Abstract
We have used cDNA microarrays to examine changes in gene expression during Arabidopsis seed development and to compare wild-type and mutant wrinkled1 (wri1) seeds that have an 80% reduction in oil. Between 5 and 13 days after flowering, a period preceding and including the major accumulation of storage oils and proteins, ∼35% of the genes represented on the array changed at least twofold, but a larger fraction (65%) showed little or no change in expression. Genes whose expression changed most tended to be expressed more in seeds than in other tissues. Genes related to the biosynthesis of storage components showed several distinct temporal expression patterns. For example, a number of genes encoding core fatty acid synthesis enzymes displayed a bell-shaped pattern of expression between 5 and 13 days after flowering. By contrast, the expression of storage proteins, oleosins, and other known abscisic acid–regulated genes increased later and remained high. Genes for photosynthetic proteins followed a pattern very similar to that of fatty acid synthesis proteins, implicating a role in CO2 refixation and the supply of cofactors for oil synthesis. Expression profiles of key carbon transporters and glycolytic enzymes reflected shifts in flux from cytosolic to plastid metabolism. Despite major changes in metabolism between wri1 and wild-type seeds, <1% of genes differed by more than twofold, and most of these were involved in central lipid and carbohydrate metabolism. Thus, these data define in part the downstream responses to disruption of the WRI1 gene.
INTRODUCTION
Most terrestrial life depends directly or indirectly on plant seeds for existence. For the survival of Spermatophyta, a seed must package together all of the genetic material and macronutrients and micronutrients needed to allow successful propagation of the species. The establishment of a new generation requires seed dispersal, germination, and mobilization of stored reserves until seedling photosynthesis can provide new material for growth. Although the means of dispersal are diverse and seeds differ greatly in size and form, one common element in all plant seeds is the storage of reserve materials. The storage compounds found in most seeds contribute up to 90% or more of the seed dry weight and consist of carbohydrates (often starch), oils (almost always triacylglycerols), and specialized storage proteins. In most agricultural field crops, these components also represent the economic value of the crop and provide the major value of seeds to humans and many other animals. Consequently, for centuries, much effort by plant breeders has focused on strategies to enhance both the quantity and the quality of seed storage reserves, and more recently, molecular genetic approaches have been used to modify seed components (Voelker et al., 1996; Mazur et al., 1999; Ye et al., 2000).
Different plant species vary greatly in the relative proportions of carbohydrate, oil, and protein stored in seeds. For example, most grain seeds contain >85% starch, whereas many oilseeds produce 50 to 70% oil, and some legumes such as soybeans contain 40% of the seed dry weight as protein. The biochemical pathways that produce these different storage components are largely known, but the factors that determine the relative proportions of the different storage components are not well understood. The accumulation of each class of storage component requires the coordination of many genes that encode the enzymes of the respective pathways. Although it can be presumed that coordinated expression of these genes occurs, relatively few studies have examined more than one or a few of the members of any pathway in seeds. To date, there have been no large-scale studies of the timing of expression of multiple genes from different biosynthetic pathways in developing seeds. Microarrays offer the possibility to study thousands of genes using RNA extracted from the same plant material, thus avoiding the complications that arise when comparing expression patterns from different studies. With the completion of the Arabidopsis genome sequence, microarray studies with Arabidopsis provide an additional opportunity to examine common elements of promoter sequences from coregulated genes.
In this study, we have examined the steady state mRNA levels of >3500 genes at different stages of Arabidopsis seed development, when the seeds undergo major changes in metabolism. During early embryogenesis, when the tissues and organelles are established, carbon and other nutrients are used mainly for rapid cell division and embryo growth. After the cessation of cell division, during the maturation phase, resources are allocated to synthesize storage compounds. In Arabidopsis, this maturation phase is characterized by a transient accumulation of starch, followed by major increases in the oil and protein contents. During the subsequent late maturation and desiccation phases, the overall biosynthetic activity decreases as the seed prepares for dormancy (Mansfield and Briarty, 1991; Harada, 1994).
As detailed in this study, the gene expression patterns associated with these developmental changes can be described as “contrapuntal,” because groups of genes change expression simultaneously in opposite directions. In addition to analyzing wild-type seeds, we also studied differences in mRNA profiles between wild-type seeds and the wrinkled1 (wri1) mutant, which is defective in the accumulation of storage oils (Focks and Benning, 1998). This study has not only identified and cataloged temporal changes in gene expression but has begun to provide insight into the structure of the primary transcriptional networks that coordinate the genome-wide response to seed developmental programs and lead to the distribution of carbon among oil, protein, and carbohydrate reserves.
RESULTS AND DISCUSSION
Accumulation of Storage Reserves
One major objective of this study was to describe the patterns of steady state mRNA abundances during the accumulation of storage reserves in developing Arabidopsis seeds. In this article, the term “gene expression” refers to these mRNA levels. Of course, changes in mRNA levels are not always reflective of changes in gene transcription, protein levels, or enzyme activities. However, in general, transcriptional regulation has been demonstrated to provide major control of countless processes involved in plant development and metabolism, and microarrays offer the opportunity to simultaneously examine changes in mRNA levels for thousands of genes.
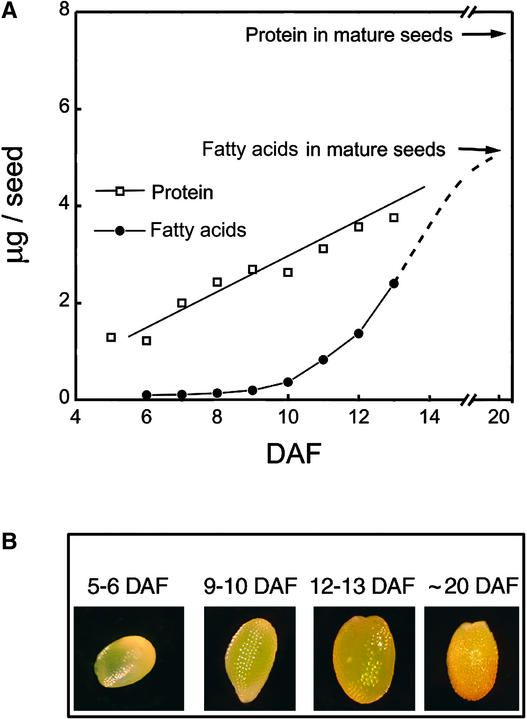
Seed development from pollination to mature desiccated seeds occurs over 18 to 21 days in Arabidopsis var Columbia plants grown in standard conditions (Koornneef and Karssen, 1994). The major storage reserves accumulate from 8 to 16 days after flowering (DAF), with maximum rates of accumulation between 9 and 13 DAF (Focks and Benning, 1998). As shown in Figure 1A, for seeds used in this study, the major accumulation of storage lipids started at 9 to 10 DAF, as indicated by an increase in total fatty acid content and the appearance of long-chain fatty acids, which are characteristic of storage oils (20:1; data not shown). Seeds at 6 to 8 DAF contained ∼0.1 μg of total fatty acids per seed. Oil content increased >20-fold to 2.4 μg per seed by 13 DAF, corresponding to 50 to 60% of the typical total lipid content of mature seeds. The total protein content was 1.25 μg per seed at 5 DAF and increased to 3.8 μg per seed at 13 DAF, corresponding to ∼50% of the total protein content of mature seeds (Mansfield and Briarty, 1991). Thus, the protein content of the developing seeds increased more steadily and less dramatically than the lipid content through the time course.
Figure 1.
Development of Arabidopsis Seeds during the Experimental Period.
(A) Time courses of total fatty acid and total protein accumulation in developing wild-type Arabidopsis seeds. Values are averages of two independent measurements, and the seed material used was the same as for the microarray experiments. The arrows indicate the typical fatty acid and protein contents of mature, dry seeds. The dotted line illustrates the typical accumulation pattern for storage oil, which reaches its maximum at ∼18 DAF (Focks and Benning, 1998). The protein content increases more steadily.
(B) Seeds at the beginning of the microarray time course (5 to 6 DAF), at the midstage (used for reference; 9 to 10 DAF), at the end of the experimental period (12 to 13 DAF), and at ∼20 DAF, when the seeds are fully mature and begin to desiccate.
We chose to focus our study on the period between 5 and 13 DAF because 5 to 7 DAF precedes the rapid increase in storage product synthesis and 8 to 13 DAF includes the major period when seed biosynthetic pathways are at maximum activity. The experimental period included the early maturation stage, when the seeds had generally attained their final size but desiccation had not yet started (Figure 1B). We further reasoned that mRNA changes involved in the accumulation of biosynthetic enzymes and the controls over this process would precede the appearance of the enzymes and their products. Because other available cDNA and oligonucleotide arrays were underrepresented in seed-expressed genes, the microarrays used in this study were designed specifically to study Arabidopsis seeds and were based on 10,500 cDNAs sequenced from an Arabidopsis developing seed library (White et al., 2000).
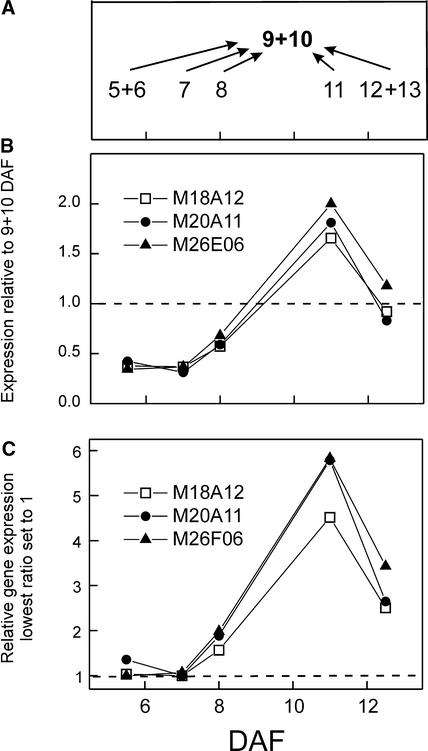
The ∼6000 DNA elements on the array were amplified from cDNA clones that mapped to ∼3500 unique Arabidopsis genes. Thus, almost 60% of the genes on the array were represented by multiple clones, and their combined analysis allowed increased reliability in expression analysis (Figures 2B and 2C). The cDNA-based microarrays were hybridized simultaneously with probes derived from two populations of mRNA, allowing accurate and reliable measurements of the ratio of expression between two mRNA samples for thousands of genes represented on the arrays. To obtain consistent comparisons of mRNA from several different time points during seed development, we chose seeds in the middle of the time course (9 and 10 DAF) as a constant reference for comparison with mRNA at other stages of development. Five time points were selected over a 9-day period between 5 and 13 DAF. As illustrated in Figure 2A, probes derived from mRNA extracted from seeds at each stage of development were cohybridized with probes from the reference 9+10-DAF mRNA.
Figure 2.
Experimental Procedure and Conversion of the Expression Ratios.
(A) Five different time points were compared with the common reference (9 to 10 DAF).
(B) Expression profiles of three clones mapped to chlorophyll a/b binding protein. The average expression ratios from two hybridizations, relative to 9+10 DAF, are shown.
(C) The lowest average expression ratio for each clone was set to 1, and the other ratios were adjusted accordingly.
General Patterns of Expression: The Majority of Genes Change Little between 5 and 13 DAF
Of the ∼6000 clones on the array, 4475 had an average signal intensity more than two times the background; these were considered reliable and were included for further analysis. Approximately 35% (1525) of these reliable clones changed more than twofold during the time course. The clones that changed significantly belonged to ∼1160 unique genes. Surprisingly, gene expression changes during the 5- to 13-DAF time course were moderate for the majority of clones, despite large changes in seed metabolism during this period. Only 29 clones changed >10-fold, 235 changed >4-fold, and the remaining 1312 changed only 2- to 4-fold during the experimental period.
In part, the reasons for the small expression differences between 5 and 13 DAF could reflect technical issues. The development of embryos within a silique is not synchronous (Bowman and Mansfield, 1994), leading to mixing of seeds of different developmental stages within the samples extracted for mRNA. In addition, array data may compress differentials (Hihara et al., 2001). Also, because ∼60% of Arabidopsis genes belong to gene families (Arabidopsis Genome Initiative, 2000), cross-hybridization between members may result in lack of discrimination by cDNA microarrays among closely related genes (Girke et al., 2000).
Nevertheless, our observations from a large number of individual genes agreed with earlier results obtained using hybridization kinetics to study different mRNA classes in developing seeds. Goldberg et al. (1981) found that a large fraction of seed transcripts were present throughout seed development and were even stored in dry seeds. Moreover, Hill et al. (2000) described gene expression of Caenorhabditis elegans thorough its life cycle using arrays that contained 10,000 open reading frames. Using oligonucleotide-based gene arrays that provide more accurate discrimination between gene family members, Hill et al. (2000) found that only 22% of all open reading frames with detectable signals changed significantly during the life cycle, a number very similar to our observations. Thus, a general conclusion of global gene expression analysis from several organisms is that the majority of genes expressed in a tissue are expressed constitutively at the mRNA level.
We previously characterized tissue-specific expression patterns of Arabidopsis genes using microarrays (Girke et al., 2000). Interestingly, in the current study, we found that genes that changed most during days 5 to 13 of Arabidopsis seed development also tended to be expressed preferentially in seeds. Among those genes that were expressed at least twofold higher in seeds than in leaves or whole plantlets, the majority (70%) changed at least twofold during the time course. By contrast, only 33% of the non-seed-specific genes were found to change more than twofold between 5 and 13 DAF (data not shown).
Lipid-Related Genes Have at Least Two Different Patterns of Expression
Although previous studies have reported changes during seed development in transcripts for enzymes of lipid metabolism, these studies generally have been limited to one or a few genes. The arrays used in this study allowed the simultaneous analysis of >100 genes involved in lipid metabolism, enabling us to assemble a broader picture of the regulation of transcripts for the pathway.
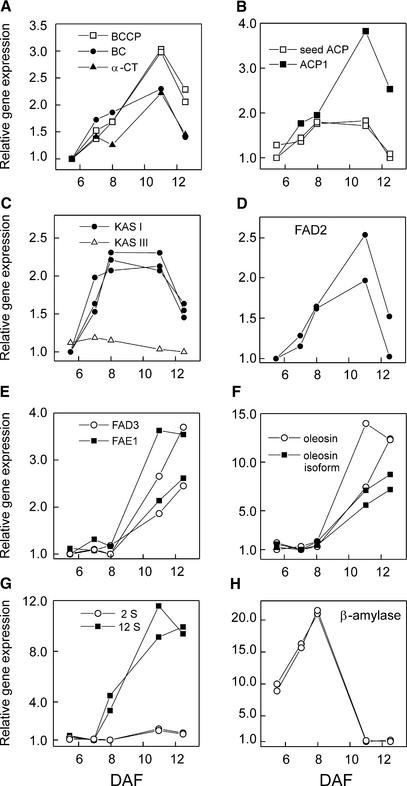
Approximately 40% of the lipid genes on the array changed more than twofold during the time course, a similar proportion to the figure for all genes. The genes that changed could be characterized by two main expression patterns. The expression in the first group followed a bell-shaped pattern that increased through the early developmental stages, peaked between 8 and 11 DAF, and then decreased. This is illustrated in Figures 3A to 3D for the expression of three subunits of plastidial acetyl-CoA carboxylase (ACCase; Figure 3A), two isoforms of acyl carrier protein (ACP; Figure 3B), 3-ketoacyl-ACP synthase I (KAS I; Figure 3C), and oleate desaturase (FAD2; Figure 3D).
Figure 3.
Expression Profiles of Storage Compound–Related Genes.
(A) Biotin carboxyl carrier protein (BCCP), biotin carboxylase (BC), and α-carboxyltransferase (α-CT) subunits of plastidial acetyl-CoA carboxylase.
(B) Acyl carrier protein1 precursor (ACP1) and seed ACP.
(C) 3-Ketoacyl-ACP synthase I (KAS I) and 3-ketoacyl-ACP synthase III (KAS III).
(D) Oleate desaturase (FAD2).
(E) Linoleate desaturase (FAD3) and fatty acid elongase1 (FAE1).
(F) Oleosin isoform and 18.5-kD oleosin.
(G) Storage proteins 12S and 2S.
(H) β-Amylase.
Expression is related to 9+10 DAF, and the ratios were rescaled by setting the lowest ratio to 1. See Methods for accession numbers.
The second group was characterized by a distinctly different profile, with maximum expression later in development. For example, the expression of linoleate desaturase (FAD3) and fatty acid elongase (FAE1) was low in young seeds and started to increase at ∼8 DAF (Figure 3E). Similarly, the expression of genes encoding different oleosin isoforms was induced strongly at ∼8 DAF (Figure 3F). Unlike the genes in the first group, the expression of genes with this pattern remained high until the end of the time course.
A third group included genes that did not change significantly during the time course. This set included some core enzymes of fatty acid synthesis (FAS), including KAS III (Figure 3C) and isoforms of ACP, that are known to be expressed in all tissues. In addition, several acyltransferases, enzymes of phospholipid metabolism, and genes for reactions later in the oil synthesis pathway changed little between 5 and 13 DAF (data not shown). Thus, although our expectation was that the entire lipid biosynthetic pathway would undergo coordinated regulation similar to that observed for flavonoid biosynthesis genes (Bruce et al., 2000; Harmer et al., 2000), this was not the case.
These microarray results extend and coincide with more limited earlier studies of developing soybeans (Heppard et al., 1996), Brassica napus embryos (for review, see Post-Beittenmiller et al., 1993), and Arabidopsis siliques (Ke et al., 2000). For example, the mRNA levels of ACCase and other FAS enzymes (such as ACP, KAS I, and enoyl-ACP reductase) tend to increase before the onset of seed oil accumulation and thus can be expected to be under similar control mechanisms. In accordance with these mRNA studies, measurements of enzyme activities and protein amounts have shown a transient increase in ACCase activity during the development of castor beans (Simcox et al., 1979) and Brassica seeds (Turnham and Northcote, 1983), with the maximum enzyme activity during the rapid phase of oil accumulation.
The microarray results also agreed with earlier evidence that the expression of some fatty acid–modifying enzymes, as well as oleosins, is under different control than the expression of the core FAS enzymes (Post-Beittenmiller et al., 1993). It is known that abscisic acid induces oleosin expression (Holbrook et al., 1991), and FAD3 and FAE1 also are regulated by abscisic acid (Finkelstein and Somerville, 1990; Zou et al., 1995; Qi et al., 1998). These functionally related genes (FAD3, FAE1, and oleosins) also had a similar temporal expression pattern (Figures 3E and 3F) that was clearly different from that of many FAS enzymes. Agreement between the microarray results described here and several previous studies using other techniques supports the validity of microarray analysis in revealing gene expression patterns.
Storage Proteins
The two major storage proteins in Arabidopsis are 12S (cruciferin-like) and 2S (napin-like) proteins. In general, these two classes behaved differently. 2S transcripts were highly abundant at all stages between 5 and 13 DAF, and their expression increased approximately twofold toward the end of the time course. In contrast, most of the 12S transcripts gave weaker signals, but their expression increased up to 10-fold in the maturing seeds between 8 and 13 DAF (Figure 3G). Similar results to 12S also were obtained for a number of transcripts encoding vicilin-like storage protein (data not shown). Again, these results complement earlier studies using RNA gel blot analysis. Guerche et al. (1990) studied four different genes from the 2S family and found that all were similarly temporally regulated such that their expression increased approximately threefold between 4 to 6 and 10 days after pollination. On the other hand, the 12S protein transcripts in Arabidopsis become abundant only during the final stages of embryo development (Heath et al., 1986).
It is known that storage protein gene expression in Brassica and Arabidopsis is under abscisic acid–mediated control (Finkelstein et al., 1985; Finkelstein and Somerville, 1990), and, according to our data, their induction and expression profiles (especially 12S and vicilin) correlated with those of the abscisic acid–regulated lipid-related genes but was clearly different from those of the core FAS enzymes. These results further emphasize that there are distinct mechanisms regulating storage oil and protein accumulation in seeds. In addition, both oleosin and most storage protein transcripts were significantly more abundant than FAS enzymes, and their expression increased severalfold more than FAS enzymes, which are involved in enzymatic catalysis rather than serving as storage products.
Starch Metabolism Genes Are Expressed Most Actively in Young Seeds
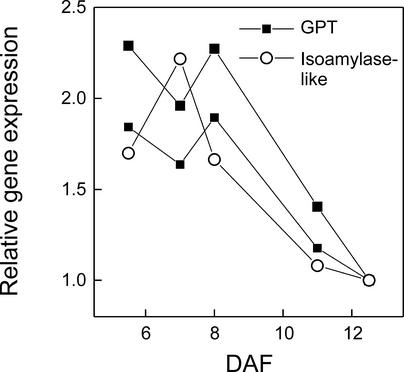
Young Arabidopsis seeds transiently accumulate starch, which later disappears and presumably is used as a carbon source for fatty acid and protein synthesis. Kang and Rawsthorne (1994) reported that in Brassica embryos, both starch synthesis and degradation were most active early in development, before the oil accumulation phase. In general, this pattern also was found in microarray analysis of Arabidopsis seeds. The expression pattern of isoamylase-like protein, which has a role in transitory starch synthesis in Arabidopsis (Zeeman et al., 1998), was highest in young seeds and then decreased (see Figure 5). In addition, the expression of the starch-degrading enzyme β-amylase peaked at 8 DAF and then decreased rapidly (Figure 3H). The close correlation between these transcript patterns and the reported changes in Arabidopsis seed starch content (Focks and Benning, 1998) suggests that in developing Arabidopsis seeds, the transient accumulation and subsequent starch breakdown is a developmental program controlled at the transcript level rather than a response to shifts in carbon import or utilization by other pathways.
Figure 5.
Coordinated Downregulation of the Import of Glc-6-P into Plastids and Starch Metabolism.
Presented are chloroplast Glc-6-P translocator (GPT; see Figure 4F) and isoamylase-like protein. See Methods for accession numbers.
Carbon Pathway from Suc to Fatty Acids
Sink organs such as developing seeds import Suc, which is cleaved by Suc synthase (SuSy) to provide carbon skeletons for all major classes of storage product. Studies with leguminous plants have established that SuSy is a marker for storage accumulation activity and that its transcripts are induced by Suc (Heim et al., 1993). However, in leguminous seeds, it is the onset of starch synthesis that is accompanied by an increase in SuSy activity. In Arabidopsis seeds, the expression of several SuSy clones increased strongly after 8 DAF (Figure 4A). Thus, in contrast to its action in legumes, SuSy induction in Arabidopsis coincided with an increase in oil and protein synthesis rather than with the intermittent starch accumulation in young seeds. A similar trend in SuSy activity was reported in developing Brassica seeds (King et al., 1997). SuSy ESTs also were found to be very abundant in developing Arabidopsis seeds (White et al., 2000) and exhibited preferential expression in seeds (Girke et al., 2000), emphasizing the essential role of SuSy in providing carbon for oilseed metabolism (King et al., 1997).
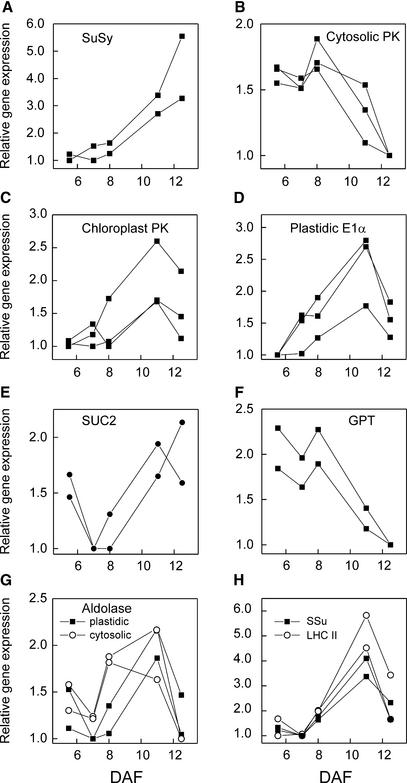
Figure 4.
Expression Profiles of Carbohydrate Metabolism–Related Genes.
(A) Suc synthase (SuSy).
(B) Cytosolic pyruvate kinase (PK).
(C) Chloroplast pyruvate kinase.
(D) Chloroplast E1α subunit of pyruvate dehydrogenase.
(E) Suc-proton transporter protein (SUC2).
(F) Chloroplast Glc-6-P translocator (GPT).
(G) Chloroplast and cytosolic aldolases.
(H) Photosystem II light-harvesting complex protein (LHC II) and small subunit of Rubisco (SSu).
Expression is related to 9+10 DAF, and the ratios were rescaled by setting the lowest ratio to 1. See Methods for accession numbers.
The products of SuSy are metabolized largely through the glycolytic pathway before utilization in subsequent reactions, such as fatty acid and amino acid biosynthesis. In addition to the cytosolic pathway, oilseeds also have a complete set of glycolytic enzymes in plastids (Kang and Rawsthorne, 1994), but it remains unclear how these two pathways are used. Eastmond and Rawsthorne (2000) reported that the activities of plastid forms of several glycolytic enzymes decreased during oil synthesis in Brassica embryos. Together with the analysis of Arabidopsis seed ESTs (White et al., 2000), these results suggest that a major route of carbohydrate conversion into oil involves cytosolic glycolysis to phosphoenolpyruvate (PEP) followed by PEP import to plastids and its conversion to pyruvate and acetyl-CoA. As described below, the microarray data generally agreed with this notion, but they also provided additional information about the temporal sequence of events of carbon metabolism and transport in the maturing seed.
The presence of both cytosolic and plastidial forms of pyruvate kinase (PK) on the arrays enabled us to compare the final reactions of the two glycolytic pathways. Significantly, the expression patterns of the two isoforms changed in opposite directions. The expression of the cytosolic PK decreased after 8 DAF (Figure 4B), whereas the plastid form increased toward the active oil accumulation period and then decreased again in the oldest seeds (Figure 4D) in the bell-shaped pattern similar to ACCase and other FAS enzymes. The coordinated channeling of carbon from PEP to acetyl-CoA inside the plastid was further reflected by the expression profiles of the E1α subunit of the plastid pyruvate dehydrogenase (PDH) (Figure 4D) as well as its E2 subunit (data not shown), which peaked at 11 DAF and then decreased. Thus, the production and consumption of acetyl-CoA inside the plastid were coordinated at the transcript level, as demonstrated by the similar expression patterns of plastid PK, PDH, and ACCase, and these patterns coincided with the active oil synthesis period.
Additional insight into this model was derived from an analysis of carbon transporters, which have been suggested to play a role in regulating the metabolic transition of young oilseed embryos from starch-storing organs to oil-storing organs (Eastmond and Rawsthorne, 2000). The expression of H+-Suc symporter increased toward seed filling (after 8 DAF), implying that both Suc delivery to the embryo and its successive catabolism by SuSy were enhanced coordinately (Figures 4A and 4E). On the other hand, the expression profile of plastid PEP/Pi translocator was similar to that of many FAS enzymes, although it changed only ∼1.5-fold (data not shown). In contrast, the expression of plastid Glc-6-P translocator (GPT) decreased rapidly after 8 DAF (Figure 4F). The capital downregulation of the plastid GPT transcripts (Figure 5) and activity (Eastmond and Rawsthorne, 2000) may limit carbon transport into plastids as hexoses and, together with the decreased expression of cytosolic PK, ensures that the main carbon flux during oil biosynthesis includes glycolysis to PEP in the cytosol followed by its uptake into plastids. The general downregulation of carbon storage as starch in seeds is evident in the parallel decreases of GPT and isoamylase gene expression (Figure 5), as well as that of β-amylase (Figure 3H), at the time of the metabolic transition to oil accumulation.
Thus, the microarray data provided an overall picture of numerous parallel and contrapuntal changes in the expression of translocators and glycolytic enzymes that support the model of primary carbon metabolism in Arabidopsis seeds shown in Figure 6. Pyruvate transport into plastids also increases during Brassica seed development (Eastmond and Rawsthorne, 2000) and may contribute to carbon flux into oil, although this route would deliver less plastidial ATP and the decrease in cytosolic PK (Figure 4B) may result in less pyruvate available for this process.
Figure 6.
Contrapuntal Patterns of Gene Expression for Proteins Involved in the Conversion of Suc to Oil in Developing Arabidopsis Seeds.
Only the major enzymes and transporters involved in the conversion of Suc to oil are shown. Microarray expression patterns for enzymes in the cytosol are shown with red lines, reactions in the plastid are shown with green lines, and membrane transporters are shown with blue lines. Gray arrows indicate reactions or transporters whose expression decreases during development; they are believed to carry less flux than the major pathway represented by brown arrows. The shift during development from starch biosynthesis and cytosolic metabolism of PEP to oil biosynthesis and plastid uptake and metabolism of PEP is reflected in the coordinated expression patterns of enzymes and transporters. Not shown are additional contributions to oil metabolism, such as dark and light reactions of photosynthesis, the oxidative pentose phosphate pathway, and others. AcCoa, acetyl-CoA; FAS, fatty acid synthesis; GPT, glc-6-P translocator; MalCoA, malonyl-CoA; Pyr, pyruvate; TAG, triacylglyceride; TP, triose phosphate; TPT, triose-P translocator.
All Glycolytic Enzymes Are Not Regulated Similarly in Developing Seeds
Based on the coordinated transcriptional regulation of several glycolytic enzymes observed in yeast during diauxic shift (DeRisi et al., 1997) and in Escherichia coli grown on different carbon sources (Oh and Liao, 2000), we expected to see groups of glycolytic enzymes with similar temporal expression patterns in the developing seeds. However, in general, this was not the case. For example, several genes encoding different aldolase isoforms, both plastidial and cytosolic forms, were represented on the arrays. Some genes changed during the time course and some did not, and in cases in which the expression changed, both cytosolic and plastid genes behaved similarly (Figure 4G). In addition, the enzymes that catalyze the interconversions of hexoses in the cytosol had different profiles. The expression of UDP-Glc pyrophosphorylase and phosphoglucomutase was highest in young seeds (5 to 7 DAF), whereas fructokinase behaved similarly to ACCase subunits, peaking at 11 DAF (data not shown).
Noncoordinated expression patterns for the glycolytic enzymes in developing seeds could be related to the multiple pathways and demands on central enzymes of intermediary metabolism. For example, plastid aldolase and glyceraldehyde 3-P dehydrogenase can participate in both glycolysis and photosynthetic carbon reduction. In addition, during embryogenesis, the active use of carbon precursors and energy from glycolysis is required for rapid cell division, and in the maturation phase, these resources may simply be redirected to storage compound synthesis. Finally, studies with Brassica embryos established that the activities of all glycolytic enzymes in plastids were well in excess of the rate of fatty acid synthesis (Eastmond and Rawsthorne, 2000) and so are not likely to have a strong regulatory role.
Role of Photosynthesis in Developing Oilseeds
Developing Brassica and Arabidopsis embryos are green and contain functional chloroplasts. However, studies on the role of photosynthesis during seed filling in Brassicacea have led to different conclusions regarding its contribution to carbon or energy metabolism in the seed (Eastmond et al., 1996; King et al., 1997). ESTs encoding chlorophyll binding proteins, as well as ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunits, were found to be highly abundant in developing Arabidopsis seeds (White et al., 2000). Our microarray analysis indicated that the expression of several genes from both photosynthetic light reactions (such as light-harvesting complex II and photosystem II oxygen-evolving complex) and dark reactions (Rubisco and phosphoribulokinase) was low in the youngest seeds, increased strongly by 11 DAF, and then decreased again, following the bell-shaped profile of several FAS enzymes (Figure 4H; see also Figure 2).
A similar temporal pattern has been reported for Rubisco activity in developing castor seeds (Simcox et al., 1979) as well as for Arabidopsis seed chlorophyll content (Focks, 1997). These profiles were very different from those of the starch metabolic enzymes and storage proteins, which were highest in young seeds and older seeds, respectively. Thus, the embryo's maximal photosynthetic capacity coincided most closely with the oil accumulation period rather than with starch or protein accumulation. These data support the hypothesis that photosynthetic activity in these seeds contributes to carbon acquisition as well as to the heavy demand by fatty acid synthesis for reductants and ATP (King et al., 1998).
The fixation of atmospheric CO2 is likely to be low within the seed as a result of the silique wall and other diffusion barriers (King et al., 1997). Therefore, why does Rubisco expression increase during seed development? In oil-storing seeds, this enzyme may be especially important in recycling the carbon released during fatty acid biosynthesis. One-third of the carbon devoted to oil synthesis is lost in the conversion of pyruvate to acetyl-CoA by the plastid pyruvate dehydrogenase. Thus, in seeds such as Arabidopsis that store oil, CO2 release represents a substantial proportion of the seed's carbon economy. The expression of Rubisco and other Calvin cycle enzymes provides a plastidial route to recapture this carbon, provided that sufficient reductant and cofactors are available.
The penetrance of ∼20% light through the silique wall (King et al., 1998), together with the expression of photosynthetic light and dark reaction pathways (Figure 4H), may provide these additional factors. Based on the microarray data, we propose that this route may be more significant than the alternative of carbon fixation through cytosolic PEP carboxylase, whose transcript levels were very low and decreased further in maturing seeds (data not shown). As suggested by Neuhaus and Emes (2000), green plastids in storage organs may operate in a “mixed carbon economy” in which heterotrophic and autotrophic pathways operate simultaneously for energy production and biosynthetic activities.
Regulatory Factors
The changes in expression of the mRNA transcripts described above for enzymes and proteins involved in storage product accumulation are regulated in large part by DNA binding transcription factors or proteins involved in RNA turnover. Protein phosphatases and kinases, in turn, provide means to transduce internal (e.g., hormones) and external (e.g., temperature) signals into transcriptional and/or chemical responses in cells. A large part of current knowledge regarding the regulation of gene expression in developing seeds has been generated from the analysis of transcription factors that function in embryo development (e.g., ABI3, FUS3, and LEC1 [for review, see Wobus and Weber, 1999]). Almost no protein phosphatases and kinases from seeds have been analyzed in similar detail. Two abscisic acid–responsive kinases, ABI1 and ABI2, have been identified (Leung et al., 1997), as have two auxin-induced mitogen-activated protein kinases that are expressed at low levels in Arabidopsis seeds (Mizoguchi et al., 1994).
The seed microarray contained >300 genes annotated as putative transcription factors or protein phosphatases/kinases. Almost none of these has been studied previously. We found that 33 putative transcription factors (of 95 with reliable signals) and 59 putative protein phosphatases or kinases (of 180 reliable genes) changed at least twofold between 5 and 13 DAF. We further compared their expression profiles with those of other genes of interest. For example, a striking feature of several core FAS genes is that their expression is downregulated strongly at the time when oil accumulation is most active.
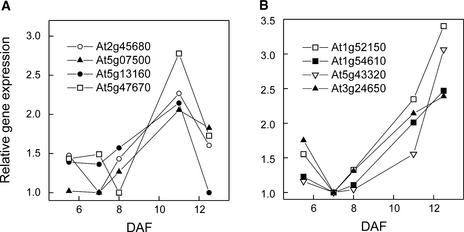
We identified several regulatory factors whose expression patterns either followed or preceded these FAS enzymes (Figure 7A) and thus are candidates for positive effectors of FAS gene expression. On the other hand, a group of regulators whose expression increased throughout the time course (Figure 7B) included ABI3, a known abscisic acid–regulated transcription factor. Therefore, this group is likely to contain genes involved in abscisic acid–mediated signal transduction as well as candidates for other abscisic acid–responsive DNA binding proteins. The temporal expression patterns of these regulatory genes thus provided an initial characterization of the regulatory networks that function in developing seeds as well as candidates for further, more detailed work, such as overexpression or gene-silencing experiments.
Figure 7.
Examples of Expression Profiles for Putative Regulatory Factors.
(A) Genes that follow the expression of ACCase-type FAS enzymes: putative PCF2-like DNA binding protein (At2g45680; open circles); zinc finger transcription factor (At5g07500; closed triangles); protein kinase–like protein (At5g13160; closed circles); and CCAAT-box binding transcription factor-like protein (At5g47670; open squares).
(B) Genes whose expression increases during active oil accumulation: putative HD-Zip protein (At1g52150; open squares); putative CRK1 protein (At1g54610; closed squares); casein kinase (At5g43320; open inverted triangles); and ABI3 (At3g24650; closed triangles).
Expression is related to 9+10 DAF, and the ratios were rescaled by setting the lowest ratio to 1.
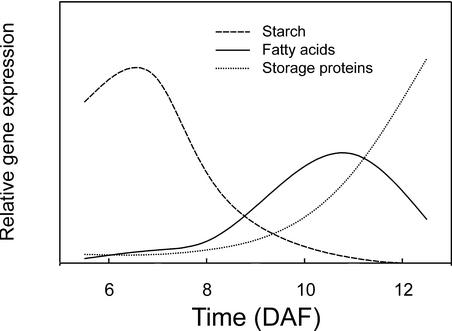
Genes Associated with Different Storage Compounds Have Different Profiles
The general patterns of gene expression associated with starch, fatty acid, and storage proteins are summarized in Figure 8. The temporal patterns of gene expression were clearly distinct for different storage compounds, suggesting that different networks of control separately regulate these pathways. For example, transcripts for oil biosynthesis were turned off before those for storage protein biosynthesis. These separately controlled pathways may explain in part why decreasing the amount of one storage compound in Arabidopsis seeds generally does not lead to a balancing increase in others. Examples of this nonplasticity of the pathways include Arabidopsis abi/aba mutants (Finkelstein and Somerville, 1990), in which storage protein content is reduced strongly but oil content does not change significantly, or the wri1 mutant (Focks and Benning, 1998), which accumulates much less oil but has similar storage protein content as wild-type seeds.
Figure 8.
Summary of Temporal Expression Patterns for Genes Involved in the Synthesis of Different Storage Compounds.
Wild-Type and wri1 Mutant Seed Differences in Gene Expression
Microarrays provide an opportunity to examine the extent of gene expression changes in mutants that are altered in metabolism. Focks and Benning (1998) described the Arabidopsis mutation wri1, which is characterized by an 80% decrease in seed oil content. Although the total protein content in the seeds is not different, the carbohydrate content of developing wri1 seeds is higher than in the wild type, with approximately twofold more starch and two to four times more soluble sugars (Suc, Glc, and Fru).
A comparison of gene expression between developing Arabidopsis wild-type and wri1 seeds at three different time points (5 to 7, 8 to 10, and 11 to 13 DAF) is summarized in Table 1 (the complete data set is available at http://www.bpp.msu.edu/Seed/SeedArray.htm). Despite the marked difference in seed metabolism between the wild type and wri1, relatively few gene expression differences were observed. Altogether, only 45 genes varied more than approximately twofold at any of the time points. Of the 45 differentially expressed genes, the large majority (40) were higher in wild-type than in wri1 seeds, perhaps reflecting the lower biosynthetic and metabolic activity of wri1 seeds. The genes that were reduced in wri1 clearly did not reflect a random selection of those on the array; instead, most encoded key carbon and lipid metabolism enzymes. In addition, most of these genes were among those that also changed in a bell-shaped pattern in wild-type seeds during the time course, such as biotin carboxyl carrier protein, KAS I, enoyl-ACP reductase, two ACP isoforms, FAD2, plastid PK, and chloroplast PDH E1α subunit.
Table 1.
Examples of Genes Whose mRNA Levels Differed between Developing Wild-Type (WT) and wri1 Seeds
| Class | Clone No. | Gene | Description | WT/wri1 5 to 7 DAF |
WT/wri1 8 to 10 DAF |
WT/wri1 11 to 13 DAF |
|---|---|---|---|---|---|---|
| Fatty acid | R64960 | At5 g15530 | Biotin carboxyl carrier protein precursor | 1.4 | 2.3 | 3.4 |
| R90100 | At2 g05990 | Enoyl-ACP reductase | 1.3 | 1.9 | 2.1 | |
| 600036066R1 | At1 g54630 | Putative seed ACP | 1.2 | 1.9 | 1.5 | |
| 600036781R1 | At5 g46290 | KAS I precursor | 1.2 | 2.3 | 1.4 | |
| L26296 | At3 g12120 | FAD2 | 1.1 | 2.0 | 1.1 | |
| Carbon metabolism | 600037175R1 | At4 g15210 | β Amylase | 2.0 | 2.2 | 1.1 |
| M46A08 | At5 g52920 | Chloroplast pyruvate kinase | 1.4 | 2.7 | 2.4 | |
| M20C09 | At1 g01090 | Plastid pyruvate dehydrogenase E1α | 1.4 | 2.6 | 2.1 | |
| 600034732r1 | At5 g49190 | Susy | 1.1 | 1.2 | 3.2 | |
| M51F10 | At3 g16950 | Plastid dihydrolipoamide dehydrogenase, putative | 1.1 | 1.6 | 2.0 | |
| Miscellaneous | M60F12 | At3 g63410 | Putative chloroplast inner envelope protein | 1.1 | 2.4 | 1.4 |
| M48A07 | At5 g13930 | Chalcone synthase | 1.0 | 2.2 | 0.85 | |
| M52G06 | At2 g34420 | Photosystem II type I chlorophyll a/b binding protein | 1.8 | 2.1 | 0.85 | |
| M36C05 | At1 g07940 | Elongation factor 1α | 1.3 | 2.0 | 0.68 | |
| M48B08 | At5 g67360 | Cucumisin-like Ser protease | 1.2 | 2.5 | 0.75 | |
| Higher in wri1 | M26E12 | At3 g54320 | Aintegumenta-like protein | 0.75 | 0.65 | 0.53 |
| 600035783R1 | At3 g58740 | Citrate synthase–like protein | 1.0 | 1.1 | 0.51 | |
| M42D09 | At5 g48180 | Putative protein, jasmonate inducible-like | 1.1 | 1.1 | 0.49 | |
| 600036894R1 | At1 g68170 | MtN21 (nodulin)-like protein | 1.1 | 0.85 | 0.52 | |
| M44A11 | At2 g38530 | Putative nonspecific lipid-transfer protein | 1.3 | 0.81 | 0.44 |
Genes with expression ratios >1 are expressed more in wild-type seeds, and genes with ratios <1 are expressed more in wri1 seeds. Values for time points at which the difference was twofold or more are shown in boldface.
In contrast, genes such as oleosins and fatty acid elongase, which are involved in lipid synthesis but do not belong to the bell-shaped expression group, were not altered in wri1. Thus, the effect of WRI1 on lipid gene expression clearly was not general. This correspondence between WRI1 effects and the grouping of a bell-shaped “regulon” reinforces the conclusion that this set of genes is regulated coordinately by the same or related signals and further implies that the WRI1 gene is involved either directly or indirectly in the regulation of this set of genes. Because 35% (∼1160) of the genes changed during 5 to 13 DAF of seed development but only 45 genes differed between the wild type and wri1, major differences in seed metabolism can occur with only minor changes at the transcript level. This result contrasts sharply with the impact of the phyA mutation, in which 10% of genes represented on an 8200-gene oligonucleotide array were altered in expression between the wild type and the mutant during greening (Tepperman et al., 2001).
Comparison between Microarray and Biochemical Data
Focks and Benning (1998) extensively analyzed developing seeds of wri1 plants, including measurements of several enzyme activities; thus, it is possible to compare the earlier biochemical data with the expression data from microarray experiments. We found almost no transcript differences between wri1 and the wild type in young seeds (5 to 7 DAF) before storage lipid accumulation starts; additionally, in these youngest seeds, the biochemical differences were smallest. Focks and Benning (1998) reported that the activity of several glycolytic enzymes was decreased in wri1 seeds. Activities of hexokinase and PPi-dependent phosphofructokinase were decreased most severely (60 to 80%). In addition, five other enzymes (fructokinase, aldolase, phosphoglycerate mutase, enolase, and PK) had decreased activities (∼60% of wild-type levels).
In contrast, the array data indicated that the transcript abundance of only a few of these enzymes differed between the wild type and wri1: cytosolic fructokinase, cytosolic phosphoglycerate mutase, and plastid PK all were approximately twofold lower in wri1. This finding suggests that the general downregulation of the glycolytic enzymes may have occurred at the biochemical or metabolic level rather than the transcript level. In addition to these few glycolytic enzymes, we found that the expression of other carbon-metabolizing enzymes, such as SuSy and β-amylase, was altered in wri1 seeds.
High Soluble Sugar Concentration Does Not Alter Gene Expression in wri1
Based on the very specific impact of WRI1 on gene expression, it is likely that the genes that were downregulated in wri1 seeds reflected specific consequences downstream of the WRI1 gene. However, a possible less direct reason for the repression of some genes seen in wri1 could be a response to sugar sensing. The wri1 seeds have twofold to threefold more Glc and Suc during development, which may lead to higher levels of hexose phosphates, which are known to be involved in the regulation of gene expression. For example, photosynthetic genes such as the chlorophyll a/b binding protein CAB1 and the Rubisco small subunit are repressed by sugars (Sheen, 1994). Of several known sugar-repressed genes, we found that only type 1 chlorophyll a/b-binding protein was downregulated in wri1 (Table 1). Furthermore, β-amylase and SuSy also were decreased in wri1, but in other studies, these genes were induced by Suc (Sheen, 1994). Thus, the effect of the wri1 mutation may have overridden any regulation by sugars. In addition, there are no previous reports of Glc repression of plant FAS enzymes; therefore, the lower expression of these genes is unlikely to be a response to sugar sensing. A few genes with potential regulatory function were found to be different between the wild type and wri1, and these might represent regulatory functions that act downstream from the WRI1 gene (Table 1).
In summary, many of the genes that were expressed at lower levels in wri1 seeds encoded central fatty acid and glycolytic enzymes. The FAS genes that were decreased belonged to the bell-shaped, ACCase-like expression group. Inspection of the time course data for FAS gene expression in the wild type, and the differences between the wild type and wri1, suggested that these enzymes were not downregulated actively in wri1; rather, the twofold to threefold induction observed in the wild type was missing in wri1 (compare the expression of biotin carboxyl carrier protein in Figure 2A and its twofold to threefold higher expression in wild-type seeds [Table 1]). Significantly, the developmental induction that caused gene expression to increase was missing only for the ACCase-type core FAS enzymes, whereas the expression of abscisic acid–regulated genes (FAE1, FAD3, oleosins, and storage proteins) was similar in both the wild type and wri1.
Conclusions
This study has provided a new data set that describes the patterns of expression of >3500 Arabidopsis genes during the period of early storage product accumulation of seed development. Full datasets are available at http://www.bpp.msu.edu/Seed/SeedArray.htm. These results have not only identified and cataloged expression patterns, but more importantly, they have begun to provide insight into the structure of the primary transcriptional networks that coordinate the metabolic responses to seed developmental programs.
Several general conclusions have emerged from these data. First, although essentially all of the cell types are established and cell division has ceased by ∼6 DAF (Koornneef and Karssen, 1994), our results emphasize that dynamic and distinct developmental patterns of gene transcription for many pathways continue throughout seed development.
Second, the timing of expression of many genes involved in central carbon transport and metabolism clearly was coordinated with changes in metabolism. Thus, these patterns are likely to control in large part the shifts from cell division to cell enlargement and from starch to oil and protein accumulation. Although this general conclusion might have been expected, some new details have been revealed. For example, gene expression changed in diametrically opposed patterns for cytosolic versus plastidial isoforms of PK and for cell membrane Suc symporter versus plastid hexose transporters (Figure 5). Therefore, these changes describe how carbon flux through central metabolism is directed from the cytosol to plastidial pathways for fatty acid and amino acid biosynthesis. A corollary conclusion is that control over the major metabolic changes in seeds is less likely to reflect post-translational biochemical controls or a changing supply of maternal factors but rather is “hard-wired” into the developmental transcriptional programs. In addition, our data suggest that the regulation of transcripts for a subset of enzymes and transporters can account for the rearrangement of metabolic fluxes through different pathways during tissue development.
Third, although groups of enzymes within a pathway showed similar regulation, not all enzymes within the lipid, carbohydrate, or other metabolic pathways behaved the same. These results help define those members of each pathway that are coregulated and presumably share common cis and trans elements involved in this control.
Fourth, analysis of a mutant with 80% reduction in storage oil indicated that a major modification in metabolism was accompanied by surprisingly few changes in gene expression. Thus, the many changes in carbon metabolism that occurred in the mutant seeds, including twofold to fourfold increased soluble carbohydrate levels, did not override the major patterns of transcription that are programmed into the scheme of Arabidopsis seed development.
Finally, this study has identified a number of potential regulatory factors whose expression patterns were undescribed previously and that correlate with both metabolic and structural gene expression patterns. These represent candidates for genes primarily or secondarily involved in the control of metabolic networks. Experiments are under way at present to determine whether overexpression or underexpression of these genes in transgenic plants can yield instructive or useful changes in seed metabolism.
METHODS
Plant Material, RNA Extraction, Probe Synthesis, and Array Hybridization
Arabidopsis thaliana plants (ecotype Columbia-2 and wri1 mutant) were grown in a growth chamber with a 16-h light period at 80 to 100 μE light intensity and day/night temperatures of 22/20°C. To harvest siliques of defined age, individual flowers were tagged with colored threads on the day of flower opening. Developing seeds were dissected from siliques between 5 and 13 days after flowering, frozen immediately with dry ice, and stored at −80°C. Because of limited quantities of developing Arabidopsis seeds of defined developmental stages, for microarray experiments, we chose a probe-labeling method that requires only 3 μg of total RNA (3DNA Submicro Expression Array Detection Kit; Genisphere, Montvale, NJ), as opposed to the 1 μg of poly(A)+ RNA needed by the conventional method (Eisen and Brown, 1999). With this probe system, we achieved essentially the same detection sensitivity (1:50,000 to 1:100,000) and specificity (measured as cross-hybridization threshold) as in previous experiments using poly(A)+-derived probes. RNA extraction, probe synthesis, array hybridization procedures, and comparison between labeling methods are described in detail elsewhere (Ruuska and Ohlrogge, 2001). Amplification of cDNAs and printing of the microarrays on aldehyde-coated glass substrates (SuperAldehyde; TeleChem International, Sunnyvale, CA) were performed as described previously (Girke et al., 2000).
The microarrays contained ∼6000 DNA elements. Of these, 2715 were previously described ESTs collected from a cDNA library of developing Arabidopsis seeds, 82 additional cDNA clones from genes of the lipid and carbohydrate pathways, and sensitivity and cross-hybridization controls (Girke et al., 2000). This clone set was supplemented with an additional 2515 clones that were selected from Arabidopsis seed EST set II (White et al., 2000). To estimate the number of unique genes on the array, the ESTs were mapped on the completed Arabidopsis genome by nucleotide BLAST searches against the June 13, 2001, version of the TIGR annotations. Because of the nature of the short EST sequences, many clones originally considered different were found to represent different regions of the same gene. The 6000 DNA elements on the array mapped to ∼3500 unique genes. Thus, almost 60% of the genes on the array were present as replicate spots, and their combined analysis allowed increased reliability in expression analysis (Figure 2). The GenBank numbers of clones used for the array and the genes to which these sequences map are available at http://www.bpp.msu.edu/Seed/SeedArray.htm.
Experimental Design
Wild-Type Time Course
To examine changes in gene expression during seed development, two RNA samples from seeds of different ages were compared. To provide a constant reference for all hybridizations, a 1:1 mixture of RNA extracted from 9- and 10-day-old seeds was selected as a reference for the wild-type time course. At this stage, the seeds are metabolically highly active, with a high proportion of genes expressed. The following five time points were compared with this reference (Figure 2A): a 1:1 mixture of RNA from 5- and 6-day-old seeds (referred to as 5+6 DAF); 7 DAF; 8 DAF; 11 DAF; and a 1:1 mixture of RNA from 12- and 13-day-old seeds (referred to as 12+13 DAF).
The expression ratios (expression at x DAF/expression at 9+10 DAF) were calculated for every clone for each of the five time points. The wild-type expression ratios are an average of two replicate hybridizations that were performed using the same RNA (technical replicates), most often with a Cy3/Cy5 dye swap. The reproducibility of the hybridizations was assessed from comparisons of the initial (nonnormalized) signal intensities of the same RNA sample from replicate arrays as described (Girke et al., 2000). For example, in the case of 8-DAF versus 9+10-DAF hybridizations, fluorescence intensities of Cy3-labeled 9+10-DAF RNA from slide one were compared with intensities of Cy5-labeled 9+10-DAF RNA from slide two. Similarly, fluorescence intensities of Cy5-labeled 8-DAF RNA from slide one were compared with intensities of Cy3-labeled 8-DAF RNA from slide two. For the technical replicates, the Pearson correlation coefficient for the intensities was always >0.9.
Wild-Type versus wri1 Mutant Time Course
Comparison between wild-type and wri1 mutant seeds was performed using a mixture of equal amounts of RNA extracted from young (5 to 7 DAF), midstage (8 to 10 DAF), and maturing (11 to 13 DAF) seeds. Each wild-type versus mutant hybridization was repeated three times using the same RNA, and the expression ratios for each clone were averaged.
Data Analysis
The arrays were scanned with Scanarray 4000 (GSI Lumonics, Billerica, MA) at a resolution of 10 μm per pixel. The mean fluorescence intensities for each cDNA clone were determined using ScanAlyze version 2.44 software (http://rana.lbl.gov/EisenSoftware.htm). We have observed that the channels in two-color microarray experiments frequently are nonlinear with respect to intensity. The data are not normalized accurately using one global factor, because the relationship between the channels changes as a function of intensity. To address this problem, we adopted a simple linear step-wise normalization procedure. First, the intensities from both Cy3 and Cy5 channels were sorted separately and divided into 10 subgroups. The average signal intensity of both channels was calculated for each subgroup, and the ratio between them was used to normalize the Cy5 intensities.
Technical and Biological Variation
To estimate technical variation, control experiments in which the same RNA was labeled with Cy3 and Cy5 were performed. Using the step-wise normalization procedure and averaging expression ratios from two hybridizations, only 9 of 5937 ESTs (0.15%) showed >2-fold variation, and 196 (3.3%) showed >1.5-fold variation. We also assessed the degree of biological variability in the seeds by comparing 9+10-DAF seeds that we used as the time-course reference with seeds of similar age harvested from plants grown ∼1 month later under the same conditions. Altogether, only 52 clones (<1%) varied more than twofold between the two seed lots.
Data Presentation
To calculate the change each clone undergoes during the time course, all five time points were compared together. This is illustrated in Figure 2B, in which the expression ratios of three separate clones encoding the same chlorophyll a/b binding protein are presented. At any time point, none of the clones differed by >2.5-fold from the reference. However, when the minimum and maximum ratios were compared, it was evident that these clones underwent 4.5- to 6-fold change during the experimental period. To illustrate this finding, we rescaled the time-course data such that the lowest expression ratio was set to 1, and the other ratios were adjusted accordingly (Figure 2C). This modification makes it easier to visualize the shape and magnitudes of the expression profiles in the figures.
A clone was deemed reliable only if its average signal intensity from both channels was more than two times the average background intensity. A change in gene expression was considered significant if overall expression changed more than twofold over the time course (however, clones that changed somewhat less than twofold are discussed in the text). To assess the internal consistency of the time course, we compared gene expression between 5+6 DAF and 12+13 DAF seeds in two ways. First, we calculated the indirect ratio from our time course as a relationship between expression ratios of 5+6 DAF versus control and 12+13 DAF versus control [(5+6 DAF/9+10 DAF)/(12+13 DAF/9+10 DAF)]. Second, we made a direct comparison by hybridizing 5+6 DAF RNA with 12+13 DAF RNA. The agreement between the two methods was very good, with a Pearson correlation coefficient for the ratios of 0.80 (data not shown).
Clone Identity and Expression Profile Verification
As discussed previously (Girke et al., 2000), cDNA-based arrays are subject to errors related to sample tracking or cross-contamination. For almost all of the data discussed in this study, we present results from genes that had at least two array elements (derived from different clones) and that had similar expression profiles. The underlying array element expression data for these key clones also was inspected manually for reliability. The identity of the majority of these clones was verified by resequencing of the DNA, which was spotted directly onto the array. In addition, real-time PCR experiments were performed on a set of genes that exhibited different temporal expression patterns. The profiles of the large majority of the genes studied in this way (85%) were identical in the two sets of experiments (for details of the validation experiment, see http://www.bpp.msu.edu/Seed/SeedArray.htm).
Lipid and Protein Analysis
The total lipid content of seeds was measured by gas chromatography–mass spectrometry after direct esterification of fatty acids (Browse et al., 1986) using triheptadecanoylglycerol as an internal standard. To avoid hydrolysis of fatty acids, frozen seeds (20 to 50) were dropped directly into a preheated (90°C) mixture of 5% H2SO4-methanol (1 mL) and toluene (0.3 mL) and incubated for 1 h at the same temperature. For total protein assays, 5 to 10 seeds were homogenized in 250 μL of extraction buffer (25 mM Tris-HCl, pH 8, 125 mM NaCl, 0.5 mM EDTA, and 0.5% [w/v] SDS). Protein content was measured from 200 μL of the crude homogenate with the Bio-Rad Detergent Compatible protein assay system using γ-globulin as a standard.
Accession Numbers
The accession numbers for the proteins described in the figures are as follows: chlorophyll a/b binding protein (At1g29920); biotin carboxyl carrier protein (BCCP; At5g15530), biotin carboxylase (BC; At5g35360), and α-carboxyltransferase (α-CT; At2g38040) subunits of plastidial acetyl-CoA carboxylase; acyl carrier protein1 precursor (ACP1; At3g05020) and seed ACP (At1g54630); 3-ketoacyl-ACP synthase I (At5g46290) and 3-ketoacyl-ACP synthase III (At1g62640); oleate desaturase (FAD2; At3g12120), linoleate desaturase (FAD3; At2g29980), and fatty acid elongase1 (FAE1; At4g34520); oleosin isoform (At3g27660) and 18.5-kD oleosin (At4g25140); storage proteins 12S (At4g28520) and 2S (At4g27140); β-amylase (At4g15210); Suc synthase (SuSy; At5g49190); cytosolic PK (At5g52920); chloroplast PK (At3g22960); chloroplast E1α subunit of pyruvate dehydrogenase (At1g01090); Suc-proton transporter protein (SUC2; At1g22710); chloroplast Glc-6-P translocator (GPT; At5g54800); chloroplast (At4g38970) and cytosolic (At3g52930) aldolases; photosystem II light-harvesting complex protein (LHC II; At1g29930) and small subunit of Rubisco (SSu; At1g67090); isoamylase-like protein (At4g09020); putative PCF2-like DNA binding protein (At2g45680); zinc finger transcription factor (At5g07500); protein kinase–like protein (At5g13160); CCAAT-box binding transcription factor-like protein (At5g47670); putative HD-Zip protein (At1g52150); putative CRK1 protein (At1g54610); casein kinase (At5g43320); and ABI3 (At3g24650).
Supplementary Material
Acknowledgments
We acknowledge the members of the Ohlrogge and Benning labs for help with plant material collection, Rob Halgren for assistance with data analysis, and the Michigan Agricultural Experiment Station for support. We also thank Ellen Wisman and Arabidopsis Functional Genomic Consortium members for advice and access to equipment. This work was supported in part by grants from the Consortium for Plant Biotechnology Research, Dow AgroSciences, and the National Science Foundation (MCB 9817882).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.000877.
Footnotes
Online version contains Web-only data.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Mansfield, S.G. (1994). Embryogenesis: Introduction. In Arabidopsis: An Atlas of Morphology and Development, J.L. Bowman, ed (New York: Springer-Verlag), pp. 351–361.
- Browse, J., McCourt, P.J., and Somerville, C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152, 141–145. [DOI] [PubMed] [Google Scholar]
- Bruce, W., Folkerts, O., Garnaat, C., Crasta, O., Roth, B., and Bowen, B. (2000). Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., Kolacna, L., and Rawsthorne, S. (1996). Photosynthesis by developing embryos of oilseed rape (Brassica napus L.). J. Exp. Bot. 47, 1763–1769. [Google Scholar]
- Eastmond, P.J., and Rawsthorne, S. (2000). Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryo. Plant Physiol. 122, 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., and Brown, P.O. (1999). DNA arrays for analysis of gene expression. Methods Enzymol. 303, 179–205. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Somerville, C.R. (1990). 3 classes of abscisic-acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 94, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Tenbarge, K.M., Shumway, J.E., and Crouch, M.L. (1985). Role of ABA in maturation of rapeseed embryos. Plant Physiol. 78, 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks, N. (1997). Charakterisierung von Mutantnen der Pflanze Arabidopsis thaliana mit Veranderter ol- oder Pigmentakkumulation in Samen. PhD dissertation (Berlin: Freie Universitat Berlin).
- Focks, N., and Benning, C. (1998). Wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate. Plant Physiol. 118, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J.O. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., Hoschek, G., Tam, S.H., Ditta, G.S., and Breidenbach, R.W. (1981). Abundance, diversity and regulation of mRNA sequence sets in soybean embryogenesis. Dev. Biol. 83, 201–217. [DOI] [PubMed] [Google Scholar]
- Guerche, P., Tire, C., de Sa, F.G., De Clercq, A., van Montagu, M., and Krebbers, E. (1990). Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 2, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, J. (1994). Seed maturation and control of dormancy. In Cellular and Molecular Biology of Plant Seed Development, B.A. Larkins, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 545–592.
- Harmer, S.L., Hogenesch, L.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Heath, J.J., Weldon, R., Monnot, C., and Meinke, D.W. (1986). Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169, 304–312. [DOI] [PubMed] [Google Scholar]
- Heim, U., Weber, H., Baumlein, H., and Wobus, U. (1993). A sucrose synthase gene of Vicia faba L.: Expression pattern in developing seeds in relation to starch synthesis and metabolic regulation. Planta 191, 394–401. [DOI] [PubMed] [Google Scholar]
- Heppard, E.P., Kinney, A.J., Stecca, K.L., and Miao, G.H. (1996). Developmental and growth temperature regulation of two different microsomal omega-6 desaturase genes in soybeans. Plant Physiol. 110, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara, Y., Kamei, A., Kanehisa, M., Kaplan, A., and Ikeuchi, M. (2001). DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13, 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A.A., Hunter, C.P., Tsung, B.T., Tucker-Kellogg, G., and Brown, E.L. (2000). Genomic analysis of gene expression in C. elegans. Science 290, 809–812. [DOI] [PubMed] [Google Scholar]
- Holbrook, L.A., van Rooijen, G.J.H., Wilen, R.W., and Moloney, M.M. (1991). Oilbody proteins in microspore-derived embryos of Brassica napus. Plant Physiol. 97, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, F., and Rawsthorne, S. (1994). Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.). Plant J. 6, 795–805. [Google Scholar]
- Ke, J.S., Wen, T.N., Nikolau, B.J., and Wurtele, E.S. (2000). Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme A carboxylase. Plant Physiol. 122, 1057–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.P., Badger, M.R., and Furbank, R.T. (1998). CO2 refixation characteristics of developing canola seeds and silique wall. Aust. J. Plant Physiol. 25, 377–386. [Google Scholar]
- King, S.P., Lunn, J.E., and Furbank, R.T. (1997). Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol. 114, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., and Karssen, C.M. (1994). Seed dormancy and germination. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 313–334.
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1991). Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can. J. Bot. 70, 151–164. [Google Scholar]
- Mazur, B., Krebbers, E., and Tingey, S. (1999). Gene discovery and product development for grain quality traits. Science 285, 372–375. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Gotoh, Y., Nishida, E., Yamaguchi-Shinozaki, K., Hayashida, N., Iwasaki, T., Kamda, H., and Shinozaki, K. (1994). Characterization of the cDNAs that encode MAP kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 5, 111–122. [DOI] [PubMed] [Google Scholar]
- Neuhaus, H.E., and Emes, M.J. (2000). Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 111–140. [DOI] [PubMed] [Google Scholar]
- Oh, M.K., and Liao, J.C. (2000). Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol. Prog. 16, 278–286. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller, D., Ohlrogge, J.B., and Somerville, C.R. (1993). Regulation of plant lipid biosynthesis: An example of developmental regulation superimposed on a ubiquitous pathway. In Control of Plant Gene Expression, D.P.S. Verma, ed (Boca Raton, FL: CRC Press), pp. 157–174.
- Qi, Q.G., Rose, P.A., Abrams, G.D., Taylor, D.C., Abrams, S.R., and Cutler, A.J. (1998). (+)-Abscisic acid metabolism, 3-ketoacyl-coenzyme A synthase gene expression, and very-long-chain monounsaturated fatty acid biosynthesis in Brassica napus embryos. Plant Physiol. 117, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska, S.A., and Ohlrogge, J.B. (2001). Protocol for small-scale RNA isolation and transcriptional profiling of developing Arabidopsis seeds. Biotechniques 31, 752–758. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1994). Feedback control of gene expression. Photosynth. Res. 39, 427–438. [DOI] [PubMed] [Google Scholar]
- Simcox, P.D., Garland, W., Deluca, V., Canvin, D.T., and Dennis, D.T. (1979). Respiratory pathways and fat synthesis in the developing castor-oil seed. Can. J. Bot. 57, 1008–1014. [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnham, E., and Northcote, D.H. (1983). Changes in the activity of acetyl-CoA carboxylase during rapeseed formation. Biochem. J. 208, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, T.A., Hayes, T.R., Cranmer, A.M., Turner, J.C., and Davies, H.M. (1996). Genetic engineering of a quantitative trait: Metabolic and genetic parameters influencing the accumulation of laurate in rapeseed. Plant J. 9, 229–241. [Google Scholar]
- White, J.A., Todd, J., Newman, T., Focks, N., Girke, T., Martínez de Ilárduya, O., Jaworski, J.G., Ohlrogge, J.B., and Benning, C. (2000). A new set of Arabidopsis expressed sequence tags from developing seeds: The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 124, 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Seed maturation: Genetic programmes and control signals. Curr. Opin. Plant Biol. 2, 33–38. [DOI] [PubMed] [Google Scholar]
- Ye, X., Salim, A.B., Klöti, A., Zhang, J., Lucca, P., Beyer, P., and Potrykus, I. (2000). Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 14, 303–305. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Umemoto, T., Lue, W.L., Au-Yeung, P., Martin, C., Smith, A.M., and Chen, J. (1998). A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10, 1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J.T., Abrams, G.D., Barton, D.L., Taylor, D.C., Pomeroy, M.K., and Abrams, S.R. (1995). Induction of lipid and oleosin biosynthesis by (+)-abscisic acid and its metabolites in microspore-derived embryos of Brassica napus L. cv Reston. Plant Physiol. 108, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.