Abstract
The tumor suppressor protein, p53, is part of the cell's emergency team that is called upon following cellular insult. How do cells sense DNA damage and other cellular stresses and what signal transduction pathways are used to alert p53? How is the resulting nuclear accumulation of p53 accomplished and what determines the outcome of p53 induction? Many posttranslational modifications of p53, such as phosphorylation, dephosphorylation, acetylation and ribosylation, have been shown to occur following cellular stress. Some of these modifications may activate the p53 protein, interfere with MDM2 binding and/or dictate cellular localization of p53. This review will focus on recent findings about how the p53 response may be activated following cellular stress.
Keywords: phosphorylation, blocked RNA polymerase II, nucleocytoplasmic shuttling, MDM2, proteasome
Introduction
The p53 gene product plays an important role in tumor suppression. The evidence for this stems from the fact that the p53 gene is the most frequently mutated gene found in human cancers [1]. Furthermore, individuals with the Li-Fraumeni syndrome, who have inherited a faulty allele of the p53 gene, or mice strains in which the p53 gene has been knocked out, are at much higher risk for contracting cancers than normal humans or normal mice. In some types of cancers, p53 inactivation occurs early in tumorigenesis [2] while in others, it is a late event [3]. Not only does functional inactivation of p53 predispose individuals to the induction of cancer, successful outcome of chemotherapy and radiotherapy has been suggested to depend on functional p53 [1,4,5]. Thus, elucidation of the function and regulation of p53 are of great importance for the understanding of the process of carcinogenesis, as well as for finding new avenues for therapeutic intervention.
The p53 Response — A Two-Edged Sword
The mechanism of tumor suppression by p53 is thought to be related to its function as a transcription activator [6]. This is based on the fact that the great majority of p53 mutations observed in human tumors are found in the DNA-binding domain of the protein. This DNA-binding domain is required for its transactivation function [7]. However, transcription-independent functions appear to be important in tumor suppression as well [8,9]. The induction of p53 involves the stabilization of the protein itself [10,11], transformation of the protein from a latent to an active form [12,13] and localization of the protein to the nucleus [14].
Some 100 genes are thought to be transactivated by p53 [15–18], but the consequences of this gene activation are not fully understood. In addition to transactivation of target genes, p53 concurrently reduces transcription of other genes [19]. This can be accomplished either by an indirect mechanism involving sequestering of transcription factors [19–22] or through a direct mechanism involving inhibition of specific genes by histone deacetylation [23]. It is well-known that the p2lWAF1 gene product, which is induced by p53, is a potent inhibitor of G1 cyclin-dependent kinases [24]. It has been suggested that it is through the p2lWAF1 protein that cells arrest at the G1/S border of the cell cycle following exposure to ionizing radiation [25]. Activated p53 also plays an important role in other protective functions by stimulating nucleotide excision repair [26–31] and participating in the induction of senescence [32] (Figure 1). Recently, we have shown that wild-type p53 function is important for the efficient recovery of mRNA synthesis following ultraviolet (UV) irradiation [33].
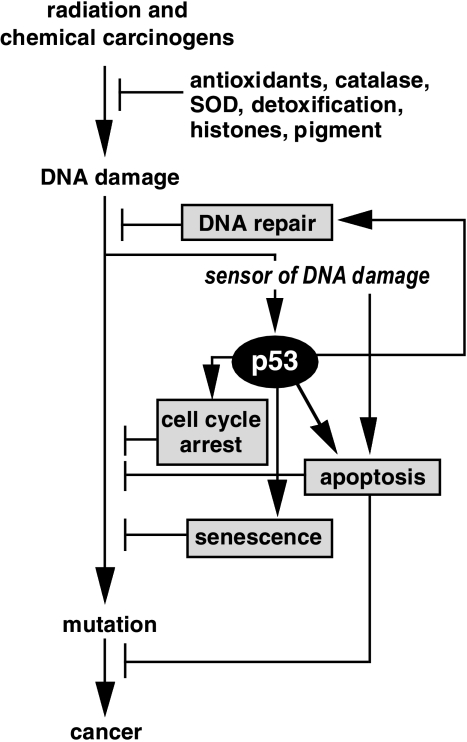
Figure 1.
Model describing some of the roles of p53 as a tumor suppressor. Cancer is a disease of multiple mutations most of which originate from unrepaired DNA damage induced by radiation or chemical carcinogens. Many defense mechanisms have evolved to safeguard the genetic material and to avoid the formation of mutations. In addition to antioxidants and DNA repair systems, mammalian cells can induce the p53 response to modify the behavior of a cell to avoid the formation of mutations. Following the induction of DNA damage, a “sensor” will trigger the induction of the p53 response that will manifest itself as cell cycle arrest, senescence, enhancement of nucleotide excision repair or apoptosis. All of these scenarios will reduce the probability of mutations arising in the exposed cells, and thus reduce the likelihood of the organism to develop cancer.
In contrast to its protective functions, the p53 protein has also been implicated in the induction of apoptosis in certain cell types following DNA damage [8,11,34–36]. The role of p53 in apoptosis, which is thought to be mediated through caspase-9 and Apaf-1 [37], may involve induction of distinct target genes such as bax [38], fas [39] and killer [40]. However, the observation that transactivation-deficient p53 mutants also appear capable of inducing cells to undergo apoptosis suggests that p53 may trigger apoptosis through both transactivation-dependent and transactivation-independent mechanisms [41]. Transactivation-independent mechanisms may involve generation of reactive oxygen species [42–44] and abrogation of mitochondrial membrane potential [44]. The p53-mediated transport of Fas receptors from the Golgi complex to the cell surface is also thought to influence the induction of apoptosis [45]. Aggregation of these receptors following UV irradiation [46,47] triggers a cascade of events leading to the activation of caspase-8 with subsequent activation of a cascade of caspases leading to apoptosis [48,49].
As outlined above, p53 can play both protective and apoptosis-promoting roles following exposure to DNA-damaging agents. The decision whether to save or eliminate the cell depends on many factors such as cell type, severity of damage, and the oncogenic status of the cell [50]. It has recently been shown that wild-type p53 plays a protective role against the induction of apoptosis following moderate doses of UV light [33]. This protection by wild-type p53 expression in human fibroblasts correlated with a p53-dependent enhancement of the recovery of mRNA synthesis following UV irradiation. Furthermore, stimulation of recovery of mRNA synthesis and the protection against apoptosis by wild-type p53 appears to require p53-mediated transactivation prior to UV irradiation (McKay et al., submitted). Thus, p53-mediated transactivation by basal levels of p53 prior to the insult confers protection, while induction of p53 following irradiation augments the UV-induced apoptotic process.
Wasteful Waiting — Suppression of the p53 Response
The p53 tumor suppressor protein negatively regulates cell growth through the induction of cell cycle arrest or apoptosis. Thus, in dividing tissue, there must be mechanisms put in place to downregulate the function of p53. It is now known that in proliferating cells, p53 is kept at a very low level by a mechanism involving proteasome-mediated degradation [51–55]. The proteasome-mediated protein degradation pathway plays an important role in the regulation of various cellular processes such as cell cycle progression, cell differentiation, signal transduction, stress responses and apoptosis [56–59]. The degradation of p53 involves a cascade of enzymatic reactions leading to the ubiquitylation and degradation of p53 by the 26S proteasome (Figure 2).
Figure 2.
Ubiquitin-dependent proteolysis of p53. In a sequential enzymatic reaction (E1–E3), ubiquitin (Ub) is activated and added onto the p53 protein. The ubiquitylated p53 protein is than targeted for proteolysis by the 26S proteasome. The proteins involved in directing ubiquitylation of p53 are the cellular proteins, MDM2 and JNK. The HPV 16 protein E6 can also target p53 for degradation by acting as a ubiquitin ligase. Following cellular stresses, proteasome-mediated degradation of p53 is stopped. The p53 protein can also accumulate by default following inhibition of proteasome activity with certain drugs.
It is well-established that the MDM2 protein is involved in the degradation of p53 [60,61]. The MDM2 protein binds tightly to the N-terminus of p53 and this interaction leads to the ubiquitylation and subsequent proteasome-mediated degradation of the p53 protein. The MDM2 protein has been shown to have ubiquitin ligase activity and probably acts as the E3 ligase for p53 [62,63]. Results suggest that in addition to the N-terminal domain to which MDM2 binds, the C-terminal domain is important for MDM2-mediated degradation [64,65].
In addition to MDM2, the jun kinase (JNK) has been shown to target p53 for ubiquitin-mediated degradation in non-stressed cells [66,67]. This targeting is dependent on the binding of JNK to the amino acids 97–155 of p53 and occurs independently of MDM2. While MDM2 is found to complex with p53 specifically in the S and G2/M phases of the cell cycle, JNK-p53 complexes are preferentially found in the G0/G1 phases [66]. This suggests that degradation of p53 by MDM2 and JNK occurs at different stages of the cell cycle and that MDM2 and JNK act independently of each other. Interestingly, cell extracts depleted of both MDM2 and JNK can still support some degradation of p53, suggesting that there may be additional factors involved in targeting p53 for degradation [66]. Some studies suggest that in addition to proteasome-mediated degradation, the p53 protein can be targeted for proteolysis by calpain proteases [68–70].
The Alarm Goes Off — Mobilization of the p53 Response
In order for p53 to accumulate in cells and to transactivate target genes, the degradation of p53 must be inhibited, the p53 protein must accumulate in the nucleus and the sequence-specific binding activity must be induced. Numerous agents have been found to cause the mobilization of p53 in cells (Figure 3). Some of these agents cause DNA damage and some do not. What sensor protein(s) does the cell use to learn about the inflicted damage or stress? How are these sensors signaling p53? Can the signals be fed through different signaling pathways depending on the type of insult inflicted and if so, does activation of different signaling pathways lead to distinct p53 modifications that tailor their function to best address the specific stress situation?
Figure 3.
The p53 response is triggered by many different stresses involving both DNA-damaging and non-DNA-damaging agents. These include ribonucleotide synthesis inhibitors resulting in perturbations of nucleotide pools [270], thymidine dinucleotides [271], media depletion [272], hypoxia [273], antioxidants [274,275], inhibition of ubiquitylation or the proteasome proteolysis pathway [53,107,108], DNA strand breaks [95], bulky DNA lesions [10,11], DNA topoisomerase inhibitors [11,276], blockage of RNA polymerase II [71–75], heat shock [273,277], cold shock [278], viral infection [217,279], pRb deregulation [216,280] and oncogene expression [214,215].
Much has been learned lately about the triggering mechanisms for the p53 response. The emerging picture is that multiple, distinct sensors and signaling pathways are triggered following exposure to different stresses [71] (Figure 4). The mechanism of p53 induction by two different DNA-damaging agents, ionizing radiation and UV light, will be contrasted below. Although these two physical agents both induce damage to DNA and trigger p53 accumulation, the types and amounts of DNA lesions induced are very different as are the cellular responses inducing the mobilization of the p53 response (Table 1). Also, certain agents appear to trigger p53 accumulation by directly interfering with the degradation pathway of p53. Finally, recent studies have implicated mismatch repair proteins in signaling p53 following induction of specific types of DNA damage.
Figure 4.
The p53 response is activated through multiple pathways. Top: UV light and cisplatin induce bulky lesions in DNA that if formed in the transcribed strand of an active gene will impede the elongation of RNA polymerases. Also, certain drugs that poison the activity of RNA polymerase II, such as actinomycin D, DRB and α-amanitin, trigger the p53 response. Right: DNA strand breaks induced by ionizing radiation may be recognized by the proteins ATM, ATR and NBS which directly or indirectly initiate p53 induction. Bottom: Inhibition of the proteasome by specific drugs, nitric oxide or the overexpression of β-catenin leads to accumulation of p53. Left: O6-methylguanine and cisplatin adducts are recognized by mismatch repair proteins resulting in the induction of p53 and p73 (see text for details).
Table 1.
The DNA-Damaging Agents Ionizing Radiation and UV Light Induce Different Types of DNA Lesions Leading to Contrasting Cellular Responses.
Stuck on the Tracks — Blockage of RNA Polymerase II by UV-Induced DNA Lesions Signals p53
It has been proposed that blockage of RNA polymerase may be the trigger for p53 accumulation following exposure to UV light [71–75]. This has been based on the findings that cells defective in the removal of UV-induced DNA lesions from the transcribed strand of active genes induce p53 at significantly lower doses than cells with proficient repair. These results suggest that persistent UV-induced lesions in the transcribed strand of active genes, but not lesions elsewhere in the genome, appear to be the triggering signal. Since UV-induced DNA lesions in the transcribed strand block elongation of RNA polymerase II [76–78], these results suggest that blockage of RNA polymerase II elongation may trigger the activation of the p53 pathway following exposure to UV light.
Recent studies have suggested that inhibition of RNA polymerase II may be sufficient for the induction of p53 [71]. The RNA synthesis inhibitors, actinomycin D, DRB, H7 and α-amanitin, were found to all induce the accumulation of p53 in the same dose range as was shown to cause inhibition of mRNA synthesis. Furthermore, the induction of p53 by these agents did not correlate to the induction of DNA strand breaks. Although these studies suggest that inhibition of mRNA synthesis is sufficient to trigger p53 accumulation, certain agents such as ionizing radiation or proteasome inhibitors were found to trigger p53 without affecting mRNA synthesis. Thus, inhibition of RNA polymerase II may be a common mechanism by which many, but not all, p53-inducing agents trigger the accumulation of p53 (Figure 4).
What proteins are involved in sensing that RNA polymerase II elongation is blocked and how is the signal transmitted to p53? There are a number of proteins that are part of the RNA polymerase II holoenzyme that could directly or indirectly modify p53 (see Tables 2–4). The Cdk-activating kinase (CAK) plays an important role in the cell cycle by regulating the activity of Cdks. As part of the transcription factor TFIIH, CAK regulates the phosphorylation of the carboxyl-terminal domain (CTD) of RNA polymerase II [79]. Recent studies have shown that CAK can directly phosphorylate p53 in vitro [80,81]. Thus, CAK could potentially be a very powerful sensor of transcription elongation activating both p53 and cell cycle arrest if transcription is inhibited. Another interesting component of the RNA polymerase II holoenzyme that potentially could regulate p53 is the TAFII250, which possesses both acetyl transferase and kinase activity [82,83]. TAFII250 may indirectly regulate p53 stability through its role in the expression of the MDM2 gene [84]. Finally, the transcription activators p300/CBP and PCAF are also proteins with the capacity to modify p53 by acetylation [85,86]. Taken together, there are many proteins associated with the transcription machinery that potentially could act as sensors of blocked RNA polymerases and could transmit (or stop transmitting) a signal resulting in p53 modifications/accumulation (Figure 5).
Table 2.
Phosphorylation of p53 by Various Kinases.
| Kinases | General | Inducer | p53 modifications | Effect on p53 | References |
| ATM | binds double-stranded DNA preferentially at DNA ends in Ku-independent manner; no affinity for UV-irradiated DNA; phosphorylates BRCA1 following IR but not UV | IR, not UV; inhibited by caffeine and wortmannin | ser15, ser37 | blocks MDM2 binding; stimulates acetylation of C-terminus; plays a role in p53-mediated apoptosis | [36,86,112–126] |
| ATR | protective function against IR and UV; involved in cell cycle check-point control | IR and UV; inhibited by caffeine but not very sensitive to wortmannin | ser15, ser37 | blocks MDM2 binding; stimulates acetylation of C-terminus; plays a role in p53-mediated apoptosis | [86,121–130] |
| DNA-PK | binds to and is activated by DNA ends; involved in double-strand break repair | IR, inhibited by wortmannin | ser15, ser37 in vitro only | not involved in p53 modification in vivo; required for efficient transactivation by p53 | [126,131–137] |
| JNK | associates with p53; direct ubiquitylation when not activated as a kinase | UV; ATM-dependent activation following IR | murine ser34, possibly ser37 on human p53 | blocks MDM2 binding | [66,104,131,138–141] |
| c-Abl | interacts with p53 and DNA-PK after IR; phosphorylates p73 but not p53; neutralizes inhibition of p53 by MDM2; phosphorylates CTD of RNA pol II | IR (ATM-dependent activation), cisplatin | tyr99 of p73 after IR | stimulates p73-mediated transactivation and apoptosis | [100–103,142–144] |
| CK I | expression may be induced by p53 | etoposide | ser4, ser6, ser9 | ? | [145] |
| CK II | strong affinity for binding to p53 | UV; constitutively phosphorylated? | ser392 | increases tetramerization and sequence-specific DNA binding; regulates p53-dependent transcriptional repression; inhibits p53-mediated renaturation of DNA strands; nuclear localization? | [13,146–151] |
| CAK | Cdk-activating kinase; phosphorylates CTD of RNA pol II | UV light downregulates CAK activity | ser33, C-terminus (ser371, 376, 378 and 392 potential targets) | enhances sequence-specific DNA binding in vitro | [80,81,131,152] |
| PK-C | stimulates apoptosis | UVA, but not UVB or UVC | ser371, ser376, ser378 in vitro only | enhances sequence-specific DNA binding in vitro | [131,153–157] |
| Cdc2/cycl in B Cdk2/cycl in A |
regulates cell cycle progression; activated by CAK | UV light downregulates Cdc2 and Cdk2 | ser315 | increases sequence-specific DNA binding in vitro; targets p53 for degradation; attenuates tetramerization; target p53 for cytoplasmic localization? | [13,131,151,158–166] |
| p38 | Interact with p53 | UV, not IR | ser33, ser46, ser392 | ser33 and ser46 phosphorylation stimulate phosphorylation of ser15 and ser37 and is important for UV-induced apoptosis; for effect of ser392 phosphorylation see above for CKII | [167,281] |
| PKR | ser/thr kinase modulating protein synthesis; requires double-stranded RNA for its activity; can bind p53 | interferon-inducible; UV?, IR? | ser392 | see above for p38 | [168,169] |
| CHK1 | may be an ATR-induced kinase | UV, IR | ser20 | blocks MDM2 interaction; plays a role in p53-mediated apoptosis | [124,170,171,282–286] |
| CHK2 | ATM-induced kinase; may be induced by ATR following UV light exposure, phosphorylates BRCA1 (ser988) following IR | UV, IR | ser20 | blocks MDM2 interaction; plays a role in p53-mediated apoptosis | [124,170,171,282,286–290] |
| Raf-1 | participates in the Ras signaling pathway initiated from the membrane | UV, IR | ser27 of mouse p53 | increases transactivation ability of p53 | [172–175] |
Table 3.
Dephosphorylation of p53 by Various Phosphatases.
| Phosphatases | General | Inducers | p53 modification | Effect on p53 | References |
| PP2A | major kinase phosphatase in eukaryotic cells | ? | ser378 | reduces DNA sequence-specific binding of p53; reduces apoptosis | [153,176,177] |
| PP5 | participates in cell cycle checkpoint control | UV light reduces PP5 mRNA | ? | reduces p53's transactivation activity | [178] |
| hCDC14A&B | birds to C-terminal domain of p53; dephosphorylate Cdc2 and Cdk2 substrates; involved in exit of mitosis in yeast | ? | ser315 | expected to favor tetramerization by removing inhibitory ser315 phosphorylation; increased stability? nuclear localization? | [151,179,283] |
| ? | ATM-dependent activation of unknown phosphatase | IR | ser376 | creating a binding site for 14-3-3 protein resulting in increased sequence-specific DNA binding | [105] |
Table 4.
Acetylation and Ribosylation of p53.
| Modifier | General | Inducer | p53 modification | Effect on p53 | References |
| P300 | acetyl transferase; acetylation of p53 favored if N-terminus of p53 is phosphorylated; functions in the apoptotic response to IR | UV, IR | acetylation of lys382 both in vitro/in vivo | increases sequence-specific binding of p53; increases stability of p53 | [85,86,180–182] |
| PCAF | acetyl transferase; acetylation of p53 favored if N-terminus of p53 is phosphorylated | UV, IR | acetylation of lys320 both in vitro/in vivo | increases sequence-specific binding of p53 | [86,181] |
| PARP | abundant chromatin-bound enzyme activated by DNA strand breaks; ribosylates itself, histones and other proteins; plays role in strand break and base excision repair; interacts with p53 | IR, not UV | ribosylates sites within central core and C-terminal domain | increases p53 stability; increases transactivation of target genes | [183–194] |
Figure 5.
Map of some of the modifications known to occur following ionizing radiation (top) and UV light (bottom). The p53 protein is divided in its three functional domains. The N-terminus contains the transactivation domain and binding sites for MDM2. The central core contains the sequence-specific binding domain. This is where most mutations are found in human tumors. The C-terminus harbors the NLS, NES and tetramerization domains. The p53-activating signal induced by IR is thought to be triggered by DNA strand breaks while UV light will induce p53 through a mechanism involving blocked RNA polymerase II at sites of DNA damage as well as from signals originating from the cell membrane (i.e., p38). The potential sensors are boxed in and mediators/effectors are listed. The dashed lines represents hypothetical pathways. Circled P represents phosphorylated residue while boxed Ac represents acetylated residue.
Inhibition of RNA polymerase II results in dramatic alterations in nuclear architecture [87,88]. The nucleus increases in size, the chromatin aggregates at the nuclear periphery and the nucleoli disintegrate. It is therefore possible that the induction of p53 following inhibition of transcription could be triggered by nuclear architectural alterations rather than direct signaling from the blocked RNA polymerase complex. Alternatively, since nuclear export is thought to be critical for the normal turnover of p53, the nuclear dismay caused by inhibition of transcription could interfere with the nuclear export machinery leading to the entrapment of p53 in the nucleus. The inhibition of transcription may also indirectly interfere with the p53 ubiquitylation process [54], or by a decrease in the cellular level of MDM2 which in turn would result in p53 accumulation [89] (see below). Further studies are needed to decipher the mechanism(s) of how inhibition of transcription leads to p53 accumulation.
Breaks in the Helix — Ionizing Radiation Induces a Rapid p53 Response
Ionizing radiation induces a number of different types of DNA lesions [90]. Among these are base damage, single- and double-strand breaks and DNA-protein cross-links. Compared to equitoxic doses of UV light, ionizing radiation induces relatively few lesions in DNA [91] (Table 1). Furthermore, the repair of radiation-induced DNA strand breaks is significantly more rapid than the removal of bulky UV-induced lesions [92,93]. It is well-established that ionizing radiation induces a rapid induction of p53 in mammalian cells [36,94] most likely triggered by radiation-induced DNA strand breaks [95]. Is it the strand break itself that is recognized by the cell or is it its effect on chromatin structure and/or function that sets off the alarm?
It has been shown that the DNA in mammalian cells contains unconstrained torsional tension [96]. Studies of the DHFR gene domain have revealed that this unconstrained negative superhelicity is localized to the promoter region of the gene when it is poised for transcription [97]. Interestingly, this localized tension can be abolished when DNA breaks are introduced, on the average, 30 kb away from the area of tension. It is tempting to speculate that the topological integrity of chromatin loops may be monitored by sensor molecules located like spiders in their webs. When tension is lost following introduction of a strand break in a domain, these sensor molecules may signal p53. One such protein may be the ATM protein, which has been shown to be chromatin-associated and able to modify interactions between DNA and the nuclear matrix [98]. Although the “spider-in-the-web” hypothesis is an attractive model for how DNA strand breaks may rapidly trigger p53 mobilization, it has been shown that microinjection of exogenous DNA with free ends into mammalian cells appears to be sufficient to activate p53 [99]. This would argue against a mechanism involving the monitoring of DNA topology for p53 activation, although such a mechanism of p53 activation following ionizing radiation cannot be ruled out.
Proteins thought to be the sensors of radiation-induced DNA strand breaks are the Ataxia Telangiectasia protein (ATM), the ATM-related protein (ATR), the Nejmeegen Breakage Syndrome (NBS) protein and poly(ADP-ribose)polymerase (PARP) (see Tables 2 and 4 and Figure 5). The ATM protein can directly (ser15) [119,120] or indirectly via CHK2 (ser20) [286–289], JNK (ser37) [104] or c-Abl [100–103] phosphorylate p53 following exposure to ionizing radiation. Furthermore, ATM activates a phosphatase that specifically dephosphorylates the ser376, which activates the sequence-specific binding of p53 by allowing for the binding of the 14-3-3 protein [105]. The p53 modifications known to be induced following ionizing radiation are summarized in Figure 5.
Staying Around — Inhibition of Proteasome-Mediated Degradation Extends p53's Half-life
As described above, the p53 tumor suppressor protein is under normal conditions rapidly degraded by the 26S proteosome [52,53,55] in a process dependent on ubiquitylation mediated by MDM2 [60–62] and/or JNK [66]. A number of peptide aldehydes that inhibit the 26S proteasome, such as lactacystin, LLnL and MG132, have been developed [106]. When treating cell cultures with these compounds, the level of the p53 protein rapidly increases [52,53]. Certain cell types have also been shown to undergo apoptosis following treatment with proteosome inhibitors [284,285].
Nitric oxide is a signal mediator that has been implicated in the induction of p53 and apoptosis in macrophages [107]. Recently, it was suggested that the mechanism by which nitric oxide induces the accumulation of p53 is linked to its inhibitory effect on the 26S proteasome [107]. Similarly, it has been suggested that overexpression of the multifunctional protein β-catenin induces p53 accumulation through inhibition of the proteasome degradation pathway [108]. Taken together, there may be a group of compounds or gene products that induce p53 accumulation through the interference of the activity of the 26S proteasome.
Tagging the Damage — Mismatch Repair Proteins Alert p53
Cisplatin adducts in DNA are efficient blocks for transcription both in vitro [109] and in vivo [110]. As discussed above, blockage of transcription is sufficient to induce p53 and apoptosis. Thus, this may be one mechanism of how cisplatin induces p53 and apoptosis in exposed cells. However, recent studies suggest that mismatch repair proteins may also be involved in mediating a signal leading to the induction of apoptosis [100]. This signal transduction pathway, which is initiated by a MLH1-dependent event, involves activation of c-Abl resulting in the phosphorylation and accumulation of the p53 family member p73.
Exposure of cells to the alkylating agent, MNNG, results in many types of DNA lesions including O6-methylguanine. This lesion has been shown to induce p53 and apoptosis in normal cells but not in cells lacking the mismatch repair protein complex MutSα [111]. Thus, it appears that the mismatch repair system recognizes this lesion and induces a signal leading to p53 activation. The mechanism for p53 induction is unknown, but it is possible that the O6-methylguanine lesion complexed with mismatch repair proteins causes an impediment for the elongating RNA polymerase II. Thus, by converting a DNA lesion from a form that most likely is bypassed by RNA polymerase II to a form that blocks elongation, the MutSα protein complex may alert the cells of the damage by using the transcription machinery to signal p53.
Regulation of p53 at the Level of Transcription and Translation
The transcription of the p53 gene is regulated by both transcriptional activators and repressors. A synergistic activation of transcription from the p53 promoter can be accomplished by the transcription factors AP-1, NF-κB and Myc/Max [195]. In addition, YY1 and NF1 [196] can bind to and stimulate p53 promoter activity. In contrast, a number of members of the Pax transcription factor family, which is implicated in the control of mammalian development, have been shown to inhibit transcription of the p53 gene [197]. In addition, the virally encoded Tax protein [198], and overexpression of c-Jun [199] leads to the repression of p53 transcription. Interestingly, the p53 protein itself has been suggested to negatively regulate its own transcriptional expression through an indirect mechanism [200] (Figure 6).
Figure 6.
A p53 autoregulatory feedback loop. Following accumulation, p53 can direct the inhibition of its own transcription by an indirect mechanism and inhibition of its own translation by binding to the 3′ UTR of the p53 mRNA.
Since the stress-inducible transcription factors AP-1 and NF-κB stimulate transcription from the p53 promoter, it is possible that this increased transcription from the p53 gene contributes to the accumulation of p53 following cellular stresses. In fact, studies using a CAT reporter plasmid assay found that a number of different genotoxic agents could stimulate expression from the human p53 promoter [201]. This effect was attributed to a novel genotoxic-responsive p53 promoter element. However, increased levels of endogenous p53 mRNA were not observed in cells exposed to ionizing radiation, UV light, cisplatin or etoposide [11,94]. Since blockage of transcription leads to p53 accumulation [71], regulation of p53 transcription is unlikely to be a universal mechanism by which p53 accumulates following cellular stresses. Thus, increased transcription of the p53 gene may play some role in the accumulation of p53 under certain circumstances but clearly is not a major mechanism by which the p53 response is launched following cellular stress.
The 3′ untranslated region (UTR) of human p53 mRNA [202] and the 5′ UTR of murine p53 mRNA [203] contain sequences that potentially could form stable secondary structures. The human 3′ UTR sequence has the ability to repress translation presumably through RNA-binding factors acting at the 3′ UTR [204]. Interestingly, the p53 protein itself has been found to bind tightly to the 5′ UTR of murine p53 mRNA resulting in the inhibition of its own translation [203]. In addition, the thymidylate synthase protein has been shown to bind to and inhibit translation from the p53 mRNA [205]. It has been hypothesized that following DNA damage-induced stress, the suppression of p53 mRNA translation is reversed. In fact, ionizing radiation, but not UV light, has been shown to partially overcome the repression exerted by the human 3′ UTR of p53 [202]. Taken together, regulation of p53 transcription and translation may contribute to p53 accumulation following certain types of stresses. Moreover, the suppressive role of p53 on its own transcription and translation sets up an interesting negative feedback loop where accumulation of p53 following cellular stress would lead to the shutdown of both its own transcription and translation (Figure 6). This would ensure that the p53 response is turned off shortly after the damage responsible for triggering the response has been repaired.
Breaking the Loop — Inhibiting MDM2- and JNK-Mediated Degradation
It has been proposed that MDM2 suppresses p53 in nonstressed cells in at least three ways [206]. First, MDM2 binds to the same region of p53 as do components of the transcription factor TFIID and thus, the ability of p53 to transactivate target genes is diminished. Second, the MDM2 protein may direct nuclear export of the p53/MDM2 complex. Third, MDM2 can act as a ubiquitin ligase targeting p53 for proteasome-mediated degradation [62]. Mutation of the nuclear export sequence (NES) of MDM2 or inhibition of the export machinery by the drug leptomycin B inhibits the nucleocytoplasmic shuttling of MDM2, resulting in nuclear accumulation of p53 [207].
To unfold the p53 response following cellular stress, the inhibitory activities of the MDM2 protein must be overcome. This could be accomplished by modifications of the p53 protein itself so that MDM2 can no longer bind to it. Alternatively, the MDM2 protein, or other proteins involved in the ubiquitylation and degradation of p53, may be modified so that they no longer can interact with p53. There is evidence suggesting that both of these scenarios may play roles in the accumulation of p53 in stressed cells (Figure 7).
Figure 7.
The p53-MDM2-ARF regulatory feedback loops. Under non-stressed conditions, the MDM2 and p53 proteins balance each other and are found in low amounts. In order for p53 to accumulate in cells, It has to escape the inhibitory action of MDM2. This can be accomplished by (A) mutations in the p53 gene resulting in a transactivation-deficient p53 protein; (B) inhibition of general transcription by e.g., UV light; (C) modifications of the MDM2 protein so that it binds p53 less efficiently; (D) induction of the p14ARF protein which reverses MDM2's inhibitory action on p53 or (E) modifications of the N-terminus of p53 leading to reduced binding to MDM2. DNA-damaging agents may cause p53 accumulation by stimulating the processes denoted with dashed lines, while solid lines denote processes repressing p53 accumulation. Following p53 accumulation, transactivation of the MDM2 gene ensures that the p53 response can be turned off soon after the signal that triggered p53 is removed.
Mutated p53 proteins found in tumors are often much more stable than wild-type p53. This has been suggested to be caused by the inability of the mutant p53 to transactivate the MDM2 gene [65]. Inhibition of MDM2 expression is also accomplished by blockage of RNA polymerase II elongation. It has been shown that following UV irradiation or incubation in the presence of the RNA polymerase II inhibitors DRB and H7, the level of MDM2 protein rapidly decreases [14,89]. Thus, this represents an indirect mechanism by which p53 may be stabilized in cells by the loss of expression of its ubiquitin ligase MDM2. However, induction of p53 can occur at very low doses of UV light in DNA repair-deficient xeroderma pigmentosum cells (XP-A) without concurrent decrease of MDM2 protein levels [73]. Although the synthesis of total mRNA was reduced by about 50% following an exposure of XP-A cells to 4 J/m2 of UV light, the protein level of MDM2 was found to be significantly increased. This would argue that induction of p53 following UV irradiation is more complex than simply resulting from the inhibition of MDM2 expression.
In addition to transcriptional regulation of MDM2, the activity of MDM2 can be regulated through posttranslational modifications. The DNA-dependent protein kinase (DNA-PK), which is activated by DNA strand breaks, can directly phosphorylate MDM2 at ser17 leading to the attenuation of the ability of MDM2 to interact with p53 [208]. Another damage-inducible protein that negatively regulates MDM2-mediated degradation of p53 is the protein tyrosine kinase, c-Abl [103]. The mechanism for how c-Abl neutralizes the inhibitory effect of MDM2 on p53 is not known but could involve blockage of MDM2-mediated nuclear export of p53 or direct interference of the p53 degradation process [103].
The ability of MDM2 to bind to and to ubiquitylate p53 can also be abrogated by expression of the tumor suppressor P14ARF [209–211]. The p14ARF protein has been shown to physically interact with MDM2 and this interaction interferes with the ability of MDM2 to act as a p53 ubiquitin ligase [212]. Expression of the p14ARF is not thought to be induced by DNA-damaging agents [213] but it is induced by oncogenes such as Myc [214], Ras [215] and E2F-1 [216], as well as the viral protein E1A [217]. However, a recent study shows that ARF-/- mouse lymphoma cells have an attenuated induction of p53 and p21WAF1 following treatment with the alkylating agent, cyclophosphamide [218]. In addition, these cells were more chemoresistant compared to corresponding wild-type ARF-expressing cells. Thus, DNA-damage signaling to p53 may in part be mediated by the p14ARF protein. Interestingly, wild-type p53 expression leads to the inhibition of p14ARF transcription, setting up a feedback loop between p53 and p14ARF [219] (Figure 7).
An important mechanism by which DNA-damaging agents may interrupt MDM2 binding to p53 is by modifications of the MDM2-binding domain of p53. Both ionizing radiation and UV light have been shown to induce phosphorylations of serines 15, -33 and -37 of p53 [170,171,220,221] (Figure 5). Ionizing radiation and UV light also induce phosphorylation of ser20, [170,282]. These modifications have been suggested to result in the abrogation of p53-MDM2 interactions leading to increased stability and activity of the p53 protein [220,221,282] (Figure 7). Phosphorylation of only serine 15 will cause inhibition of p53 transactivation activity by abolishing the interaction between p53 and the TATA-binding protein TBP [125], but phosphorylation of both serines 15 and 37 restores transactivation but abrogates the interaction between MDM2 and p53 [125].
JNK has also been shown to mediate ubiquitylation and degradation of p53 in nonstressed cells [66]. JNK can directly interact with p53 within a region spanning amino acids 97–155 of p53. JNK may be an adapter molecule that links p53 to an ubiquitin-ligase complex [66]. Complexes of JNK-p53 are preferentially observed in the G0/G1 phases of the cell cycle, while MDM2-p53 complexes are found preferentially in S/G2/M. Following cellular stresses that induce the activation of JNK, the JNK-mediated ubiquitylation and degradation of p53 are abolished [140]. Activated JNK actually phosphorylates p53 leading to the attenuation of the interaction between p53 and MDM2. Thus, kinases upstream of JNK may be critical regulators of p53 stability by controlling both JNK- and MDM2-mediated ubiquitylation of p53 (Figure 8).
Figure 8.
The regulation of p53 by JNK. In addition to MDM2, JNK can direct ubiquitylation of p53. Following exposure to agents that induce JNK kinase activity, the JNK no longer can direct degradation of p53 but rather, phosphorylates p53 at the N-terminus. This phosphorylation leads to inhibition of MDM2 binding to p53. Thus, by activating the JNK, UV light knocks out two systems that normally direct degradation of p53.
The Cellular Postal Office — Regulating Nucleocytoplasmic Shuttling
The functions of many cell cycle regulators and transcription factors are regulated by nucleocytoplasmic shuttling. To be allowed access into the nucleus, a protein needs to carry a specific zip code, or nuclear localization signal (NLS). Similarly, proteins to be exported need to display a nuclear export signal (NES) or piggyback onto other proteins containing NES. The p53 protein has been shown to shuttle between the cytoplasmic and nuclear compartments in a cell-cycle-dependent fashion [222–224].
The nuclear import of p53 is an active process involving the association of the importin complex with the NLS of p53. In addition to a functional NLS, additional sites on p53 contribute to nuclear import. Mutation of a single lysine or arginine residue (Iys305 or Arg306), or changing the positioning of these two amino acids relative to the NLS, has been shown to result in cytoplasmic localization of the p53 protein [225–227]. The importin complex brings proteins to be imported in contact with the nuclear envelope, and translocation through the nuclear pores is facilitated by the small and abundant GTPase protein, Ran [228–230]. In the cytoplasm. Ran is bound to GDP, but following completion of nuclear import of a protein, the GDP is exchanged for GTP. For nuclear export, Ran-GTP will complex with the exportins and their cargo proteins to facilitate nuclear export. Nuclear export of p53 is thought to be carried out by the export receptor, CRM1 [231]. The export of p53 can be blocked by the drug leptomycin B, resulting in the accumulation of p53 in the nucleus [207,231].
In certain tumors, such as breast cancer and neuroblastoma, p53 is commonly found localized to the cytoplasm [232,233]. This is not due to mutations in the p53 gene. In fact, mutant p53 is most frequently found in the nucleus of cancer cells. The nuclear exclusion of p53 in these cancer cells was initially interpreted as being the result of an inability of p53 to translocate to the nucleus [233]. However, it was recently shown that the cytoplasmic “sequestering” of p53 could be reversed following inhibition of the nuclear export machinery with leptomycin B [231]. Thus, the cytoplasmic localization of p53 in these cells is not due to the inability of p53 to enter the nucleus, but rather is the result of an hyperactive nuclear export of p53.
How is nucleocytoplasmic shuttling of p53 regulated? First, p53 may be anchored in the cytoplasm by specific cytoplasmic proteins or structures. In order for p53 to be imported to the nucleus, the interaction with anchoring proteins must be overcome [227]. Second, the accessibility of the NLS or NES of the p53 protein may be regulated by proteins that bind to this region and block the interaction with the importin complex. It has been shown that the apoptosis antagonist, Bcl-2, in combination with c-Myc, appears to interfere with the nuclear import of p53 although the mechanism for this is unclear [234,235]. Third, the NLS, NES or adjacent sequences of p53 may be modified by phosphorylation, acetylation, ribosylation or other modifications that either stimulate or inhibit nuclear import [236].
Many proteins that shuttle between the cytoplasm and nucleus are regulated by phosphorylation mediated by CK II or Cdc2/cyclin B at sites near their NLS and NES domains [236]. While phosphorylation of these proteins by CKII is associated with nuclear location, phosphorylation by Cdc2/cyclin B is associated with cytoplasmic localization (see Table 2). The C-terminal domain of p53 has both Cdc2/cyclin B and CK II phosphorylation sites. Phosphorylation of ser392 by CK II has been shown to stimulate tetramerization of p53 [151]. Since tetramerization of p53 has been suggested to hide the NES of p53 and thus block nuclear export [231], phosphorylation of p53 by CK II may stimulate nuclear accumulation of p53 by blocking p53's export. In contrast, phosphorylation induced by Cdc2/cyclin B is associated with cytoplasmic localization of many different types of proteins. It has been shown that the tetramerization-stimulating effect of ser392 phosphorylation by CK II can be blocked by phosphorylation of ser315 by Cdc2/cyclin B [151]. Thus, it is possible that the nucleocytoplasmic shuttling of p53 could be regulated, in part, by these two kinases where CK II may stimulate nuclear import and Cdc2/cyclin B may stimulate nuclear export (Figure 9).
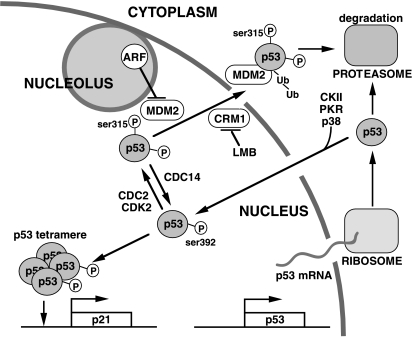
Figure 9.
Nucleocytoplasmic shuttling of p53. Following its synthesis, the cytoplasmic p53 protein may be directly degraded by the proteasome or imported into the nucleus. Phosphorylation of ser392 may stimulate nuclear localization by favoring tetramerization. p53 is also subject to nuclear export via the CRM1 nuclear export protein complex, a process that can be inhibited by the drug leptomycin B (LMB). The nuclear export process may be stimulated by CDK-mediated phosphorylation of ser315 and by the MDM2 protein. The phosphorylation of ser315, which can be reversed by the Cdc14 phosphatases, inhibits p53 tetramerization. The p14ARF protein may interfere with the shuttling of both MDM2 and p53 by sequestering MDM2. Following export, the p53 protein is directed to the 26S proteasome by MDM2. Whether some of the exported p53 are allowed to re-enter the nucleus is not clear.
In addition to its potential role in cytoplasmic localization of p53, phosphorylation of ser315 may stimulate degradation of the p53 protein [163]. Whether this increased degradation is due to the phosphorylation itself or its stimulation of nuclear export is not clear. Unexpectedly, studies in which single or combinational mutations have been engineered in the p53 gene to eliminate phosphorylation sites showed no evidence for a role of any of the phosphorylation sites in regulating the stability of p53 [237–239]. Interestingly, deletion of the tetramerization domain of p53 was found to result in a p53 form that was not further stabilized by cellular stresses such as UV light. This suggests that the ability to form tetramers may be important for the stabilization of p53 following cellular stress [237–239]. However, these tetramerization-defective p53 mutants were expressed to higher levels than wild-type p53 in transient transfection assays, even in unirradiated cells. The high level of protein expression of these mutants may be due to diminished MDM2 binding since it has been suggested that an intact tetramerization domain is essential for the binding of MDM2 to the N-terminus of p53 [13]. Thus, the results from these studies regarding the role of the tetramerization domain in stabilizing p53 following stress are difficult to interpret.
The recently identified human protein phosphatases Cdc14A and Cdc14B, which were cloned by homology to the yeast Cdc14 [179], have been found to dephosphorylate p53 [283]. These phosphatases bind to the C-terminus of p53 and specifically dephosphorylate the ser315 site of p53. In yeast, Cdc14 specifically dephosphorylates proteins that are substrates for the yeast Cdc2 homologue, Cdc28/clb. One example of a protein that is subject to phosphorylation by Cdc28/clb and dephosphorylation by Cdc14 is the transcription factor, Swi5. Phosphorylation of Swi5 by Cdc28/clb stimulates its nuclear export, while Cdc14-mediated dephosphorylation of the same site leads to its nuclear localization [240]. By analogy to the yeast Cdc28/clb and Cdc14, the human Cdc2/cyclin B and Cdc14 may dictate the cellular localization of p53 by regulating the phosphorylation status of the ser315 site of p53 (Figure 9). Whether the activity or cellular localization of the human Cdc14 phosphatases is altered following certain cellular stresses is currently being investigated.
Activation of DNA-Binding Activity and the Battle for Cofactors
As discussed above, stabilization of the p53 protein and nuclear localization are important mechanisms required for the activation of the p53 response. However, to achieve efficient transactivation of target genes, two more events need to take place. First, the sequence-specific DNA-binding domain needs to be activated, and second, the p53 protein needs to interact with cofactors to stimulate transcription of the target genes.
The p53 protein is thought to contain a cryptic central DNA-binding domain negatively regulated by the C-terminal domain [12]. It has been shown that antibodies or small peptides directed against the C-terminal domain, inhibitors of molecular chaperones, as well as phosphorylation or acetylation of the C-terminal domain, result in the activation of the sequence-specific activity of the DNA-binding domain [13,241]. However, recent findings show that nuclear accumulation of p53 per se may be sufficient for transactivation of target genes [207,242]. In these studies, both p21WAF1 and MDM2 proteins accumulated in the nucleus of cells treated with the nuclear export inhibitor, leptomycin B. Furthermore, the accumulated p53 proteins isolated from cells incubated with proteasome inhibitors were fully capable of binding DNA in a sequence-dependent manner [242]. It is possible that the “latent” form of p53 has some residual sequence-specific DNA-binding activity, that if present in large enough amounts in the nucleus will result in the activation of target genes. Alternatively, accumulation of p53 in the nucleus favors the p53 tetramerization which is thought to lead to activation of the DNA-binding domain by induced conformational alterations [13]. It is possible that some of the “activating” modifications of the C-terminus, such as phosphorylation of ser392, are related to stimulation of tetramerization which in turn activates the DNA-binding domain [148]. Association of p53 with noncovalent binding activators such as REF-1 and HMG-1, which are abundant nuclear proteins, may lead to the activation of the DNA-binding properties of p53 without the need for covalent modifications of its C-terminus [131,243,244]. Thus, induction of p53-mediated transactivation may be achieved either by specific modifications of the C-terminus leading to the induction of the sequence-specific DNA-binding activity, or in a less sophisticated manner, by brute force accumulation of p53 proteins in the nucleus.
To stimulate transcription of its target genes, p53 needs to interact with various transcription cofactors in order to stimulate transcription. It has been shown that the acetyl transferase CBP, which stimulates transcription by acetylating histones, plays an important role in p53-mediated transactivation [245–248]. Other proteins that p53 interacts with that could stimulate transcription are transcription factors TBP [20,249,250], TFIIH [251,252], TAFs [253], Sp1 [254], the p300 cofactor JMY [255] and DNA topoisomerase I [256]. Since these enzymes and factors must be shared between many different genes, activated p53 and p53-inducible genes must compete for these factors with other transcription activators and genes. As a result of p53 accumulation following cellular stress, transcription factors will be sequestered by p53 leading to the activation of genes containing p53-binding sequences, while transcription from genes lacking p53-binding sequences will be attenuated [19–22]. In addition to the nonspecific inhibition of many genes by the sequestering transcription factors, p53 has recently been shown to specifically repress certain genes by bringing a histone deacetylase complex to these genes [23]. The deacetylase activity will reverse histone acetylation resulting in the compaction of the chromatin and repression of transcription.
Various stresses that induce accumulation of p53, such as UV light, concurrently induce the transcription factors NF-κB and AP-1 [257]. NF-κB binds, like p53, to the histone acetyl transferase CBP. It has been suggested that the battle between p53 and NF-κB for CBP sets up a transcriptional cross-talk between these two stress pathways [258,259]. The survival-promoting functions of NF-κB could perhaps be due, in part, to the inhibition of the apoptosis-promoting function of p53 by the sequestering of CBP [258,259]. Viral proteins, such as the adenovirus E1A protein, can bind to and suppress the activity of p300/CBP and PCAF, thereby interfering with p53 function [260,261]. To favor interactions between p53 and CBP following stress, stress-induced modifications of the p53 protein may increase the binding affinity of CBP for p53. In fact, phosphorylation of ser15 of p53 has been found to stimulate CBP-binding to p53 [123]. Binding of CBP to the N-terminus will also be favored by phosphorylations that exclude MDM2 binding [220,245]. Taken together, viral proteins and parallel stress signaling pathways may limit the transactivation activity of p53 by inactivating or competing for transcriptional cofactors while stress-induced modifications of the p53 protein may stack the cards in favor of p53.
Conclusions and Future Directions
It is not difficult to appreciate that a protein that has been bestowed so much power over the fate of a cell must adhere to intricate and rigid regulation. Its role in transactivation is regulated by protein accumulation, nucleocytoplasmic shuttling, induction of its sequence-specific DNA-binding activity and through the competition for transcription cofactors. Relying predominantly on inhibition of its own degradation for induction, p53-mediated apoptosis can be achieved even after insults that severely limit the ability of the cell to perform transcription. If a damaged cell survives long enough to fully repair itself, multiple feedback systems are in place to eliminate p53 so that the repaired cell can re-enter the cell cycle. Following UV irradiation, e.g., the cell has to completely remove all UV-induced transcription blocks in the MDM2 gene before the p53 response can be turned off. Thus, the monitoring of the recovery of MDM2 mRNA synthesis is a way for the cell to assess the severity of the insult and determine whether apoptosis would be an appropriate solution.
It would be of great interest to better understand the alternative mechanisms used by tumor cells to inactivate their p53 function in addition to mutations in the p53 gene. What regulates the hyperactive nuclear export and cytoplasmic localization of “wild-type” p53 in cells of many tumor types [231]? What is different about the regulation of p53 in epithelial cells compared to fibroblasts [262–264]? How important are the inhibitory effects of p53 function by environmental pollutants such as arsenic [265] and cadmium [266] for human health?
More than 15,000 papers featuring p53 have been published since its discovery in 1979 [267–269]. About 10 papers are currently published daily. With this productivity in both basic and clinical research, there are good prospects that our further understanding of the regulation and function of p53 soon will lead to fruitful new efforts in both the prevention and treatment of cancer.
Acknowledgements
I would like to apologize to everyone whose work I have overlooked while writing this review. I thank current and former members of my research group for their contributions to this review and Sara Ljungman, Bruce McKay, Jiayuh Lin and Mohamed Khan for proofreading this manuscript. The p53 work in my laboratory is supported by a grant from the National Institute of Health (CA82376-01).
References
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 3.Baker S, Preisinger A, Jessup J, Paraskeva C, Markowitz S, Wilson J, Hamilton S, Vogelstein B. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 4.Fisher DE. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78:539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 5.Harris CC. Structure and function of the p53 tumor suppressor gene: clues for rational cancer therapeutic strategies. J Natl Cancer Inst. 1996;88:1442–1455. doi: 10.1093/jnci/88.20.1442. [DOI] [PubMed] [Google Scholar]
- 6.Levine A, Perry M, Chang A, Silver A, Dittmer D, Wu M, Welsh D. The 1993 Walter Hubert lecture: the role of the p53 tumour-suppressor gene in tumorigenesis. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 8.Canman C, Kastan M. Role of p53 in apoptosis. Adv Pharmacol. 1997;41:429–460. doi: 10.1016/s1054-3589(08)61068-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhou XL, Wang XW, Xu LX, Hagiwara K, Nagashima M, Wolkowicz R, Zurer I, Rotter V, Harris CC. COOH-terminal domain of p53 modulates p53-mediated transcriptional transactivation, cell growth, and apoptosis. Cancer Res. 1999;59:843–848. [PubMed] [Google Scholar]
- 10.Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 12.Hupp T, Meek D, Midgley C, Lane D. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 13.Hupp TR. Regulation of p53 protein function through alterations in protein-folding pathways. Cell Mol Life Sci. 1999;55:88–95. doi: 10.1007/s000180050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman DA, Levine AJ. Regulation of the p53 protein by the MDM2 oncoprotein — thirty-eighth G.H.A. Clowes Memorial Award lecture. Cancer Res. 1999;59:1–7. [PubMed] [Google Scholar]
- 15.Smith ML, Fornace AJ. p53-mediated protective responses to UV irradiation. Proc Natl Acad Sci USA. 1997;94:12255–12257. doi: 10.1073/pnas.94.23.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amundson SA, Bittner M, Chen YD, Trent J, Meltzer P, Fornace AJ. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 17.Zambetti G, Bargonetti J, Walker K, Prives C, Levine A. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 18.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 19.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can downmodulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seto E, Usheva A, Zambetti G, Momand J, Horikoshi N, Weinmann R, Levine A, Shenk T. Wild-type p53 binds to the TATA-binding protein and repress transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Berk AJ. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 23.Murphy M, Ahn J, Walker K, Hoffman W, Evans R, Levine A, George D. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harper J, Adami G, Wei N, Keyormarsi K, Elledge S. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 25.Dulic V, Kaufmann WK, Wilson SJ, TIsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 26.Smith ML, Chen IT, Zha QM, Oconnor PM, Fornace AJ. Involvement of the p53 tumor suppressor in repair of UV-type DNA damage. Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 27.Ford JM, Hanawalt PC. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 29.McKay BC, Francis MA, Rainbow AJ. Wild-type p53 is required for heat shock and ultraviolet-light-enhanced repair of a UV-damaged reporter gene. Carcinogenesis. 1997;18:245–249. doi: 10.1093/carcin/18.2.245. [DOI] [PubMed] [Google Scholar]
- 30.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKay BC, Ljungman M, Rainbow AJ. Potential roles for p53 in nucleotide excision repair. Carcinogenesis. 1999;20:1389–1396. doi: 10.1093/carcin/20.8.1389. [DOI] [PubMed] [Google Scholar]
- 32.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16(INK4a) Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 33.McKay B, Ljungman M. Role for p53 in the recovery of transcription and protection against apoptosis induced by ultraviolet light. Neoplasia. 1999;1:276–284. doi: 10.1038/sj.neo.7900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe S, Schmitt E, Smith S, Osborne B, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 35.Clarke A, Purdie C, Harrison D, Morris R, Bird C, Hooper M, Wyllie A. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 36.Lu X, Lane DP. Differential induction of transcriptionally active p53 following UV or ionizing radiation — defects in chromosome instability syndromes? Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 37.Soengas MS, Alarcón RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, Lowe SW. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 38.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 39.Owen-Schaub LB, Zhang W, Cusack JC, Angelo LS, Santee SM, Fujiwara T, Roth JA, Deisseroth AB, Zhang WW, Kruzel E, et al. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu GS, Burns TF, McDonald ER, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, ElDeiry WS. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 41.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by transactivation-deficient p53. Gene Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 42.Johnson TM, Yu ZX, Ferrans VJ, Lowenstein RA, Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci USA. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 44.Li P-F, Dietz R, von Harsdorf R. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 1999;18:6027–6036. doi: 10.1093/emboj/18.21.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett M, Macdonald K, Chan SW, Luzio JP, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- 46.Rehemtulla A, Hamilton CA, Chinnaiyan AM, Dixit VM. Ultraviolet-radiation-induced apoptosis is mediated by activation of CD-95 (Fas/APO-1) J Biol Chem. 1997;272:25783–25786. doi: 10.1074/jbc.272.41.25783. [DOI] [PubMed] [Google Scholar]
- 47.Hill LL, Ouhtit A, Loughlin SM, Kripke ML, Ananthaswamy HN, Owen-Schaub LB. Fas ligand: a sensor for DNA damage critical in skin cancer etiology. Science. 1999;285:898–900. doi: 10.1126/science.285.5429.898. [DOI] [PubMed] [Google Scholar]
- 48.Chinnaiyan A. The apoptosome: heart and soul of the cell death machine. Neoplasia. 1999;1:5–15. doi: 10.1038/sj.neo.7900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nuñez G, Benedict M, Hu Y, Inohara N. Caspases: the proteases of the apoptotic pathway. Oncogene. 1998;17:3237–3245. doi: 10.1038/sj.onc.1202581. [DOI] [PubMed] [Google Scholar]
- 50.Sionov R, Haupt Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 51.Scheffner M, Werness B, Huibregtse J, Levine A, Howley P. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 52.Chowdary D, Dermody J, Jha K, Ozer H. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 54.Maki CG, Howley PM. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown J, Pagano M. Mechanism of p53 degradation. Biochim Biophys Acta. 1997;1332:O1–O6. doi: 10.1016/s0304-419x(96)00048-0. [DOI] [PubMed] [Google Scholar]
- 56.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 57.Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 58.Koepp D, Harper J, Elledge S. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 59.Coux O, Tanaka K, Goldberg A. Structure and functions of the 20S and 26S proteosomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 60.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 61.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 62.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 63.Fuchs SY, Adler V, Buschmann T, Wu XW, Ronai Z. Mdm2 association with p53 targets its ubiquitination. Oncogene. 1998;17:2543–2547. doi: 10.1038/sj.onc.1202200. [DOI] [PubMed] [Google Scholar]
- 64.Kubbutat M, Ludwig R, Ashcroft M, Vousden K. Regulation of Mdm2-directed degradation by the C-terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 66.Fuchs SY, Adler V, Buschmann T, Yin ZM, Wu XW, Jones SN, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuchs SY, Fried VA, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 68.Kubbutat MHG, Vousden KH. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol Cell Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pariat M, Carillo S, Molinari M, Salvat C, Debussche L, Bracco L, Milner J, Piechaczyk M. Proteolysis by calpains: a possible contribution to degradation of p53. Mol Cell Biol. 1997;17:2806–2815. doi: 10.1128/mcb.17.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonen H, Shkedy D, Barnoy S, Kosower NS, Ciechanover A. On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 1997;406:17–22. doi: 10.1016/s0014-5793(97)00225-1. [DOI] [PubMed] [Google Scholar]
- 71.Ljungman M, Zhang FF, Chen F, Rainbow AJ, McKay BC. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 72.Yamaizumi M, Sugano T. UV-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene. 1994;9:2775–2784. [PubMed] [Google Scholar]
- 73.Ljungman M, Zhang F. Blockage of RNA polymerase as a possible trigger for UV light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 74.Dumaz N, Duthu A, Ehrhart JC, Drougard C, Appella E, Anderson CW, May P, Sarasin A, DayaGrosjean L. Prolonged p53 protein accumulation in trichothiodystrophy fibroblasts dependent on unrepaired pyrimidine dimers on the transcribed strands of cellular genes. Mol Carcinogen. 1997;20:340–347. [PubMed] [Google Scholar]
- 75.McKay BC, Ljungman M, Rainbow AJ. Persistent DNA damage induced by ultraviolet light inhibits p21 (waf1) and bax expression: implications for DNA repair, UV sensitivity and the induction of apoptosis. Oncogene. 1998;17:545–555. doi: 10.1038/sj.onc.1201963. [DOI] [PubMed] [Google Scholar]
- 76.Sauerbier W, Hercules K. Gene and transcription mapping by radiation effects. Annu Rev Genet. 1978;12:329–363. doi: 10.1146/annurev.ge.12.120178.001553. [DOI] [PubMed] [Google Scholar]
- 77.Selby G, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 78.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JHJ, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 80.Ko LJ, Shieh SY, Chen XB, Jayaraman L, Tamai K, Taya Y, Prives C, Pan ZQ. p53 is phosphorylated by CDK7-cyclin H in a p36(MAT1)-dependent manner. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu H, Fisher RP, Bailey P, Levine AJ. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol Cell Biol. 1997;17:923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:81–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 83.Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, OwenHughes T, Workman J, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 84.Léveillard T, Wasylyk B. The MDM2 C-terminal region binds to TAF(II)250 and is required for MDM2 regulation of the cyclin A promoter. J Biol Chem. 1997;272:30651–30661. doi: 10.1074/jbc.272.49.30651. [DOI] [PubMed] [Google Scholar]
- 85.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 86.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brasch K. Drug and metabolite-induced perturbations in nuclear structure and function: a review. Biochem Cell Biol. 1990;68:408–426. doi: 10.1139/o90-059. [DOI] [PubMed] [Google Scholar]
- 88.Haaf T, Ward DC. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp Cell Res. 1996;224:163–173. doi: 10.1006/excr.1996.0124. [DOI] [PubMed] [Google Scholar]
- 89.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog Nucleic Adds Res Mol Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 91.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and repairability. Prog Nucleic Adds Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 92.Ljungman M. Repair of radiation-induced DNA strand breaks does not occur preferentially in transcriptionally active DNA. Radiat Res. 1999;152:444–449. [PubMed] [Google Scholar]
- 93.Bohr VA, Smith CL, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 94.Kastan M, Onyekwere O, Sidransky D, Vogelstein B, Craig R. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 95.Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ljungman M, Hanawalt PC. Localized torsional tension in the DNA of human cells. Proc Natl Acad Sci USA. 1992;89:6055–6059. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ljungman M, Hanawalt P. Presence of negative torsional tension in the promoter region of the transcriptionally poised dihydrofolate reductase gene in vivo. Nucleic Acids Res. 1995;23:1782–1789. doi: 10.1093/nar/23.10.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smilenov LB, Dhar S, Pandita TK. Altered telomere nuclear matrix interactions and nucleosomal periodicity in ataxia telangiectasia cells before and after ionizing radiation treatment. Mol Cell Biol. 1999;19:6963–6971. doi: 10.1128/mcb.19.10.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang LC, Clarkin KG, Wahl GM. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G(1) arrest. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Levrero M, Wang JYJ. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 101.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73 alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 102.Yuan ZM, Shioya H, Ishiko T, Sun XG, Gu JJ, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D. P73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 103.Sionov RV, Moallem E, Berger M, Kazaz A, Gerlitz O, Ben-Neriah Y, Oren M, Haupt Y. c-Abl neutralizes the inhibitory effect of Mdm2 on p53. J Biol Chem. 1999;274:8371–8374. doi: 10.1074/jbc.274.13.8371. [DOI] [PubMed] [Google Scholar]
- 104.Shafman TD, Saleem A, Kyriakis J, Weichselbaum R, Kharbanda S, Kufe DW. Defective induction of stress-activated protein kinase activity in ataxia telangiectasia cells exposed to ionizing radiation. Cancer Res. 1995;55:3242–3245. [PubMed] [Google Scholar]
- 105.Waterman M, Stavridi E, Waterman J, Halazonetis T. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 106.Rock K, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A. Inhibitors of the proteosome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 107.Glockzin S, von Knethen A, Scheffner M, Brune B. Activation of the cell death program by nitric oxide involves inhibition of the proteasome. J Biol Chem. 1999;274:19581–19586. doi: 10.1074/jbc.274.28.19581. [DOI] [PubMed] [Google Scholar]
- 108.Damalas A, Ben-Ze'ev A, Simcha I, Shtutman N, Leal JFN, Zhurinsky J, Geiger B, Oren M. Excess beta-catenin promotes accumulation of transcriptionally active p53. EMBO J. 1999;18:3054–3063. doi: 10.1093/emboj/18.11.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Corda Y, Anin MF, Leng M, Job D. Spectrum of DNA-platinum adduct recognition by prokaryotic and eukaryotic DNA-dependent RNA polymerases. Biochemistry. 1993;32:8582–85884. doi: 10.1021/bi00084a027. [DOI] [PubMed] [Google Scholar]
- 110.Mello JA, Lippard SJ, Essigmann JM. DNA adducts of cis-diamminedichloroplatinum (II) and its trans-isomer inhibit RNA polymerase II differentially in vivo. Biochemistry. 1995;34:14783–14791. doi: 10.1021/bi00045a020. [DOI] [PubMed] [Google Scholar]
- 111.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kastan MB, Zhan QM, Eldeiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 113.Jongmans W, Vuillaume M, Chrzanowska K, Smeets D, Sperling K, Hall J. Nijmegen breakage syndrome cells fail to induce the p53-mediated DNA damage response following exposure to ionizing radiation. Mol Cell Biol. 1997;17:5016–5022. doi: 10.1128/mcb.17.9.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carney J, Maser R, Olivares H, Davis E, Le Beau M, Yates J, III, Hays L, Morgan W, Petrini J. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 115.Khanna KK, Lavin MF. Ionizing radiation and UV induction of p53 by different pathways in ataxia telangiectasia cells. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 116.Cortez D, Wang Y, Qin J, Elledge S. Requirement of ATM-dependent phosphorylation of Brca-1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 117.Smith GCM, Cary RB, Lakin ND, Hann BC, Teo SH, Chen DJ, Jackson SP. Purification and DNA binding properties of the ataxia telangiectasia gene product ATM. Proc Natl Acad Sci USA. 1999;96:11134–11139. doi: 10.1073/pnas.96.20.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Suzuki K, Kodama S, Watanabe M. Recruitment of ATM protein to double strand DNA irradiated with ionizing radiation. J Biol Chem. 1999;274:25571–25575. doi: 10.1074/jbc.274.36.25571. [DOI] [PubMed] [Google Scholar]
- 119.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 120.Banin S, Moyal L, Shieh SY, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATN in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 121.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Tay Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 122.Blasina A, Price BD, Turenne GA, McGowan CH. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 123.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 124.Unger T, Sionov RV, Moallem E, Yee CL, Howley PM, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 125.Pise-Masison C, Radonovich M, Sakaguchi K, Appella E, Brady J. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J Virol. 1998;72:6348–6355. doi: 10.1128/jvi.72.8.6348-6355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sarkaria J, Tibbetts R, Busby E, Kennedy A, Hill D, Abraham R. Inhibition of phosphoinostide 3-kinase related kinases by the radiosenzitizing agent, wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 127.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wright J, Keegan K, Herendeen D, Bentley N, Carr A, Hoekstra M, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lakin N, Hann B, Jackson S. The ataxia telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 131.Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kachnic LA, Wu B, Wunsch H, Mekeel KL, DeFrank JS, Tang W, Powell SN. The ability of p53 to activate downstream genes p21WAF1/cip1 and MDM2, and cell cycle arrest following DNA damage is delayed and attenuated in scid cells deficient in the DNA-dependent protein. J Biol Chem. 1999;274:13111–13117. doi: 10.1074/jbc.274.19.13111. [DOI] [PubMed] [Google Scholar]
- 133.Abraham J, Spaner D, Benchimol S. Phosphorylation of p53 protein in response to ionizing radiation occurs at multiple sites in both normal and DNA-PK deficient cells. Oncogene. 1999;18:1521–1527. doi: 10.1038/sj.onc.1202454. [DOI] [PubMed] [Google Scholar]
- 134.Araki R, Fukumura R, Fujimori A, Taya Y, Shiloh Y, Kurimasa A, Burma S, Li GC, Chen DJ, Sato K, Hoki Y, Tatsumi K, Abe M. Enhanced phosphorylation of p53 serine 18 following DNA damage in DNA-dependent protein kinase catalytic subunit-deficient cells. Cancer Res. 1999;59:3543–3546. [PubMed] [Google Scholar]
- 135.Jimenez GS, Bryntesson F, Torres-Arzayus MI, Priestley A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo PA, Taccioli GE, Wahl GM, Hubank M. DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature. 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- 136.Huang LC, Clarkin KC, Wahl GM. P53-dependent cell cycle arrests are preserved in DNA-activated protein kinase-deficient mouse fibroblasts. Cancer Res. 1996;56:2940–2944. [PubMed] [Google Scholar]
- 137.Take Y, Kumano M, Teraoka H, Nishimura S, Okuyama A. DNA-dependent protein kinase inhibitor (OK-1035) suppresses p21 expression in HCT116 cells containing wild-type p53 induced by adriamycin. Biochem Biophys Res Commun. 1996;221:207–212. doi: 10.1006/bbrc.1996.0575. [DOI] [PubMed] [Google Scholar]
- 138.Hu MCT, Qiu WR, Wang YP. JNK1, JNK2 and JNK3 are p53 N-terminal serine 34 kinases. Oncogene. 1997;15:2277–2287. doi: 10.1038/sj.onc.1201401. [DOI] [PubMed] [Google Scholar]
- 139.Milne DM, Campbell LE, Campbell DG, Meek DW. p53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- 140.Fuchs SY, Adier V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 142.Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan MB, Wang JYJ. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 143.Shafman T, Kanna KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, Egerton M, Shiloh Y, Kharbanda S, Kufe D, Lavine MF. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 144.Kharbanda S, Pandey P, Jin SF, Inoue S, Bharti A, Yuan ZM, Weichselbaum R, Weaver D, Kufe D. Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature. 1997;386:732–735. doi: 10.1038/386732a0. [DOI] [PubMed] [Google Scholar]
- 145.Knippschild U, Milne DM, Campbell LE, DeMaggio AJ, Christenson E, Hoekstra MF, Meek DW. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene. 1997;15:1727–1736. doi: 10.1038/sj.onc.1201541. [DOI] [PubMed] [Google Scholar]
- 146.Meek D, Simon S, Kikkawa U, Eckhart W. The p53 tumor suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hall SR, Campbell LE, Meek DW. Phosphorylation of p53 at the casein kinase II site selectively regulates p53-dependent transcriptional repression but not transactivation. Nucleic Acids Res. 1996;24:1119–1126. doi: 10.1093/nar/24.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hupp TR, Lane DP. Two distinct signaling pathways activate the latent DNA binding function of p53 in a casein kinase II-independent manner. J Biol Chem. 1995;270:18165–18174. doi: 10.1074/jbc.270.30.18165. [DOI] [PubMed] [Google Scholar]
- 149.Filhol O, Baudier J, Chambaz EM, Cochet C. Casein kinase 2 inhibits the renaturation of complementary DNA strands mediated by p53 protein. Biochem J. 1996;316:331–335. doi: 10.1042/bj3160331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Appel K, Wagner P, Boldyreff B, Issinger O-G, Montenarh M. Mapping of the interaction sites of the growth suppressor protein p53 with the regulatory b-subunit of protein kinase CK2. Oncogene. 1995;11:1971–1978. [PubMed] [Google Scholar]
- 151.Sakaguchi K, Sakamoto H, Lewis MS, Anderson CW, Erickson JW, Appella E, Xie D. Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry. 1997;36:10117–10124. doi: 10.1021/bi970759w. [DOI] [PubMed] [Google Scholar]
- 152.Adamczewski J, Rossignol M, Tassan J-P, Nigg E, Moncollin V, Egly J-M. MAT1, cdk7 and cyclin H form a complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 153.Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- 154.Youmell M, Park S, Basu S, Price B. Regulation of the p53 protein by protein kinase Cα and protein kinase Cξ. Biochem Biophys Res Commun. 1998;245:514–518. doi: 10.1006/bbrc.1998.8471. [DOI] [PubMed] [Google Scholar]
- 155.Rusnak JM, Lazo JS. Downregulation of protein kinase C suppresses induction of apoptosis in human prostatic carcinoma cells. Exp Cell Res. 1996;224:189–199. doi: 10.1006/excr.1996.0127. [DOI] [PubMed] [Google Scholar]
- 156.Milne DM, Mckendrick L, Jardine LJ, Deacon E, Lord JM, Meek DW. Murine p53 is phosphorylated within the PAb421 epitope by protein kinase C in vitro, but not in vivo, even after stimulation with the phorbol ester o-tetradecanoylphorbol 13-acetate. Oncogene. 1996;13:205–211. [PubMed] [Google Scholar]
- 157.Matsui M, DeLeo V. Induction of protein kinase C activity by ultraviolet radiation. Carcinogenesis. 1990;11:229–234. doi: 10.1093/carcin/11.2.229. [DOI] [PubMed] [Google Scholar]
- 158.Bischoff J, Friedman P, Marshak D, Prives C, Beach D. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc Natl Acad Sci USA. 1990;87:4766–4770. doi: 10.1073/pnas.87.12.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stürzbecher H-W, Maimets T, Chumakov P, Brain R, Addison C, Simanis V, Rudge K, Philp R, Grimaldi M, Court W, Jenkins J. p53 interacts with p34cdc2 in mammalian cells: implications for the cell cycle control and oncogenesis. Oncogene. 1990;5:795–801. [PubMed] [Google Scholar]
- 160.Price BD, Hughesdavies L, Park SJ. cdk2 kinase phosphorylates serine 315 of human p53 in vitro. Oncogene. 1995;11:73–80. [PubMed] [Google Scholar]
- 161.Wagner P, Fuchs A, Gotz C, Nastainczyk W, Montenarh M. Fine mapping and regulation of the association of p53 with p34(Cdc2) Oncogene. 1998;16:105–111. doi: 10.1038/sj.onc.1201510. [DOI] [PubMed] [Google Scholar]
- 162.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 163.Lin W-C, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 164.Herzinger T, Funk J, Hillmer K, Eick D, Wolf D, Kind P. Ultraviolet B irradiation-induced G2 cell cycle arrest in human keratinocytes by inhibitory phosphorylation of the cdc2 cell cycle kinase. Oncogene. 1995;11:2151–2156. [PubMed] [Google Scholar]
- 165.Poon RYC, Jiang W, Toyoshima H, Hunter T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage. J Biol Chem. 1996;271:13283–13291. doi: 10.1074/jbc.271.22.13283. [DOI] [PubMed] [Google Scholar]
- 166.Poon RYC, Chau MS, Yamashita K, Hunter T. The role of Cdc2 feedback loop control in the DNA damage checkpoint in mammalian cells. Cancer Res. 1997;57:5168–5178. [PubMed] [Google Scholar]
- 167.Huang CS, Ma WY, Maxiner A, Sun Y, Dong ZG. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J Biol Chem. 1999;274:12229–12235. doi: 10.1074/jbc.274.18.12229. [DOI] [PubMed] [Google Scholar]
- 168.Cuddihy AR, Li SY, Tam NWN, Wong AHT, Taya Y, Abraham N, Bell JC, Koromilas AE. The double-stranded RNA activated protein kinase PKR enhances transcriptional activation by tumor suppressor p53. Mol Cell Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cuddihy AR, Wong AHT, Tam NWN, Li SY, Koromilas AE. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- 170.Shieh SY, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Unger T, Juven-Gershon T, Moallem E, Berger M, Sionov RV, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jamal S, Ziff EB. Raf phosphorylates p53 in vitro and potentiates p53-dependent transcriptional transactivation in vivo. Oncogene. 1995;10:2095–2101. [PubMed] [Google Scholar]
- 173.Kasid U, Suy S, Dent P, Ray S, Whiteside TL, Sturgill TW. Activation of Raf by ionizing radiation. Nature. 1996;382:813–816. doi: 10.1038/382813a0. [DOI] [PubMed] [Google Scholar]
- 174.Radler-Pohl A, Sachsenmaier C, Gebel S, Auer HP, Bruder JT, Rapp U, Angel P, Rahmsdorf HJ, Herrlich P. UV-induced activation of AP-1 involves obligatory extranuclear steps including Raf-1 kinase. EMBO J. 1993;12:1005–1012. doi: 10.1002/j.1460-2075.1993.tb05741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Devary Y, Gottlieb R, Smeal T, Karin M. The mammalian ultraviolet response is triggered by activation of Src tyrosine kinase. Cell. 1992;71:1081–1091. doi: 10.1016/s0092-8674(05)80058-3. [DOI] [PubMed] [Google Scholar]
- 176.Yan Y, Shay JW, Wright WE, Mumby MC. Inhibition of protein phosphatase activity induces p53-dependent apoptosis in the absence of p53 transactivation. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- 177.Millward T, Zolnierowicz S, Hemmings B. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–191. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- 178.Zuo Z, Dean N, Honkanen R. Serine/threonine protein phosphatase type 5 acts upstream of p53 to regulate the induction of p21WAF1/Cip1 and mediate growth arrest. J Biol Chem. 1998;273:12250–12258. doi: 10.1074/jbc.273.20.12250. [DOI] [PubMed] [Google Scholar]
- 179.Li L, Ernsting B, Wishart M, Lohse D, Dixon J. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J Biol Chem. 1997;272:29403–29406. doi: 10.1074/jbc.272.47.29403. [DOI] [PubMed] [Google Scholar]
- 180.Yuan ZM, Huang YY, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Yokoyama K, Weichselbaum R, Shi Y, Kufe D. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- 181.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yuan ZM, Huang YY, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Shi Y, Weichselbaum R, Kufe D. Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene. 1999;18:5714–5717. doi: 10.1038/sj.onc.1202930. [DOI] [PubMed] [Google Scholar]
- 183.Wesierska-Gadek J, Bugajskaschretter A, Cerni C. ADP-ribosylation of p53 tumor suppressor protein: mutant but not wild-type p53 is modified. J Cell Biochem. 1996;62:90–101. doi: 10.1002/(sici)1097-4644(199607)62:1<90::aid-jcb10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 184.Wesierska-Gadek J, Schmid G, Cerni C. ADP-ribosylation of wild-type p53 in vitro: binding of p53 protein to specific p53 consensus sequence prevents its modification. Biochem Biophys Res Commun. 1996;224:96–102. doi: 10.1006/bbrc.1996.0990. [DOI] [PubMed] [Google Scholar]
- 185.Ahnström G, Ljungman M. Effects of 3-aminobenzamide on the rejoining of DNA-strand breaks in mammalian cells exposed to methyl methanesulphonate; role of poly(ADP-ribose) polymerase. Mutat Res. 1988;194:17–22. doi: 10.1016/0167-8817(88)90052-1. [DOI] [PubMed] [Google Scholar]
- 186.Althaus FR, Kleczkowska HE, Malanga M, Müntener CR, Pleschke JM, Ebner M, Auer B. Poly ADP-ribosylation: a DNA break signal mechanism. Mol Cell Biochem. 1999;193:5–11. [PubMed] [Google Scholar]
- 187.Jeggo PA. DNA repair: PARP — another guardian angel? Curr Biol. 1997;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 188.Kumari SR, Mendoza-Alvarez H, Alvarez-Gonzalez R. Functional interactions of p53 with poly(ADP-ribose) polymerase (PARP) during apoptosis following DNA damage: covalent poly (ADP-ribosyl)ation of p53 by exogenous PARP and noncovalent binding of p53 to the Mr 85,000 proteolytic fragment. Cancer Res. 1998;58:5075–5078. [PubMed] [Google Scholar]
- 189.Venkatachalam S, Denissenko M, Wani AA. Modulation of (±)-anti-BPDE mediated p53 accumulation by inhibitors of protein kinase C and poly (ADP-ribose) polymerase. Oncogene. 1997;14:801–809. doi: 10.1038/sj.onc.1200890. [DOI] [PubMed] [Google Scholar]
- 190.Vaziri H, West MD, Allsopp RG, Davison TS, Wu YS, Arrowsmith CH, Poirier GG, Benchimol S. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the posttranslational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wesierska-Gadek J, Wang ZQ, Schmid G. Reduced stability of regularly spliced but not alternatively spliced p53 protein in PARP-deficient mouse fibroblasts. Cancer Res. 1999;59:28–34. [PubMed] [Google Scholar]
- 192.Simbulan-Rosenthal CM, Rosenthal DS, Luo RB, Smulson ME. Poly(ADP-ribosyl)ation of p53 during apoptosis in human osteosarcoma cells. Cancer Res. 1999;59:2190–2194. [PubMed] [Google Scholar]
- 193.Wang XJ, Ohnishi K, Takahashi A, Ohnishi T. Poly(ADP-ribosyl)ation is required for p53-dependent signal transduction induced by radiation. Oncogene. 1998;17:2819–2825. doi: 10.1038/sj.onc.1202216. [DOI] [PubMed] [Google Scholar]
- 194.Whitacre CM, Hashimoto H, Tsai ML, Chatterjee S, Berger SJ, Berger NA. Involvement of NAD-poly(ADP-ribose) metabolism in p53 regulation and its consequences. Cancer Res. 1995;55:3697–3701. [PubMed] [Google Scholar]
- 195.Kirch HG, Flaswinkel S, Rumpf H, Brockmann D, Esche H. Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-kappaB and Myc/Max. Oncogene. 1999;18:2728–2738. doi: 10.1038/sj.onc.1202626. [DOI] [PubMed] [Google Scholar]
- 196.Furlong EEM, Rein T, Martin F. YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element, and YY1 and E1A cooperate to amplify p53 promoter activity. Mol Cell Biol. 1996;16:5933–5945. doi: 10.1128/mcb.16.10.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stuart E, Haffner R, Oren M, Gruss P. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J. 1995;14:5638–5645. doi: 10.1002/j.1460-2075.1995.tb00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Uittenbogaard MN, Giebler HA, Reisman D, Nyborg JK. Transcriptional repression of p53 by human T-cell leukemia virus type I tax protein. J Biol Chem. 1995;270:28503–28506. doi: 10.1074/jbc.270.48.28503. [DOI] [PubMed] [Google Scholar]
- 199.Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian JM, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53-dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hudson JM, Frade R, Bareli M. Wild-type p53 regulates its own transcription in a cell-type specific manner. DNA Cell Biol. 1995;14:759–766. doi: 10.1089/dna.1995.14.759. [DOI] [PubMed] [Google Scholar]
- 201.Sun XG, Shimizu H, Yamamoto KI. Identification of a novel p53 promoter element involved in genotoxic stress-inducible p53 gene expression. Mol Cell Biol. 1995;15:4489–4496. doi: 10.1128/mcb.15.8.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fu LN, Benchimol S. Participation of the human p53 3′ UTR in translational repression and activation following gamma-irradiation. EMBO J. 1997;16:4117–4125. doi: 10.1093/emboj/16.13.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fu LN, Minden MD, Benchimol S. Translational regulation of human p53 gene expression. EMBO J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 205.Ju JF, Pedersen-Lane J, Maley F, Chu E. Regulation of p53 expression by thymidylate synthase. Proc Natl Acad Sci USA. 1999;96:3769–3774. doi: 10.1073/pnas.96.7.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 209.Pomerantz J, SchreiberAgus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, CordonCardo C, DePinho RA. The Ink4a tumor suppressor gene product, p19(Arf), interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 210.Zhang YP, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 211.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kamijo T, Zindy F, Roussel M, Quelle D, Downing J, Ashum R, Grosveld G, Sherr C. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 214.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 216.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 217.de Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Schmitt C, McCurrach M, de Stanchina E, Wallace-Brodeur R, Lowe S. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and downregulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 221.Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Shaulsky G, Ben-Ze'ev A, Rotter V. Subcellular distribution of the p53 protein during the cell cycle of Balb/c 3T3 cells. Oncogene. 1990;5:1707–1711. [PubMed] [Google Scholar]
- 223.David-Pfeuty T, Chakrani F, Ory K, Nouviandooghe Y. Cell cycle-dependent regulation of nuclear p53 traffic occurs in one subclass of human tumor cells and in untransformed cells. Cell Growth Differ. 1996;7:1211–1225. [PubMed] [Google Scholar]
- 224.Komarova EA, Zelnick CR, Chin D, Zeremski M, Gleiberman AS, Bacus SS, Gudkov AV. Intracellular localization of p53 tumor suppressor protein in gamma-irradiated cells is cell cycle regulated and determined by the nucleus. Cancer Res. 1997;57:5217–5220. [PubMed] [Google Scholar]
- 225.Liang SH, Hong D, Clarke MF. Cooperation of a single lysine mutation and a C-terminal domain in the cytoplasmic sequestration of the p53 protein. J Biol Chem. 1998;273:19817–19821. doi: 10.1074/jbc.273.31.19817. [DOI] [PubMed] [Google Scholar]
- 226.Liang SH, Clarke MF. The nuclear import of p53 is determined by the presence of a basic domain and its relative position to the nuclear localization signal. Oncogene. 1999;18:2163–2166. doi: 10.1038/sj.onc.1202350. [DOI] [PubMed] [Google Scholar]
- 227.Liang S-H, Clarke M. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J Biol Chem. 1999;274:32699–32703. doi: 10.1074/jbc.274.46.32699. [DOI] [PubMed] [Google Scholar]
- 228.Nigg E. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 229.Talcott B, Moore M. Getting across the nuclear pore complex. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- 230.Adam S. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- 231.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Moll U, Riou G, Levine A. Two distinct mechanism alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci USA. 1992;92:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Moll U, LaQuaglia M, Bernard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci USA. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ryan J, Prochowink E, Gottlieb C, Apel I, Merino R, Nuñez G, Clarke M. c-myc and bcl-2 modulate p53 function by altering p53 subcellular trafficking during the cell cycle. Proc Natl Acad Sci USA. 1994;91:5878–5882. doi: 10.1073/pnas.91.13.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Beham A, Marin MC, Fernandez A, Herrmann J, Brisbay S, Tari AM, LopezBerestein G, Lozano G, Sarkiss M, McDonnell TJ. Bcl-2 inhibits p53 nuclear import following DNA damage. Oncogene. 1997;15:2767–2772. doi: 10.1038/sj.onc.1201464. [DOI] [PubMed] [Google Scholar]
- 236.Vandromme M, Guuthier-Rouviére C, Lamb F, A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biol Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 237.Hengstermann A, Whitaker NJ, Zimmer D, Zentgraf H, Scheffner M. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene. 1998;17:2933–2941. doi: 10.1038/sj.onc.1202282. [DOI] [PubMed] [Google Scholar]
- 238.Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 239.Ashcroft M, Kubbutat MHG, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;9:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Visintin R, Craig K, Hwang E, Prinz S, Tyers M, Amon A. The phosphotase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 241.Hupp T, Sparks A, Lane D. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 242.Lain S, Midgley C, Sparks A, Lane EB, Lane DP. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 243.Jayaraman L, Murthy KGK, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 244.Jayaraman L, Moorthy NC, Murthy KGK, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by GBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 246.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 247.Scolnick DM, Chehab NH, Stavridi ES, Lien MC, Caruso L, Moran E, Berger SL, Halazonetis TD. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 248.Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 249.Chen XB, Farmer G, Zhu H, Prywes R, Prives C. Cooperative DNA binding of p53 with TFIID (TBP) — a possible mechanism for transcriptional activation. Genes Dev. 1993;7:1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- 250.Martin D, Munoz R, Subler M, Deb S. p53 binds to the TATA-binding protein-TATA complexes. J Biol Chem. 1993;268:13062–13067. [PubMed] [Google Scholar]
- 251.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier JL, Triezenberg SJ, Reinberg D, Flores O, Ingles CJ, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Léveillard T, Andera L, Bissonnette N, Schaeffer L, Bracco L, Egly JM, Wasylyk B. Functional interactions between p53 and the TFIIH complex are affected by tumour-associated mutations. EMBO J. 1996;15:1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 253.Farmer G, Colgan J, Nakatani Y, Manley JL, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gualberto A, Baldwin AS. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J Biol Chem. 1995;270:19680–19683. doi: 10.1074/jbc.270.34.19680. [DOI] [PubMed] [Google Scholar]
- 255.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 256.Gobert C, Skladanowski A, Larsen AK. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc Natl Acad Sci USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tyrell R. Activation of mammalian gene expression by the UV component of sunlight — from models to reality. BioEssays. 1996;18:139–148. doi: 10.1002/bies.950180210. [DOI] [PubMed] [Google Scholar]
- 258.Webster GA, Perkins ND. Transcriptional cross talk between NF-kappaB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wadgaonkar R, Phelps KM, Haque Z, Williams AJ, Silverman ES, Collins T. CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J Biol Chem. 1999;274:1879–1882. doi: 10.1074/jbc.274.4.1879. [DOI] [PubMed] [Google Scholar]
- 260.Chakravarti D, Ogryzko V, Kao H-Y, Nash A, Chen H, Nakatani Y, Evans R. A viral mechanism for inhibition of p300 and PCAF acetyl transferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 261.Hamamori Y, Sartorelli V, Ogryzko V, Puri P, Wu H-Y, Wang Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 262.Girinsky T, Koumenis C, Graeber TG, Peehl DM, Giaccia AJ. Attenuated response of p53 and p21 in primary cultures of human prostatic epithelial cells exposed to DNA-damaging agents. Cancer Res. 1995;55:3726–3731. [PubMed] [Google Scholar]
- 263.Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 264.Meyer KM, Hess SM, TIsty TD, Leadon SA. Human mammary epithelial cells exhibit a differential p53-mediated response following exposure to ionizing radiation or UV light. Oncogene. 1999;18:5795–5805. doi: 10.1038/sj.onc.1202977. [DOI] [PubMed] [Google Scholar]
- 265.Hamadeh HK, Vargas M, Lee E, Menzel DB. Arsenic disrupts cellular levels of p53 and mdm2: a potential mechanism of carcinogenesis. Biochem Biophys Res Commun. 1999;263:446–449. doi: 10.1006/bbrc.1999.1395. [DOI] [PubMed] [Google Scholar]
- 266.Méplan C, Mann K, Hainaut P. Cadmium induces conformational modifications of wild-type p53 and suppresses p53 response to DNA damage in cultured cells. J Biol Chem. 1999;44:31663–31670. doi: 10.1074/jbc.274.44.31663. [DOI] [PubMed] [Google Scholar]
- 267.Kress M, May E, Cassingena R, May P. Simian virus-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J Virol. 1979;31:472–483. doi: 10.1128/jvi.31.2.472-483.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lane D, Crawford L. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 269.Linzer D, Levine A. Characterization of a 54K dalton cellular SV40 tumour antigen present in SV40 transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 270.Linke SP, Clarkin KC, Dileonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G(0)/G(1) cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 271.Eller MS, Maeda T, Magnoni C, Atwal D, Gilchrest BA. Enhancement of DNA repair in human skin cells by thymidine dinucleotides: evidence for a p53-mediated mammalian SOS response. Proc Natl Acad Sci USA. 1997;94:12627–12632. doi: 10.1073/pnas.94.23.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhan Q, Carrier F, Fornace A. Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol Cell Biol. 1993;13:4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Graeber TG, Peterson JF, Tsai M, Monica K, Fornace AJ, Giaccia AJ. Hypoxia induces accumulation of p53 protein, but activation of a G(1)-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Plaumann B, Fritsche M, Rimpler H, Brandner G, Hess RD. Flavonoids activate wild-type p53. Oncogene. 1996;13:1605–1614. [PubMed] [Google Scholar]
- 275.Liu M, Pelling J, Ju J, Chu E, Brash D. Antioxidant action via p53-mediated apoptosis. Cancer Res. 1998;58:1723–1729. [PubMed] [Google Scholar]
- 276.Tishler R, Calderwood S, Coleman C, Price B. Increases in sequence specific DNA binding by p53 following treatment with chemotherapeutic and DNA damaging agents. Cancer Res. 1993;53:2212–2216. [PubMed] [Google Scholar]
- 277.Matsumoto H, Shimura M, Omatsu T, Okaichi K, Majima H, Ohnishi T. p53 proteins accumulated by heat stress associate with heat shock proteins HSP72/HSC73 in human glioblastoma cells lines. Cancer Lett. 1994;87:39–46. doi: 10.1016/0304-3835(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 278.Ohnishi T, Wang X, Ohnishi K, Takahashi A. p53-dependent induction of WAF1 by cold shock in human glioma cells. Oncogene. 1998;16:1507–1511. doi: 10.1038/sj.onc.1201663. [DOI] [PubMed] [Google Scholar]
- 279.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 280.Hsieh J-K, Chan F, O'Connor D, Mittnacht S, Zhong S, Lu X. Rb regulates the stability and the apoptotic function of p53 via MDM2. Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 281.Bulavin D, Saito S, Hollander M, Sakaguchi K, Anderson C, Appella E, Fornace A. Phosphorylation of human p53 by p38 kinase coordinates N-terminal Phosphorylation and apoptosis in response to UV irradiation. EMBO J. 1999;18:6845–6854. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li L, Ljungman M, Dixon J. The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53. J Biol Chem. 2000;275:2410–2414. doi: 10.1074/jbc.275.4.2410. [DOI] [PubMed] [Google Scholar]
- 284.Shinohara K, Tomioka M, Nakano H, Tone S, Ito H, Kawashima S. Apoptosis induction resulting from proteasome inhibition. Biochem J. 1996;317:385–388. doi: 10.1042/bj3170385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Drexler HCA. Activation of the cell death program by inhibition of proteasome function. Proc Natl Acad Sci USA. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases chk1 and cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 287.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 288.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 289.Tominaga K, Morisaki H, Kaneko Y, Fujimoto A, Tanaka T, Ohtsubo M, Hirai M, Okayama H, Ikeda K, Nakanishi M. Role of human Cds1 (Chk2) kinase in DNA damage checkpoint and its regulation by p53. J Biol Chem. 1999;274:31463–31467. doi: 10.1074/jbc.274.44.31463. [DOI] [PubMed] [Google Scholar]
- 290.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]