrow minor groove that is characteristic of A:T-rich sequences. The most notable A:T-rich binding motif is the AT-hook, several copies of which are typically found in A:T-rich binding proteins such as HMG-I (31, 33). The fission yeast protein Orc4p binds to replication origin DNA via multiple AT-hooks (10), and the AT-hook protein D1 has been implicated in position effect variegation in Drosophila melanogaster (2).
Results of several experiments support the idea that an AT-hook type protein may recognize methylation signals. Specifically, we found that distamycin A, which competes with the AT-hook proteins for binding to the narrow minor groove of A:T tracts (30), suppresses DNA methylation and gene silencing in Neurospora; that a purified human AT-hook protein, HMG-I, preferentially bound to our most potent methylation signals and showed differential sensitivity to the orientation of G:C pairs, as we found in our in vivo methylation assay; and that an activity in Neurospora extracts binds with similar specificity and its binding is inhibited by distamycin.
AT-hooks are thought to require tracts of at least four A:T pairs for optimal binding (51). Similarly, A/T tracts longer than 4 bp did not appear to further promote methylation. Although not as powerful as most sequences with tracts of at least four A:T pairs, arrays of TAA or AAT separated by C:G pairs induced substantial methylation so long as the sequence did not include CAT trinucleotides (Fig. 4A). Remarkably, even sequences with just two contiguous A:T pairs, (CTA)n and (GATC)n, can trigger methylation. It is noteworthy that the latter sequence is 50% G:C. These observations do not indicate that an AT-hook protein is not involved in DNA methylation, however. Short A:T tracts can be bound, although weakly, by AT-hook proteins (data not shown). Moreover, different classes of AT-hook proteins may have distinctive DNA-binding patterns. This may account for our observation that the presence of TpA dinucleotides inhibited binding of HMG-I, which contrasts with its apparent effect in signaling DNA methylation.
It is interesting that the methylated relics of RIP in N. crassa have functional and structural features in common with heterochromatin in other eukaryotes. Methylated DNA in N. crassa and characterized heterochromatin in other systems are both marked by gene silencing and methylation of lysine-9 of histone H3 (17, 26, 35, 53). In addition, heterochromatic regions and products of RIP are both typically associated with A:T-rich, repeated sequences that are prone to methylation in organisms that show this modification. Finally, distamycin A interferes with SAR protein interactions (18) and with DNA methylation in relics of RIP. It is also noteworthy that a multi-AT hook protein, MATH alleviates position effect variegation associated with heterochromatin in Drosophila (13). It seems likely that signaling DNA methylation in N. crassa involves recognition of the minor groove of short stretches of mixed A:T base pairs by one or more proteins sporting an AT-hook or a functionally similar motif. The striking similarity of our best two methylation signals [(TAAA)n and (TTAA)n] to proposed nucleosome-positioning sequences (20) raises the possibility that nucleosomes may also play an active role in reading signals for de novo DNA methylation.
Acknowledgments
We greatly appreciate the gift of HMG-I from Raymond Reeves and his stimulating discussions and encouragement. We also thank Michael Freitag for designing ζ−η primers, Raymond Reeves and Michael Freitag for comments on the manuscript, and former and present members of our laboratory for advice. H.T. thanks Laura Coffin for inspiration.
This study was supported by a grant from the National Institutes of Health (GM35690).
REFERENCES
- 1.Adachi, Y., E. Kas, and U. K. Laemmli. 1989. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 8:3997-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aulner, N., C. Monod, G. Mandicourt, D. Jullien, O. Cuvier, A. Sall, S. Janssen, U. K. Laemmli, and E. Kas. 2002. The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation. Mol. Cell. Biol. 22:1218-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bestor, T. H., G. Gundersen, A.-B. Kolstø, and H. Prydz. 1992. CpG islands in mammalian gene promoters are inherently resistant to de novo methylation. Genet. Anal. Technol. Appl. 9:48-53. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321:209-213. [DOI] [PubMed] [Google Scholar]
- 5.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 6.Burge, C., A. M. Campbell, and S. Karlin. 1992. Over- and under-representation of short oligonucleotides in DNA sequences. Proc. Natl. Acad. Sci. USA 89:1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cambareri, E. B., B. C. Jensen, E. Schabtach, and E. U. Selker. 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244:1571-1575. [DOI] [PubMed] [Google Scholar]
- 8.Cambareri, E. B., M. J. Singer, and E. U. Selker. 1991. Recurrence of repeat-induced point mutation (RIP) in Neurospora crassa. Genetics 127:699-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, X., and R. J. Roberts. 2001. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29:3784-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang, R. Y., and T. J. Kelly. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA 96:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foss, H. M., C. J. Roberts, K. M. Claeys, and E. U. Selker. 1993. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science 262:1737-1741. [DOI] [PubMed] [Google Scholar]
- 12.Freitag, M., R. L. Williams, G. O. Kothe, and E. U. Selker. 2002. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 99:8802-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard, F., B. Bello, U. K. Laemmli, and W. J. Gehring. 1998. In vivo analysis of scaffold-associated regions in Drosophila: a synthetic high-affinity SAR binding protein suppresses position effect variegation. EMBO J. 17:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant, S. G., and V. M. Chapman. 1988. Mechanisms of X-chromosome regulation. Annu. Rev. Genet. 22:199-233. [DOI] [PubMed] [Google Scholar]
- 15.Grayburn, W. S., and E. U. Selker. 1989. A natural case of RIP: degeneration of DNA sequence in an ancestral tandem duplication. Mol. Cell. Biol. 9:4416-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irelan, J. T., and E. U. Selker. 1997. Cytosine methylation associated with repeat-induced point mutation causes epigenetic gene silencing in Neurospora crassa. Genetics 146:509-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 18.Käs, E., E. Izaurralde, and U. K. Laemmli. 1989. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA) · oligo(dT) tracts. J. Mol. Biol. 210:587-599. [DOI] [PubMed] [Google Scholar]
- 19.Kipp, M., F. Gohring, T. Ostendorp, C. M. van Drunen, R. van Driel, M. Przybylski, and F. O. Fackelmayer. 2000. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 20:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiyama, R., and E. N. Trifonov. 2002. What positions nucleosomes? A model. FEBS Lett. 523:7-11. [DOI] [PubMed] [Google Scholar]
- 21.Kouzminova, E. A., and E. U. Selker. 2001. dim-2 encodes a DNA-methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 20:4309-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macleod, D., J. Charlton, J. Mullins, and A. P. Bird. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 22a.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Margolin, B. S., M. Freitag, and E. U. Selker. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44:34-36. [Google Scholar]
- 24.Miao, V. P., M. Freitag, and E. U. Selker. 2000. Short TpA-rich segments of the zeta-eta region induce DNA methylation in Neurospora crassa. J. Mol. Biol. 300:249-273. [DOI] [PubMed] [Google Scholar]
- 25.Miao, V. P. W., M. J. Singer, M. R. Rountree, and E. U. Selker. 1994. A targeted replacement system for identification of signals for de novo methylation in Neurospora crassa. Mol. Cell. Biol. 14:7059-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Natale, D. A., A. E. Schubert, and D. Kowalski. 1992. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc. Natl. Acad. Sci. USA 89:2654-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natale, D. A., R. M. Umek, and D. Kowalski. 1993. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 21:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paluh, J. L., M. J. Orbach, T. L. Legerton, and C. Yanofsky. 1988. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc. Natl. Acad. Sci. USA 85:3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radic, M. Z., M. Saghbini, T. S. Elton, R. Reeves, and B. A. Hamkalo. 1992. Hoechst 33258, distamycin A, and high mobility group protein I (HMG-I) compete for binding to mouse satellite DNA. Chromosoma 101:602-608. [DOI] [PubMed] [Google Scholar]
- 31.Reeves, R. 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277:63-81. [DOI] [PubMed] [Google Scholar]
- 32.Reeves, R., and L. Beckerbauer. 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519:13-29. [DOI] [PubMed] [Google Scholar]
- 33.Reeves, R., and M. S. Nissen. 1990. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265:8573-8582. [PubMed] [Google Scholar]
- 34.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089-1093. [DOI] [PubMed] [Google Scholar]
- 35.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 36.Russell, P. J., K. D. Rodland, E. M. Rachlin, and J. A. McCloskey. 1987. Differential DNA methylation during the vegetative life cycle of Neurospora crassa. J. Bacteriol. 169:2902-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Selker, E. U. 1999. Gene silencing: repeats that count. Cell 97:157-160. [DOI] [PubMed] [Google Scholar]
- 39.Selker, E. U. 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24:579-613. [DOI] [PubMed] [Google Scholar]
- 40.Selker, E. U. 1998. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95:9430-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selker, E. U., E. B. Cambareri, B. C. Jensen, and K. R. Haack. 1987. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell 51:741-752. [DOI] [PubMed] [Google Scholar]
- 42.Selker, E. U., M. Freitag, G. O. Kothe, B. S. Margolin, M. R. Rountree, C. D. Allis, and H. Tamaru. 2002. Induction and maintenance of non-symmetrical DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 99:16485-16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selker, E. U., D. Y. Fritz, and M. J. Singer. 1993. Dense non-symmetrical DNA methylation resulting from repeat-induced point mutation (RIP) in Neurospora. Science 262:1724-1728. [DOI] [PubMed] [Google Scholar]
- 44.Selker, E. U., and P. W. Garrett. 1988. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 85:6870-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selker, E. U., B. C. Jensen, and G. A. Richardson. 1987. A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science 238:48-53. [DOI] [PubMed] [Google Scholar]
- 46.Selker, E. U., G. A. Richardson, P. W. Garrett-Engele, M. J. Singer, and V. Miao. 1993. Dissection of the signal for DNA methylation in the ζ-η region of Neurospora. Cold Spring Harbor Symp. Quant. Biol. 58:323-329. [DOI] [PubMed] [Google Scholar]
- 47.Selker, E. U., and J. N. Stevens. 1985. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc. Natl. Acad. Sci. USA 82:8114-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selker, E. U., and J. N. Stevens. 1987. Signal for DNA methylation associated with tandem duplication in Neurospora crassa. Mol. Cell. Biol. 7:1032-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singer, J., J. Roberts-Ems, F. W. Luthardt, and A. D. Riggs. 1979. Methylation of DNA in mouse early embryos, teratocarcinoma cells and adult tissues of mouse and rabbit. Nucleic Acids Res. 7:2369-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singer, M. J., B. A. Marcotte, and E. U. Selker. 1995. DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol. Cell. Biol. 15:5586-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solomon, M. J., F. Strauss, and A. Varshavsky. 1986. A mammalian high mobility group protein recognizes any stretch of six A · T base pairs in duplex DNA. Proc. Natl. Acad. Sci. USA 83:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strick, R., and U. K. Laemmli. 1995. SARs are cis DNA elements of chromosome dynamics: synthesis of a SAR repressor protein. Cell 83:1137-1148. [DOI] [PubMed] [Google Scholar]
- 53.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 54.Turker, M. S. 1999. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin. Cancer Biol. 9:329-337. [DOI] [PubMed] [Google Scholar]
- 55.Vogel, H. J. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435-446.
- 56.Wegner, M., and F. Grummt. 1990. Netropsin, distamycin and berenil interact differentially with a high-affinity binding site for the high mobility group protein HMG-I. Biochem. Biophys. Res. Commun. 166:1110-1117. [DOI] [PubMed] [Google Scholar]
- 57.Yates, P. A., R. W. Burman, P. Mummaneni, S. Krussel, and M. S. Turker. 1999. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 274:36357-36361. [DOI] [PubMed] [Google Scholar]
- 58.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, K., E. Käs, E. Gonzalez, and U. K. Laemmli. 1993. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in Hi-depleted chromatin. EMBO J. 12:3237-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]