Abstract
Microtubules interact strongly with the viral movement protein (MP) of Tobacco mosaic virus (TMV) and are thought to transport the viral genome between plant cells. We describe a functionally enhanced DNA-shuffled movement protein (MPR3) that remained bound to the vertices of the cortical endoplasmic reticulum, showing limited affinity for microtubules. A single amino acid change was shown to confer the MPR3 phenotype. Disruption of the microtubule cytoskeleton in situ with pharmacological agents, or by silencing of the α-tubulin gene, had no significant effect on the spread of TMV vectors expressing wild-type MP (MPWT) and did not prevent the accumulation of MPWT in plasmodesmata. Thus, cell-to-cell trafficking of TMV can occur independently of microtubules. The MPR3 phenotype was reproduced when infection sites expressing MPWT were treated with a specific proteasome inhibitor, indicating that the degradation of MPR3 is impaired. We suggest that the improved viral transport functions of MPR3 arise from evasion of a host degradation pathway.
INTRODUCTION
The mechanism of cell-to-cell movement of plant viruses has been the subject of intense investigation (for reviews, see Citovsky and Zambryski, 1991; Zambryski, 1995; Carrington et al., 1996; Lazarowitz, 1999; Tzfira et al., 2000). Most plant viruses encode one or more movement proteins (MPs) that facilitate the passage of the viral genome through the plasmodesmata, the small pores that connect higher plant cells. The most extensively studied viral MP is that of Tobacco mosaic virus (TMV). This MP binds single-stranded RNA (Citovsky et al., 1990), accumulates in plasmodesmata during viral infection (Tomenius et al., 1987; Oparka et al., 1997; Roberts et al., 2001), and increases the size exclusion limit of plasmodesmata to allow cell-to-cell passage of the viral genome, a phenomenon referred to as “gating” (Wolf et al., 1989; Waigmann et al., 1994; Oparka et al., 1997). Most models of TMV movement support the view that the intercellular movement complex comprises an unfolded and elongated viral ribonucleoprotein complex, the size and structure of which is compatible with the gated plasmodesmal pore (Citovsky and Zambryski, 1991; Carrington et al., 1996; Lazarowitz, 1999; Reichel and Beachy, 1999; Tzfira et al., 2000; Aaziz et al., 2001).
It is clear that cytoplasmic host cell factors must interact with the TMV MP to facilitate the transfer of the viral genome from its subcellular site of synthesis to plasmodesmata. Recently, both F-actin (McLean et al., 1995) and microtubules (MT) (Heinlein et al., 1995, 1998; Padgett et al., 1996; Mas and Beachy, 1999; Boyko et al., 2000a, 2000b) have been shown to interact with MP during the viral replication cycle, leading to the view that elements of the cytoskeleton play a crucial role in targeting the viral RNA (vRNA) to plasmodesmata (Heinlein et al., 1995, 1998; Zambryski, 1995; Lazarowitz and Beachy, 1999; Boyko et al., 2000a, 2000b; Aaziz et al., 2001).
Because MT have been shown to function in the intracellular transport of several mRNAs (Bassel and Singer, 1997; Hazelrigg, 1998; Jansen, 1999; Aaziz et al., 2001), the targeting of MP-vRNA complexes to plasmodesmata along MT has emerged as an attractive hypothesis to explain the intracellular trafficking of plant viral genomes (for review, see Reichel and Beachy, 1999; Aaziz et al., 2001). Recently, Boyko et al. (2000a) identified a conserved tobamovirus MP sequence exhibiting similarity to a tubulin motif and postulated that this conserved region mediates the association of MP with MT during the cell-to-cell movement process. Viruse that display point mutations in the putative tubulin binding domain of the MP showed reduced cell-to-cell spread and did not label MT, suggesting that the spread of vRNA is linked closely to the ability of MP to interact with MT (Boyko et al., 2000a).
Although the association of MP with MT has led to models that implicate MT in cell-to-cell transport of the viral genome (Boyko et al., 2000a, 2000b), some reports suggested that the association of MP with MT may represent an intracellular pathway involved in targeting the MP for degradation during the later stages of viral infection (Padgett et al., 1996; Reichel and Beachy, 1998; Mas and Beachy, 1999; for review, see Tzfira et al., 2000). In studies with protoplasts, it was shown that the TMV MP associates first with cortical elements of the endoplasmic reticulum (ER) at sites that also contain the viral replicase (Mas and Beachy, 1999). From these sites, the MP is transferred subsequently from the ER to the MT (Heinlein et al., 1998; Reichel and Beachy, 1998; Mas and Beachy, 1999). In studies of expanding viral infection sites of the tobamovirus Ob, only cells deeper in the infection site showed labeling with MP–green fluorescent protein (GFP), consistent with the view that binding of MP to MT occurs late in the infection process (Padgett et al., 1996). However, in a recent study of TMV movement, it was shown that increasing the temperature of infection sites from 22 to 32°C caused MT labeling to appear in cells at the infection front, rather than in older infected cells, consistent with a role for MT in trafficking of vRNA (Boyko et al., 2000b).
We showed recently that cell-to-cell movement of TMV-based vectors could be enhanced through DNA shuffling of the MP gene (Toth et al., 2002). Three rounds of shuffling yielded a clone, R3, that moved up to fivefold faster than the progenitor GFP-expressing clone. The MPR3 gene contained three silent nucleotide changes (in the codons for amino acids Asn-10, Thr-104, and Gly-184) and a nucleotide change that resulted in an amino acid substitution L72V. The first two silent changes and the coding change were shown to enhance viral movement independently.
In this study, we examined the basis of the enhanced cell-to-cell movement functions of MPR3 by studying facets of the TMV infection process known specifically to involve viral MP: MT association, plasmodesmal gating, and vRNA trafficking. To facilitate the colocalization of MP with associated subcellular structures, we made translational fusions of wild-type MP (MPWT) and MPR3 with the red fluorescent protein DsRed (Matz et al., 1999) and subsequently examined these on transgenic Nicotiana benthamiana plants expressing GFP targeted to the endoplasmic reticulum (erGFP) or as a translational fusion to α-tubulin (tua-GFP).
We show that during early infection, both MPWT and MPR3 accumulate on the vertices of the cortical ER network. Subsequently, MPWT is transferred from the cortical ER onto underlying MT. However, MPR3 showed restricted association with MT and remained on the cortical ER during the infection process. When the MT cytoskeleton was disrupted with pharmacological agents, or by viral silencing of the tua gene, TMV vectors expressing MPWT moved unimpeded through the treated cells. The MPR3 localization pattern was reproduced when infection sites expressing MPWT were treated with a specific proteasome inhibitor, indicating that the degradation of MPR3 is impaired. We conclude that the cell-to-cell movement of TMV does not require MT and suggest that the improved viral trafficking functions of MPR3 occur by evasion of a host degradation pathway.
RESULTS
MPR3 Confers Improved Viral Transport Functions
To examine the subcellular localization of MPWT and MPR3, a range of viral vectors expressing these MPs were constructed (Figures 1A to 1D), together with a series of plasmids in which the different MPs were expressed from a 35S promoter (Figures 1E to 1H). In keeping with previous studies, the viral vectors used for subcellular localization studies lacked the coat protein gene, allowing a study of the nonencapsidated viral genome (Figures 1A and 1B) (Heinlein et al., 1995, 1998; Padgett et al., 1996; Boyko et al., 2000a, 2000b).
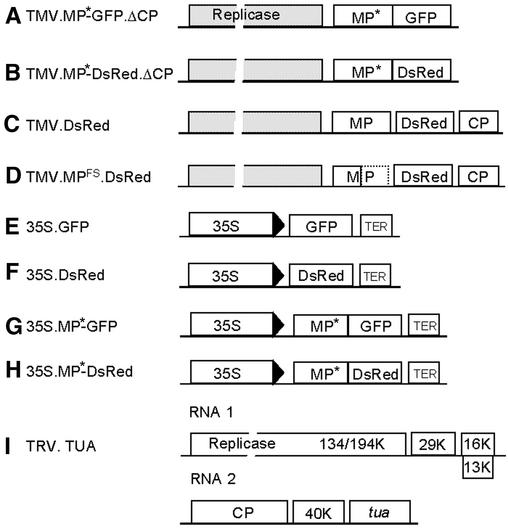
Figure 1.
Viral Vector and Transient Expression Constructs.
Schemes of the constructs used in these studies. The schemes are not drawn to scale.
(A) to (D) In the TMV vector constructs, replicase indicates the 126/183-kD open reading frame required for replication, and the box representing the open reading frame is broken to indicate that its relative length is greater than shown. CP, coat protein.
(E) to (H) Transient expression constructs were based on the plant expression vector pRTL2. The fluorescent proteins GFP and DsRed were cloned in front of the CaMV 35S promoter (35S) alone and as fusions to the TMV MP. TER, 35S polyadenylation signal.
(I) In the TRV vector construct for VIGS of α-tubulin (tua) expression, the Arabidopsis α-tubulin cDNA was cloned into the TRV RNA 2 cDNA in place of the 33-kD protein. The RNA 1 cDNA clone carrying the replication and movement functions also was required for infection. Other viral open reading frames are represented by the approximate sizes of their protein products in kDa (K).
The asterisks following MP in TMV vector and transient expression constructs ([A], [B], [G], and [H]) indicate that these constructs were made with different forms of MP that are indicated in the text by superscripts (e.g., 35S.MPWT-GFP and 35S.MPR3-GFP). Only the wild-type MP form of TMV.DsRed (C) was used. TMV.MPFS.DsRed (D) contained a frameshift mutation that rendered the MP nonfunctional.
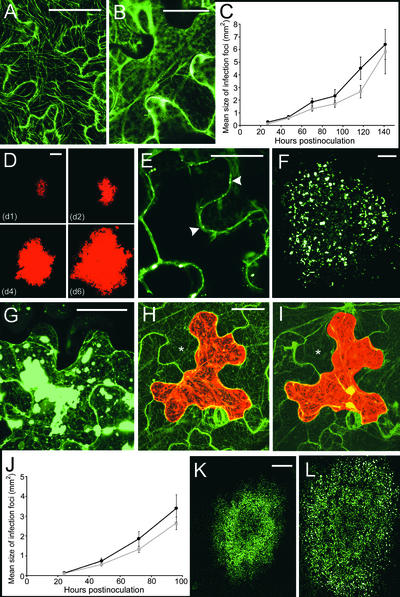
We first examined whether two known functions of viral MPs, plasmodesmal gating and vRNA trafficking, were improved for MPR3 compared with MPWT. As a convenient measure of plasmodesmatal gating, we examined the movement of cytoplasmic GFP from source leaf cells in the presence of either MPR3 or MPWT. We bombarded a plasmid expressing GFP biolistically (molecular mass, 27 kD) under the control of the 35S promoter of Cauliflower mosaic virus (CaMV) (35S.GFP; Figure 1E) into source leaf epidermal cells of N. benthamiana with or without plasmids expressing MPWT or MPR3 fused to the red fluorescent protein DsRed (35S.MPWT-DsRed or 35S.MPR3-DsRed; Figure 1H). MP-DsRed fusion proteins were used in preference to unfused MPs to determine, unequivocally, the locations of single cobombarded cells. These data are shown in Figures 2A to 2D).
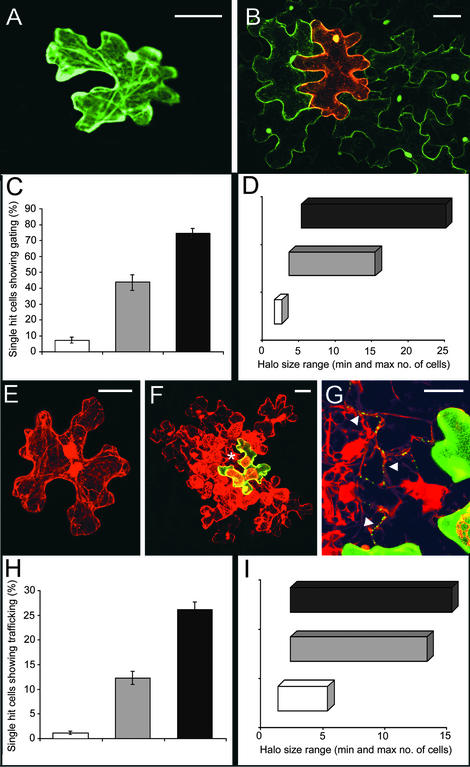
Figure 2.
Effect of Viral MPs on Plasmodesmal Gating and Viral RNA Trafficking in Source Leaves.
(A) Control. Bombardment of 35S.GFP results in fluorescence that is restricted largely to single epidermal cells. Bar = 20 μm.
(B) Cobombardment of 35S.GFP plus 35S.MPR3-DsRed results in plasmodesmal gating and movement of GFP to several surrounding cells. Bar = 20 μm.
(C) Measurement of cells showing plasmodesmal gating (movement of cytoplasmic GFP out of the hit cell). 35S.GFP cobombarded with 35S.MPWT-DsRed (gray bar) and 35S.MPR3-DsRed (black bar) (±se) relative to control bombardment with 35S.GFP alone (white bar).
(D) Range of cell numbers in a halo after biolistic bombardment of 35S.GFP (white bar) with and without 35S.MPWT-DsRed (gray bar) or 35S.MPR3-DsRed (black bar).
(E) Bombardment of an infectious transcript shows that the viral vector TMV.MPFS.DsRed is restricted to single cell infections. Bar = 20 μm.
(F) Cobombardment of TMV.MPFS.DsRed and 35S.MPR3-GFP results in viral RNA trafficking from the cobombarded cell (yellow) to several surrounding cells. Bar = 20 μm.
(G) Detail of the region marked by the asterisk in (F) showing labeling of plasmodesmata by MPR3-GFP in cells surrounding the bombarded cell. Bar = 5 μm.
(H) Comparison of viral RNA trafficking capacities of 35S.MPWT-GFP (gray bar) and 35S.MPR3-GFP (black bar) plasmids cobombarded with transcript of TMV.MPFS.DsRed relative to control (white bar; TMV.MPFS.DsRed transcript alone).
(I) Range of cell numbers in a halo after biolistic bombardment of TMV.MPFSDsRed transcript (white bar) with or without 35S.MPWT-GFP (gray bar) and 35S.MPR3-GFP (black bar) plasmids.
In control leaves (bombardments of 35S.GFP alone), GFP showed restricted trafficking to neighboring cells (Figures 2A and 2C). This result confirmed the relatively low size exclusion limit of source leaf plasmodesmata (Oparka et al., 1999). In cobombardments of 35S.GFP and 35S.MPWT-DsRed, plasmodesmal gating was increased significantly (P < 0.001) compared with the control, as shown by the increased number of bombarded cells showing outward movement of GFP (Figure 2C). The number of cells showing outward movement of GFP in the presence of MPR3-DsRed was approximately double that observed in the presence of MPWT-DsRed, indicating substantial improvement in the gating function of the viral MP (Figures 2B and 2C). In addition, the number of surrounding cells into which the GFP moved (defined as a “halo”; Oparka et al., 1999) was significantly greater (P < 0.001) in the presence of MPR3-DsRed than MPWT-DsRed (Figure 2D).
To examine the capacity of MPWT and MPR3 to traffic vRNA, we constructed a replicating, movement-defective viral vector that expressed DsRed as a cytosolic protein (TMV.MPFS.DsRed; Figure 1D) (Roberts et al., 2001). We bombarded transcript of this vector into source epidermal cells of N. benthamiana with or without 35S.MPWT-GFP or 35S.MPR3-GFP plasmids (Figure 1G). Biolistic bombardment was used to maximize the number of cells that received both the viral vector transcript and the plasmid DNA. As expected, TMV.MPFS.DsRed alone failed to move from cell to cell on source leaves (Figures 2E and 2H). In cells cobombarded with 35S.MPWT-GFP or 35S.MPR3-GFP, vRNA trafficking was rescued, that is, replicating virus was detected in epidermal cells several cells away from the cobombarded cell (an example of TMV.MPFS.DsRed with 35S.MPR3-GFP is shown in Figure 2F).
In the case of cobombardments of TMV.MPFS.DsRed with 35S MPR3-GFP, rescue of vRNA trafficking was observed from a significantly greater (P < 0.001) number of cobombarded cells, and over a significantly greater (P < 0.001) range of cell boundaries, than from cobombardments of TMV.MPFS.DsRed with 35S MPWT-GFP (Figures 2F, 2H, and 2I). In all bombardment experiments using MP-GFP fusions, MP was detected within plasmodesmata in cells beyond the originally bombarded cell (shown for MPR3-GFP in Figure 2G), indicating that MP had trafficked between cells after viral rescue.
MPR3 Is Not Degraded at the Center of Expanding Infection Sites
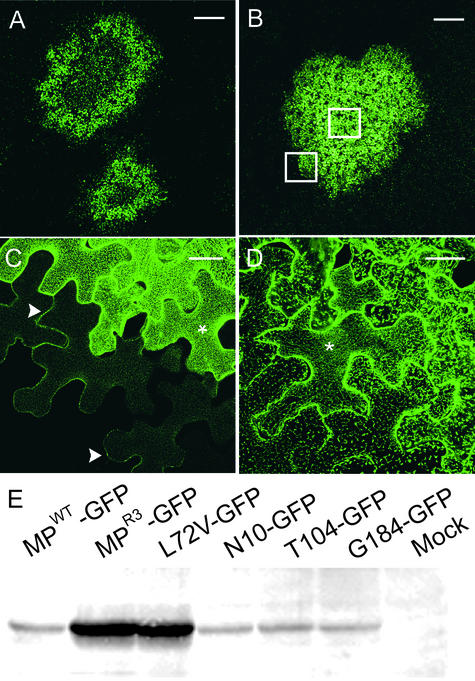
We found that infection sites of TMV.MPWT-GFP had a characteristic ring-like appearance attributable to a cycle of MP synthesis and degradation (Figure 3A), a feature reported previously for TMV (Heinlein et al., 1995, 1998; Padgett et al., 1996; Boyko et al., 2000b). In these infection sites, cells at the leading edge showed prominent labeling of plasmodesmata and punctate cortical bodies, those at the trailing edge of the fluorescent ring had predominantly MT labeling, and cells at the darker center of the infection displayed only labeled plasmodesmata (for details, see Heinlein et al., 1995, 1998; Padgett et al., 1996). In contrast, infection sites of TMV.MPR3-GFP were labeled more uniformly throughout (Figure 3B), suggesting that degradation of MPR3 had not occurred to the same extent as degradation of MPWT.
Figure 3.
Appearance of MPWT and MPR3 Infection Sites.
(A) TMV.MPWT-GFP infection sites have a characteristic ring shape as a result of a cycle of MP synthesis and degradation. Bar = 1 mm.
(B) TMV.MPR3-GFP infection sites are uniformly fluorescent. Bar = 1 mm.
(C) and (D) Detail of boxed regions in (B). At the leading edge of infection (C), plasmodesmata are labeled (arrowheads). Note punctate localization of MPR3-GFP in the cell cortex (asterisks). At the center of infection (D), the cortical punctae are retained. Bars = 20 μm.
(E) Protein gel blot of protein extracts from Bright Yellow 2 protoplasts transfected with viral vectors expressing GFP fusions to MP variants.
(G) At the leading edge, some MPWT-DsRed punctae are not associated with MT (arrowheads), although cells deeper in the infection show strong colabeling of MPWT and MT. Bar = 25 μm.
(H) to (J) Details of the association of MPWT-DsRed ([H], red) with MT ([I], green). Merged image (J) shows that not all MT are labeled with MPWT-DsRed. Bar = 25 μm.
(K) and (L) MPR3-DsRed fails to associate with MT.
(K) Recently infected cells showing MPR3-DsRed punctae (arrowheads). Unlike MPWT-DsRed (cf. [G]), MPR3-DsRed is not transferred onto MT in cells deeper in the infection site. Bars = 25 μm.
(L) Detail of an infected cell showing the lack of MPR3-DsRed association with underlying MT. Bar = 10 μm.
At the leading edge of TMV.MPR3-GFP infection sites, plasmodesmata were labeled strongly, and regularly spaced punctae were apparent in the cell cortex (Figure 3C). These punctae became brighter in cells deeper within the infection site (Figure 3C). At the center of the infection site, the enlarged cortical punctae remained labeled with MPR3-GFP (Figure 3D). The single amino acid change L72V was found to be responsible for the punctate localization characteristic of MPR3 (data not shown). When the plasmids expressing MPWT and MPR3 fluorescent protein fusions (Figures 1G and 1H) were bombarded individually into single epidermal cells of N. benthamiana, the subcellular phenotypes described above were maintained (data not shown), suggesting that the subcellular localizations of MPWT and MPR3 are determined by intrinsic properties of the respective MPs, not by additional viral factors.
MPR3 Is Retained on the Cortical ER
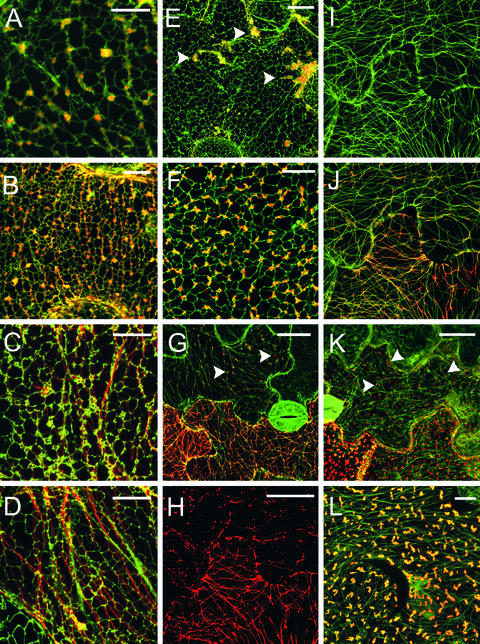
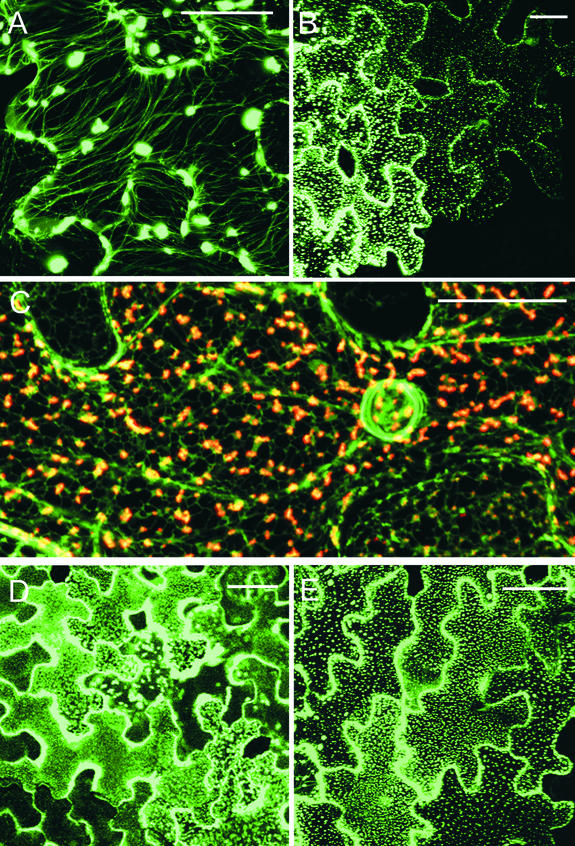
To examine the relationship of MP with the cortical ER, we infected transgenic N. benthamiana plants expressing erGFP by manual inoculation with transcripts of viral vectors expressing MPWT and MPR3 fused to DsRed (Figures 1B and 4). The cortical ER of higher plant cells is dynamic and composed of a polygonal network of tubules connected by short regions of cisternal ER (Quader et al., 1987; Allen and Brown, 1988; Hepler et al., 1990; Cantrill et al., 1999). In TMV.MPWT-DsRed infections, the initial DsRed fluorescence was localized to the cortical ER network. At these sites, the MPWT-DsRed formed aggregates on small regions of cisternal ER (Figure 4A). After this, MPWT-DsRed moved outward from these sites along ER tubules (Figure 4B).
Figure 4.
Subcellular Localization of MPWT-DsRed and MPR3-DsRed Fusion Proteins on GFP-ER– and GFP-MT–Labeled Transgenic Plants.
The images are orientated so that the leading edge of the infection site is toward the top of each image and to the top of each vertical sequence.
(A) to (D) Sequence of MPWT-DsRed association with cortical ER (green) across an infection site.
(A) At the leading edge, MPWT-DsRed punctae are associated with the vertices of the cortical ER network. Bar = 5 μm.
(B) MPWT-DsRed demonstrates spread along the cortical ER network, which shows linear distortions in places. Bar = 10 μm.
(C) MPWT-DsRed is transferred onto underlying MT (red). Bar = 5 μm.
(D) Deeper in the infection site, the ER restores its polygonal structure. All MPWT-DsRed has been transferred from the ER to MT at this stage. Bar = 5 μm.
(E) and (F) Association of MPR3-DsRed with the cortical ER.
(E) Recently infected cell showing MPR3 punctae on the cortical ER network (arrowheads). Bar = 15 μm.
(F) MPR3-DsRed punctae remain associated with the vertices of the cortical ER throughout infection. Bar = 5 μm.
(G) to (J) Sequence of MPWT-DsRed association with MT (green) across an infection site.
Concurrent with this stage, the cortical ER became stretched, distorting the polygonal network in places, although MPWT-DsRed remained associated with the ER (Figure 4B). With increasing distance from the leading edge, the cortical ER became entwined with underlying linear structures (Figure 4C), and at this stage, most of the MPWT-DsRed was transferred from the distended ER tubules onto the linear structures. After transfer of MPWT-DsRed, the cortical ER resumed its characteristic polygonal structure (Figure 4D). In cells infected with TMV.MPR3-DsRed, fluorescence was associated initially with the vertices of the cortical ER network (Figure 4E). With time, these sites enlarged to form punctae that remained bound to the ER (Figure 4F). In contrast to the results with MPWT, MPR3-DsRed fluorescence remained associated with the ER throughout the infection site, and the cortical ER did not undergo the deformation found within TMV.MPWT-DsRed–infected cells (data not shown).
MPR3 Shows Restricted Association with MT
To study more closely the relationship between MPWT and MT, we constructed transgenic N. benthamiana plants that expressed tua-GFP (Ueda et al., 1999). We infected these plants with transcripts of the TMV.MPWT-DsRed or TMV.MPR3-DsRed vector (Figures 1A and 1B). As anticipated, at 28°C, MPWT-DsRed became strongly associated with MT during virus infection. At the infection front, MPWT-DsRed was associated initially with punctae that did not colocalize with underlying MT (Figure 4G). However, in cells immediately behind the infection front, MPWT-DsRed fluorescence was transferred from the punctae onto MT. These MT showed precise colabeling with both GFP and DsRed (Figures 4H to 4J), confirming that the DsRed-labeled linear structures found in infected erGFP plants (see above) were MT.
However, not all MT within infected cells became decorated with MPWT-DsRed (Figure 4J). In TMV.MPR3-DsRed infection sites, the initial pattern of fluorescence in newly infected cells was similar to that observed with MPWT-DsRed, with numerous punctae appearing at the cell periphery (Figure 4K). With time (increasing distance behind the infection front), the punctae became larger and more regularly spaced (Figure 4L), but transfer of MPR3-DsRed onto MT was reduced greatly compared with that of MPWT-DsRed. When the temperature was increased to 32°C, some MT labeling became apparent at the trailing edge of TMV.MPR3-DsRed infection sites, although the cortical punctae described above were retained throughout the infection site (data not shown).
The L72V Coding Change Causes MPR3 to Accumulate in Protoplasts
The restricted transfer of MPR3 fusions from the ER to MT (Figures 4K and 4L), together with the lack of MP degradation at the center of TMV.MPR3-GFP infection sites (Figure 3B), suggested that levels of MPR3 might be increased in infected cells. To test this, we examined MP expression in protoplasts transfected with transcript of either TMV.MPWT-GFP or TMV.MPR3-GFP (Figure 1A). When we examined MP levels in protoplasts infected with viral clones expressing the coding and silent changes independently, increased levels of MP were detected only in protoplasts infected with virus expressing the amino acid change L72V. In protoplasts infected with viruses expressing MPs containing the silent nucleotide changes in codons Asn-10, Tyr-104, or Gly-184 (Toth et al., 2002), MP levels were similar to those in MPWT (Figure 3E).
Disruption of MT with Pharmacological Agents
Because MT have been suggested to play a role in TMV cell-to-cell movement (Boyko et al., 2000a), we were surprised that MPR3 seldom associated with MT during the infection process. Therefore, we infiltrated the MT-depolymerizing drug colchicine into intact leaves of N. benthamiana plants to determine whether TMV movement could occur in the absence of MT. In transgenic plants expressing tua-GFP, 0.1 mM colchicine was found to completely depolymerize the MT cytoskeleton without affecting cytoplasmic streaming (Figures 5A and 5B). We manually inoculated colchicine-treated, nontransgenic leaves with infectious transcripts of TMV.MPWT-DsRed or TMV.MPWT-GFP (Figures 1A and 1B). These viruses moved extensively through colchicine-treated cells at rates similar to those observed with water-infiltrated tissues (Figures 5C and 5D; data not shown for MPR3 versions of the constructs).
Figure 5.
Treatment of Leaves with MT Inhibitors.
(A) Control epidermal cell from a transgenic plant expressing tua-GFP shows an intact MT cytoskeleton. Bar = 30 μm.
(B) Epidermal cell expressing tua-GFP after treatment with 0.1 mM colchicine. MT are absent, although the cells displayed strong cytoplasmic streaming. Bar = 20 μm.
(C) Growth rates of TMV.DsRed infection sites in leaves infiltrated with colchicine (black circles) or water (gray squares).
(D) Growth of a single TMV.DsRed infection site over a 6-day period (day indicated at bottom left) on colchicine. Bar = 1 mm.
(E) Plasmodesmata (arrowheads) are labeled with MPWT-GFP in colchicine-treated infected cells. Bar = 20 μm.
(F) Large MPWT-GFP aggregates accumulate in colchicine-treated cells (see also [G]). Bar = 500 μm.
(G) Detail of a single cell at the leading edge of a colchicine-treated TMV.MPWT-GFP infection site. Note the massive aggregates of MPWT-GFP. Bar = 20 μm.
(H) and (I) A TMV.MPFS.DsRed transcript bombarded into single epidermal cells of tua-GFP transgenic plants fails to move from cell to cell. The same cell was imaged before (H) and after (I) treatment with 0.1 mM colchicine. Note the depolymerization of MT in cells surrounding the bombarded cell (asterisks). Bars = 20 μm.
(J) Growth rates of TMV.DsRed infection sites in leaves infiltrated with oryzalin (black circles) or water (gray squares).
(K) Appearance of a 3-day-old TMV.MPWT-GFP infection site before the addition of oryzalin. Bar = 1 mm.
(L) Appearance of the infection site shown in (K) 1 day after treatment with oryzalin. Note the formation of MPWT-GFP aggregates within infected cells. Bar = 1 mm.
Colchicine-treated cells infected with TMV.MPWT-GFP showed no labeling of MT, although plasmodesmata were labeled strongly in these cells (Figure 5E). In colchicine-treated tissue, the infected cells showed intense labeling of large MPWT-GFP aggregates distributed across the infection site (Figures 5F and 5G). To confirm that MT depolymerization had persisted in the colchicine-treated leaves, leaves of tua-GFP transgenic plants were infiltrated with colchicine and monitored in parallel with virus-infected tissue. MT labeling was not observed in these leaves. Cytoplasmic streaming was observed throughout the time courses of these experiments, indicating that the cells remained alive.
Colchicine Does Not Affect Plasmodesmatal Permeability
Because inhibitors of the actin cytoskeleton have been shown to increase the size exclusion limit of plasmodesmata (Ding, 1996), we investigated whether the treatment of leaf cells with colchicine had an effect on the permeability of plasmodesmata to both nonspecific protein movement and virus movement. To assess protein movement, we biolistically bombarded a plasmid expressing DsRed (molecular mass, 27 kD) under the control of the CaMV 35S promoter (35S.DsRed; Figure 1F) into individual source leaf epidermal cells. Colchicine was applied by infiltration immediately after plasmid bombardment. Of 109 bombardments, none showed DsRed movement into adjacent cells at 48 h after bombardment (data not shown).
To assess whether virus movement was affected by colchicine treatment, we bombarded transcripts of the movement-defective TMV.MPFS.DsRed vector (Figure 1D) into source leaf epidermal cells. To monitor the disassembly of MT, we used tua-GFP plants for these experiments. Colchicine was applied immediately after viral transcript bombardment, and leaves were infiltrated subsequently with colchicine as described above. TMV.MPFS.DsRed failed to move out of the bombarded cells (306 bombardments), although MT disassembly was observed immediately after treatment with colchicine (Figures 5H and 5I). Cytoplasmic streaming continued throughout the treatment of leaf tissues with colchicine. We conclude that colchicine did affect the basic permeability properties of source leaf epidermal plasmodesmata.
In subsequent experiments, leaf tissue was infiltrated before infection with oryzalin (5 to 20 μM), an inhibitor of MT polymerization (Hugdahl and Morejohn, 1993). In leaves expressing tua-GFP, the response to oryzalin was rapid (<10 min), and the labeled MT became broken into short fragments, followed by the gradual disappearance of MT from the cells (data not shown) (Ueda et al., 1999). Viral cell-to-cell movement in oryzalin occurred at comparable rates in control leaves (Figure 5J).
When either colchicine or oryzalin was added to established infection sites (Figure 5K), the growth of the infection site continued (cf. Figures 5K and 5L), but MT labeling was eliminated. In the infected cells, the MPWT-GFP was found in large aggregates dispersed throughout the cytoplasm (Figure 5L). In young infection sites (1 to 3 days after inoculation), oryzalin depolymerized the MT cytoskeleton completely. However, in older infection sites established before the addition of oryzalin (>5 days after inoculation), depolymerization of MT was incomplete, and many of the infected cells retained short MT fragments (data not shown). In contrast, colchicine depolymerized the MT cytoskeleton completely in all infection sites.
Virus-Induced Gene Silencing of tua
In subsequent experiments, we used a virus-induced gene silencing (VIGS) strategy to inhibit MT assembly with a viral vector of Tobacco rattle virus (TRV) (Ratcliff et al., 2001) that expressed a tua sequence homologous with that present in tua-GFP transgenic plants (Figure 1I). Transcripts of TRV.TUA (Figure 1I) were inoculated manually onto tua-GFP N. benthamiana plants. The vector moved systemically from source to sink leaves within 6 days of inoculation, with the first signs of tua-GFP silencing appearing in emerging leaves at 10 days after inoculation.
In young sink leaves, in which guard cells were still coupled symplastically to epidermal cells, tua silencing occurred in all cells of the leaf, and MT could not be detected with the confocal microscope. In older sink leaves, in which the guard cells were isolated symplastically before the arrival of virus, the guard cells did not become silenced (Figure 6A). At 20 days after inoculation, leaves displaying tua-GFP silencing were inoculated manually with TMV.DsRed transcript (Figure 1C). Within 3 days, expanding TMV.DsRed infection sites had become established on the silenced leaves (Figures 6B and 6C). With continued growth of the silenced plants (>20 days after inoculation with respect to the TRV infections), the new leaves showed abnormal growth patterns, consistent with the elimination of tua expression in these leaves (data not shown).
Figure 6.
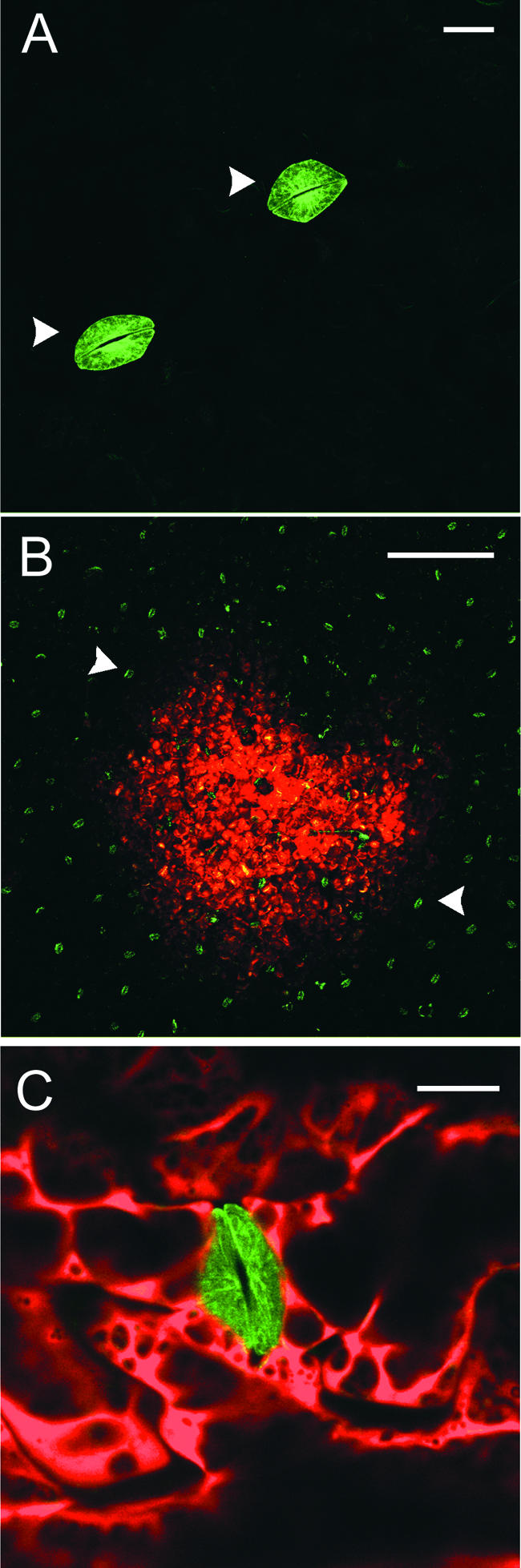
Infection of tua-Silenced Leaves with TMV.DsRed.
(A) After systemic silencing of tua-GFP in transgenic plants infected with TRV vector (TRV.TUA; see Figure 1I), only symplastically isolated guard cells remained fluorescent (arrowheads). Bar = 25 μm.
(B) Establishment of a TMV.DsRed infection site (3 days after inoculation) on a tua-silenced leaf. Note the numerous tua-GFP–expressing guard cells (arrowheads). Bar = 5 mm.
(C) Detail of TMV.DsRed-infected, tua-silenced epidermal cells surrounding a stomatal complex. Bar = 25 μm.
Inhibition of the 26S Proteasome Causes MPWT to Accumulate on the Cortical ER
Because degradation of the TMV MP is mediated by the 26S proteasome (Reichel and Beachy, 2000), we tested, in situ, the effects of the proteasome inhibitor clasto-lactacystin β-lactone on TMV.MPWT-GFP infection sites. In treated leaves, the inhibitor eliminated MT labeling and induced numerous cortical punctae to form (cf. Figures 7A and 7B), a phenotype strongly resembling that of the untreated TMV.MPR3-GFP (cf. Figures 4J and 4L). By infecting erGFP transgenic plants with TMV.MPWT-DsRed, it was shown that the punctate aggregates, produced after treatment with proteasome inhibitor, were associated with the vertices of the cortical ER network (Figure 7C). Treatment of TMV.MPR3-GFP infection sites with proteasome inhibitor had little or no effect on the MPR3-GFP subcellular localization phenotype, and the punctate appearance of cells was retained throughout (cf. Figures 7D and 7E).
Figure 7.
Treatment of TMV.MP-GFP Infection Sites with Proteasome Inhibitors.
(A) TMV.MPWT-GFP infection site showing MT labeling at the trailing edge of infection. Bar = 20 μm.
(B) TMV.MPWT-GFP infection site after treatment with clasto-lactacystin β-lactone. Note the punctate appearance of MPWT-GFP in infected cells. Bar = 20 μm.
(C) TMV.MPWT-DsRed infection site on an erGFP transgenic plant after treatment with clasto-lactacystin β-lactone. MPWT-DsRed is localized to the vertices of the cortical ER network. Bar = 15 μm.
(D) and (E) TMV.MPR3-GFP infection site before (D) and after (E) treatment with clasto-lactacystin β-lactone. Bar = 20 μm.
DISCUSSION
MT Are Not Required for Cell-to-Cell Movement of TMV
TMV is the type member of the tobamoviruses, a group of viruses suggested to use the MT cytoskeleton to transport the viral genome within and between plant cells (Heinlein et al., 1995, 1998; Lazarowitz and Beachy, 1999; Boyko et al., 2000a, 2000b; Tzfira et al., 2000; Aaziz et al., 2001). The recent identification of a MT binding motif on the MPWT has lent credence to the view that MP–MT interactions form an integral part of the viral cell-to-cell transport process (Boyko et al., 2000a). However, although the viral MP has been shown to interact strongly with both vRNA (Citovsky et al., 1990) and MT (Heinlein et al., 1995, 1998; Padgett et al., 1996; Boyko et al., 2000a, 2000b), direct evidence for cell-to-cell trafficking of vRNA-MP complexes along MT is lacking.
In agreement with previous reports of TMV infection, we found that MPWT was transferred from the cortical ER onto underlying MT in cells at the trailing edge of infection foci (Heinlein et al., 1995, 1998; Padgett et al., 1996). By contrast, at the same temperature (28°C), MPR3 (Toth et al., 2002) showed reduced association with MT and remained bound to the vertices of the cortical ER throughout the infection process. The single amino acid coding change L72V was shown to confer the MPR3 phenotype; other silent changes present in MPR3 clones (Toth et al., 2002) did not affect MP localization.
The improved cell-to-cell movement functions of MPR3 made us question whether MT association is a necessary step in the TMV movement process. Viral vectors expressing MPWT or MPR3 moved unimpeded through cells in which the MT cytoskeleton was disrupted with colchicine or oryzalin, confirming the notion that intact MT are not required for the cell-to-cell spread of vRNA. The inability of the movement-defective virus TMV.MPFS.DsRed, and also of free DsRed, to spread from cell to cell in the presence of colchicine confirmed that MT disassembly had no effect on the basic permeability of plasmodesmata. Significantly, the targeting of plasmodesmata by MP and the formation of large, dispersed MP aggregates were unaffected by MT-destabilizing agents, suggesting that both of these phenomena can occur independently of the MT cytoskeleton.
Although colchicine depolymerized MT completely, some infection sites (>5 days after inoculation) treated with oryzalin were partially resistant to MT degradation. The TMV MP interacts with MT nucleation sites and contains a motif that is similar to a region in tubulin that is proposed to mediate lateral contacts between MT protofilaments (Boyko et al., 2000a). A number of factors have been identified that noncompetitively inhibit the binding of oryzalin to tubulin dimers (Hugdahl and Morejohn, 1993), and we suggest that the tubulin binding motif of the MP (Boyko et al., 2000a) may mask oryzalin binding sites on tubulin subunits, protecting the MT cytoskeleton from further disassembly.
As an alternative to chemical inhibitors, we used a VIGS strategy to inhibit MT assembly, using TRV to silence the tua gene in systemically infected leaves. TRV has been shown previously to induce effective and persistent gene silencing in N. benthamiana plants (Ratcliff et al., 2001). When tua-silenced leaves were infected with TMV.DsRed, cell-to-cell movement of virus occurred in the absence of MT. Collectively, our observations are consistent with previous data showing that MPWT interacts strongly with MT during infection. However, they do not support the view that MT are required for the cell-to-cell movement of vRNA.
MP Degradation
It has been suggested that the association of MP with MT could occur as part of a targeted protein degradation pathway in plant cells (Padgett et al., 1996; Reichel and Beachy, 1998; Mas and Beachy, 1999). The transient expression of MPWT in plant cells (Watanabe et al., 1984) is reflected by a temporal modification of plasmodesmal gating in newly infected cells (Oparka et al., 1997) that may be regulated by MP phosphorylation (Waigmann et al., 2000). Subsequent MP localizations, such as ER and MT associations, are lost as the infection front advances. In cells at the center of an infection, most of the MP is degraded, although some MP remains associated with plasmodesmata (Heinlein et al., 1998; Padgett et al., 1996; Oparka et al., 1997). By contrast, MPR3 remained on the vertices of the cortical ER throughout infection, with reduced transfer to MT. As a consequence, infection sites expressing MPR3 fusions were labeled uniformly throughout. These observations suggest a lack, or reduction, of MP degradation in viral vectors expressing MPR3.
It was shown recently that in tobacco protoplasts, TMV MP is degraded by the 26S proteasome (Reichel and Beachy, 2000). Specific proteasome inhibitors enhanced the accumulation of high-molecular-mass forms of MP, leading to increased stability of the MP. Furthermore, MP-GFP fusions accumulated preferentially on the cisternal ER (Reichel and Beachy, 2000). Protein degradation by the 26S proteasome is a ubiquitin-dependent pathway in which polyubiquitinated proteins become targeted selectively for degradation (Vierstra, 1996; Von Kampen et al., 1996). We found that the MPR3 phenotype could be reproduced when viral infection sites expressing MPWT were treated with a specific proteasome inhibitor, indicating that the MPR3 phenotype might arise from impairment of the ubiquitin-proteasome degradation pathway.
In many mammalian cells, inhibition of proteasome activity leads to the accumulation of high-molecular-mass, multiubiquitinated proteins. Such proteins accumulate on the ER and later associate with MT before being transported to the perinuclear region of the cell, where they form large protein aggregates termed “aggresomes” (Johnson et al., 1998). In the model described above, MT function to transport aberrant proteins away from their site of synthesis on the ER when the ubiquitin-proteasome system is saturated (Johnson et al., 1998).
MP–ER Interactions
We have shown that during infection by TMV expressing MPWT fusions to fluorescent proteins, the cortical ER is “pulled” into close proximity with the MT cytoskeleton to permit the direct transfer of MPWT onto MT. During infection with TMV, the cortical ER has been shown to undergo transitions from tubular to cisternal forms, reverting to tubular forms in late stages of infection, changes that parallel the accumulation and degradation of viral MP (Reichel and Beachy, 1998). The hypothesis that the vertices of the cortical ER might act as viral replication “factories” (Schaad et al., 1997) has gained support from the demonstration that both the vRNA and replicase are associated with these sites (Mas and Beachy, 1999).
In the case of MPR3, which cannot be dislocated readily from the ER, MT showed reduced labeling with viral MP. Significantly, in the presence of proteasome inhibitors, MPWT remained on the cortical ER and did not associate with MT. At present, the reason why MPR3 remains on the vertices of the cortical ER throughout infection is unknown. However, we suggest that the L72V coding change might inhibit the transfer of MPR3 to the ubiquitin-proteasome pathway for degradation. When we increased the temperature from 28 to 32°C, some MT labeling became apparent at the trailing edge of MPR3 infection sites. In agreement with the model of Johnson et al. (1998), we suggest that at high temperatures, the ubiquitin-proteasome pathway can become saturated, inducing the transfer of limited amounts of MPR3 to MT.
Are Improvements in Virus Movement Caused by Evasion of MP Degradation?
When we examined MPR3 for the enhancement of known viral transport functions, both plasmodesmal gating and vRNA trafficking were improved significantly over these functions in MPWT. It is unlikely that MPR3 conferred unique improvements in both of these functions, because the L72V change that contributes to improved movement (Toth et al., 2002) does not occur in regions of the MP known to affect either of these cell-to-cell transport functions (Berna et al., 1991; Waigmann et al., 1994; Kahn et al., 1998). Furthermore, the L72V change does not occur in the putative tubulin binding domain identified by Boyko et al. (2000a).
We hypothesize that the observed improvements in virus movement were brought about indirectly by an increase of MP levels that arose from a block in MP degradation. This model is supported by two observations: (1) MPR3 clones expressing the L72V change accumulated to greater levels than MPWT in isolated protoplasts; and (2) infection sites expressing MPR3 did not display the degradation pattern (ring phenotype) characteristic of MPWT infection. The inability of the host cell to degrade MPR3 early in the viral replication cycle could lead to the accumulation of stable MP on the cortical ER (Reichel and Beachy, 2000), providing the invading virus with an enhanced pool of nonubiquitinated MP with which to transport vRNA between cells.
Conclusions
Despite a strong association of the TMV MP with MT during the infection process, we conclude that MT are not essential for trafficking of the viral genome between cells. Instead, our data are consistent with a role for MT in the degradation of MP (Padgett et al., 1996; Reichel and Beachy, 1998; Mas and Beachy, 1999). Alternative models now are required to explain how vRNA is transported within and between plant cells. From the viral replication factories on the vertices of the ER, vRNA must be transported to, and through, plasmodesmata. In the absence of a role for intact MT in cell-to-cell movement, the cortical ER emerges as a candidate for transporting the viral genome between cells (Reichel and Beachy, 1998).
In higher plants, the ER is continuous through plasmodesmata via the desmotubule, a central structure composed of tightly appressed ER membranes (Overall and Blackman, 1996). To move between cells, the MP-vRNA complex may traffic along ER tubules (Reichel and Beachy, 1998) or, alternatively, become bound to an already motile cortical ER system. It is well established that the cortical ER system is dynamic (Allen and Brown, 1988; Hepler et al., 1990; Grabski, 1993; Cantrill et al., 1999), and actin has been implicated in cortical ER movements (Quader et al., 1987) as well as in providing a “scaffold” for trafficking ER-associated Golgi stacks (Boevink et al., 1996). Inhibitors of the actin cytoskeleton increase the size exclusion limit of plasmodesmata (Ding, 1996), and actin and myosin have been immunolocalized to plasmodesmata (White et al., 1994; Radford and White, 1998; Baluska et al., 2000). Therefore, we suggest that future studies address the role of actin as a potential cytoskeletal element for transporting ER-bound vRNA complexes to, and through, higher plant plasmodesmata.
METHODS
Plant Material
Nicotiana benthamiana plants were grown from seed and maintained at 28°C in a Snijder Climatic Cabinet (Snijder, Tilburg, The Netherlands) with a photoperiod of 16 h. Light intensity was ∼400 μE·m−2·s−1 PAR. Plants were used for experiments when they were between 15 and 60 days old. N. benthamiana plants expressing a fusion of green fluorescent protein (GFP) to Arabidopsis α-tubulin (tua-GFP) were produced by transformation with 35S::GFP-TUA (Ueda et al., 1999) as described previously (Benvenuto et al., 1991).
Construction of Viral Vectors
Standard molecular techniques were used for DNA manipulation (Sambrook et al., 1989). Receptor clones for the construction of C-terminal fusions of the different movement protein (MP) gene sequences with fluorescent proteins were made by amplifying the cycle 3 GFP and DsRed sequences from 30B-based vectors (Shivprasad et al., 1999) using 5′ primers incorporating an AvrII site and a 3′ primer downstream from the unique BsiWI site in Tobacco mosaic virus (TMV). The amplification products, encompassing the fluorescent protein genes and TMV pseudoknot sequences, were digested with AvrII and BsiWI and cloned between the same sites of 30B.GFPe (Toth et al, 2002). MP gene sequences were cloned into the resulting receptor clones by HiFidelity (Expand High Fidelity PCR system; Roche, Mannheim, Germany) amplification of the MP sequence with a 5′ primer covering the EagI site and a 3′ primer incorporating an AvrII site.
To prevent the production of a subgenomic mRNA encoding free fluorescent protein, the subgenomic mRNA transcriptional start site was modified (g5703c) to prevent initiation, and the initiation codons of the fluorescent proteins were removed. The coat protein genes were deleted from the resulting clones. Single nucleotide mutations in the MP gene sequence were generated by overlap PCR with mutagenic primers and subsequent cloning back into the appropriate receptor clone. TMV.DsRed was constructed by exchanging the PacI-KpnI fragment of a DsRed-containing 30B derivative for the same fragment in 30B. Construction of TMV.MPFS.DsRed was described previously (Roberts et al., 2001). Clones were checked by sequencing.
Preparation of Infectious Transcripts and Plant Inoculation
Runoff transcripts were synthesized from viral vectors using a T7 transcription kit (Ambion, Austin, TX). For manual inoculation, transcripts were reassembled with TMV coat protein (Sleat et al., 1986) and then rubbed onto aluminum oxide–dusted N. benthamiana leaves.
Construction of Transient Expression Vectors
35S promoter plasmid constructs for bombardments were constructed in the plant expression vector pRTL2 (Restrepo et al., 1990). pRTL2.GFP was a kind gift of Biao Ding (Ohio State University, Columbus, OH). pRTL2.DsRed was constructed by amplifying the DsRed sequence from pDsRed (Clontech, Palo Alto, CA) using primers containing NcoI and KpnI sites. The PCR product was digested with NcoI and KpnI and cloned into similarly digested pRTL2. For the pRTL2 MP–fluorescent protein fusion constructs, the MP–fluorescent protein sequences were amplified by HiFidelity PCR from viral vector fusion protein clones using a 5′ primer that primed from the second codon of the MP gene and 3′ fluorescent protein primers that incorporated a KpnI site. The amplification products were digested with KpnI and cloned into pRTL2 between the NcoI site (which had been end filled to regenerate an initiation codon for the MP) and the KpnI site. Correct clones were confirmed by sequencing.
Microprojectile Bombardment
Plasmid DNAs and transcripts from viral constructs were introduced into N. benthamiana source leaf epidermal cells by particle bombardment. The bombardment method was essentially as described by Gal-On et al. (1997). Briefly, 5 μL of ethanol was added to ∼2 μg of plasmid DNA or RNA transcript. This was mixed with 0.55 mg of 1-μm gold microcarrier (Bio-Rad Laboratories, Hercules, CA) in ethanol to make a final volume of 18 μL. Two microliters of the nucleic acid–gold mixture was loaded onto a discharge assembly (13-mm Plastic Swinney Filter Holder; Pall Gelman Laboratory, Ann Arbor, MI). The ethanol was allowed to evaporate before bombardments were started. Unless stated otherwise, bombarded leaves were observed with the confocal laser scanning microscope after 24 h. In cobombardment experiments, equal amounts of the plasmids and RNA transcript (2 μg total) were mixed before the addition of the gold particles.
Plasmodesmal Gating Experimental Design and Statistical Analysis
In all experiments described, two intact source leaves per plant were bombarded (usually the second and third true leaves). Each shot was taken as a replicate, and there were 20 replicates per treatment (giving ∼250 to 500 transformed “hit” cells), with five shots per leaf and four leaves per treatment. Statistical analyses were performed using the Minitab Release 10.2 (Minitab, State College, PA) and GenStat Release 4.2 (5th edition; Lawes Agricultural Trust, Rothamsted, UK) statistical packages for Windows. Data were checked for normality using an Anderson-Darling normality test and then subjected to analysis of variance (fixed effects model); where applicable, separation of means was performed using Tukey's test. Count data (e.g., number of cells per infection foci) were analyzed using the nonparametric Kruskal-Wallis test.
Transfection of Protoplasts and Protein Gel Blot Analysis
Protoplasts were prepared from tobacco Bright Yellow 2 suspension culture cells and infected with in vitro transcripts by electroporation essentially as described elsewhere (Lewandowski and Dawson, 1998) with minor modifications. For each electroporation, 5 × 105 protoplasts were mixed with 150 μL of cold electroporation buffer consisting of 10% mannitol, 6 mM CaCl2, and 80 mM KCl. The products from a 5-μL transcription reaction were added just before transferring to a 2-mm-gap cuvette. Electroporated protoplasts were resuspended in 1 mL of culture medium and cultured in 24-well culture dishes at 25°C in the light. Protoplasts were harvested at 40 h after infection by grinding in 50 μL of PBS per 5 × 105 protoplasts and centrifuging at 10,000 rpm for 2 min. The supernatant was retained for standardization by ELISA, and 50 μL of 1 × SDS loading dye was added to the pellet fraction for protein gel blot analysis.
Accumulation of TMV coat protein in transfected protoplasts was determined by indirect triple-antibody sandwich ELISA essentially as described by Torrance (1992). ELISA plate wells were coated with polyclonal antibody against TMV coat protein raised in goats (a kind gift of M.H.V. van Regenmortel). After incubation with protoplast supernatant samples, bound coat protein was detected using rabbit polyclonal antiserum (Hwang et al., 1994) followed by alkaline phosphatase–conjugated anti-rabbit IgG antibody (Sigma, St. Louis, MO). All antibodies were used at a dilution of 1:2000. Levels of coat protein were calculated using Biolinx software (Dynatech Laboratories, Chantilly, VA) with TMV strain U5 as a standard. Aliquots of the pellet fraction that corresponded to equal levels of viral replication were subjected to SDS-PAGE and protein gel blot analysis. Protein gel blot analysis was performed as described elsewhere (Reichel and Beachy, 1998). Polyclonal antiserum raised against GFP (Santa Cruz et al., 1996) was used at a dilution of 1:2000 at 4°C overnight. Alkaline phosphatase–conjugated antibody raised against rabbit IgG was used at a dilution of 1:2000 for 2 to 3 h at room temperature.
Treatment with Inhibitors
A stock solution of clasto-lactacystin β-lactone (Calbiochem-Novabiochem UK Ltd., Nottingham, UK) was prepared in DMSO and diluted 1:1000 in water to produce a final inhibitor concentration of 20 μM. Colchicine (Sigma) solutions were prepared directly in water at 0.1 mM. Stock solutions of oryzalin (a kind gift from C. Hawes) were made up in ethanol. These were diluted in water to produce a working solution with a final inhibitor concentration of 20 μM. Leaf tissue was syringe-infiltrated with inhibitor solutions through stomata on the abaxial leaf surface. The infiltrated tissue then was floated adaxial surface down on the inhibitor solution under a light source for the duration of the treatment. Tissue samples were imaged between 2 and 24 h after infiltration with clasto-lactacystin β-lactone and up to 5 days after infiltration with the microtubule inhibitors.
Imaging of Intact Leaves
Intact leaves or pieces of leaf tissue infected with TMV vectors were either mounted on glass microscope slides in distilled water under a glass cover slip or stuck to the slides using double-sided adhesive tape (Sellotape GB Ltd., Dunstable, UK). The tissue was imaged for GFP or DsRed using a Leica TCS SP (Leica Microsystems, Heidelberg, Germany) spectral confocal laser scanning microscope.
Acknowledgments
The authors thank T. Hashimoto for the kind gift of TUA6-GFP. This work was supported by the Scottish Executive Environment and Rural Affairs Department and by Biosource Genetics Corporation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002303.
References
- Aaziz, R., Dinant, S., and Epel, B.L. (2001). The cytoskeleton and plasmodesmata: For better or for worse. Trends Plant Sci. 6, 326–330. [DOI] [PubMed] [Google Scholar]
- Allen, N.S., and Brown, D.T. (1988). Dynamics of the endoplasmic reticulum in living onion epidermal cells in relation to microtubules, microfilaments and intracellular particle movement. Cell Motil. Cytoskeleton 10, 153–163. [Google Scholar]
- Baluska, F., Barlow, P.W., and Volkmann, D. (2000). Actin and myosin VIII in developing root cells. In Actin: A Dynamic Framework for Multiple Plant Cell Functions, C.J. Staiger, F. Baluska, D. Volkmann, and P.W. Barlow, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 457–476.
- Bassel, G., and Singer, R.H. (1997). mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 9, 109–115. [DOI] [PubMed] [Google Scholar]
- Benvenuto, E., Ordas, R.J., Tavazza, R., Ancora, G., Biocca, S., Cattaneo, A., and Galeffi, P. (1991). ‘Phytoantibodies’: A general vector for expression of immunoglobulin domains in transgenic plants. Plant Mol. Biol. 17, 865–874. [DOI] [PubMed] [Google Scholar]
- Berna, A.R., Gafny, S., Wolf, S., Lucas, W.J., Holt, C.A., and Beachy, R.N. (1991). The TMV movement protein: Role of the carboxyl-terminal 73 amino acids in subcellular localisation and function. Virology 182, 682–689. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Santa Cruz, S., Hawes, C., Harris, N., and Oparka, K.J. (1996). Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J. 10, 935–941. [Google Scholar]
- Boyko, V., Ferralli, J., Ashby, J., Schellenbaum, P., and Heinlein, M. (2000. a). Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2, 826–832. [DOI] [PubMed] [Google Scholar]
- Boyko, V., Ferralli, J., and Heinlein, M. (2000. b). Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J. 22, 315–325. [DOI] [PubMed] [Google Scholar]
- Cantrill, L.C., Overall, R.L., and Goodwin, P.B. (1999). Cell-to-cell communication via plant endomembranes. Cell Biol. Int. 23, 653–661. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V., Knorr, D., Schuster, G., and Zambryski, P. (1990). The P30 movement protein of tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell 60, 637–647. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., and Zambryski, P. (1991). How do plant virus nucleic acids move through intercellular connections? Bioessays 13, 373–379. [DOI] [PubMed] [Google Scholar]
- Ding, B. (1996). Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 10, 157–164. [Google Scholar]
- Gal-On, A., Meiri, E., Elman, C., Gray, D.J., and Gaba, V. (1997). Simple hand-held devices for the efficient infection of plants with viral-encoding constructs by particle bombardment. J. Virol. Methods 64, 103–110. [DOI] [PubMed] [Google Scholar]
- Grabski, S. (1993). Endoplasmic reticulum forms a dynamic continuum for lipid diffusion between contiguous soybean root cells. Plant Cell 5, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg, T. (1998). The destinies and destinations of RNAs. Cell 95, 451–460. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., Epel, B.L., Padgett, H.S., and Beachy, R.N. (1995). Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270, 1983–1985. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., Padgett, H.S., Gens, J.S., Pickard, B.G., Casper, S.J., Epel, B.L., and Beachy, R.N. (1998). Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., Palevitz, B.A., Lancelle, S.A., McCauley, M.M., and Lichtscheidl, I. (1990). Cortical endoplasmic reticulum in plants. J. Cell Sci. 96, 355–373. [Google Scholar]
- Hugdahl, J.D., and Morejohn, L.C. (1993). Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin of Zea mays L. Plant Physiol. 102, 725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, D.J., Roberts, I.M., and Wilson, T.M. (1994). Expression of Tobacco mosaic virus coat protein and assembly of pseudovirus particles in Escherichia coli. Proc. Natl. Acad. Sci. USA 91, 9067–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R.P. (1999). RNA-cytoskeletal association. FASEB J. 13, 455–466. [PubMed] [Google Scholar]
- Johnson, J.A., Ward, C.L., and Kopito, R.R. (1998). Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, T.W., Lapidot, M., Heinlein, M., Reichel, C., Cooper, B., Gafny, R., and Beachy, R.N. (1998). Domains of the TMV movement protein involved in subcellular localisation. Plant J. 15, 15–25. [DOI] [PubMed] [Google Scholar]
- Lazarowitz, S.G. (1999). Probing plant cell structure with viral movement proteins. Curr. Opin. Plant Biol. 2, 332–338. [DOI] [PubMed] [Google Scholar]
- Lazarowitz, S.G., and Beachy, R.N. (1999). Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski, D.J., and Dawson, W.O. (1998). Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 251, 427–437. [DOI] [PubMed] [Google Scholar]
- Mas, P., and Beachy, R.N. (1999). Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intercellular distribution of viral RNA. J. Cell Biol. 147, 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz, M.V., Fradkov, A.F., Labas, Y.A., Savitsky, A.P., Zaraisky, A.G., Markelov, M.L., and Lukyanov, S.A. (1999). Fluorescent proteins from nonbioluminescent Anthazoa species. Nat. Biotechnol. 17, 969–973. [DOI] [PubMed] [Google Scholar]
- McLean, B.G., Zupan, J., and Zambryski, P. (1995). Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco plants. Plant Cell 7, 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J., Prior, D.A.M., Santa Cruz, S., Padgett, H.S., and Beachy, R.N. (1997). Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus. Plant J. 12, 781–789. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I.M., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B.E. (1999). Simple, but not branched, plasmodesmata allow the non-specific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. [DOI] [PubMed] [Google Scholar]
- Overall, R.L., and Blackman, L.M. (1996). A model of the macromolecular structure of plasmodesmata. Trends Plant Sci. 1, 307–311. [Google Scholar]
- Padgett, H.S., Epel, B.L., Kahn, T.W., Heinlein, M., Watanabe, Y., and Beachy, R.N. (1996). Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J. 10, 1079–1088. [DOI] [PubMed] [Google Scholar]
- Quader, H., Hoffman, A., and Schnepf, E. (1987). Shape and movement of the endoplasmic reticulum in onion bulb cells: Possible involvement of actin. Eur. J. Cell Biol. 44, 17–26. [Google Scholar]
- Radford, J.E., and White, R.G. (1998). Localization of a myosin-like protein to plasmodesmata. Plant J. 19, 555–567. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Reichel, C., and Beachy, R.N. (1998). Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 95, 11169–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel, C., and Beachy, R.N. (1999). The role of the ER and cytoskeleton in plant viral trafficking. Trends Plant Sci. 4, 458–462. [DOI] [PubMed] [Google Scholar]
- Reichel, C., and Beachy, R.N. (2000). Degradation of tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74, 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, I.M., Boevink, P., Roberts, A.G., Sauer, N., Reichel, C., and Oparka, K.J. (2001). Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218, 31–44. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Santa Cruz, S., Chapman, S., Roberts, A.G., Roberts, I.R., Prior, D.A.M., and Oparka, K.J. (1996). Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. USA 93, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C., Jensen, P.E., and Carrington, J.C. (1997). Formation of plant RNA virus replication complexes on membranes: Role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivprasad, S., Pogue, G.P., Lewandowski, D.J., Hidalgo, J., Donson, J., Grill, L.K., and Dawson, W.O. (1999). Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255, 312–323. [DOI] [PubMed] [Google Scholar]
- Sleat, D.E., Turner, P.C., Finch, J.T., Butler, P.J.G., and Wilson, T.M.A. (1986). Packaging of recombinant RNA molecules into pseudovirus particles directed by the origin-of-assembly sequence from tobacco mosaic virus RNA. Virology 155, 299–308. [DOI] [PubMed] [Google Scholar]
- Tzfira, T., Rhee, Y., Chen, M.-H., Kunik, T., and Citovsky, V. (2000). Nucleic acid transport in plant-microbe interactions: The molecules that walk through the walls. Annu. Rev. Microbiol. 54, 187–219. [DOI] [PubMed] [Google Scholar]
- Tomenius, K., Clapham, D., and Meshi, T. (1987). Localization, by immunogold cytochemistry, of the virus-coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 160, 363–371. [DOI] [PubMed] [Google Scholar]
- Torrance, L. (1992). Serological methods to detect plant viruses: production and use of monoclonal antibodies. In Techniques for the Rapid Detection of Plant Pathogens, J.M. Duncan and L. Torrance, eds (Oxford: Blackwell Scientific Publications), pp. 7–33.
- Toth, R.L., Pogue, G.P., and Chapman, S. (2002). Improvement of a plant virus based vector through DNA shuffling. Plant J. 30, 593–600. [DOI] [PubMed] [Google Scholar]
- Ueda, K., Matsuyama, T., and Hashimoto, T. (1999). Visualisation of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206, 201–206. [Google Scholar]
- Vierstra, R.D. (1996). Proteolysis in plants: Mechanisms and functions. Plant Mol. Biol. 32, 275–302. [DOI] [PubMed] [Google Scholar]
- Von Kampen, J., Wettern, M., and Schulz, M. (1996). The ubiquitin system in plants. Physiol. Plant. 97, 618–624. [Google Scholar]
- Waigmann, E., Chen, M.-H., Bachmaier, R., Goshroy, S., and Citovsky, V. (2000). Regulation of plasmodesmal transport by phosphorylation of the tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19, 4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann, E., Lucas, W.J., Citovsky, V., and Zambryski, P. (1994). Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc. Natl. Acad. Sci. USA 91, 1433–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y.Y., Emori, I., Ooshika, T., Meshi, T., Ohno, T., and Okada, Y. (1984). Synthesis of TMV-specific RNAs and proteins at the early stage of infection in tobacco protoplasts: Transient expression of the 30K protein and its mRNA. Virology 133, 18–24. [DOI] [PubMed] [Google Scholar]
- White, R.G., Badelt, K., Overall, R.L., and Vesk, M. (1994). Actin associated with plasmodesmata. Protoplasma 180, 169–184. [Google Scholar]
- Wolf, S., Deom, C.M., Beachy, R.N., and Lucas, W.J. (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246, 377–379. [DOI] [PubMed] [Google Scholar]
- Zambryski, P. (1995). Plasmodesmata: Plant channels for molecules on the move. Science 270, 1943–1944. [DOI] [PubMed] [Google Scholar]