Abstract
Domestic, low-level exposure to radon gas is considered a major environmental lung-cancer hazard involving DNA damage to bronchial cells by α particles from radon progeny. At domestic exposure levels, the relevant bronchial cells are very rarely traversed by more than one α particle, whereas at higher radon levels—at which epidemiological studies in uranium miners allow lung-cancer risks to be quantified with reasonable precision—these bronchial cells are frequently exposed to multiple α-particle traversals. Measuring the oncogenic transforming effects of exactly one α particle without the confounding effects of multiple traversals has hitherto been unfeasible, resulting in uncertainty in extrapolations of risk from high to domestic radon levels. A technique to assess the effects of single α particles uses a charged-particle microbeam, which irradiates individual cells or cell nuclei with predefined exact numbers of particles. Although previously too slow to assess the relevant small oncogenic risks, recent improvements in throughput now permit microbeam irradiation of large cell numbers, allowing the first oncogenic risk measurements for the traversal of exactly one α particle through a cell nucleus. Given positive controls to ensure that the dosimetry and biological controls were comparable, the measured oncogenicity from exactly one α particle was significantly lower than for a Poisson-distributed mean of one α particle, implying that cells traversed by multiple α particles contribute most of the risk. If this result applies generally, extrapolation from high-level radon risks (involving cellular traversal by multiple α particles) may overestimate low-level (involving only single α particles) radon risks.
Domestic exposure to radon gas in homes generally is considered to be the single largest naturally occurring environmental hazard (1). The basic mechanism involves DNA damage to bronchioepithelial cells by α particles emitted by radon progeny. The most recent report (1) from the U.S. National Academy of Sciences on the health effects of environmental exposure to radon gas (BEIR VI) estimated that 10–14% of all lung cancer deaths—amounting to central estimates of about 15,400–21,800 per year in the United States—are linked to radon gas exposure from the environment.
The BEIR VI (1) estimates (and others) of the risks of domestic radon exposure were made by extrapolating risks from underground miners who received radon exposures that were, on average, many times larger than those of people in most homes. The problem inherent in this extrapolation is that, at these high exposures, the cells at risk in the bronchial epithelium of miners may be traversed by several α particles during a short period, whereas for individuals exposed in homes at normal domestic radon levels, it is unlikely that any cell at risk will be traversed by more than one α particle in a lifetime (1).
Even in the laboratory, there has until now been no direct way of measuring the oncogenic transforming effects of exactly one α particle without the confounding effects of a significant fraction of exposed cells being subject to multiple α-particle traversals, and this has led to significant uncertainty in low-dose radon risk estimates (2).
In recent years, several groups have developed charged-particle microbeams, in which cells on a dish are individually irradiated by a predefined exact number of α particles, thus allowing the effects of exactly one (or more) α particle traversals to be assessed (3–8). However, earlier microbeam irradiation systems have been too slow to measure oncogenic transformation frequencies, because the low probabilities involved require that many cells (≈105) be individually irradiated. Specifically, the current overall irradiation throughput for the microbeam experiments described here is ≈3,000 cells per hr, which may be compared with earlier microbeam system throughputs of ≈120 cells per hr (6). In practice, the earlier low throughput precluded the irradiation of the ≈105 cells necessary for measurements of oncogenic transformation frequencies. This increased microbeam throughput, made possible by developments in both hardware and software, now permits sufficient numbers of cells to be irradiated, and we report here the direct measurement of the oncogenic risk of exactly one α particle.
The goal of this study was to investigate the oncogenic effects produced by exactly one α particle traversing cell nuclei. Specifically, we have investigated whether the oncogenic effects of exactly one α particle are similar to the effects of a (Poisson) mean of one α particle, with the latter’s attendant proportion of cells traversed by more than one α particle. Because it is much less likely that there would be a difference between the effects of, say, exactly four and a Poisson-mean of four α particles, we have made such comparisons to act as positive controls. Equality in these positive controls for multiple α-particle traversals would imply that any differences seen between the effects of exactly one α particle and a Poisson-mean of one α particle are not artifacts of any differences in irradiation conditions between the microbeam and the broad-beam systems—although we have striven to make the irradiation conditions as similar as possible.
The layout and irradiation procedure of the Columbia University microbeam have been described in detail elsewhere (3, 8). Briefly, each cell attached in a monolayer to the thin polypropylene base of a cell culture dish is identified and located by using an image analysis system, and its coordinates are stored in a computer; the cell dish is then moved under computer control such that the centroid of each cell nucleus in the dish is in turn positioned over a highly collimated shuttered beam of α particles. The nucleus of each cell is exposed to a predetermined exact number of α particles having an energy that simulates the emission from radon progeny, and a particle detector above the cell signals to close the accelerator shutter when the desired number of particles (e.g., 1) are recorded, after which the next cell is moved under the beam.
In parallel with these microbeam studies, we have conducted “broad beam” α-particle exposures by using different fluences of α particles such that cell nuclei were traversed by mean numbers of 0, 1, 2, 4, 6, and 8 particles. In this case, Poisson statistics determine the probability that any given cell nucleus is traversed by a given number of α particles. For example, if the fluence is chosen such that the mean number of particles traversing nuclei is 1, then 37% are not traversed at all, 37% are traversed once, and the remaining 26% are traversed by 2 or more α particles. Groups of cell nuclei traversed by a given exact number of α particles, and by the same (Poisson-distributed) mean number of α particles can be considered to have received the same average dose.
C3H10T½ cells were used in this study, which can be quantitatively assayed for in vitro oncogenic transformation (9). Transformed cells can be identified by their altered morphology, and extensive prior studies have shown that such transformed cells produce tumors when injected into animals (10).
METHODS AND MATERIALS
Microbeam Irradiations.
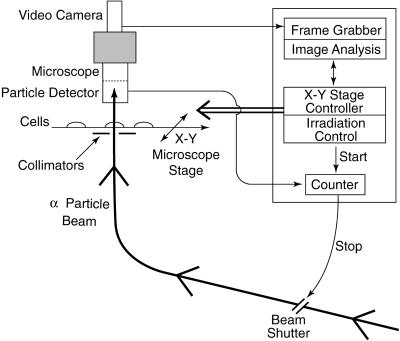
A schematic of the microbeam system is shown in Fig. 1. α particles accelerated by a 4-MV Van de Graaff accelerator to an energy of 5.3 MeV (1 eV = 1.602 × 10−19 J; stopping power 90 keV/μm) were used for the microbeam irradiations. As described below, the cells to be irradiated were attached to the bottom of thin-bottomed plastic dishes and left for sufficient time in exponential growth to ensure that they were completely asynchronous. Individual nuclei (including mitotic cell nuclei) were identified and located with an optical image analysis system, which detects the fluorescent staining pattern with 366-nm UV light. For each dish, a computer/microscope-based image analysis system first automatically locates the positions of all the cell nuclei on the dish (Fig. 2). Next, the dish is moved under computer control such that the first cell nucleus is positioned over a highly collimated α-particle beam. The beam shutter is opened until the required number of α particles are detected (with a solid state detector located above the cell) to have passed though the nucleus. The shutter is then closed, and the next cell is moved under the beam. The overall spatial precision of the beam, including positioning and beam spread, was about ±3.5 μm. By using Monte-Carlo simulations, we estimated—based on the measured morphometric characteristics of exponentially growing C3H10T½ cells (11)—that the particle beam would miss the targeted nucleus at a rate of <0.3%. Because of the lag time of shutter closure, about 0.2% of irradiated nuclei would have received 1 more particle than expected.
Figure 1.
Schematic of microbeam system.
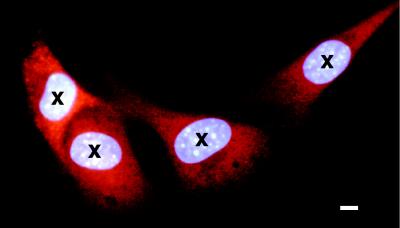
Figure 2.
Detail of C3H10T½ cells attached to the polypropylene surface of the mini-well, as detected by the automated microbeam image analysis system. The cells were stained with a low concentrations of Hoechst 33342 fluorescent dye as described in the text, which is preferentially taken up by the cell nucleus. The cell dish is moved under computer control such that the centroid of every cell nucleus as determined by the image analysis system (marked by the image analysis system as crosses) is sequentially situated under the collimated microbeam for irradiation with a predetermined number of α particles. For illustrative purposes, the cells shown here also were stained with orange fluorescent probe (CellTracker orange CMTMR, Molecular Probes), which is preferentially taken up in the cytoplasm and is used when microbeam irradiation of only the cytoplasm is required. The bar illustrates the overall spatial precision of the α particle microbeam of ±3.5 μm. (Bar = 7 μm.)
Including all handling times, the average time to locate and irradiate a single cell nucleus in the microbeam is typically around 1 sec. Overall, each dish was out of the incubator for a total of about 10–15 min for irradiation, after which the cells were replated as described below for subsequent incubation and assessment of oncogenic transformation rates. In these microbeam studies, a total of more than 260,000 cells were individually imaged, positioned, and irradiated, corresponding (after accounting for cell killing and plating efficiency) to around 110,000 viable cells.
Broad-Beam Irradiations.
To ensure compatibility with the microbeam irradiations, cells were stained with the same low concentration of Hoechst dye as described above and exposed to the same low fluence of 366-nm photons. The same energy α particles (5.3 MeV) as was used for the microbeam irradiations were directed vertically through the dishes. The cell dishes were mounted in a specimen wheel that was rotated horizontally under computer control, allowing each dish to pass over a slot-shaped α-particle beam at a rate determined by the required fluence and the instantaneous fluence rate of the beam. Based on the morphometry of the target-cell nuclei (11), the broad-beam irradiations were such that an average of 1, 2, 4, 6, or 8 α particles passed through each nucleus. In these broad-beam studies, a total of >500,000 viable cells, including sham-irradiated controls, were analyzed.
Cell Culture.
Before irradiation, C3H10T½ mouse fibroblast cells from passages 8–14 were grown in Eagle’s basal medium supplemented with 10% heat-inactivated fetal bovine serum with added iron and 25 μg/ml gentamycin at 37.5°C in a 95% air/5% CO2 atmosphere.
For the microbeam exposures, 300–800 exponentially growing C3H10T½ cells were plated into each of a series of 6.3-mm diameter mini-wells 24 hr before exposure. Each mini-well consists of a 6.3-mm hole in the center of a standard 60-mm cell culture dish, covered on the bottom with a thin (3.8 μm) polypropylene film. The DNA of attached cells was stained with an extremely low concentration (50 nM) of Hoechst 33342 dye for 30 min, enabling individual nuclei to be identified and located with the enhanced optical image analysis system, which detects the fluorescent staining pattern with 366-nm light. To enhance cell attachment and spreading, the polypropylene film was coated with Cell-Tak (Becton Dickinson), an adhesive protein.
For the broad-beam exposures, 44 hr before radiation exposure, exponentially growing C3H10T½ cells were plated at a density of 200,000 cells per dish onto thin-bottomed (6-μm mylar) 35-mm diameter stainless steel dishes. To ensure compatibility with the microbeam irradiations, cells were stained with the same low concentration of Hoechst dye as described above and exposed to the same low fluence of 366-nm photons. It is very unlikely that the difference in preirradiation growth times (24 vs. 44 hr) would be significant, because adequate time in either case is available for the exponentially growing cells to become completely asynchronous.
Irradiation times for each dish (which was effectively the time that the cells were in a pH-controlled environment) were 10–15 min both for the microbeam and the broad-beam irradiations, after which the treatment protocols for both types of irradiations were identical. Specifically, the cells were trypsinized from the irradiation container (recovery rates both from the microbeam and the broad-beam irradiation dishes were ≈70%) and replated into 100-mm diameter cell culture dishes. To assess radiation-induced oncogenic transformation, the replated cell densities after both microbeam and broad-beam irradiations were such that 200–400 viable cells per dish would survive the cell plating and radiation exposure. These low cell numbers per dish were chosen to avoid artifacts that have been reported at higher cell densities (12). Cells were incubated for 7 wk with fresh culture medium every 12 days before being fixed and stained to identify morphologically transformed types II and III foci as described elsewhere (9).
In parallel studies, dishes were plated with about 30 viable cells that had been subject to exactly the same conditions both as the microbeam- and the broad-beam-irradiated cells and incubated for 2 wk, after which the cells were stained to determine plating efficiencies and surviving fractions of control vs. irradiated cells.
RESULTS AND DISCUSSION
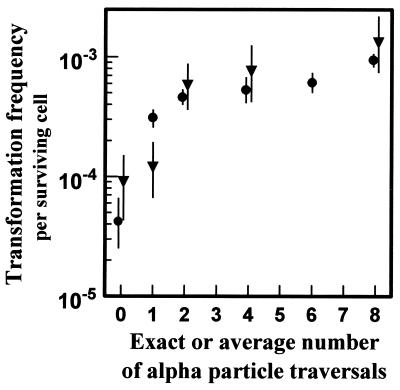
Table 1 and Fig. 3 (with detail in Fig. 4) shows the measured transformation incidence per surviving cell as a function of the exact number of α particles traversing the cell nucleus (microbeam irradiation). Results from the corresponding broad-beam irradiations also are shown where graded doses of α particles were used (calculated to yield the same mean number of α-particle traversals), but with a nucleus-to-nucleus Poisson distribution in the actual number of α-particle traversals. Control rates for the microbeam and broad-beam irradiations were not significantly different (P = 0.28, using a likelihood ratio test). In addition, as shown in Fig. 3, the data from the microbeam and the broad-beam irradiations coincide for 2, 4, and 8 α particles, indicating that the dosimetry and the biological protocols for the two irradiation systems are compatible.
Table 1.
Clonogenic survival rates, numbers of viable cells exposed in transformation studies, number of transformed clones produced, and transformation frequencies for microbeam (exact number of α particles) and broad-beam (Poisson-distributed number of α particles) irradiations
| Irradiation | Exact or mean no. of α particles | Clonogenic surviving fraction (plating efficiency) | Number* of viable cells exposed/104 | Number of transformants produced | Transformation frequency/ 104 surviving cells |
|---|---|---|---|---|---|
| Microbeam | 0 (control) | 0.60 | 4.62 | 4 | 0.86 |
| 1 | 0.83 | 4.27 | 5 | 1.2 | |
| 2 | 0.64 | 1.22 | 7 | 5.8 | |
| 4 | 0.41 | 0.66 | 5 | 7.6 | |
| 8 | 0.16 | 0.38 | 5 | 13.2 | |
| Broad-beam | 0 (control) | 0.33 | 14.37 | 6 | 0.42 |
| 1 | 0.85 | 12.42 | 38 | 3.1 | |
| 2 | 0.77 | 11.06 | 51 | 4.6 | |
| 4 | 0.46 | 3.76 | 20 | 5.3 | |
| 6 | 0.28 | 5.06 | 31 | 6.1 | |
| 8 | 0.18 | 7.05 | 66 | 9.4 |
Estimated, accounting for plating efficiency and clonogenic survival.
Figure 3.
Yield per surviving C3H10T½ cell of oncogenically transformed cells produced by nuclear traversals by 5.3-MeV α particles. ▾ represents exposure of cell nuclei to exact numbers of α-particle traversals by using the microbeam system. • represents exposure to a Poisson-distributed number of α-particles traversals with a given average traversal number. Standard errors (±1 SD) were estimated by assuming an underlying Poisson distributed number of transformed cells (13).
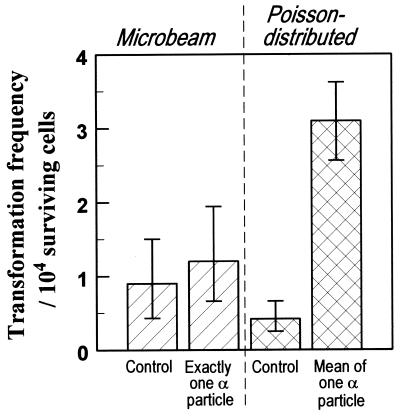
Figure 4.
Yield per surviving C3H10T½ cell of oncogenically transformed cells produced by nuclear traversals by exactly one (microbeam irradiation) or a Poisson mean of one (broad-beam irradiation) 5.3-MeV α particle. Standard errors (±1 SD) were estimated assuming an underlying Poisson distributed number of transformed cells (13).
Having established with these positive control results that the dosimetry and biological protocols for the microbeam and the broad-beam irradiations are compatible, we show in Fig. 4 a comparison of the oncogenic effects of exactly one α particle and a Poisson distribution of α particles with a mean of one (in which 26% of the cells will be traversed by more than one α particle). Cells whose nuclei were all traversed by exactly one α particle show significantly lower oncogenic risks (P = 0.02 using a likelihood ratio test) than cells whose nuclei were traversed by a mean (from a Poisson distribution) of one α particle. Nuclear traversal by exactly one α particle is not only significantly less effective than traversal by an average of one α particle but also is not significantly more effective than a zero-dose (sham) irradiation.
If single α-particle traversals produce either zero or only a small risk of oncogenic transformation in this system, the majority of the yield of transformed cells obtained after traversal by a (Poisson) mean of one α particle must come from the minority of cells whose nuclei were subject to multiple α-particle traversals.
If it were generally the case that traversal of cell nuclei by a single α particle does not produce significant oncogenic risks, this result would have important implications for the standard method of assessing the risks of exposure to low levels of radon in homes. Because these low-exposure risks cannot be epidemiologically assessed with sufficient precision, they have generally been quantitatively established by extrapolating from high-exposure radon risks assessed by epidemiological studies of uranium miners (1). If traversal by a single α particle does not significantly raise the risk of oncogenic transformation, a linear extrapolation to low exposures of risks that were estimated in situations in which bronchial cells can receive multiple α-particle traversals could significantly overestimate the risk at low (i.e., domestic) radon exposure levels where essentially no target cell is traversed by more than a single α particle.
Finally, it is important to note that these initial results have, of necessity, been generated with the C3H10T½ mouse fibroblast model system, because no quantitative oncogenic transformation systems involving normal human cells have yet been developed. Consequently, caution should be used in applying the conclusions to radon risk estimation in humans. On the other hand, this quantitative model system has, in the past, been quite reliable in predicting trends relevant to the radon problem in humans (14, 15).
Acknowledgments
The Columbia University Radiological Research Accelerator Facility (RARAF) is a National Institutes of Health Supported Resource Center. This work was supported by Grants P41 RR-11623 (NCRR) and CA-37967 (NCI) from the National Institutes of Health.
References
- 1.National Research Council. Health Effects of Exposure to Radon: BEIR VI. Washington, DC: Natl. Acad. Press; 1998. [PubMed] [Google Scholar]
- 2.Brenner D J. In: Low Dose Radiation: Biological Bases of Risk Assessment. Baverstock K F, Stather J W, editors. London: Taylor & Francis; 1989. pp. 477–480. [Google Scholar]
- 3.Geard C R, Brenner D J, Randers-Pehrson G, Marino S A. Nucl Instrum Meth Phys Res B. 1991;54:411–416. [Google Scholar]
- 4.Michael B D, Folkard M, Prise K M. Int J Radiat Biol. 1994;65:503–508. doi: 10.1080/09553009414550581. [DOI] [PubMed] [Google Scholar]
- 5.Folkard M, Vojnovic B, Prise K M, Bowey A G, Locke R J, Schettino G, Michael B D. Int J Radiat Biol. 1997;72:375–385. doi: 10.1080/095530097143158. [DOI] [PubMed] [Google Scholar]
- 6.Nelson J M, Brooks A L, Metting N F, Khan M A, Buschbom R L, Duncan A, Miick R, Braby L A. Radiat Res. 1996;145:568–574. [PubMed] [Google Scholar]
- 7.Cholewa M, Saint A, Legge G J F, Kamiya T. Nucl Instrum Meth Phys Res B. 1997;130:275–279. [Google Scholar]
- 8.Hei T K, Wu L J, Liu S X, Vannais D, Waldren C A, Randers-Pehrson G. Proc Natl Acad Sci USA. 1997;94:3765–3770. doi: 10.1073/pnas.94.8.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reznikoff C A, Bertram J S, Brankow D W, Heidelberger C. Cancer Res. 1973;33:3239–3249. [PubMed] [Google Scholar]
- 10.Kennedy A R, Fox M, Murphy G, Little J B. Proc Natl Acad Sci USA. 1980;77:7262–7266. doi: 10.1073/pnas.77.12.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geard C R, Randers-Pehrson G, Hei T K, Jenkins G J, Miller R C, Wu L J, Brenner D J, Hall E J. In: Microdosimetry, an Interdisciplinary Approach. Goodhead D T, O’Neill P, Menzel H G, editors. Cambridge, U.K.: R. Soc. Chem.; 1997. pp. 327–330. [Google Scholar]
- 12.Terzaghi M, Little J B. Cancer Res. 1976;36:1367–1374. [PubMed] [Google Scholar]
- 13.Brenner D J, Quan H. Int J Radiat Biol. 1990;57:1031–1046. doi: 10.1080/09553009014551141. [DOI] [PubMed] [Google Scholar]
- 14.Brenner D J, Miller R C, Huang Y, Hall E J. Radiat Res. 1995;142:61–69. [PubMed] [Google Scholar]
- 15.Brenner D J. Health Phys. 1994;67:76–79. doi: 10.1097/00004032-199407000-00010. [DOI] [PubMed] [Google Scholar]