Abstract
The chloroplast Albino3 (Alb3) protein is a chloroplast homolog of the mitochondrial Oxa1p and YidC proteins of Escherichia coli, which are essential components for integrating membrane proteins. In vitro studies in vascular plants have revealed that Alb3 is required for the integration of the light-harvesting complex protein into the thylakoid membrane. Here, we show that the gene affected in the ac29 mutant of Chlamydomonas reinhardtii is Alb3.1. The availability of the ac29 mutant has allowed us to examine the function of Alb3.1 in vivo. The loss of Alb3.1 has two major effects. First, the amount of light-harvesting complex from photosystem II (LHCII) and photosystem I (LHCI) is reduced >10-fold, and total chlorophyll represents only 30% of wild-type levels. Second, the amount of photosystem II is diminished 2-fold in light-grown cells and nearly 10-fold in dark-grown cells. The accumulation of photosystem I, the cytochrome b6f complex, and ATP synthase is not affected in the ac29 mutant. Mild solubilization of thylakoid membranes reveals that Alb3 forms two distinct complexes, a lower molecular mass complex of a size similar to LHC and a high molecular mass complex. A homolog of Alb3.1, Alb3.2, is present in Chlamydomonas, with 37% sequence identity and 57% sequence similarity. Based on the phenotype of ac29, these two genes appear to have mostly nonredundant functions.
INTRODUCTION
The biogenesis of the photosynthetic complexes of the thylakoid membrane depends on the concerted interactions of the chloroplast and nuclear genetic systems. Genetic and biochemical studies in Chlamydomonas, maize, and Arabidopsis have revealed a large number of nucleus-encoded factors that are involved in transcriptional and post-transcriptional steps of chloroplast gene expression (Barkan and Goldschmidt-Clermont, 2000). Once proteins of the photosynthetic apparatus have been synthesized, they need to be targeted to the thylakoid membrane, inserted into or translocated through the membrane, and assembled into functional complexes.
At least four pathways for thylakoid membrane insertion and translocation have been characterized that display unique energy and stromal protein requirements (Keegstra and Cline, 1999). Each of these pathways targets a distinct subset of proteins to the thylakoid. The Sec pathway, which requires ATP and cpSecA, is related to the Sec system of the bacterial export machinery. The presence of this system in chloroplasts is not surprising in light of the generally accepted view that chloroplasts evolved from a prokaryotic endosymbiont. Another pathway, related to the bacterial Tat pathway, uses a transthylakoid pH gradient as its sole energy source to transport proteins and does not require any stromal factor (Robinson and Bolhuis, 2001).
The light-harvesting complex protein (LHCP) integration pathway shares several features with the GTP-dependent signal recognition particle (SRP) system of the endoplasmic reticulum and bacteria (Li et al., 1995). The chloroplast SRP contains a homolog of SRP54. However, it differs from other SRPs because it lacks any SRP RNA, contains a novel 43-kD subunit, and interacts with substrates post-translationally (Schuenemann et al., 1998; Klimyuk et al., 1999). It is believed generally that the solubility of the hydrophobic LHCP is maintained in the stroma by its binding to SRP to form a targeting intermediate of 120 kD termed the transit complex.
Together with GTP and the chloroplast homolog FtsY, an Escherichia coli protein necessary for E. coli SRP function, the transit complex promotes efficient integration of LHCP into the thylakoid membrane (Tu et al., 1999). However, the fact that LHCP can insert into thylakoid membranes in the absence of functional SRP (Amin et al., 1999) suggests that alternative targeting pathways exist. Finally, some thylakoid proteins are able to insert into the thylakoid membrane without the requirement of protein factors.
Several factors specific for the assembly of photosynthetic complexes have been identified. They include BtpA (Bartsevich and Pakrasi, 1997; Zak et al., 1999; Zak and Pakrasi, 2000), Ycf3 and Ycf4 (Wilde et al., 1995; Boudreau et al., 1997; Ruf et al., 1997), which are required specifically for the accumulation of photosystem I (PSI), and Hcf 136, which is required for the assembly of photosystem II (PSII) (Meurer et al., 1998). However, the action of these factors at the molecular level remains poorly understood.
Studies with yeast mitochondrial protein export have shown that the inner membrane protein Oxa1p is an essential component for integrating a subset of inner membrane proteins encoded by nuclear and mitochondrial DNA (Bonnefoy et al., 1994; Hell et al., 1998, 2001). Oxa1p is homologous with the YidC protein of E. coli that also acts as a mediator of membrane protein assembly (Luirink et al., 2001). YidC operates either as a separate unit or in connection with the Sec YEG translocon, depending on the substrate protein that is integrated into the membrane. A homolog of Oxa1p, termed Alb3, has been found in chloroplasts (Sundberg et al., 1997). A mutant of Arabidopsis deficient in Alb3 displays an albino phenotype, indicating that this protein is required for thylakoid biogenesis (Sundberg et al., 1997).
Recently, it was shown that treatment of thylakoid membranes with an Alb3 antibody interferes with the integration of Lhcb1, the major light-harvesting chlorophyll binding protein (Moore et al., 2000). The same treatment also blocks the insertion of two other chlorophyll binding proteins, Lhcb4.1 and Lhcb5 (Woolhead et al., 2001). By contrast, preincubation of thylakoids with anti-Alb3 antibodies does not affect the thylakoid Sec or Delta pH pathways (Moore et al., 2000), nor does it block the insertion of PsbS, PsbX, PsbW, and PsbY (Woolhead et al., 2001). The latter three proteins contain signal peptides, and their insertion into the thylakoid membrane is SRP independent. These in vitro studies reveal a strict correlation between the requirements for Alb3 and SRP.
To test the role of Alb3 in vivo, we have characterized a mutant of Chlamydomonas, ac29-3, in which the Alb3.1 gene is inactivated. This mutant is principally deficient in LHC. The level of PSII also is reduced, especially in dark-grown cells. The Alb3.1 protein appears to form two distinct complexes, a lower molecular mass complex of a size similar to that of LHC and a high molecular mass complex of unknown function. Chlamydomonas contains a homolog of Alb3.1 that appears to play only a modest role in the integration of LHC in the thylakoid membrane, based on the phenotype of the ac29 mutant.
RESULTS
Characterization of ac29 Mutants
The first two mutants affected at the AC29 locus of Chlamydomonas, CC-44 and CC-45, were identified as yellow, leaky, acetate-requiring mutants (Levine and Goodenough, 1970). This locus is linked tightly to that of the MATING-TYPE (MT), which was cloned and characterized previously (Ferris and Goodenough, 1994; Ferris, 1995). Several new mutants affected at the AC29 locus were isolated during a search for large deletions in the MT locus (P. Ferris, unpublished results).
A diploid having a prototrophic green mt− phenotype was constructed from CC-1336 (nic7 ac29-2 mt− pf14) and CC-1067 (++ mt+ +). Gamma irradiation of the diploid generated yellow and/or nicotinamide-requiring mutant strains carrying new mutations at these loci. Six yellow nicotinamide prototrophs (acy9, acy10, acy20, acy25, acy27, and acy32) were obtained that have DNA rearrangements in the region marked by the ac29-2 allele in Figure 1. DNA gel blot analysis of four of these mutants and their diploid parental strain is shown in Figure 2A.
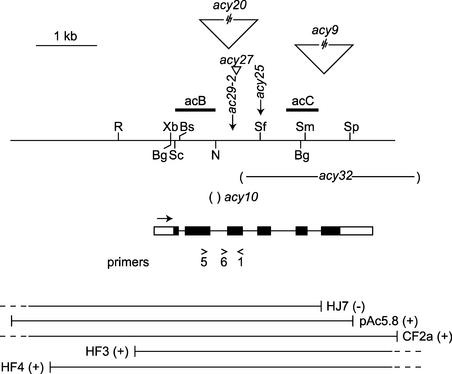
Figure 1.
Map of the AC29 Locus.
The probes acB and acC are shown. The approximate location of the point mutations, insertions (wedges), and deletions (−) of the acy mutants are marked. The exons and introns of the Alb3 gene are indicated with black bars and lines, respectively. Untranslated regions are shown as open bars. Transcription proceeds from left to right. The primers used are indicated. Plasmids and phages used in transformations are shown at bottom. Rescue of the mutant phenotype is indicated by +. Restriction sites are indicated at top: Bg, BglII; Bs, BstEII; N, NsiI; R, EcoRI; Sc, SacI; Sf, SfiI; Sm, SmaI; Sp, SpeI; Xb, XbaI.
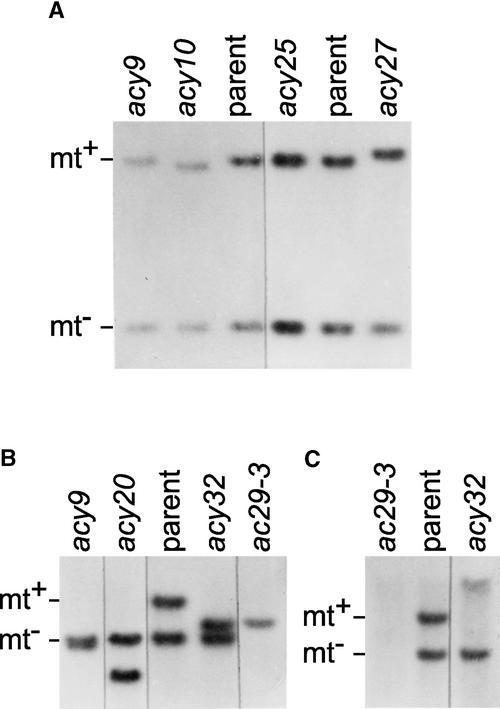
Figure 2.
DNA Gel Blot Analysis of ac29 Mutants.
DNA was isolated from the diploid parent (nic7 ac29-2 mt− pf14/ ++ mt+ +) and the diploid acy9, acy10, acy20, acy25, and acy32 mutant strains and from the haploid ac29-3 mutant containing the mt+ chromosome from acy32. The DNAs of the blots were digested with BglII (A) or SacI ([B] and [C]) and probed with the acB probe ([A] and [B]) or the acC probe (C).
The DNAs were digested with BglII and probed with the acB probe (Figure 1). The probe hybridizes to two fragments of the parent, one (1.3 kb) from the ac29-2 mt− chromosome and the other (2.2 kb) from the AC29 mt+ chromosome. The acy9 strain shows no change within the BglII fragments but has an insertion larger than 10 kb farther downstream (Figure 1 and data not shown). The acy27 and acy10 strains carry a small insertion and a deletion, respectively, within the 2.2-kb BglII fragment. These changes in the two mutants localize between the BstEII and SfiI sites (Figure 1 and data not shown). The only change detectable in the acy25 strain is the loss of the SfiI site.
Figure 2B shows DNA gel blots obtained after digestion with SacI from the parent and the acy9, acy20, and acy32 mutants. The mt− and mt+ chromosomes yield fragments of 7.2 and 10.8 kb, respectively. In the mutants, the mt+ fragment has been replaced by smaller fragments. Additional DNA gel blots revealed that acy9 and acy20 contain large insertions (>10 kb) with several internal SacI sites, resulting in SacI fragments smaller than in the original, whereas acy32 carries a 3-kb deletion (data not shown). The approximate locations of the insertions and the deletion are indicated in Figure 1.
Identification and Structure of the AC29 (Alb3) Gene
The sequence of 4654 bp proceeding to the right from the EcoRI site in Figure 1 was determined from the genomic copy of AC29. A corresponding cDNA was isolated, and its sequence was determined. Comparison of the two sequences revealed that this gene consists of six exons and five introns of 113, 313, 230, 440, and 213 nucleotides. A BLAST search (in April 2002) of the database with the protein of 495 residues predicted from the cDNA had significant sequence identity with the chloroplast Albino3 protein of Arabidopsis (46% identity and 65% similarity; Figure 3). Accordingly, the gene is called Alb3.1.
Figure 3.
Sequence Comparison of the Alb3 Protein with YidC and Oxa1p.
The protein sequences were aligned using ClustalW (Thompson et al., 1994) and Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Alb3.1 and Alb3.2 are from Chlamydomonas; atAlb3 (Sundberg et al., 1997) and atAlb3.2 are from Arabidopsis; YidC is from E. coli (Luirink et al., 2001); Oxa1p is from S. cerevisiae (Bonnefoy et al., 1994). The sites corresponding to the introns are indicated by open and closed wedges for Alb3.1 of Chlamydomonas and atAlb3 of Arabidopsis, respectively. In the phylogenetic tree at bottom, the numbers of amino acid substitutions per alignment site are indicated on the branches. The phylogenetic analysis was performed using the PHYLO_WIN program (Galtier and Gouy, 1996).
Chlamydomonas ESTs indicate that Alb3.1 mRNAs are polyadenylated at multiple locations. The position of Alb3.1 is consistent with previous transformation data (Ferris, 1995). Thus, the green phenotype of the ac29-2 mutant was restored by transformation with the pAc5.8 plasmid, which terminates within the 3′ untranslated region, but not by transformation with the HJ7 phage, which lacks the final exon (Figure 1). The location of Alb3.1 also is consistent with the observation that phages HF3 and CF2a were able to rescue the ac29-2 mutant by transformation (Ferris, 1995).
Further evidence that the AC29 locus corresponds to the Alb3.1 gene is provided by the ac29-2 mutation in the third exon of Alb3.1. Restriction fragment length polymorphism analysis had revealed polymorphic BglII-XbaI tandem sites in the ac29-2 allele of CC-44. A PCR product spanning the mutant site obtained with primers 1 and 5 (Figure 1) was cloned and sequenced. The mutation consists of a single C-to-T base change that produces a stop codon and creates two novel restriction sites, BglII and XbaI (Figure 4).
Figure 4.
Sequence Analysis of the Region of Alb3.1 Affected in the ac29-2 Mutant and Three Revertant Strains.
Only the sequence surrounding the BglII site of ac29-2 is shown.
It is possible that some other changes in the ac29-2 allele also are responsible for the phenotype, because the entire allele was not sequenced. However, examination of three green revertants by DNA gel blot analysis revealed that they had retained the BglII site but had lost the XbaI site. The only change in the sequence of the revertants relative to ac29-2 was within the XbaI site that restored the Alb3.1 open reading frame (Figure 4). Analysis of the acy27 mutant revealed that it contains a 78-bp insertion that apparently resulted from the duplication of adjoining sequences in exon 3 (data not shown).
To determine whether the acy32 deletion (referred to as the ac29-3 allele) confers a more stringent acetate-requiring phenotype than the ac29-2 nonsense mutation, it was first necessary to create a haploid strain for the NIC7 ac29-3 mt+ chromosome. Such an ac29-3 strain was constructed by crossing the acy32 diploid to a green nic7 mt+ strain and identifying a prototrophic yellow mt+ strain among the progeny. DNA gel blot analysis (Figure 2B) demonstrated that the strain carried only the ac29-3 chromosome and failed to hybridize with the acC probe predicted to lie in the deleted region (Figures 1 and 2C).
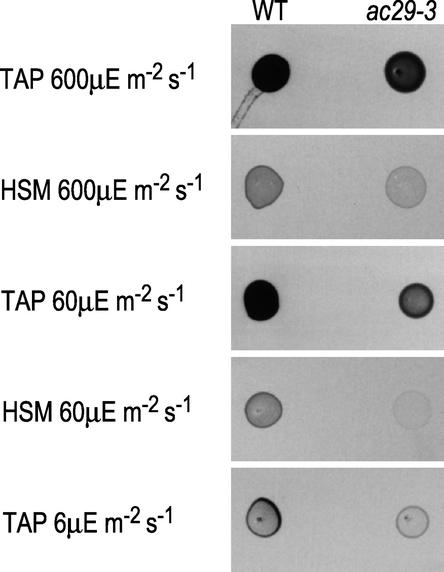
The haploid ac29-3 strain, like ac29-2, is not strictly acetate requiring. It grows normally on Tris-acetate phosphate (TAP) medium under high light. However, its growth on minimal medium or on TAP medium under low light is retarded significantly (Figure 5). It grows more slowly on high-salt medium (HSM) plates than ac29-2 (data not shown). Transformation of ac29-3 was performed with linearized pAc5.8. Only 2 × 106 cells were transformed, and they were spread onto 18 HSM plates. Several green sectors were isolated, and ultimately, 11 independent green transformants were isolated after subcloning. These results indicate that the yellow color of ac29-3 is caused by the deletion in the AC29 locus. This mutant was used for further studies.
Figure 5.
Growth Patterns of the Wild Type (WT) and ac29-3.
Five microliters from an exponentially growing culture was spotted on TAP or HSM (minimal) under different light conditions as indicated.
The N-terminal part of the Alb3.1 protein of Chlamydomonas is rich in Ser, Thr, and Arg, a typical feature of chloroplast transit peptides, and is predicted by the Chloro P program to contain a chloroplast transit sequence (Emanuelsson et al., 1999). This is consistent with the location of the Arabidopsis Alb3 protein in the chloroplast (Sundberg et al., 1997) and with the finding that the Alb3.1 protein of Chlamydomonas is associated with thylakoid membranes (see below).
Besides its similarity to the Alb3 protein of Arabidopsis, two regions of the Chlamydomonas protein, residues 128 to 232 and 284 to 358, show significant sequence identity to the Oxa1p mitochondrial protein of Saccharomyces cerevisiae and to the inner membrane YidC protein of E. coli (Figure 3). The C-terminal region contains a long stretch of Ala residues and a short highly positively charged region of seven residues.
Figure 3 also indicates the positions of the Alb3 residues of Chlamydomonas and Arabidopsis that correspond to the intron insertion sites. It can be seen that among the five introns of Chlamydomonas and the nine introns of Arabidopsis, only intron 2 is inserted at the same site in both proteins. This finding suggests that, in contrast to the other introns, the second intron of Alb3 was present before the divergence of plants and green algae during evolution.
Localization of Alb3.1
Attempts to produce an antiserum against Alb3.1 were unsuccessful. Therefore, to localize the Alb3.1 protein within the chloroplast, the protein was tagged with an epitope. This was achieved by inserting the coding sequence of the triple hemagglutinin (HA) epitope at the 3′ end of the Alb3.1 cDNA fused to the promoter and the first exon of the Alb3.1 gene. This construct was inserted into the nuclear genome of the ac29-3 mutant by cotransformation using the Cry1 gene, which confers resistance to emetine, as a selectable marker (Nelson et al., 1994).
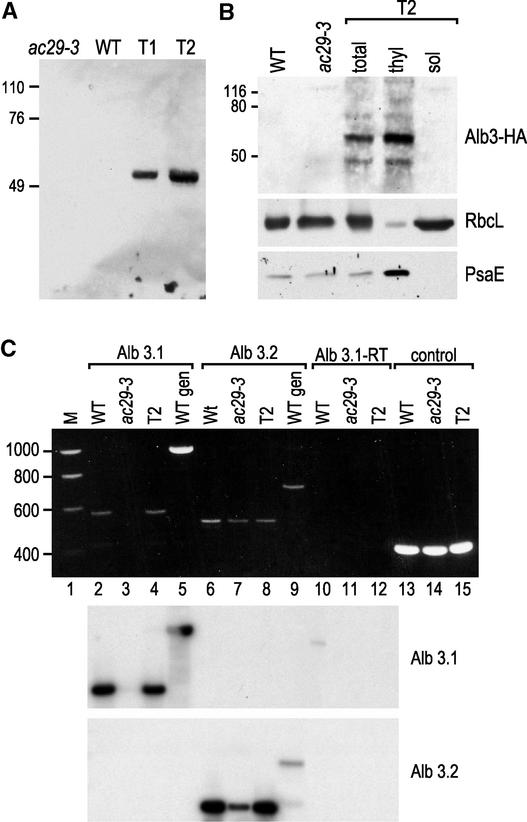
The drug-resistant transformants were screened for expression of the tagged protein by immunoblotting with an HA antibody. Among 15 transformants tested, two (T1 and T2) expressed the Alb3-HA protein to detectable levels (Figure 6A). As expected, the tagged Alb3.1 protein was found in the thylakoid membrane fraction (Figure 6B). The purity of the fractions was tested with antibodies against RbcL and PsaE.
Figure 6.
Localization of the Alb3-HA Protein and Expression of the Alb3.1 and Alb3.2 Genes.
(A) Immunoblot analysis of total cell extracts from the ac29-3 mutant, the wild type (WT), and two ac29-3 transformants (T1 and T2) rescued with the HA epitope–tagged Alb3 gene. The blot was decorated with HA antibody.
(B) Cells from the T2 transformant were lysed and fractionated into a thylakoid membrane fraction (thyl) and a soluble fraction (sol).
(C) At top, total RNA from wild-type, ac29-3, and T2 transformants was subjected to RT-PCR, and the products were fractionated by agarose gel electrophoresis using primers specific for Alb3.1 (lanes 2 to 4), Alb3.2 (lanes 6 to 8), and the G-protein β-subunit gene (Schloss, 1990) (control; lanes 13 to 15). The same reaction with the Alb3.1 primers without reverse transcriptase did not yield any signal (lanes 10 to 12). The same primers for Alb3.1 and Alb3.2 were used for PCR on wild-type genomic DNA (lanes 5 and 9, respectively). Lane 1 contained molecular mass markers (M). At bottom, the DNA fragments were blotted onto filters and hybridized with probes specific for Alb3.1 and Alb3.2 as indicated.
To determine the amount of Alb3.1 mRNA in the wild type and the rescued ac-29 strain, total RNA was isolated and examined by RNA gel blot analysis. Because the amount of RNA was undetectable by this method, reverse transcriptase–mediated (RT) PCR was performed using appropriate Alb3.1-specific oligonucleotides (see Methods). A fragment of the expected size was present in similar amounts in the wild-type and rescued strains and was absent, as expected, in the ac-29 mutant (Figure 6C, lanes 2 to 4). However, because the RT-PCR method used is semiquantitative, we were not able to measure accurately the relative levels of expression in the two strains.
The Alb3 Protein Is Required for the Normal Accumulation of LHC
The ac29 mutant has a pale phenotype, indicating reduced accumulation of chlorophyll. The amount of chlorophyll in light-grown ac29-3 mutant cells is 31% relative to wild-type cells; in dark-grown cells, the corresponding value is only 16% (Table 1). The reduced accumulation of chlorophyll in the dark is attributable to the ac29-3 mutation, because mutant cells rescued with the wild-type copy of the Alb3.1 gene by transformation accumulated similar chlorophyll levels in both the light and the dark (Table 1). The rescued cells accumulated only 60% of the chlorophyll that wild-type cells accumulated, possibly because the HA-tagged version of Alb3.1 lacks introns, which are known to be required in most cases for efficient gene expression in Chlamydomonas (Fischer and Rochaix, 2001). Another possibility is that the insertion of the HA tag diminishes the stability of the Alb3.1 protein or affects its function.
Table 1.
Chlorophyll Accumulation in ac29-3
| Strain | Chlorophyll Concentration (μg/μg protein) |
ac29-3 Concentration (%) |
Chlorophyll a/b Ratio |
|---|---|---|---|
| Wild-type L | 0.091 ± 0.001 | 100 | 2.4 |
| Wild-type D | 0.077 ± 0.016 | 84 | 2.2 |
| ac29-3 L | 0.028 ± 0.004 | 31 | 4.6 |
| ac29-3 D | 0.015 ± 0.001 | 16 | 5.1 |
| ac29-3-HA L | 0.072 ± 0.007 | 78 | 2.3 |
| ac29-3-HA D | 0.057 ± 0.015 | 62 | 2.3 |
In all cases, the percentage was determined by comparison of the mutant with the wild-type cells grown under the same light or dark conditions. Chlorophyll concentrations were determined according to Porra et al. (1989). Measurements were performed five times, and the average is indicated with the standard deviation. D, darkness; L, light (60 μE·m−2·s−1).
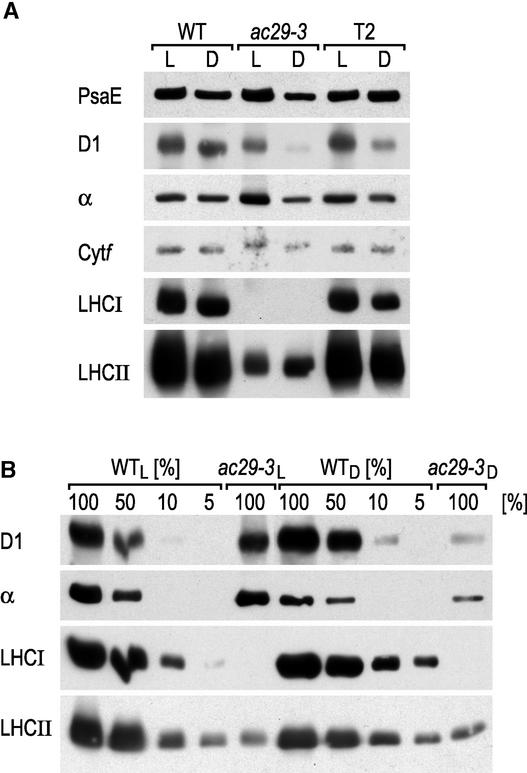
To study the role of the Alb3.1 protein in the biogenesis of the photosynthetic apparatus, the levels of the different photosynthetic complexes in the thylakoid membranes of the wild type and the ac29-3 mutant were determined. Figure 7 shows the results obtained by immunoblotting of thylakoid membrane extracts and reacting the blots with antisera raised against specific subunits of PSI (PsaE), PSII (D1), the ATP synthase complex (α), and the cytochrome b6f complex (Cytf). In all of these cases, no significant difference was observed between mutant and wild-type cells grown in the light, with the exception of the level of PSII, which was reduced twofold in ac29-3 cells.
Figure 7.
The Abundance of LHCI and LHCII Is Reduced in the ac29-3 Mutant.
(A) Thylakoid proteins from the wild type (WT), ac29-3, and ac29-3 rescued with a HA epitope–tagged Alb3 gene (T2) were separated by PAGE, immunoblotted, and decorated with antibodies against PSI (PsaE), PSII (D1), ATP synthase (α), cytochrome b6f complex (Cytf), LHCI, and LHCII.
(B) Dilutions of the protein extracts shown in (A) were used to estimate the amount of protein accumulated in ac29-3.
D, darkness; L, light.
The effect of the ac29-3 mutation on PSII was more pronounced in dark-grown cells, in which this complex was reduced nearly 10-fold. The major effect of the loss of Alb3.1 was the strong reduction of the LHC polypeptides (Figure 7). Quantitative immunoblot analysis with the LHCI-specific P14.1 and the LHCII-specific P11 antibodies indicated that LHCI accumulates to <5% and that LHCII accumulates to 5 to 10% of wild-type levels in light-grown mutant cells, in agreement with the chlorophyll measurements. This observed decrease of LHC is compatible with the chlorophyll remaining in ac29-3, 30% relative to the wild type.
The chlorophyll antennae from PSII and PSI of Chlamydomonas have been estimated at 320 and 290 chlorophyll molecules, respectively (Polle et al., 2000). If one takes into account the chlorophylls that are bound to the core complexes of PSII (36 molecules) and PSI (100 molecules), one can estimate that a 90% reduction of total LHC would lead to a 70% reduction of total chlorophyll. The chlorophyll a/b ratio was increased significantly in the ac29-3 mutant, as expected from a deficiency in LHC (Table 1).
Alb3.1-HA Is Associated with Two Complexes
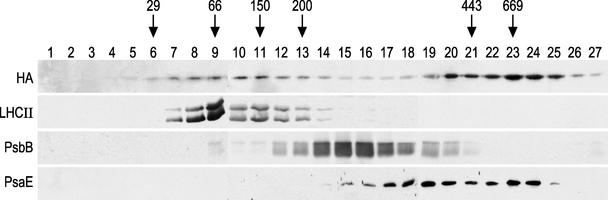
To determine whether Alb3.1 is associated with a large molecular mass complex, purified thylakoid membranes were solubilized with dodecyl maltoside and fractionated by Suc density gradient centrifugation. An equal aliquot of each fraction was separated by PAGE and used for immunoblot analysis with antibodies against HA, LHCII, the PSII subunit PsbB, and the PSI subunit PsaE. Figure 8 shows that the Alb3-HA protein fractionates in two distinct regions of the gradient.
Figure 8.
The Alb3-HA Protein Is Associated with Two Complexes.
Thylakoid membranes from the T2 transformant were solubilized with dodecyl maltoside and fractionated on a Suc density gradient. Fractions of 1 mL were collected, and 32-μL aliquots were used for PAGE and immunoblotting. The antibodies used are indicated at left. Antibodies specific for LHCII, PSII (PsbB), and PSI (PsaE) were used. Masses are indicated in kD and were determined by comparison with molecular mass standards provided with a molecular marker kit (Sigma). HA, HA-tagged Alb3 protein.
The first peak of Alb3.1, with a size in the 60- to 80-kD range, cofractionates with LHCII, and the second peak corresponds to a high molecular mass complex, in the 600- to 700-kD range, that is slightly larger than the PSI complex. However, because the multiple forms of these complexes have been observed with the HA-tagged Alb3.1 protein, and because the tag could alter the properties of Alb3.1, it remains to be determined whether the untagged protein also is associated with two complexes.
Chlamydomonas Contains Two Alb3-Related Genes
Screening of the Chlamydomonas ESTs with the Alb3.1 sequence revealed a second Alb3-related gene named Alb3.2 that encodes a protein of 422 amino acids and that shows 37% sequence identity and 54% sequence similarity to Alb3.1 over its entire length (Figure 3). As for Alb3.1, the Alb3.2 mRNA is poorly expressed and was detected only by RT-PCR (Figure 6C, lanes 6 to 8). Because the method used is not quantitative, we have not been able to measure the relative levels of these two transcripts. The central region of both proteins, comprising ∼260 residues, is significantly more conserved (46% identity and 65% similarity).
Alb3.2 displays 53% sequence identity and 71% sequence similarity to the Alb3 protein of Arabidopsis. The relatedness between Alb3.1 and the Arabidopsis protein is slightly smaller (46% identity and 65% similarity). These three Alb3 proteins have significantly more sequence similarity between themselves than to the yeast mitochondrial Oxa1p protein (19% sequence identity and 38% sequence similarity to Alb3.1, 18% sequence identity and 37% sequence similarity to both Alb3.2 and Alb3 of Arabidopsis). The relatedness to the bacterial YidC protein also is similar for Alb3.1 (27% sequence identity and 47% sequence similarity), Alb3.2 (31% sequence identity and 50% sequence similarity), and Alb3 of Arabidopsis (30% sequence identity and 48% sequence similarity).
The relationship between these different proteins is illustrated by the dendrogram at the bottom of Figure 3, which shows that Alb3.2 is related more closely to Alb3 of Arabidopsis than to Alb3.1. The Alb3 proteins of Chlamydomonas and Arabidopsis contain very similar hydrophobicity profiles, with five common putative transmembrane domains (data not shown). However, Alb3.1 appears to contain an additional transmembrane domain in its C-terminal region (data not shown). The Alb3.2 protein is likely to be a chloroplast protein, based on the prediction of the ChloroP program (Emanuelsson et al., 1999), although this assignment remains to be tested with appropriate cellular localization studies.
In spite of their similar structure, it is likely that the functions of Alb3.1 and Alb3.2 differ significantly, because the Alb3.1-deficient ac29-3 mutant is affected drastically in the accumulation of LHCI and LHCII. However, because small amounts of LHCII still are present in this mutant, it is possible that Alb3.2 also plays some role in the assembly of this complex. Besides Alb3, a BLAST search for Alb3 homologs in Arabidopsis revealed another gene encoding a protein, named atAlb3.2, of 1013 residues that is related to Alb3 in its C-terminal half (45% identity and 67% similarity). Based on its N-terminal sequence, the atAlb3.2 protein is not predicted to be localized within the chloroplast, although the phlyogenetic tree indicates that its C-terminal part is closely related to Alb3 (Figure 3).
DISCUSSION
The AC29 Locus Encodes Alb3.1
In this study, we have shown that the AC29 locus encodes the Alb3.1 protein. This assignment is based on the physical characterization of several ac29 mutants with insertions and deletions at the AC29 locus, on the molecular characterization of pseudorevertants of the ac29-2 nonsense mutation that restore the open reading frame of Alb3.1, and on the rescue of the mutant phenotype by the transformation of ac29 mutants with the genomic Alb3.1 clone or with its cDNA.
The Alb3.1 protein shows a sequence identity of 42% to the Arabidopsis Alb3 protein. Furthermore, two regions of the protein display significant sequence identity to mitochondrial Oxa1p of yeast and the YidC inner membrane protein of E. coli. These proteins appear to be evolutionarily conserved mediators of membrane protein assembly in chloroplasts, mitochondria, and bacteria (Luirink et al., 2001; Jiang et al., 2002).
Inactivation of Alb3.1 in Chlamydomonas
The inactivation of Alb3.1 in Chlamydomonas leads to a drastic reduction of the LHCII and LHCI polypeptides. It is interesting that in this case, LHCI is more affected than LHCII, which accumulate to <5% and nearly 10% of wild-type levels, respectively. Whereas PSI, cytochrome b6f, and ATP synthase are largely unaffected by the loss of Alb3.1, PSII is reduced twofold in light-grown cells. Surprisingly, this complex is reduced nearly 10-fold in dark-grown cells. This dark effect appears to be restricted to PSII, because the accumulation of LHCI and LHCII is the same in the light and the dark. It is interesting that ribosome nascent chain complexes of the PSII reaction center polypeptide D1 with SRP have been detected (Nilsson et al., 1999).
Together, these results agree with the idea derived from in vitro experiments that Alb3 is connected closely to the SRP pathway (Woolhead et al., 2001). The enhanced effect of the ac29-3 mutation on PSII in the dark cannot be attributed to an alteration in the chlorophyll synthesis pathway in the dark because the rescued mutant accumulates chlorophyll to the same level in dark- and light-grown cells. This finding indicates that the Alb3.1 gene influences chloroplast metabolism in the dark in a manner that remains unknown.
Role of Alb3 in LHC Assembly
Moore et al. (2000) recently demonstrated that Alb3 is required for the post-translational integration of the light-harvesting chlorophyll binding proteins into thylakoid membranes, based on the observation that thylakoids treated with an anti-Alb3 antibody are unable to integrate LHCP. The same treatment did not affect protein transport by the thylakoid Sec or Delta pH pathway. Suc density gradient fractionation of solubilized thylakoid membranes reveals that a portion of Alb3.1 forms a complex of 60 to 80 kD that cofractionates with the LHC complex (Figure 8). The cofractionation could imply an interaction between LHC and Alb3.1, but direct demonstration of such an interaction by coimmunoprecipitation was not possible.
The major portion of Alb3.1 is contained within a high molecular mass (600 to 700 kD) complex that is slightly larger than PSI. The current accepted model is that LHCP bound to chloroplast SRP in a soluble transit complex represents the LHCP targeted to the thylakoid membrane. LHCP integration requires the SRP receptor homolog cpFtsY, which partitions between stroma and thylakoids (Tu et al., 1999). One possible reason for this effect is that the high molecular mass Alb3.1 complex of Chlamydomonas participates in the recruitment of FtsY to the thylakoids. Another possible explanation is that the high molecular mass Alb3.1 complex includes the Sec translocon. In this respect, it is interesting that the E. coli YidC protein can act independently as a translocon, but it also has been found to be associated with the Sec translocon (Valent et al., 1998; Houben et al., 2000). Likewise, yeast Oxa1p is part of an oligomeric complex (Hell et al., 1998). This complex of 170 to 180 kD is homooligomeric and forms an essential part of the mitochondrial export translocase of Neurospora crassa (Nargang et al., 2002).
Although our data support the view that Alb3.1 is part of a translocon system that is used specifically to integrate LHCII and LHCI into thylakoid membranes, as much as 10% of LHCII still accumulates in the absence of Alb3.1. Thus, an alternative pathway must exist for LHC integration into the thylakoid membrane. In this respect, it is interesting to note the existence of Alb3.2, a homolog of Alb3.1 that belongs clearly to the Alb3 family rather than to the Oxa1p group (Figure 3). Although this Alb3 homolog has not yet been characterized fully, it is possible that it also plays a role in LHC assembly and that it is partially redundant with Alb3.1. This could explain why LHC accumulation is not blocked fully in the absence of Alb3.1. However, it is clear that the functions of these two Alb3 homologs differ considerably. The ac29-3 mutant still is able to grow photoautotrophically, although at a much reduced rate.
A similar mutant with a disruption of the Alb3 gene has been described in Arabidopsis with a phenotype that is significantly more severe (Sundberg et al., 1997). The leaves of the mutant are yellowish to white, and the overall chlorophyll content is only 5% of that of wild-type plants. There are only a few thylakoid membranes, and the mutant dies at the seedling stage. Thus, the loss of Alb3 leads to a much milder phenotype in Chlamydomonas than in Arabidopsis. In particular, the polytopic membrane proteins of PSI and PSII that are synthesized in the chloroplast are much less affected by the mutation. Because of its specific effect on LHC, this algal system offers promising possibilities for elucidating the exact function of Alb3.
METHODS
Strains and Growth Conditions
Wild-type and mutant strains of Chlamydomonas reinhardtii were maintained on solid Tris-acetate phosphate (TAP) medium supplemented with 4 μg/mL nicotinamide or 100 μg/mL Arg as needed. Sueoka's high-salt medium was used whenever an acetate-deficient medium was required (Harris, 1989). Crosses were performed using standard protocols (Harris, 1989). Transformations were performed using the glass beads/vortex protocol (Kindle, 1990).
DNA Gel Blot Analysis
Genomic DNA and DNA gel blots were prepared as described by Ferris (1989). Probes were derived as follows: probe acB is a 0.8-kb NsiI-SacI fragment from pAc5.8, and probe acC is a 0.6-kb SstII fragment from pAc5.8.
cDNA Cloning
The pAc5.8 plasmid was used as a probe for screening a cDNA library from Chlamydomonas as described (Perron et al., 1999).
DNA Sequencing
The genomic sequence of a 4654-bp region beginning at the EcoRI site and spanning the Alb.3 gene was determined from plasmid subclones of the λEMBL3 chromosome walk from strain CC-621 (mt−) (Ferris and Goodenough, 1994). The bulk of the DNA sequencing was performed on single-stranded DNA using the Sequenase kit (United States Biochemical) after cloning restriction fragments into pUC118 or pUC119.
The segment of the ac29-2 allele from strain CC-44 that carries the BgIII-XbaI polymorphism was isolated as follows. PCR was performed on 25 ng of genomic DNA from the ac29-2 mutant using Vent DNA polymerase (New England Biolabs, Beverly, MA) primers 1 and 6 under the following conditions: 30 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min. The resulting fragment was gel purified and ligated into HincII-digested pUC118 and sequenced.
Three potential revertants of ac29-2 were analyzed. Revertant 1 was a culture of CC-2663 received from the Chlamydomonas Genetics Center (Durham, NC) that was green, rather than the expected yellow, at the time of receipt. Revertant 2 was a culture of ac29-2 that has been in use in our laboratory for many years and had acquired a green color. Revertant 3 was a green colony isolated during an attempt to transform ac29-2 with a wild-type Alb3-1 genomic clone; it appeared not to contain any transforming DNA and therefore was assumed to be a revertant.
All three revertants, when examined by genomic DNA gel blot analysis, had retained the BglII polymorphism of the ac29-2 mutation but had lost the XbaI polymorphism. A PCR product was cloned from each strain (twice in the case of revertant 1) by the same protocol used for ac29-2 (except that primers 1 and 5 were used, with an annealing temperature of 66°C), and a segment of ∼100 bp spanning the mutant site in ac29-2 was sequenced. The only change in the sequence relative to ac29-2 was within the XbaI site.
The insertion in mutant acy27 was characterized as follows. PCR amplification was performed using 25 ng of acy27 genomic DNA and primers 1 and 5 under the following conditions: 30 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min. This resulted in the formation of two products, one of 558 bp (from the ac29-2 allele on the mt− chromosome) and one of 636 bp (from the new ac29 allele on the mt+ chromosome). The larger fragment was gel purified and ligated into HincII-digested pUC118, and the sequence of ∼250 bp that includes the insertion was determined. PCR amplification and cloning were performed twice and yielded the same result. In addition to the insertion, a base change relative the wild-type genomic sequence was noted at position 1851 (A→C). This is within the second intron and represents a strain difference rather than a mutational change.
Primers
The primers used were as follows: primer 1, 5′-ATGGTGGCCAGTGTTGG-3′; primer 5, 5′-CACGTGCCCTACTCCTAC-3′; and primer 6, 5′-TCGGAAGCTATCCTGGC-3′. Note that primer 6 has 1 bp deleted relative to the actual genomic sequence.
Generation of New Mutants
A diploid was constructed (Ebersold, 1967) by mating CC-1067 (arg2 nr-u-2-1 mt+) to CC-1336 (nic7 ac29-2 act2 pf14 msr1 mt−), plating the cells onto TAP plus 3-acetylpyridine, and incubating in the light. Because 3-acetylpyridine is toxic to nicotinamide-requiring cells, only diploids and prototrophic recombinant meiotic progeny will survive, and the diploids tend to mature sooner. In addition, because NIC7 is linked tightly to mt+, the surviving meiotic progeny almost always will be mt+, whereas the diploids will mate as mt−. A strain mating as minus was selected, and diploidy was confirmed by showing the presence of mt-linked restriction fragment length polymorphisms from both parents on DNA gel blots.
The diploid strain was plated onto TAP at low density (∼50 colonies on an 85-mm plastic Petri dish). When the colonies were 1 to 2 mm in diameter, the entire plate was irradiated at 422 rad/min for 12 min from a 137Cs source (provided by William Wright, Washington University, St. Louis, MO). Thirty-six colonies were chosen from the irradiated plate and replated onto TAP plus nicotinamide. Mutants isolated from different colonies represent independent events. The plates were replica-plated to TAP plus 3-acetylpyridine to identify nicotinamide-requiring mutants. Any colonies that appeared yellow also were chosen and tested for nicotinamide requirement.
Transformation
Cells of ac29-3 were transformed with the linearized pAc5.8 plasmid, and green sectors on the high-salt medium plates were isolated, from which individual transformants were obtained after subcloning. Alternatively, the cells were cotransformed with a plasmid containing the hemagglutinin-tagged Alb3.1 gene and a plasmid containing the Cry1 gene that confers resistance to emetine (Nelson et al., 1994). Drug-resistant transformants were screened for expression of the tagged protein by immunoblot analysis with a hemagglutinin antiserum.
Protein Extracts and Immunoblotting
Thylakoid membranes were isolated as described (Fischer et al., 1997). These membranes were solubilized with β-dodecyl maltoside and fractionated on a 0.1 to 1 M Suc density gradient as described (Fischer et al., 1997). Immunoblot analysis was performed using the chemiluminescence detection system (Supersignal; Pierce).
Isolation and Characterization of the Alb3.2 cDNA
A BLAST search of the Chlamydomonas EST library (http://www.biology.duke.edu/chlamy_genome/EST.html) for homologs of Alb3.1 identified the EST clone BE122079.1. Total RNA was extracted from a wild-type Chlamydomonas strain grown in TAP to midlogarithmic phase using TriReagent from Sigma, and the RNA was treated with RQ-DNase (Promega, Madison, WI) to remove residual DNA. Reverse transcription was primed with random hexamers. Primers for the amplification of a 358-bp fragment were derived from EST BE122079.1 (forward 1, 5′-CAGGTGGAGTCGACGCTG-3′; forward 2, 5′-TGGAGTCGACGCTGTCGCT-3′; reverse 1, 5′-CTGGCGTCTTGGCTCGCC-3′; and reverse 2, 5′-AGCACCGGCATCACCAGGTA-3′).
The resulting PCR fragment was A-tailed (elongated with dATP; 2 mM dATP and 1 unit of TaqPol) for cloning into pDRIVE (Qiagen, Valencia, CA). The cloned fragment was verified by sequencing. A 380-bp EcoRI fragment from this plasmid was used as a probe to screen a cDNA library. Screening of 800,000 plaque forming units resulted in four positive clones containing inserts of 1.5, 1.8, 2.1, and 2.1 kb. Inserts were cut out with EcoRI, cloned into pBluescript KS−, and sequenced.
Procedures for the preparation of recombinant plasmids and DNA sequencing were performed as described by Sambrook et al. (1989).
RNA Analysis
The amount of Alb3.1 and Alb3.2 RNA was measured using reverse transcriptase–mediated PCR. The primers used were 5′-GTTATG-TCGAGCTCCATGTG-3′ and 5′-CGAAACGGTCCTTGATC-3′ for Alb3.1 and 5′-GATGCTGTGCACCACG-3′ and 5′-ACCACCAAT-GGTGGTTG-3′ for Alb3.2. The transcript of the gene encoding a G-protein β-subunit–like polypeptide of Chlamydomonas (Schloss, 1990) was used for reverse transcriptase–mediated PCR with primers 5′-GACGTCATCCACTGCCTGTG-3′ and 5′-CGACGCATCCTC-AACACACC-3′.
Five micrograms of RNA from the wild type was reverse transcribed with 50 units of Superscript reverse transcriptase, 0.5 μg of oligo(dT), 0.5 mM deoxynucleotide triphosphate, 5 mM MgCl2, 10 mM DTT, 20 mM Tris-HCl, pH 8.4, and 50 mM KCl for 50 min at 50°C in a total reaction volume of 50 μL. Three micrograms were used for PCR with Taq polymerase under the following conditions: 94°C for 4 min, then 30 cycles of 94°C for 1 min, 54°C for 45 s, and 72°C for 30 s, followed by 72°C for 4 min.
Sequence Analysis
Sequence alignments were performed using ClustalW (Thompson et al., 1994) and version 3.21 of Boxshade, written by K. Hofmann and M. Baron (http://www.ch.embnet.org/software/BOX_form.html). Potential transmembrane domains were identified using the transmembrane prediction program TMHMM 2.0 (Krogh et al., 2001) and TopPred2 (http://bioweb.pasteur.fr/seqanal/tmp/toppred/). Phylogenetic analysis was performed using the neighbor-joining method (Saitou and Nei, 1987). Distances were corrected for multiple substitutions using the percentage accepted mutations matrix of Dayhoff. The PHYLO_WIN program was used (Galtier and Gouy, 1996; http://evolution.genetics.washington.edu/phylip/software.html).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession number for the Alb3.1 gene is AF492768 and for the Alb3.2 cDNA is AF514291.
Acknowledgments
We thank M. Goldschmidt-Clermont for helpful comments, L. Small for technical assistance, J. Pawlowski for help with the phylogenetic analysis, and N. Roggli for preparing the figures. This work was supported by a fellowship from EMBO and the Danish Natural Science Foundation to H.N., by National Science Foundation Grant MCB-9904667 to P.F., and by Grant 3100-050895.97 from the Swiss National Fund to J.-D.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003442.
References
- Amin, P., Sy, D.A., Pilgrim, M.L., Parry, D.H., Nussaume, L., and Hoffman, N.E. (1999). Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 121, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Bartsevich, V.V., and Pakrasi, H.B. (1997). Molecular identification of a novel protein that regulates biogenesis of photosystem I, a membrane protein complex. J. Biol. Chem. 272, 6382–6387. [DOI] [PubMed] [Google Scholar]
- Bonnefoy, N., Chalvet, F., Hamel, P., Slonimski, P.P., and Dujardin, G. (1994). OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes, controls cytochrome oxidase biogenesis. J. Mol. Biol. 239, 201–212. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., Takahashi, Y., Lemieux, C., Turmel, M., and Rochaix, J.D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16, 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersold, W.T. (1967). Chlamydomonas reinhardtii: Heterozygous diploid strains. Science 157, 447–449. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J. (1989). Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher plants. Genetics 122, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J. (1995). Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141, 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., and Goodenough, U.W. (1994). The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Fischer, N., and Rochaix, J.D. (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265, 888–894. [DOI] [PubMed] [Google Scholar]
- Fischer, N., Setif, P., and Rochaix, J.D. (1997). Targeted mutations in the psaC gene of Chlamydomonas reinhardtii: Preferential reduction of FB at low temperature is not accompanied by altered electron flow from photosystem I to ferredoxin. Biochemistry 36, 93–102. [DOI] [PubMed] [Google Scholar]
- Galtier, N., and Gouy, M. (1996). SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comp. Appl Biosci. 12, 543–548. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Hell, K., Herrmann, J.M., Pratje, E., Neupert, W., and Stuart, R.A. (1998). Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl. Acad. Sci. USA 95, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell, K., Neupert, W., and Stuart, R.A. (2001). Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, E.N., Scotti, P.A., Valent, Q.A., Brunner, J., de Gier, J.L., Oudega, B., and Luirink, J. (2000). Nascent Lep inserts into the Escherichia coli inner membrane in the vicinity of YidC, SecY and SecA. FEBS Lett. 476, 229–233. [DOI] [PubMed] [Google Scholar]
- Jiang, F., Moore, M., Chen, M., Rohl, T., van Wijk, K., de Gier, J.-W., Henry, R., and Dalbey, R. (2002). Chloroplast YidC homologue Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bac-terial and chloroplast thylakoid proteins. J. Biol. Chem. 277, 19281–19288. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle, K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk, V.I., Persello-Cartieaux, F., Havaux, M., Contard-David, P., Schuenemann, D., Meiherhoff, K., Gouet, P., Jones, J.D., Hoffman, N.E., and Nussaume, L. (1999). A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Levine, R.P., and Goodenough, U.W. (1970). The genetics of photosynthesis and of the chloroplast in Chlamydomonas reinhardtii. Annu. Rev. Genet. 4, 397–408. [DOI] [PubMed] [Google Scholar]
- Li, X., Henry, R., Yuan, J., Cline, K., and Hoffman, N.E. (1995). A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc. Natl. Acad. Sci. USA 92, 3789–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink, J., Samuelsson, T., and de Gier, J.W. (2001). YidC/Oxa1p/Alb3: Evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501, 1–5. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plucken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M., Harrison, M.S., Peterson, E.C., and Henry, R. (2000). Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275, 1529–1532. [DOI] [PubMed] [Google Scholar]
- Nargang, F.E., Preuss, M., Neupert, W., and Herrmann, J.M. (2002). The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J. Biol. Chem. 277, 12846–12853. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Savereide, P.B., and Lefebvre, P.A. (1994). The CRY1 gene in Chlamydomonas reinhardtii: Structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14, 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, R., Brunner, J., Hoffman, N.E., and van Wijk, K.J. (1999). Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.D. (1999). A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18, 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle, J.E., Benemann, J.R., Tanaka, A., and Melis, A. (2000). Photosynthetic apparatus organization and function in the wild type and a chlorophyll b-less mutant of Chlamydomonas reinhardtii: Dependence on carbon source. Planta 211, 335–344. [DOI] [PubMed] [Google Scholar]
- Porra, R., Thompson, W., and Kriedemann, P. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Robinson, C., and Bolhuis, A. (2001). Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2, 350–356. [DOI] [PubMed] [Google Scholar]
- Ruf, S., Kossel, H., and Bock, R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schloss, J.A. (1990). A Chlamydomonas gene encodes a G protein β subunit-like polypeptide. Mol. Gen. Genet. 221, 443–452. [DOI] [PubMed] [Google Scholar]
- Schuenemann, D., Gupta, S., Persello-Cartieaux, F., Klimyuk, V.V., Jones, J.D.G., Nussaume, L., and Hoffman, N.E. (1998). A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. USA 95, 10312–10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg, E., Slagter, J.G., Fridborg, I., Cleary, S.P., Robinson, C., and Coupland, G. (1997). ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C.J., Schuenemann, D., and Hoffman, N.E. (1999). Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J. Biol. Chem. 274, 27219–27224. [DOI] [PubMed] [Google Scholar]
- Valent, Q.A., Scotti, P.A., High, S., de Gier, J.W., von Heijne, G., Lentzen, G., Wintermeyer, W., Oudega, B., and Luirink, J. (1998). The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17, 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, A., Hartel, H., Hubschmann, T., Hoffmann, P., Shestakov, S.V., and Borner, T. (1995). Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhead, C.A., Thompson, S.J., Moore, M., Tissier, C., Mant, A., Rodger, A., Henry, R., and Robinson, C. (2001). Distinct Albino3-dependent and -independent pathways for thylakoid membrane protein insertion. J. Biol. Chem. 276, 40841–40846. [DOI] [PubMed] [Google Scholar]
- Zak, E., Norling, B., Andersson, B., and Pakrasi, H.B. (1999). Subcellular localization of the BtpA protein in the cyanobacterium Synechocystis sp. PCC 6803. Eur. J. Biochem. 261, 311–316. [DOI] [PubMed] [Google Scholar]
- Zak, E., and Pakrasi, H.B. (2000). The BtpA protein stabilizes the reaction center proteins of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803 at low temperature. Plant Physiol. 123, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]