Abstract
The ZNF198-FGFR1 fusion gene arises as a result of the t(8;13)(p11;q12) in the 8p11 myeloproliferative syndrome. To determine the transforming properties of this chimeric protein we transfected ZNF198-FGFR1 into the interleukin (IL)-3 dependent cell line Ba/F3. Growth factor independent subclones were obtained in which ZNF198-FGFR1, STAT1, and STAT5 were constitutively tyrosine phosphorylated, as determined by immunoprecipitation and Western blot analysis. To test the hypothesis that constitutive activation of ZNF198-FGFR1 tyrosine kinase activity is a result of self-association of the fusion protein, we in vitro transcribed and translated ZNF198-FGFR1 and a derivative construct, ZNF198-FGFR1 ΔC-myc, in which the C-terminal FGFR1 epitope was replaced by a c-myc tag. As expected, an anti-FGFR1 antibody immunoprecipitated ZNF198-FGFR1 but not ZNF198-FGFR1 ΔC-myc. However when both products were translated together, both were coimmunoprecipitated by anti-FGFR1 antisera. Similar results were obtained by using an anti-myc antibody and demonstrated a physical interaction between the two proteins. Analysis of COS-7 cells transfected with ZNF198-FGFR1 demonstrated that the fusion gene, in contrast to normal FGFR1, is located in the cytoplasm. We conclude that ZNF198-FGFR1 is a cytoplasmic protein that self-associates and has constitutive transformation activity. These data suggest that ZNF198-FGFR1 plays a primary role in the pathogenesis of the t(8;13) myeloproliferative syndrome and is the first report to implicate STAT proteins in FGFR1-mediated signaling.
Keywords: ZNF198, FGFR1, t(8;13), tyrosine kinase
Introduction
The 8p11 myeloproliferative syndrome is a rare, aggressive condition associated with chromosomal rearrangements of the short arm of chromosome 8 [1]. The most common cytogenetic abnormality in this syndrome is the t(8;13) (p11;q12), but strikingly similar clinical features have also been described in patients with a t(8;9)(p11;q34) or a t(6;8)(q27;p11). We and others have recently cloned the t(8;13) and found a fusion between a novel gene, ZNF198/RAMP/FIM (hereafter referred to as ZNF198), at 13q12 and the fibroblast growth factor receptor-1 (FGFR1) gene at 8p11 [2–5]. FISH analysis has indicated that FGFR1 is also disrupted by the t(6;8) and t(8;9) [6].
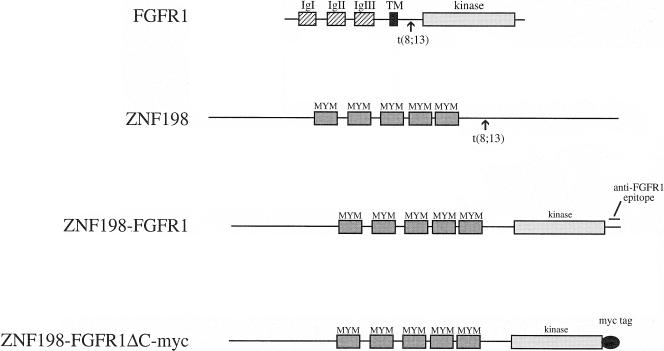
ZNF198 is a widely expressed gene of unknown function that is predicted to encode a 1377 amino acid protein of 155 kDa. The most prominent feature of ZNF198 is that it contains five MYM domains [2,3], a novel octocysteine (C8), zinc finger-related motif that is conserved in two related genes, ZNF261 (DXS6673E/KIAA0385) and ZNF262 (KIAA0425). In all t(8;13) cases thus far examined, ZNF198 exon 17 is spliced to FGFR1 exon 9 and the fusion mRNA is predicted to be translated into a chimeric protein that retains the five MYM domains of ZNF198 and virtually the entire intracellular part of FGFR1, including the tyrosine kinase domain (Figure 1).
Figure 1.
Schematic representation of ZNF198, FGFR1, the ZNF198-FGFR1 fusion and the myc-tagged, C-terminal-deleted fusion construct ZNF198-FGFR1ΔC-myc. Arrows indicate the sites of fusion immunoglobulin I–III, extracellular immunoglobulin-like domains. (TM, transmembrane region; kinase, tyrosine kinase domain; MYM, MYM domains).
Deregulation of specific nonreceptor and receptor tyrosine kinases as a result of chromosomal translocation and gene fusion has emerged as a consistent theme in myeloproliferative disorders. The first example of such a chimeric gene was BCR-ABL in chronic myeloid leukemia [7,8], but subsequently several other fusions have been identified, such as TEL-ABL [9,10] and TEL-JAK2 [11] in Philadelphia-chromosome negative/atypical chronic myeloid leukemia, TEL-PDGRF/β [12] and HIP1-PDGRFβ [13] in chronic myelomonocytic leukemia, and, as just described, ZNF198-FGFR1 in the 8p11 myeloproliferative syndrome. Functional analysis has demonstrated that BCR-ABL, TEL-PDGRFβ, and TEL-JAK2 possess transforming activity in vitro and in vivo as a result of the constitutive activation of their tyrosine kinase moieties [14–17]. Constitutive activity arises by partner gene-dependent dimerization or multimerization of the fusion proteins and thus mimics the normal process of receptor tyrosine kinase signaling after binding of their cognate ligands. In addition, the partner gene may relocalize the tyrosine kinase to a different cellular compartment from which it normally resides, thus enabling it to phosphorylate novel substrates, such as components of the focal adhesion complex in the case of BCR-ABL [18].
In this study we have analyzed the transforming activity of ZNF198-FGFR1. We show that this fusion protein is capable of self-association and that it transforms the IL-3-dependent cell line Ba/F3 to factor independence. Transformation is accompanied by constitutive high level tyrosine phosphorylation of STAT 1 and STAT 5. These are the first data to demonstrate the transforming activity of ZNF198-FGFR1 and to implicate STAT proteins in FGFR1-mediated signaling.
Experimental Procedures
Constructs
Because only partial cDNA clones were generated during the characterization of the t(8;13), the entire sequence of ZNF198-FGFR1 was reconstructed by using reverse transcriptase-polymerase chain reaction (RT-PCR). High fidelity PCR (Boehringer Mannheim, Lewes, UK) was used to amplify three fragments which were subsequently assembled to give the entire coding sequence. All positions refer to Genbank accession numbers AJ224901 (ZNF198) and X52833 (FGFR1). Fragment 1 (ZNF198 positions 30–1390) was amplified from normal peripheral blood leukocyte cDNA and introduced a BamHI site at ZNF198 position 33. Fragment 2 (ZNF198 position 998 to FGFR1 position 1495) was amplified from t(8;13) patient cDNA. Fragment 3 (FGFR1 positions 1290–2603) was amplified from an FGFR1 cDNA clone and introduced a BamHI site at FGFR1 position 2600. The complete ZNF198-FGFR1 cDNA was assembled into the BamHI site of pUC19 by using the internal restriction sites EcoRI (ZNF198 position 1379) and NheI (FGFR1 position 1455) to give plasmid pZF1. This clone was entirely sequenced and matched exactly to that expected, except that it lacked the 46-bp noncoding ZNF198 exon 3. An expression construct was generated by cloning the ZNF198-FGFR1 cDNA into the BamHI site of pcDNA3.1 (Invitrogen, UK) to give pcDNA3.1/ZNF198-FGFR1.
To generate the pCDNA3.1/ZNF198-FGFR1ΔC-myc construct, the 3.5 kb pZF1 BamHI/EagI fragment was subcloned into pcDNA3.1/Myc-His B (Invitrogen, Groningen, Holland) digested with BamHI and NotI. The resulting plasmid ZNF198-FGFR1ΔC-myc encodes a ZNF198-FGFR1 fusion in which the C-terminal 163 amino acids of FGFR1 are replaced by a c-myc epitope.
Cell Lines and Transfections
Ba/F3 cells were maintained in IL-3 medium (RPMI 1640 medium with 10% fetal calf serum [FCS] and 5% conditioned medium from the IL-3-producing WEHI-3B cell line). For electroporation, 1x107 Ba/F3 cells were washed in phosphate-buffered saline (PBS) and incubated for 10 minutes at room temperature with 20 µg of plasmid DNA in PBS. Cells were electroporated at 350 V/975 mF in a Bio-Rad Gene Pulser II. After a 10-minute incubation at room temperature, the cells were plated in 10 ml of IL-3-containing medium for 48 hours and then selected and subcloned by limiting dilution (2500 or 250 cells per well) in IL-3 medium plus 1 mg/mL G418. After 12 days, resistant subclones were washed and plated out in medium without IL-3. For growth curves, 2x106 cells from three subclones were plated in 5 mL of medium with or without IL-3, and viable cells counted on each day by trypan blue exclusion.
COS7 cells were maintained Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS, and transfections were carried out with Transfast (Promega, Southampton, UK) according to the manufacturer's instructions.
In Vitro Transcription/Translation
In vitro transcription/translation was carried out by using a rabbit reticulocyte lysate kit (TNT T7 quick coupled system; Promega) as recommended by the manufacturer. [35S]methionine incorporation was used to label the proteins. Half the reaction was diluted to 0.5 mL in lysis buffer (150 mmol/L NaCl, 50 mmol Tris-HCl, 1% Triton X-100 plus protease, and phosphatase inhibitors) and proteins immunoprecipitated as described in the next section. Immunoprecipitates and total in vitro transcribed/translated products were resolved on a 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and labeled proteins visualized by fluorography using Amplify (Amersham, St. Alban's, UK) according to the manufacturer's instructions.
Immunoprecipitation and Western Blotting
PBS washed cells were lysed in lysis buffer for 30 minutes on ice. Immunoprecipitation was carried out by incubating 1 mg of total cell lysate or in vitro transcribed/translated products in 0.5 mL lysis buffer with the appropriate primary antibody for 2 hours at 4°C. Fifty milliliters of 10% protein A-Sepharose (Pharmacia, St. Alban's, UK) made up in lysis buffer was then added for an additional 1 hour, and immunoprecipitates were washed twice in lysis buffer before boiling in loading buffer for 5 minutes and resolving by SDS-PAGE. Blocking of Western blots was carried out in either 5% dry milk in TBST (0.1% Tween-20/0.01 mol/L Tris-HCL, pH 7.6/150 mmol NaCl) or 3% dry milk in TBST for antiphosphotyrosine blots. Blots were washed in TBST and incubated for 45 minutes with primary antibody before further TBST washes and incubation with horseradish peroxidase-conjugated secondary antibody (Amersham) for a further 45 minutes. Visualization was carried out with an enhanced chemiluminescence system (Pierce, Aylesbury, UK) Antibodies used were a polyclonal rabbit antibody to residues 802–822 at the C-terminus of FGFR1, anti-STAT 1 and 5 (all from Santa Cruz Biotechnology, Santa Cruz, CA); antiphosphotyrosine (4G10; Upstate Biotechnology, Lake Placid, NY) and anti-myc (Invitrogen).
Immunofluorescence
COS7 cells transfected with pcDNA3.1/ZNF198-FGFR1 were immunostained by using the anti-FGFR1 antibody as previously described [19]. Briefly, transfected COS7 cells were grown on glass coverslips and fixed with 4% paraformaldehyde in PBS for 30 minutes at room temperature. The coverslip was then washed three times for 5 minutes in L15 medium +10% FCS at room temperature and incubated for 30 minutes in the same mixture. After incubation for 1 hour in rabbit anti-FGFR1 antibody (5 µg/mL), cells were washed in L15/FCS as before and then incubated with goat anti-rabbit antibody linked to fluoresceinisothiocyanate (1:1000) for an additional 1 hour at room temperature. After a final set of L15/FCS washes, the cells were counterstained with DAPI and images captured on a Zeiss Axioplan microscope (Zeiss, Welwyn Garden City, UK).
Results
ZNF198-FGFR1 Transforms Ba/F3 Hematopoietic Cells to Interleukin 3 Independence
The transforming activity of ZNF198-FGFR1 was tested in the murine hematopoietic cell line Ba/F3, which is dependent on IL-3 for growth and expresses no detectable endogenous FGFR1 [20]. Ba/F3 cells were transfected by electroporation with the control vector, pcDNA3.1, or pcDNA3.1/ZNF198-FGFR1 containing the full length fusion cDNA. Cells were recovered in IL-3-containing medium for 48 hours and then plated out in medium containing G418 plus IL-3 at either 2500 or 250 cells per well in 96 well microtitre plates. After transfection with pcDNA3.1/ZNF198-FGFR1 and 12 days of incubation, growth was seen in 95/96 (99%) and 22/96 (23%) of wells at the higher and lower concentration of cells, respectively. For the pcDNA3.1 vector control, 70/96 (73%) of wells grew at the higher concentration and 5/96 (5%) grew at the lower concentration. The proportions of surviving wells and the fact that all grew from a single focal point suggested that many of positive wells were clonal, at least at the lower dilutions. G418-resistant subclones were washed free of IL-3 and plated out in medium lacking added growth factor. After 12 days, none of the G418 resistant, pcDNA3.1-transfected wells had grown in the absence of IL-3. For the ZNF198-FGFR1 transfected cells, factor independent growth was seen for 67/95 (71%) of subclones from the high concentration plate and for 15/22 (68%) of subclones from the low concentration plate. The proportion of factor-independent, G418-resistant clones is comparable to that seen with BCR-ABL and TEL-ABL [21].
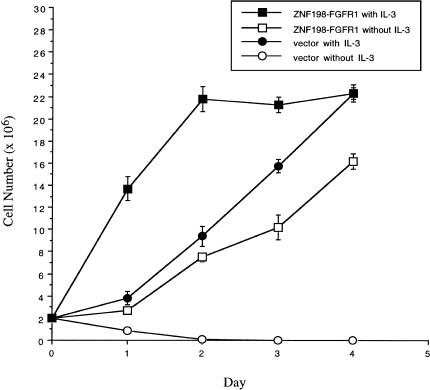
Growth curves were performed on three separate subclones for both ZNF198-FGFR1 and pcDNA3.1 transfected cells. Ba/F3 cells transfected with ZNF198-FGFR1 grew at a slightly slower rate in the absence of IL-3 compared with the Ba/F3-pcDNA3.1 control cells in the presence of IL-3. However, addition of IL-3 to the Ba/F3 cells transfected with ZNF198-FGFR1 resulted in a further stimulation of growth, indicating a cooperative effect (Figure 2). All three ZNF198-FGFR1 transformed subclones grew at a similar rate in the presence or absence of IL-3 and all expressed similar levels of the fusion protein (data not shown).
Figure 2.
ZNF198-FGFR1 transforms Ba/F3 cells to IL-3 independence. Ba/F3 cells were transfected with pcDNA3.1 or pcDNA3.1/ZNF198-FGFR1. Cells were initially selected for G418 resistance before plating in media lacking IL-3. IL-3-independent cells (pcDNA3.1/ZNF198-FGFR1) or G418-resistant cells (pcDNA3.1) were washed free of IL-3 and plated in the absence of presence of IL-3. Viable cells were counted on each day, and results shown are the average of three independent clones.
Normal Ba/F3 cells exhibited no growth in conditioned medium harvested by centrifugation and filtration from a confluent culture of Ba/F3-ZNF198-FGFR1 cells grown in the absence of IL-3 (data not shown). This suggests that the transformation of Ba/F3 cells by ZNF198-FGFR1 is not an autocrine effect of induction of expression of endogenous IL-3 or other growth factors.
ZNF198-FGFR1 is Constitutively Tyrosine-Phosphorylated in Transformed Ba/F3 Cells
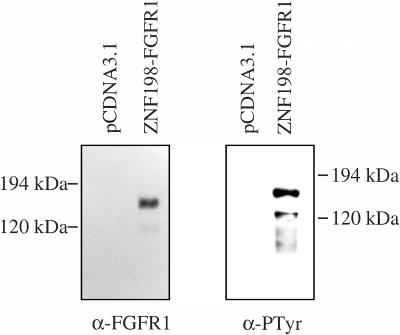
Ba/F3-ZNF198-FGFR1 cells grown in medium lacking IL-3 and Ba/F3-pcDNA3.1 cells grown with IL-3 were lysed and proteins precipitated with an anti-FGFR1 antisera. Immunoprecipitates were separated by SDS-PAGE and blotted onto nitrocellulose and then probed with either anti-FGFR1 antisera or an antiphosphotyrosine antibody (Figure 3). A tyrosine-phosphorylated product of approximately 150 kDa, the expected size of the ZNF198-FGFR1 fusion protein, was observed in the transformed Ba/F3 cells, whereas no such product was seen with either antibody in control cells transfected with vector alone. An additional tyrosine phosphorylated product of approximately 130 kDa was seen occasionally and probably corresponds to a breakdown product of ZNF198-FGFR1.
Figure 3.
ZNF198-FGFR1 is expressed and constitutively tyrosine-phosphorylated in transformed Ba/F3-ZNF198-FGFR1 cells. Ba/F3 cells were transfected with pcDNA3.1 or pcDNA3.1/ZNF198-FGFR1. All cells were selected in 1 mg/mL G418, and pcDNA3.1/ZNF198-FGFR1 cells were selected further for growth in the absence of IL-3. Proteins released from cells by lysis were immunoprecipitated with an antibody to human FGFR1, separated by SDS/PAGE and transferred to nitrocellulose. Western blotting was carried out by using the anti-FGFR1 antisera and antiphosphotyrosine antibody.
ZNF198-FGFR1 Induces Tyrosine Phosphorylation of STAT 1 and STAT 5
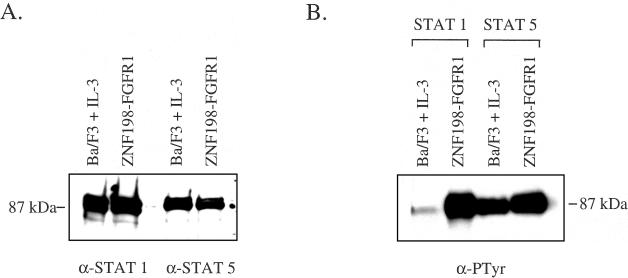
We sought to determine if STAT 1 and STAT 5 might play a role in ZNF198-FGFR1-mediated transformation of Ba/F3 cells because these proteins have been implicated in the signal transduction cascades initiated by growth factors and constitutively activated tyrosine kinases. STAT 1 and 5 were expressed at equivalent levels in wild-type parental Ba/F3 cells grown in the presence of IL-3 and ZNF198-FGFR1-transformed cells grown in the absence of IL-3 (Figure 4A). Immunoprecipitation with anti-STAT antibodies followed by anti-phosphotyrosine blotting showed that STAT 1 was weakly phosphorylated on tyrosine in parental Ba/F3 cells grown in the presence of IL-3 but strongly phosphorylated in cells transformed with ZNF198-FGFR1. STAT 5 was phosphorylated in Ba/F3 cells stimulated with IL-3 but was more strongly phosphorylated in cells transformed with ZNF198-FGFR1 (Figure 4B).
Figure 4.
ZNF198-FGFR1 induces tyrosine phosphorylation of STAT 1 and STAT 5. (A) Immunoprecipitation and Western blot of 0.5 mg total protein with STAT 1 or STAT 5 antibodies demonstrating equal levels of expression in Ba/F3 cells grown in the presence of IL-3 or transfected with ZNF198-FGFR1 and grown in the absence of IL-3. (B) Cell lysates immunoprecipitated with STAT 1 or STAT 5 antibodies and probed with antiphosphotyrosine.
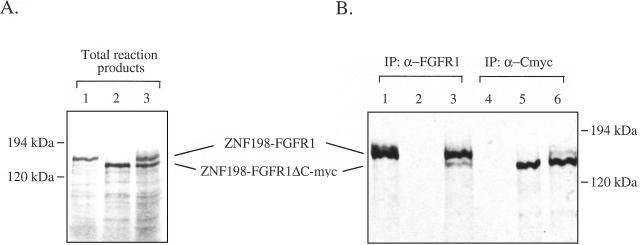
ZNF198-FGFR1 Oligomerizes In Vitro
In common with other receptor tyrosine kinases, activation of FGFR1 results from ligand-induced receptor dimerization. It is, likely, therefore that the constitutive phosphorylation of ZNF198-FGFR1 and transformation of Ba/F3 cells results from dimer/oligomerization of the ZNF198-FGFR1 fusion protein via the ZNF198 moiety. To test this hypothesis we in vitro transcribed and translated ZNF198-FGFR1 and a derivative construct, ZNF198-FGFR1ΔC-myc, in which the C-terminal epitope for the anti-FGFR1 antisera was replaced by a c-myc tag (Figure 5A). As expected, anti-FGFR1 antisera immunoprecipitated ZNF198-FGFR1 (Figure 5B, lane 1) but not ZNF198-FGFR1ΔC-myc (Figure 5B, lane 2). However when both products were translated together, both were coimmunoprecipitated by anti-FGFR1 antisera (Figure 5B, lane 3). Similarly, anti-myc antibody recognized ZNF198-FGFR1ΔC-myc but not ZNF198-FGFR1 but when both were cotranslated the anti-myc antibody immunoprecipitated both proteins (Figure 5B, lanes 4–6). Whilst these results do not prove an in vivo self-association, they do suggest that ZNF198-FGFR1 and ZNF198-FGFR1ΔC-myc interact, thereby providing evidence for dimer/oligomerization of the fusion product.
Figure 5.
ZNF198-FGFR1 oligomerises in vitro. (A) pcDNA3.1/ZNF198-FGFR1 (lane 1), pcDNA3.1/ZNF198-FGFR1ΔC-myc (lane 2), or a mixture of the two (lane 3) were in vitro transcribed and translated in the presence of [35S] methionine. (B) One half the reaction mixture was immunoprecipitated by using either an antisera to the C-terminus of human FGFR1 (lanes 1–3) or an anti-myc epitope antibody (lanes 4–6). Total reaction products or immunoprecipitates were separated by SDS/PAGE, the gel fixed, treated with Amplify (Amersham), dried, and exposed to film at -70°C.
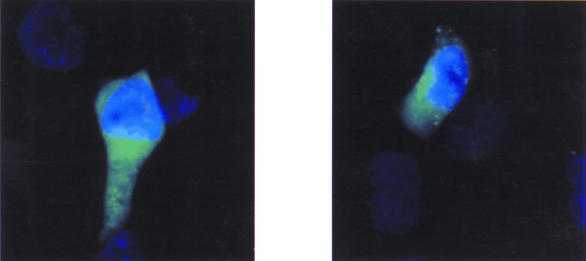
Immunolocalization of ZNF198-FGFR1
To identify the cellular location of the fusion protein, pcDNA3.1/ZNF198-FGFR1 was transfected into COS7 cells. Immunostaining of transiently transfected cells with the anti-FGFR1 antibody revealed that ZNF198-FGFR1 is located exclusively in the cytoplasm (Figure 6), unlike wild-type FGFR1 protein which is a plasma membrane protein that, at least in some cell types, shuttles transiently in the nucleus on binding of ligand [23]. Untransfected cells showed no staining.
Figure 6.
Cytoplasmic localization of ZNF198-FGFR1 COS7 cells were transiently transfected with pcDNA3.1/ZNF198-FGFR1, immunostained with anti-FGFR1, and visualized by fluorescence microscopy. Transfected cells showed diffuse cytoplasmic staining, whereas untransfected cells were unstained.
Discussion
The 8p11 myeloproliferative syndrome, most commonly associated with the t(8;13)(p11;q12), is a rare condition that has obvious clinicopathological parallels with chronic myeloid leukemia [1]. The t(9;22) in this disease fuses BCR to the ABL tyrosine kinase, and several lines of evidence, notably the induction of leukemia in mice [15], have demonstrated that BCR-ABL plays a primary role in the pathogenesis of this disease. It was of great interest, therefore, to find that the t(8;13) also disrupts a tyrosine kinase, FGFR1, by fusion to ZNF198. Here we have shown that ZNF198-FGFR1, like BCR-ABL and other related chimeric genes such as TEL-PDGFRβ, TEL-JAK2, and TEL-ABL [16,17,21,24,25], transforms the IL-3 dependent cell line Ba/F3 to factor independence. In common with these other tyrosine kinase fusion genes, ZNF198-FGFR1 self-associates, and this is presumably responsible for its constitutive tyrosine kinase activity, by mimicking the normal process of ligand-induced receptor dimerization. An obvious candidate for the self-association motif are the five ZNF198 MYM domains, zinc finger-related structures that resemble other motifs believed to mediate protein-protein interactions such as the RING, LAP/PHD, and LIM domains [2,3,26]. However it is possible that the MYM domains mediate heterotypic interactions and that the self-association motif lies elsewhere.
The normal role of FGFR1 in hemopoiesis has not been clearly defined. Current evidence suggests that FGFs are concentrated in the extracellular matrix of bone marrow stromal cells and that bFGF synergizes with SCF and GM-CSF to directly augment the growth of myeloid colony forming cells [27,28]. In contrast, bFGF appears to have no discernible effect on hemopoietic stem cells, and FGFR mRNA, including FGFR1, is undetectable by RT-PCR in cells with a primitive phenotype [28,29]. The fact that both lymphoid and myeloid cells are affected in the 8p11 myeloproliferative syndrome is indicative of a stem cell disorder, and it is likely, therefore, that fusion to ZNF198 constitutively activates FGFR1 tyrosine kinase in a cell type in which FGFR1 is not normally expressed.
The signaling pathways that are normally activated by FGFR1 have been only partly elucidated. FGFR1 activates the RAS/MAPK pathway via FRS2, a protein that is phosphorylated on tyrosine in response to FGF stimulation and subsequently forms a complex with GRB2, SOS, and SHP2 [30,31]. A total of seven tyrosine residues become autophosphorylated after binding of ligand [32] and one of these, Y766, serves as a high affinity binding site for the SH2 domain of PLCγ1 [22]. Although phosphorylation of Y766 is essential for hydrolysis of phosphatidylinositol, this function is not required for FGF-induced DNA synthesis of myeloblasts or differentiation of the neuronal cell line PC12 [33,34]. Whether these interactions are required for ZNF198-FGFR1-mediated transformation of Ba/F3 cells remains to be determined.
The role of BCR-ABL in leukemia has been studied in considerable detail. We have found that ZNF198-FGFR1, as is the case with BCR-ABL [35], is localized in the cytoplasm, and therefore these two fusion proteins may interact with a common set of substrates. However unlike BCR-ABL, ZNF198-FGFR1 does not appear to specifically associate with actin filaments or focal adhesion complexes. Signaling by BCR-ABL is highly complex and involves the phosphorylation and/or recruitment of more than 20 proteins, with concomitant activation of the RAS, JNK/SAPK, PI3 kinase and JAK/STAT signaling pathways (reviewed in [36]). STAT 5 is constitutively phosphorylated in BCR-ABL-transformed cells [37], and the finding that this protein is also phosphorylated in Ba/F3 cells transfected with ZNF198-FGFR1 or TEL-JAK2 [17] shows that activation of STAT 5-mediated pathways are a common feature of deregulated tyrosine kinases in leukemia. STAT 5 is known to play a role in the response of hematopoietic cells to diverse cytokines, which suggests that transformation by tyrosine kinase fusion proteins is mediated, at least in part, by activation or subversion of pathways employed by normal growth factors. However we found that STAT 1 is also highly phosphorylated on tyrosine in ZNF198-FGFR1-transformed Ba/F3 cells, indicating that this fusion gene employs distinct signaling pathways. To our knowledge, these are the first data to implicate STAT proteins in FGFR1 signal transduction. STAT 1 is also activated by FGFR3 mutants associated with achondroplasia [38], which suggests that STAT activation may be a common feature of FGFR-mediated signaling.
Acknowledgements
This work was supported by the Kay Kendall Leukemia Fund, the Leukemia Research Fund, and The Medical Research Council.
References
- 1.Macdonald D, Aguiar RCT, Mason PJ, Goldman JM, Cross NCP. A new myeloproliferative disorder associated with chromosomal translocations involving 8p11: a review. Leukemia. 1995;9:1628–1630. [PubMed] [Google Scholar]
- 2.Reiter A, Sohal J, Kulkarni S, Chase A, Macdonald DHC, Aguiar RCT, Goncalves C, Hernandez JM, Jennings BA, Goldman JM, Cross NCP. Consistent fusion of ZNF198 to the fibroblast growth factor receptor-1 in the t(8;13)(p11;q12) myeloproliferative syndrome. Blood. 1998;92:1735–1742. [PubMed] [Google Scholar]
- 3.Smedley D, Hamoudi R, Clark J, Warren W, Abdul Rauf M, Somers G, Venter D, Fagan K, Cooper C, Shipley J. The t(8;13)-(p11;q11-12) rearrangement associated with an atypical myeloproliferative disorder fuses the fibroblast growth factor receptor 1 gene to a novel gene RAMP. Hum Mol Genet. 1998;7:637–642. doi: 10.1093/hmg/7.4.637. [DOI] [PubMed] [Google Scholar]
- 4.Popovici C, Adelaide J, Ollendorff V, Chaffanet M, Guasch G, Jacrot M, Leroux D, Birnbaum D, Pebusque MJ. Fibroblast growth factor receptor 1 is fused to FIM in stem-cell myeloproliferative disorder with t(8;13) Proc Natl Acad Sci USA. 1998;95:5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 6.Chaffanet M, Popovici C, Leroux D, Jacrot M, Adelaide J, Dastugue N, Gregoire MJ, Hagemeijer A, Lafage Pochitaloff M, Birnbaum D, Pebusque MJ. t(6;8), t(8;9) and t(8;13) translocations associated with stem cell myeloproliferative disorders have close or identical breakpoints in chromosome region 8p11-12. Oncogene. 1998;16:945–949. doi: 10.1038/sj.onc.1201601. [DOI] [PubMed] [Google Scholar]
- 7.Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph′ translocation. Nature. 1985;315:758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- 8.Shtivelman E, Lifshit B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 9.Andreasson P, Johansson B, Carlsson M, Jarlsfelt I, Fioretos T, Mitelman F, Hoglund M. BCR/ABL negative chronic myeloid leukemia with ETV6/ABL fusion. Genes Chromosomes Cancer. 1997;20:299–304. [PubMed] [Google Scholar]
- 10.Papadopoulos P, Ridge SA, Boucher CA, Stocking C, Wiedemann LM. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. [PubMed] [Google Scholar]
- 11.Peeters P, Raynaud SD, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 12.Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 13.Ross TS, Bernard OA, Berger R, Gilliland DG. Fusion of Huntington interacting protein 1 to platelet-derived growth factor beta receptor (PDGFbetaR) in chronic myelomonocytic leukemia with t(5;7)(q33;q11.2) Blood. 1998;91:4419–4426. [PubMed] [Google Scholar]
- 14.McLaughlin J, Chianese E, Witte ON. In vitro transformation of immature hematopoietic cells by the P210 BCR/ABL oncogene product of the Philadelphia chromosome. Proc Natl Acad Sci USA. 1987;84:6558–6562. doi: 10.1073/pnas.84.18.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 16.Carroll M, Tomasson MH, Barker GF, Golub TR, Gilliland DG. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc Natl Acad Sci USA. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwaller J, Frantsve J, Aster J, Williams IR, Tomasson MH, Ross TS, Peeters P, Van Rompaey L, Van Etten RA, Ilaria R, Marynen P, Gilliland DG. Transformation of hematopoietic cell lines to growth factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 genes. EMBO J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salgia R, Sattler M, Pisick E, Li JL, Griffin JD. p210BCR/ABL induces formation of complexes containing focal adhesion proteins and the protooncogene product p120c-Cbl. Exp Hematol. 1996;24:310–313. [PubMed] [Google Scholar]
- 19.Aster JC, Robertson ES, Hasserjian RP, Turner JR, Kieff E, Sklar J. Oncogenic forms of NOTCH 1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Mohammadi M, Rodrigues GA, Schlessinger J. Reduced activation of RAF-1 and MAP kinase by a fibroblast growth factor receptor mutant deficient in stimulation of phosphatidylinositol hydrolysis. J Biol Chem. 1995;270:5065–5072. doi: 10.1074/jbc.270.10.5065. [DOI] [PubMed] [Google Scholar]
- 21.Hannemann JR, McManus DM, Kabarowski JHS, Wiedemann LM. Haemopoietic transformation by the TEL/ABL oncogene. Br J Haematol. 1998;102:475–485. doi: 10.1046/j.1365-2141.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi M, Honegger AM, Rotin D, Fischer R, Bellot F, Li W, Dionne CA, Jaye M, Rubinstein M, Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Fig) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stachowiak EK, Maher PA, Tucholski J, Mordechai E, Joy A, Moffett J, Coons S, Stachowiak MK. Growth factor regulation of cell growth and proliferation in the nervous system. A new intracrine nuclear mechanism. Oncogene. 1997;14:2201–2211. doi: 10.1038/sj.onc.1201057. [DOI] [PubMed] [Google Scholar]
- 24.McWhirter JR, Wang JY. An actin-binding function contributes to transformation by the Bcr-Abl oncoprotein of Philadelphia chromosome-positive human leukemias. EMBO J. 1993;12:1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 26.Mackay JP, Crossley M. Zinc fingers are sticking together. Trends in Biochemical Sciences. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 27.Gabrilove JL, White K, Rahman Z, Wilson EL. Stem cell factor and basic fibroblast growth factor are synergistic in augmenting committed myeloid progenitor cell growth. Blood. 1994;83:907–910. [PubMed] [Google Scholar]
- 28.Berardi AC, Wang A, Abraham J, Scadden DT. Basic fibroblast growth factor mediates its effects on committed myeloid progenitors by direct action and has no effect on hematopoietic stem cells. Blood. 1995;86:2123–2129. [PubMed] [Google Scholar]
- 29.Ratajczak MZ, Ratajczak J, Skorska M, Marlicz W, Calabretta B, Pletcher CH, Moore J, Gewirtz AM. Effect of basic (FGF-2) and acidic (FGF-1) fibroblast growth factors on early haemopoietic cell development. Br J Haematol. 1996;93:772–782. doi: 10.1046/j.1365-2141.1996.d01-1736.x. [DOI] [PubMed] [Google Scholar]
- 30.Kouhara H, Hadari YR, Spivak Kroizman T, Schilling J, Bar Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 31.Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammadi M, Dionne CA, Li W, Li N, Spivak T, Honegger AM, Jaye M, Schlessinger J. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature. 1992;358:681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- 34.Peters KG, Marie J, Wilson E, Ives HE, Escobedo J, Del Rosario M, Mirda D, Williams LT. Point mutation of an FGF receptor abolishes phosphatidylinositol turnover and Ca2+ flux but not mitogenesis. Nature. 1992;358:678–681. doi: 10.1038/358678a0. [DOI] [PubMed] [Google Scholar]
- 35.Wetzler M, Talpaz M, Van Etten RA, Hirsh Ginsberg C, Beran M, Kurzrock R. Subcellular localization of Bcr, Abl, and Bcr-Abl proteins in normal and leukemic cells and correlation of expression with myeloid differentiation. J Clin Invest. 1993;92:1925–1939. doi: 10.1172/JCI116786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyers CL. Signal transduction pathways involved in BCR-ABL transformation. Baillieres Clin Haematol. 1997;10:223–231. doi: 10.1016/s0950-3536(97)80004-2. [DOI] [PubMed] [Google Scholar]
- 37.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers CL. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 38.Su WC, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton WA, Fu XY. Activation of Stati by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]