Abstract
Deletion of any one of three subunits of the yeast Mediator of transcriptional regulation, Med2, Pgd1 (Hrs1), and Sin4, abolished activation by Gal4–VP16 in vitro. By contrast, other Mediator functions, stimulation of basal transcription and of TFIIH kinase activity, were unaffected. A different but overlapping Mediator subunit dependence was found for activation by Gcn4. The genetic requirements for activation in vivo were closely coincident with those in vitro. A whole genome expression profile of a Δmed2 strain showed diminished transcription of a subset of inducible genes but only minor effects on “basal” transcription. These findings make an important connection between transcriptional activation in vitro and in vivo, and identify Mediator as a “global” transcriptional coactivator.
Mediator was discovered as an activity in a crude yeast fraction able to relieve activator inhibition (1) and required for an activator response in a partially reconstituted RNA polymerase II transcription system (2). Mediator was initially resolved to homogeneity (3) by displacement from a complex with polymerase II (“holoenzyme”), and proved to contain the products of three groups of genes: SRBs, recovered from a genetic screen for CTD-interacting proteins (4, 5); the SIN4/RGR1 group, whose founding members were obtained from screens for mutations affecting repression (5, 6); and the MED genes, not previously identified in any screen (7, 8). Functional analysis of purified Mediator in a transcription system reconstituted from essentially homogeneous proteins revealed three biochemical activities, stimulation of basal transcription, support of activated transcription, and stimulation of CTD phosphorylation by TFIIH (3, 8). Activated transcription occurred in the absence of TATA boxing-binding protein associated factors (TAFs), consistent with the lack of a TAF requirement for regulation of most yeast promoters in vivo (9–10). The outstanding question regarding Mediator has been whether it too might prove to be dispensable for regulation in vivo, or whether it plays a general role in activated transcription in vivo, in keeping with the biochemical results.
Work done to date has begun to address the physiologic relevance of Mediator and the relationship between its functions in vivo and in vitro. Cells harboring a temperature-sensitive mutation in SRB4 ceased transcription of all promoters analyzed at the restrictive temperature, indicating a widespread requirement for Mediator, though not distinguishing between roles in basal and activated transcription (11). A temperature-sensitive mutation in MED6 was shown to diminish activation by Gal4 in vivo and by VP16 in vitro, but because two different activators were used, the effects could not be correlated (7). Finally, CTD truncation has been shown to impair activation in vivo (12) and in vitro (8); the CTD interacts with Mediator in vitro (8, 13), enabling a correlation, but only an indirect one.
Here we use multiple activators and Mediator mutants in a combined biochemical and genetic analysis. The results define a consistent pattern of structure–function relationships, establish the fidelity of transcription control in the yeast system in vitro, and identify Mediator as an important conduit of regulatory information from enhancers to RNA polymerase II promoters in vivo.
MATERIALS AND METHODS
Protein Purification.
Approximately 400 g of cells from yeast strains BJ926 (Mata/Matα trp1/TRP1 Prc1–126/Prc1–126 pep4–3/pep4–3 prp1–1122/prp1–1122 can1/can1), MG107 (MATa ade2–1 can 1–100 his 3–11 15 leu 2–3 112 trp 1–1 ura 3–1 med2Δ1∷TRP1), SSAB-4A (MATa ura 3 ade2 his 3 leu 2-k 112 hrs1Δ∷LEU2), or DY1707 (MATα Δsin4∷URA3 ade2–1 can1–100 his3–11, 15 leu 2–3, 112 trp 1–1) were used to purify wild-type, Δmed2, Δpgd1, or Δsin4 RNA polymerase II holoenzymes, respectively. The wild-type and mutant holoenzymes were purified through the Mono-Q fractionation step as described (14).
Biochemical Assays.
The stimulation of basal transcription was measured in the system reconstituted with purified yeast proteins (8) by comparing a transcription reaction containing only core polymerase to a reaction in which 25% of the polymerase activity (measured in nonspecific assays) was supplied by holoenzyme. For measurement of activated transcription, purified Gal4–VP16 (2.5 ng) or Gcn4 (10 ng) was added to reactions containing two DNA templates (3) and either core polymerase or a mixture of core polymerase and holoenzyme as described above. The stimulation of TFIIH kinase activity was measured by comparing phosphorylation of core polymerase and holoenzyme, in amounts based on polymerase activity in nonspecific assays.
Assays of β-Galactosidase Activity in Vivo.
Appropriate yeast strains were transformed with the pLGSD5 GAL-lacZ reporter (2 μm, URA3) and GAL4 fusion effector plasmids (ARS-CEN, LEU2) by using the lithium acetate procedure (15). Cells were grown in synthetic complete-Ura-Leu medium (16), and β-galactosidase assays were carried out by permeabilizing whole cells with chloroform and SDS (17).
Northern Analysis of Gene Induction.
Total RNA from wild-type MG106 (MATa ade2–1 can 1–100 his 3–11 15 leu 2–3 112 trp 1–1 ura 3–1) and Δmed2 MG107 mutant strain was prepared by hot phenol extraction (18). RNA samples (7 mg) were subjected to electrophoresis in 1% agarose-Mops-formaldehyde gels and blotted onto nylon membranes (Hybond-N, Amersham) as described (19). Prehybridization and hybridization were performed in 0.25 M sodium phosphate (pH 7.0), 1 mM EDTA, 7% SDS, and 1% BSA at 65°C. The membranes were washed twice with 2× SSC plus 0.1% SDS for 15 min followed by a wash in 0.5× SSC plus 0.1% SDS for 15 min. DNA probes for the genes of interest were generated by PCR using the following synthetic oligonucleotides: GAL1, 5′-dGGCCGGCCATGGTCGTCAACACTAAAGCCCTG-3′ and 5′-dCCGGCCGGATCCTCCTTCTGTGTCGGACTGGT-3′; DED1, 5′-dGGCCGGCCATGGCCAAATGTTGGATATCAGCGG-3′ and 5′- dCCCCGAGGATCCAAATTTCC-3′; HIS4, 5′-dTGCCTTCTTGAACAACGGAG-3′ and 5′-dTCTAACAATGCAGAGTCGTTG-3′; and ACT1, 5′-dATGGATTCTGAGGTTGCTGC-3′ and 5′-dTTAGAAACACTTGTGGTGAA-3′. The probes were labeled by random priming (United States Biochemical). Results were analyzed on a PhosphoImager (Molecular Dynamics).
DNA Microarray Analysis of Gene Expression.
For gene expression analysis under galactose-inducing conditions, two total RNA samples were prepared from wild-type MG106 and Δmed2 MG107 mutant strains as described above for RNA blot analysis. Poly(A)+ mRNA was prepared from the total RNA by using an Oligotex mRNA Kit (Qiagen) according to the manufacturer’s protocols. The two mRNA samples were labeled individually, cohybridized to a single yeast whole genome microarray, and analyzed as described (20). For gene expression analysis under heat shock induction conditions, the wild-type MG106 and Δmed2 MG107 mutant strains were grown to OD600 = 0.6 at 23°C in yeast extract/peptone/dextrose media, warmed quickly to 37°C in a 42°C water bath, grown an additional 20 min at 37°C, and harvested. The two heat shock mRNA samples were prepared and analyzed by using a microarray as described above. Complete data sets and array images for both the galactose and heat shock experiments are available on the Internet at http://cmgm.stanford.edu/pbrown/med2.
RESULTS
Mediator Protein Mutations Specific for Activated Transcription in Vitro.
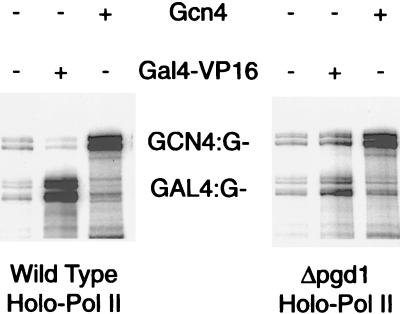
We sought to identify Mediator mutations impairing one or more of the three activities measured in vitro. Deletions of the nonessential MED2, PGD1, and SIN4 genes proved effective in this regard. RNA polymerase II holoenzymes isolated from the three mutant strains were unresponsive to the activator Gal4–VP16 in transcription reconstituted with essentially pure transcription proteins (Table 1, Fig. 1). By contrast, stimulation of basal transcription and of TFIIH kinase activity remained within a factor of 2–3 of wild-type levels. Evidently the role of Mediator in transcriptional activation is distinct from those in basal transcription and CTD phosphorylation.
Table 1.
Functional analysis of wild-type and mutant Mediators in the purified yeast transcription system
| Core Pol II | Wild-type holoenzyme | Δmed2 holoenzyme | Δpgd1 holoenzyme | Δsin4 holoenzyme | |
|---|---|---|---|---|---|
| Activation by VP16 (fold) | 1.7 | 31 | 1.7 | 1.8 | 1.1 |
| Activation by Gcn4 (fold) | 1.3 | 8.2 | 6.4 | 6.9 | 1.1 |
| Stimulation of basal transcription (fold) | — | 18 | 6.9 | 6.6 | 9.2 |
| Stimulation of TFIIH CTD-kinase activity (fold) | — | 31 | 17 | 9 | 29 |
Fold activation by VP16 and Gcn4 was the ratio of full-length transcripts in presence of activator from a template bearing the appropriate activator-binding sequence (UAS) to transcripts in the absence of the activator (see Fig. 1 for example of primary data). This ratio was normalized by division by the ratio obtained from a second template lacking the appropriate UAS. Stimulation of basal transcription was measured by the ratio of transcripts produced by holoenzyme and core polymerase under identical reaction conditions. Stimulation of kinase activity was measured by the ratio of RPB1 CTD phosphorylation in a reaction containing TFIIH and holoenzyme to CTD phosphorylation in a reaction containing TFIIH and core polymerase. Stimulation of basal transcription and TFIIH kinase activity was highly dependent on the ratio of Mediator to core polymerase in the holoenzyme fraction. This ratio, and thus the stimulatory effect, varied a few-fold for different holoenzyme preparations, even from the same strain. In contrast, VP16- and Gcn4-activated transcription was relatively unaffected by the ratio of Mediator to core polymerase.
Figure 1.
Transcription assays of wild-type and Δpgd1 holoenzymes. Transcription was performed with highly purified transcription factors and DNA templates containing binding sites for Gcn4 (GCN4:G−) and Gal4 (GAL4:G−). Gal4–VP16 activation (31-fold for wild-type holoenzyme, 1.8-fold for Δpgd1) was quantitated by comparing transcription in the presence and absence of the activator on the GAL4:G− template and dividing the ratio by any change in transcription of GCN4:G− template. Gcn4 activation (8.2-fold for wild-type holoenzyme, 6.9-fold for Δpgd1) was measured with the GCN4:G− template in a similar manner.
Two observations indicated that the effect of the Δmed2 mutation was exerted through Mediator and not by an alteration of polymerase, such as a modification, or by another indirect mechanism. First, results obtained with either the naturally occurring Mediator-RNA polymerase II complex prepared from wild-type and mutant strains (Table 1), or with a complex formed from separately isolated Mediator and polymerase, were essentially the same (not shown). Second, the addition of isolated Δmed2 Mediator to reactions containing wild-type Mediator inhibited activated transcription, showing a dominant effect of the mutant protein and its capacity to compete for polymerase interaction.
Different Requirements for Responsiveness to Two Acidic Activators.
Yeast Gcn4 protein is, along with VP16, a founding member of the family of “acidic” activator proteins (21). It has been thought that these proteins function by a common mechanism. In keeping with this idea, Gcn4 was shown, like Gal4–VP16, to require Mediator for activation in a fully reconstituted transcription system (3). We therefore were surprised to find that Gcn4 required different Mediator subunits than did Gal4–VP16 in the reconstituted system (Table 1). Only the purified Δsin4 holoenzyme was defective for the response to Gcn4. The Δmed2 and Δpgd1 holoenzymes, which also failed to support Gal4–VP16 activation, were fully functional with Gcn4. Evidently the two acidic activators contact different members of the Mediator complex or function through Mediator by different mechanisms.
Structure–Function Relationships of Yeast Mediator.
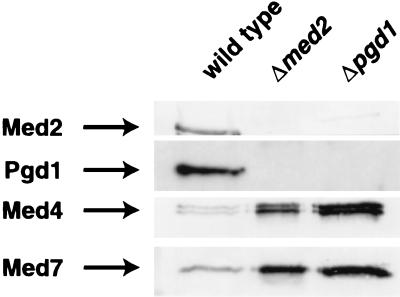
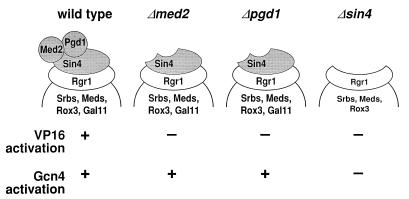
Pgd1 is associated with Mediator through Sin4, as shown by the loss of Pgd1 from holoenzymes isolated from SIN4 deletion and RGR1 truncation mutants (6, 8). Pgd1 was also absent from the Δmed2 holoenzyme isolated here (Fig. 2). Conversely, Med2 was not retained in the purified Δpgd1 holoenzyme (Fig. 2). Med2 and Pgd1 therefore must interact, either directly or indirectly, to stabilize their mutual association with the holoenzyme. SDS/PAGE and silver staining (not shown) revealed the presence of all other Mediator polypeptides in the mutant holoenzymes (the presence of Rox3 and Gal11 could not be conclusively confirmed because they comigrate exactly with Med8 and Rpb2, respectively), so Med2 and Pgd1 are likely to occupy peripheral locations. The picture of Mediator subunit organization that emerges conforms well with the results of functional studies (Fig. 3). The mutual association of Pgd1 and Med2 with the holoenzyme is reflected in their joint requirement for Gal4–VP16 activation. The interaction of these two proteins through Sin4 leads to the requirement for Sin4 as well. Finally, the peripheral location of Pgd1 and Med2 explains why they may be dispensable for Gcn4 activation whereas Sin4 is not.
Figure 2.
Immunobloting analysis of wild-type and mutant holoenzymes. Mono-Q fractions of wild-type, Δmed2, and Δpgd1 holoenzymes were subjected to immunoblot analysis by using antibodies directed against Mediator components Med2, Pgd1, Med4, and Med7 (8). The amounts of the Δmed2 and Δpgd1 holoenzymes loaded on the gel were approximately three times greater than the amount of wild-type holoenzyme, to demonstrate the absence of Med2 and Pgd1 subunits.
Figure 3.
Structure–function relationships of wild-type and mutant RNA polymerase II holoenzymes. The subunit organization of the Sin4/Rgr1 module of Mediator is based on Fig. 2 and the Results in the text. This model, however, does not preclude the existence of weak interactions among Med2, Pgd1, Sin4, and other subunits of holopolymerase that do not withstand the rigors of purification. The functional consequences of the various Mediator mutations are from Table 1.
Similar Mediator Mechanism in Vitro and in Vivo.
Having identified Mediator mutations specific for transcriptional activation in vitro, we investigated the effects of the same mutations on transcription in vivo. The Δmed2 mutation diminished Gal4–VP16 activation of a lacZ reporter gene downstream of Gal4-binding sites by ≈10-fold (Table 2). This effect clearly involved VP16, because activation with a VP16 mutant was lower and was similarly impaired by the Δmed2 mutation (Table 2). As noted above, activation by Gal4–VP16 of transcription in vitro was diminished by the Δmed2 mutation (Table 1), also by an order of magnitude, establishing a parallel between effects of Mediator mutations in vivo and in vitro.
Table 2.
Gal4-VP16 activation in wild-type, Δmed2, and Δsin4 strains
| Strain | Vector | Gal4–VP16 | Gal4–VP16FA |
|---|---|---|---|
| MED2 wild type (MG106) | <1 | 1156 | 568 |
| Δmed2 (MG107) | <1 | 138 | 35 |
| SIN4 wild type (DY1880) | <1 | 2806 | 660 |
| Δsin4 (DY1876) | <1 | 1763 | 342 |
Levels of β-galactosidase activity were assayed in strains with an ARS-CEN plasmid containing Gal4–VP16 or Gal4–VP16 bearing the Phe-442 to Ala mutation (Gal4–VP16FA) under control of the ADH1 promoter, and the pLGSD5 GAL (lacZ) reporter plasmid (23). The units of activity are normalized to cell OD600 and results shown are the means from at least three replicate assays.
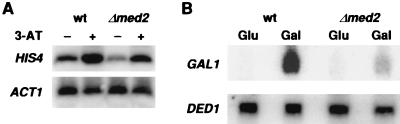
The effect of the Δmed2 mutation on Gcn4 activation in vivo was also consistent with the results obtained in vitro. The Δmed2 strain displayed wild-type levels of Gcn4-dependent HIS4 transcription (Fig. 4A), in keeping with the lack of requirement of Med2 protein for Gcn4 activation in vitro (Table 1). A similar parallel can be drawn for Sin4, because a Δsin4 strain previously was shown to be defective in the activation of HIS4 transcription (24) and, as mentioned, Sin4 is essential for Gcn4 activation in the reconstituted transcription system. The correlation breaks down, however, for HIS3, whose level of Gcn4-dependent transcription increases in a Δsin4 strain (24). Various mechanisms, mostly indirect, may be considered to account for this discrepancy, but the actual basis remains to be determined.
Figure 4.
RNA blot analysis of HIS4 and GAL1 induction in wild-type and Δmed2 strains. (A) Wild-type (MG106) and Δmed2 (MG107) mutant cells transformed with pRS313 (HIS3) (22) were grown in synthetic minimal medium (16) supplemented with 0.2 mM inositol, 2.0 mM leucine, 0.5 mM isoleucine, 0.5 mM valine, 0.4 mM tryptophan, 0.25 mM arginine, 0.1 mM adenine, 0.2 mM lysine, and 0.2 mM uracil to OD600 = 0.8. For starvation conditions, 3-aminotriazol (3-AT) was added to 100 mM, and the cultures were harvested 3 hr later. The RNA blot was hybridized to radioactively labeled probes for HIS4 and ACT1. (B) Wild-type (MG106) and Δmed2 (MG107) mutant cells were grown in yeast extract/peptone/raffinose medium overnight, washed with water and transferred to yeast extract/peptone/glucose (Glu) or yeast extract/peptone/galactose (Gal) media at a density of OD600 = 0.15, followed by harvest at OD600 = 0.6. The RNA blot was hybridized to radioactively labeled probes for GAL1 and DED1.
Effects of Mediator mutations on transcriptional activation in vivo also were manifest in cell growth phenotypes. A haploid med2 deletion strain failed to grow on galactose medium containing the respiration inhibitor antimycin A, although it was suc+, raf+, gly+, and neither UV-sensitive nor temperature-sensitive at 37°C. Blot hybridization of total RNA revealed a 7-fold decrease in the induction of GAL1 transcription in the mutant upon shifting the carbon source from glucose to galactose (Fig. 4B). In contrast, expression of the constitutively transcribed DED1 and ACT1 genes was unaffected. A Δpgd1 strain also exhibited a gal− phenotype and was defective for GAL gene induction (25), in keeping with the joint requirement for Med2 and Pgd1 components of Mediator in vitro noted above. Gal4 protein responsible for GAL gene induction, evidently, specifically requires Med2 and Pgd1 protein for transcriptional activation.
A second deviation from consistency of the data reported here is that a Δsin4 strain remains gal+ (Y. W. Jiang and D. J. Stillman, personal communication). This result is surprising because, as already mentioned, loss of Sin4 from a purified holoenzyme results in loss of Med2 and Pgd1 as well (Fig. 3). Various explanations may be advanced for this behavior. For example, Δsin4 strains exhibit derepression of GAL genes (26) that may offset the loss of activation by Sin4-associated proteins such as Med2/Pgd1. Alternatively, Med2/Pgd1 may make interactions with other Mediator components sufficient for retention in a Δsin4 mutant in vivo, but not through the course of fractionation in vitro. Two experimental observations are consistent with this idea: first, Med2 and Pgd1 are observed to comigrate with the other mediator components over the first three chromatographic steps in the purification of the Δsin4 mediator (data not shown); and second, activation by Gal4–VP16 in a Δsin4 strain was only 2-fold less than wild type, compared with the 10-fold decrease observed in the Δmed2 strain (Table 2).
Whole Genome Analysis of MED2-Dependent Transcription.
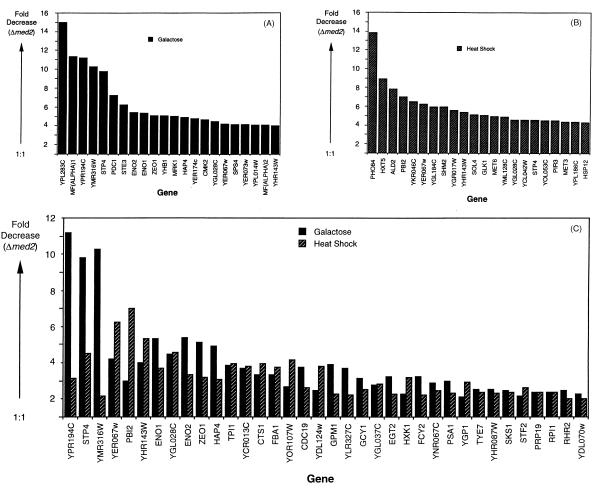
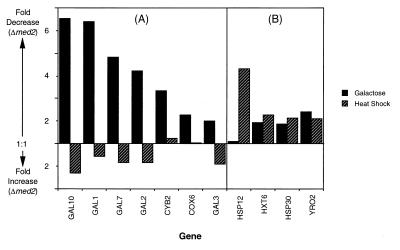
We used whole genome DNA microarrays (20) to investigate the generality of the Med2 requirement for activated transcription. Differences in specific transcript levels between Δmed2 and wild-type strains were determined under galactose and heat shock growth conditions (Fig. 5). Approximately 200 of the ≈6,000 genes analyzed showed a >2-fold decrease in expression in the Δmed2 strain grown in galactose. A similar number of genes showed a >2-fold decrease in expression in the mutant strain under heat shock induction conditions. On examination of the specific genes affected, certain patterns emerged. First, the genes most dependent on Med2 were, in general, highly transcribed under the conditions tested. The majority of abundant transcript levels did not change, however, showing specificity of the Med2 effect. Second, although there was some overlap between the sets of genes dependent on Med2 under the two different growth conditions, these sets were largely distinct. For example, deletion of MED2 diminished expression of MFα1, STE3, CDC19, and MFα2, which share common regulatory sequences and which were shown previously to require the Mediator components Med6 (7) and Gal11 (27) for optimal transcription. Of these four genes, only CDC19 was Med2-dependent under both galactose induction and heat shock growth conditions. Third, two cell cycle-related genes, CTS1 and EGT2, were less well transcribed under both growth conditions, which may relate to neither galactose nor heat shock induction, but rather reflect a requirement of Med2 for temporal induction of transcription. Defective expression of CTS1 also has been noted in a Δsin4 strain (28), consistent with the structure–function relationships described above (Fig. 3). Fourth, as anticipated from the results of blot hybridization (Fig. 4B), galactose induction of GAL genes was defective in the mutant strain, and transcription of some heat shock promoters, including HSP12, HXT6, HSP30, and YRO2, decreased as well (Fig. 6). GAL4 and GAL80 transcript levels were unchanged in the mutant, arguing against secondary effects on GAL gene transcription arising from altered expression of these regulatory proteins. An almost 2-fold defect in expression of GAL3, however, could have played a role, because gal3 mutants show a diminished rate of galactose induction of transcription (29). Finally, the effect of the MED2 deletion on galactose induction was selective: transcript levels of several genes shown by previous microarray analysis to be induced by galactose (30), such as COX12, QCR6, COR1, PET9, COX8, ATP5, ATP3, COX9, and MCR1, were essentially unchanged.
Figure 5.
Genes that display defects in transcription under galactose and heat shock growth conditions in a Δmed2 strain. Genes are identified by gene name or ORF designation (as listed in the Stanford Genome Database). (A) The 22 genes that suffer the greatest transcription defects in the Δmed2 strain under galactose induction conditions are shown. The GAL genes shown in Fig. 6A are not included in this plot. (B) The 22 genes that suffer the greatest transcription defects in the Δmed2 strain under heat shock growth conditions (excluding genes described in Fig. 6B) are shown. (C) All genes that suffer a >2-fold defect in the Δmed2 strain under both galactose and heat shock growth conditions are shown. The deleted gene, MED2, was not included on the above lists. “Fold Decrease (Δmed2)” refers to the ratio (wild type/Δmed2) of normalized transcript levels.
Figure 6.
Galactose- and heat shock-induced genes that display transcription defects in a Δmed2 strain. (A) Well characterized galactose-induced genes that suffer a >2-fold defect in expression in the Δmed2 strain grown on galactose medium are shown. For comparison, fold differences in expression of these same genes under heat shock conditions (in glucose) also are indicated. (B) Well characterized heat shock-induced genes that suffer a >2-fold defect in expression in the Δmed2 strain under heat shock (37°C) growth conditions are shown. For comparison, fold differences in expression of these same genes under galactose growth conditions (30°C) also are indicated. “Fold Decrease (Δmed2)” and “Fold Increase (Δmed2)” refer to ratios (wild-type/Δmed2 and Δmed2/wild-type, respectively) of normalized transcript levels.
The picture that emerges from the microarray analysis is one of gene-specific involvement of Med2 in transcriptional induction in vivo. There appears to be no general requirement of the protein for constitutive (“basal”) transcription. Although the array analysis also revealed increased expression of some genes in the Δmed2 strain, these increases seem likely to reflect an adaptive response rather than a direct consequence of the genetic deficiency.
DISCUSSION
The chief import of this work lies in the validation of transcriptional activation in the yeast system in vitro and the implications for the role of Mediator in vivo. The work must be considered in the context of other efforts to elucidate transcriptional activation mechanisms. Dissection of human and Drosophila systems led to the discovery of TAFs and to evidence for their requirement for activation in vitro. It now appears that the function of TAFs is to augment the sequence specificity of TATA-binding protein, rather than to facilitate enhancer–promoter communication (31). Other factors, similar or equivalent to yeast Mediator, are more important than TAFs for activation in the human system in vitro (32). The same questions arise for yeast Mediator, whether its requirement for activation in vitro holds true in vivo, and whether it conveys the regulatory influence of enhancers in vivo.
Pursuit of these questions was facilitated by our finding of Mediator mutations that abolish activated transcription in vitro with only minimal effects (at most 2- to 3-fold) on basal transcription or TFIIH kinase activity. Study of the same mutations in vivo revealed selective effects on activated transcription as well. The number of inducible genes whose expression was diminished by the mutations should be regarded as a lower estimate, because only one Med protein mutation and two inducing conditions have so far been investigated by microarray analysis. Selectivity was shown by a lack of effect of the mutations on DED1 and ACT1 transcript levels in RNA blots and by a lack of effect on expression of the majority of yeast ORFs in microarrays. Although the cellular equivalent of basal transcription in vitro has not been defined, the diminished expression of inducible genes in the med2 deletion strain stands out against the broad background of genes that are unaffected. It can be said that Med2 protein plays a positive role in the transcription of many genes but that it is not generally required, so its function in vivo is in transcriptional activation. Evidence that this role is direct and relates to upstream regulatory sequences comes from the results obtained for induction of GAL gene transcription and for Gal4–VP16 activation of a Gal4-binding reporter construct. Mediator thus provides a functional connection between upstream sequences and promoters. Evidence for related complexes in mammalian cells (33, 34) suggests the Mediator connection is conserved across species from yeast to man.
Previous work of others showed the involvement of Sin4 and Rgr1 in the negative regulation of many genes, leading to their designation as “global” repressors (35). The discovery of Mediator united these diverse molecules in a common biochemical entity (3, 6, 8). It was further shown that Sin4, Rgr1, Gal11, and Pgd1(Hrs1) interact in the same subcomplex of the Mediator assembly, thus accounting for their involvement in regulation of the same set of genes (6). Our findings extend the characterization of this Sin4/Rgr1 module in two respects. First, Med2 is identified as an additional component of this module, occupying a peripheral location, interdependent in its association with Pgd1 (Hrs1). Second, the module is required for activation of many genes in vitro and in vivo. Using previously defined terminology, Mediator may be described as a global coactivator and corepressor.
It is noteworthy that the involvement of Mediator in both activation and repression is brought about by the same subcomplex of the larger assembly. Biochemical and genetic findings thus converge on the notion of a common activation/repression mechanism. This idea is nicely compatible with a suggestion that repression occurs through the same complex of RNA polymerase II and general factors as the initiation of transcription (36).
Our finding that VP16 and Gcn4 differ in their Mediator subunit requirements for activation was unexpected in view of the common classification of these proteins in a single group of “acidic” activators. It will be instructive to determine the Mediator subunit requirements for other “acidic” activators. Genetic analyses of VP16 (37) and Gcn4 (38), examining the consequences of amino acid substitutions in the activation domains of these proteins for function in vivo, have questioned the importance of acidic amino acids, and our findings indicate that the two activators may differ in regard to mechanism.
Ultimately, activator proteins must be categorized on the basis of mechanism. Although this study does not directly address the activation mechanism, some findings are pertinent. Binding studies in vitro and functional studies in vivo have suggested that a direct physical interaction between the activation domain of Gal4 and Srb4, a mediator component, is critical for Gal4-stimulated transcription (39). Holoenzymes purified from the Δmed2, Δpgd1, and med6ts (7) strains all retain Srb4 but support neither Gal4 activation in vivo nor VP16 activation in vitro. Despite the presence of Srb4, preliminary studies indicate a diminished interaction between the Δmed2 and Δpgd1 holoenzymes and both VP16 and Gcn4 (L.C.M., C.M.G., and R.D.K., unpublished results). We investigated the possibility of activator–Med2 or –Pgd1 interaction with the use of recombinant proteins and obtained only negative results (data not shown). Others have reported that Med6 also is required for a VP16 response in vitro, that Med6 alone is lost from isolated med6ts Mediator, and that no activator–Med6 interaction can be detected (7). The separate requirements for Med2/Pgd1 and Med6, and the lack of activator interaction, are not immediately compatible with the “recruitment model” (40) of activation, but no definitive statement regarding the activation mechanism can be made at the present time.
Acknowledgments
We thank B. Cairns for assistance in the phenotypic analysis of the Δmed2 mutant and D. Bushnell for purified proteins. We also thank Y.-W. Jiang for the Δsin4 strain and helpful discussions. Whole genome yeast DNA microarrays were kindly provided by J. DeRisi and V. Iyer. The Δpgd1(Δhrs1) strain and anti-Pgd1(Hrs1) antibodies were gifts from A. Aguilera and the DY1880 and DY1876 (Δsin4) strains were gifts from D. J. Stillman. L.C.M. was recipient of a Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship, DRG-1361. C.M.G. was a recipient of a Swedish Cancer Society postdoctoral fellowship. This research was supported by Swedish Cancer Society Grant 3947 (to C.M.G.) and National Institutes of Health Grant GM36659 (to R.D.K.).
ABBREVIATIONS
- CTD
C-terminal domain of RNA polymerase II
- TAF
TATA boxing-binding protein associated factor
Footnotes
A Commentary on this article begins on page 2.
References
- 1.Kelleher R J, III, Flanagan P M, Kornberg R D. Cell. 1990;61:1209–1216. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 2.Flanagan P M, Kelleher R J, III, Sayre M H, Tschochner H, Kornberg R D. Nature (London) 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 4.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 5.Carlson M. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Bjorklund S, Jiang Y W, Kim Y-J, Lane W S, Stillman D J, Kornberg R D. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y C, Min S, Gim B S, Kim Y-J. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 10.Walker S S, Reese J C, Apone L M, Green M. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 11.Thompson C M, Young R A. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. Nature (London) 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 13.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Bjorklund S, Kim Y-J, Kornberg R D. Methods Enzymol. 1996;273:172–175. doi: 10.1016/s0076-6879(96)73017-3. [DOI] [PubMed] [Google Scholar]
- 15.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 16.Sherman F, Fink G R, Lawrence C W. Methods of Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 18.Wise J A. Methods Enzymol. 1991;194:405–423. doi: 10.1016/0076-6879(91)94031-7. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 21.Hope I A, Mahadevan S, Struhl K. Nature (London) 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski R S, Hieter P. Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y W, Stillman D J. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piruat J I, Chavez S, Aguilera A. Genetics. 1997;147:1585–1592. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, West R W, Jr, Johnson S L, Gans H, Kruger B, Ma J. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishizawa M, Suzuki Y, Nogi Y, Matsumoto K, Fukasawa T. Proc Natl Acad Sci USA. 1990;87:5373–5377. doi: 10.1073/pnas.87.14.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y W, Stillman D J. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yano K, Fukasawa T. Proc Natl Acad Sci USA. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen W-C, Green M R. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 32.Oelgeschläger T, Tao Y, Kang Y K, Roeder R G. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y W, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway J W, Conaway R C, Kornberg R D. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y W, Dohrmann P R, Stillman D J. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 37.Regier J L, Shen F, Triezenberg S J. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson B M, Drysdale C M, Natarajan K, Hinnebusch A G. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 40.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]