Abstract
Nuclear factor κB (NF-κB) is one of the key regulators of transcription of a variety of genes involved in immune and inflammatory responses. NF-κB activity has long been thought to be regulated mainly by IκB family members, which keep the transcription factor complex in an inactive form in the cytoplasm by masking the nuclear localization signal. Nowadays, the importance of additional mechanisms controlling the nuclear transcription potential of NF-κB is generally accepted. We show that the mitogen-activated protein kinase inhibitors SB203580 and PD98059 or U0126, as well as a potent mitogen- and stress- activated protein kinase-1 (MSK1) inhibitor H89, counteract tumor necrosis factor (TNF)-mediated stimulation of p65 transactivation capacity. Muta tional analysis of p65 revealed Ser276 as a target for phosphorylation and transactivation in response to TNF. Moreover, we identified MSK1 as a nuclear kinase for p65, since MSK1 associates with p65 in a stimulus-dependent way and phosphorylates p65 at Ser276. This effect represents, together with phosphorylation of nucleosome components such as histone H3, an essential step leading to selective transcriptional activation of NF-κB-dependent gene expression.
Keywords: gene activation/MSK1/NF-κB/phosphorylation/TNF
Introduction
Nuclear factor κB (NF-κB) encompasses an important family of inducible transcription factors, critical for regulation of gene expression in response to injury and inflammatory stimuli. Members of this family share a 300 amino acid region, known as the Rel homology domain (RHD), which is responsible for DNA binding and dimerization. To date, eight members of the NF-κB/Rel family have been cloned and characterized: NFKB1 (p50/p105), NFKB2 (p52/p100), cRel, RelA (p65), RelB and the Drosophila proteins Dorsal, Dif and Relish (reviewed in Ghosh and Karin, 2002; Li and Verma, 2002). The most abundant form of the transcription factor is a heterodimer that contains a p50 and p65 subunit, the latter comprising a powerful transcriptional activation domain. In uninduced cells, NF-κB is kept inactive in the cytoplasm through binding of an inhibitory protein IκB. After stimulation by a variety of inducers, such as tumor necrosis factor (TNF), interleukin-1 (IL-1) or lipopolysaccharide (LPS), the IκB protein becomes phosphorylated, ubiquitylated and degraded by the 26S proteasome. NF-κB subsequently is released and translocates to the nucleus to activate expression of various target genes.
Phosphorylation is a rapid and reversible enzymatic reaction frequently used as a molecular mechanism in several signal transduction pathways. As such, it has a number of advantages for regulating transcription factor activity. It is very effective at integrating information from various incoming signals, whereas a single kinase can differentially affect multiple transcription factors. More over, depending on the amino acid residue of the target protein modified, phosphorylation may influence different aspects of transcription factor function (Jackson, 1992). In the case of NF-κB, phosphorylation is involved in release of NF-κB from its inhibitor, nuclear transport, processing of NF-κB precursors, stabilization of NF-κB dimerization and DNA binding, kinetics of NF-κB turnover and NF-κB transactivation properties (Karin and Ben-Neriah, 2000; Schmitz et al., 2001; Vermeulen et al., 2002). In contrast to IκB phosphorylation, p65 phosphorylation has not yet been fully characterized and seems to vary depending on the stimulus, the time point or the cell type tested (Vanden Berghe et al., 2000). As far as TNF-mediated stimulation is concerned, Ser529 phosphorylation by casein kinase II was necessary for full transactivation in mouse embryo fibroblasts (MEFs) (Wang and Baldwin, 1998; Wang et al., 2000), whereas Ser536 was phosphorylated by co-expressed IKKs; in addition, TNF-induced phosphorylation of overexpressed p65 also occurs at this position (Sakurai et al., 1999). Another mechanism of cytoplasmic p65 phosphorylation, in response to LPS, was proposed previously by Zhong et al. (1997), who identified an IκB-associated protein kinase A catalytic subunit (PKAc) by co-immunoprecipitation and pointed to phosphorylation of p65 at Ser276. This might lead to a conformational change of p65, allowing efficient recruitment of the cofactor CBP (CREB-binding protein) (Zhong et al., 1998). This position was, however, found to be constitutively phosphorylated in endothelial cells (Anrather et al., 1999).
Here we focus on the nuclear phosphorylation of the NF-κB p65 subunit as the driving force for gene activation. In previous studies, we demonstrated that p38 and ERK mitogen-activated protein kinases (MAPKs) are absolutely required for NF-κB-driven gene transcription in response to TNF (Vanden Berghe et al., 1998). We report that mitogen- and stress-activated protein kinase-1 (MSK1) targets p65 for direct phosphorylation at Ser276 as an essential element for gene induction. MSK1 is a recently identified nuclear CREB and histone H3 kinase that responds to both mitogen- and stress-activated protein kinases (Deak et al., 1998; Thomson et al., 1999).
Results
Effect of kinase inhibitors on endogenous IL-6 gene expression
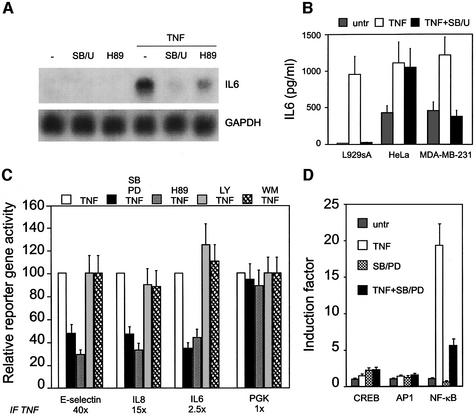
In mammalian species, MAPKs are involved in all aspects of immune responses (Pearson et al., 2001; Dong et al., 2002). Previously, we have shown that p38 and ERK MAPK are absolutely required for NF-κB-driven gene transcription in response to TNF. Here we demonstrate that endogenous IL-6 mRNA levels induced by TNF are severely affected in the presence of inhibitors of the p38 and ERK MAPK pathways together, i.e. SB203580 and U0126, or in the presence of an inhibitor, such as H89, of the dually regulated kinase MSK1 (Figure 1A). The nuclear kinase MSK1 is activated by both ERK and p38 MAPK. Interestingly, MAPK-dependent IL-6 expression seems to be cell type specific, as it occurs in L929 mouse fibroblasts and human breast cancer cells MDA-MB-231 but not in HeLa cells (Figure 1B).
Fig. 1. Regulation of NF-κB-dependent gene expression. (A) L929sA cells were left untreated or were treated with 2000 IU/ml TNF, either in combination with 10 µM H89 or following a 2 h pre-treatment with 10 µM SB203559 and 10 µM U0126, and mRNA was isolated and subjected to northern blot analysis. (B) IL-6 production in various cell lines was tested after 6 h treatment with 2000 IU/ml TNF, pre-treated or not with 10 µM SB203580 and 10 µM U0126. (C) L929sA cells were stably transfected with reporter plasmids for either the E-selectin, the IL-8 or the IL-6 promoter, and pPGKβgeobpA. Transfectants were left untreated or were treated with 2000 IU/ml TNF for 6 h, either in combination with 10 µM H89 or following a 2 h pre-treatment with 10 µM SB203580 and 10 µM PD98059, with 20 µM LY294002 or with 100 nM wortmannin. The induction factor is defined as the amount of luciferase produced in TNF-treated cells after normalization for β-galactosidase expression, compared with the non-induced state. (D) Similar experiments were performed with L929sA cells, stably transfected with synthetic CREB-, AP1- or NF-κB-dependent reporter constructs.
Effect of kinase inhibitors on NF-κB-regulated promoters
Next, we investigated the effect of these kinase inhibitors on the regulation of different NF-κB-driven reporter gene constructs, such as the E-selectin, the IL-8 and the IL-6 promoters, coupled to luciferase. As previously identified, NF-κB is the main transcription factor regulating these natural promoters in response to TNF (Vanden Berghe et al., 1998, 1999). As shown in Figure 1C, TNF causes a strong induction of the various NF-κB-driven promoters, which is counteracted by SB203580 and PD98059 together, inhibitors of the p38 and ERK MAPK pathways, respectively, or by the potent MSK1 inhibitor H89. Specific inhibitors of phosphatidylinositol 3-kinase signaling (LY294002 and wortmannin) clearly had no effect. Strong variations in TNF inducibility are due to promoter configuration, including the presence of multiple NF-κB-responsive elements (E-selectin) (Schindler and Baichwal, 1994), composition of the κB motif (Kunsch and Rosen, 1993) and/or the possible synergy with adjacent transcription factors (IL-6 and IL-8) (Mukaida et al., 1990). In parallel, we have also tested synthetic reporter gene constructs with various responsive elements preceding a minimal TATA box sequence. Only the NF-κB-responsive promoter could be stimulated by TNF and inhibited by the compounds used (Figure 1D). Remarkably, repression obtained at the promoter level is less drastic than that observed at the IL-6 mRNA and protein levels, suggesting a further potentiation of MAPK effects at the post-transcriptional and translational level (Kracht and Saklatvala, 2002).
MAPK inhibitors and H89 impede p65 transactivation, but not DNA binding
To investigate if reduced expression of the reporter genes is due to inhibition of the NF-κB DNA-binding capacity, an electrophoretic mobility shift assay (EMSA) was performed. Total lysates of L929sA cells, treated with TNF and/or various kinase inhibitors, were analyzed for their NF-κB-binding activity. Figure 2A clearly shows that pre-treatment with MAPK inhibitors and/or with H89 has no effect on TNF-induced p50/p65 DNA-binding activity, indicating that p65 transactivation is affected by the inhibitors used.
Fig. 2. Effect of kinase inhibitors on TNF-induced NF-κB binding, p65 phosphorylation and transactivation. (A) L929sA cells were left untreated or were treated with 2000 IU/ml TNF for 30 min, either in combination with 10 µM H89 and/or following a 2 h pre-treatment with 10 µM SB203580 and/or 10 µM PD98059. Total cell lysates were incubated with a 32P-labeled IL-6 κB site-containing probe. Complexes formed were analyzed by EMSA. Loading of equal amounts of protein was verified by comparison with the binding activity of the repressor molecule RBP-Jκ (Plaisance et al., 1997). (B) Pools of L929sA cells stably expressing the Gal4 DNA-binding domain, Gal4-VP16, Gal4-p65 or serine-mutated variants thereof were transiently transfected with p(gal4)2hu.IL6-luc+. At 48 h post-transfection, cells were left untreated or were treated with 2000 IU/ml TNF for 6 h. The pools stably expressing Gal4-p65 were also co-treated with 10 µM H89 or pre-treated for 2 h with 10 µM SB203580 and 10 µM PD98059. (C) [32P]orthophosphate-labeled L929sA cells were stimulated with 5000 IU/ml TNF for 15 min in the absence or presence of 10 µM H89 at 2 h before TNF. Total cell lysates were immunoprecipitated with an anti-p65 antibody, analyzed by 10% SDS–PAGE and visualized using PhosphorImager technology. (D) L929sA mouse fibroblasts were starved for 24 h in 0.5% serum. Quiescent cells were treated for 20 min with 2500 IU/ml TNF alone or following a 2 h pre-treatment with 10 µM H89. Top panels, DAPI-stained nuclei; bottom panels, corresponding signals obtained with anti-phospho Ser276 p65 antibody.
To study the effect of TNF-induced phosphorylation on p65 transactivation, we used the Gal4 ‘one-hybrid’ system. Briefly, the expression plasmids pGal4, pGal4-p65 and pGal4-VP16 were transfected in L929sA cells and stable pools were expanded. The transactivation capacity of the chimeric Gal4 proteins was scored by measuring gene activity of a transiently transfected Gal4-driven reporter construct p(gal4)250hu.IL6-luc+. This system has the advantage that the Gal4 transactivator fusion proteins are exclusively nuclear, and thus regulated independently of IκB (Schmitz and Baeuerle, 1991).
TNF is able specifically to potentiate p65 transactivation (Figure 2B) and does not show any effects on the Gal4 DNA-binding domain alone or on the chimeric protein Gal4-VP16. Pre-treatment with SB203580 and PD98059 significantly decreases p65 transactivation. Due to some basal levels of activated p38 and/or ERK MAPK, partial inhibition of the basal Gal4-p65 activity was observed. Interestingly, treatment with H89 also inhibits Gal4-p65 transactivation. Notably, TNF-enhanced p65 transactivation cannot be abolished entirely, which probably originates from some residual TNF-induced c-Jun N-terminal kinase (JNK) activity (Krause et al., 1998; Holtmann et al., 1999). Nevertheless, none of these treatments showed any effect on the binding capacity of Gal4-p65 to a Gal4 DNA-binding sequence (see Supplementary figure 1, available at The EMBO Journal Online). Taken together, these results suggest a role for multiple phosphorylation-dependent pathways in regulating p65 transactivation.
H89, a potent MSK1 inhibitor, blocks TNF-induced phosphorylation of p65 in vivo
We focused on the effect of TNF and the MSK1 inhibitor H89 on in vivo phosphorylation of p65. L929sA cells were labeled with [32P]orthophosphate and stimulated with TNF for 15 min. After cells were harvested, whole-cell extracts were subjected to immunoprecipitation with anti-p65 antibody. Proteins were separated by SDS–PAGE and visualized using PhosphorImager software. The results demonstrate that p65 is a phosphoprotein under non-induced conditions and that it can be phosphorylated further after treatment with TNF (Figure 2C). Interest ingly, H89 completely reverses TNF-induced phosphorylation to the basal p65 phosphorylation level. This result points to a role for an H89-sensitive kinase in direct phosphorylation of p65.
Identification of Ser276 as a crucial residue for TNF-mediated transactivation of p65
By analogy with the MSK phosphorylation motif at CREB Ser133 (Deak et al., 1998), we first directed our attention to the highly homologous phosphorylation motif in the RHD of p65 at Ser276. Therefore, Ser276 was mutated in the Gal4-p65 construct to eliminate the phosphorylation site. As C-terminal phosphorylation of p65 in response to TNF was also reported at Ser529 and Ser536 (Wang and Baldwin, 1998; Sakurai et al., 1999), these residues were also mutated separately, and the transactivation properties of the various mutants were compared. Figure 2B shows that, upon mutation of Ser276 to cysteine (which resembles more the native serine residue), the TNF-induced response is almost completely abrogated, whereas responses obtained with the Ser529 or Ser536 mutants are nearly identical to those for wild-type (wt) p65. This points to a crucial role for Ser276 in TNF-induced transactivation of p65.
Furthermore, phosphorylation of endogenous p65 on Ser276 was examined by using a Ser276 phospho-specific antibody. Specificity of the phospho-specific antibody was confirmed by phosphopeptide competition (Supplementary figure 2) and by transfection assays with wt p65 and Ser276-mutated expression vectors (Supplementary figure 3). Serum-starved L929 cells were treated with TNF for 20 min, in either the presence or absence of H89. Upon TNF stimulation, the amount of Ser276-phosphorylated p65 is clearly enhanced (Figure 2D). Note that the phosphorylated p65 is mainly nuclear. Pre-treatment with H89 significantly reduces the phosphorylation state of p65, as also observed in the in vivo phosphorylation assay (Figure 2C) and in the western blot with the phospho-specific antibody (Supplementary figure 3).
Mutation of p65 Ser276 selectively eliminates cofactor-mediated acetylase effects
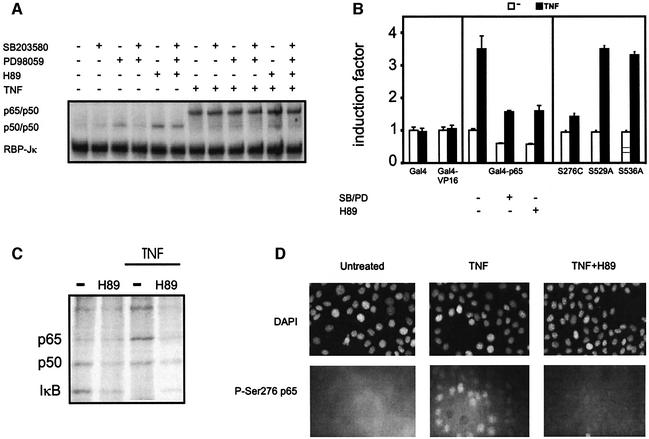
Previously, we demonstrated the crucial role of the NF-κB p65 subunit in engaging CBP/p300 and histone acetyltransferase (HAT) activity for transcriptional activation of the IL-6 promoter (Vanden Berghe et al., 1999). In this respect, we investigated the ability of p300 to synergize with wt p65 or the respective mutants S276C, S276A and S529A for transcriptional activation. Subsaturating amounts of expression plasmids of p65 (wt, S276C, S276A or S529A) and/or p300 (wt or ΔHAT) were transiently transfected to human embryonic kidney HEK293 cells together with the IL-6 reporter gene construct p1168hu.IL6P-luc+. As is clear from Figure 3, NF-κB-dependent gene expression is enhanced after overexpression of p300 together with either wt p65 or the mutant S529A, but not after co-expression of p300ΔHAT. In contrast, mutation of Ser276 to either alanine or cysteine completely destroys this synergistic effect, no difference between the two mutations being apparent. Moreover, nuclear expression levels (Figure 3) as well as the DNA-binding capacity (data not shown) of the Ser276 mutants are similar to those of wt p65. These results demonstrate the absolute requirement for Ser276 in order to establish a functional synergism between p65 and p300.
Fig. 3. Ser276 is a crucial residue for TNF-mediated transactivation of p65. HEK293 cells were transiently transfected with a combination of expression plasmids, i.e 180 ng of p1168hu.IL6-luc+, 20 ng of pPGKβgeobpA, 2 ng of pRcRSVp65 (or the respective mutant versions) and 100 ng of p300 expression plasmid (pCIp300 or pCIp300ΔHAT). The total amount of DNA was kept constant in all set ups by supplementing empty vector DNA. Cells were lysed 42 h after transfection. Luciferase levels were determined and corrected for transfection efficiency by normalization to β-galactosidase levels. In addition, HEK293 cells were transfected with an increasing amount of pRcRSVp65 or pRcRSVp65 S576C (30–900 ng in a total amount of 12 µg per medium Petri dish). Cytoplasmic (c) and nuclear (n) protein extracts were determined for p65 expression by western blot analysis.
TNF activates MSK1 in vivo
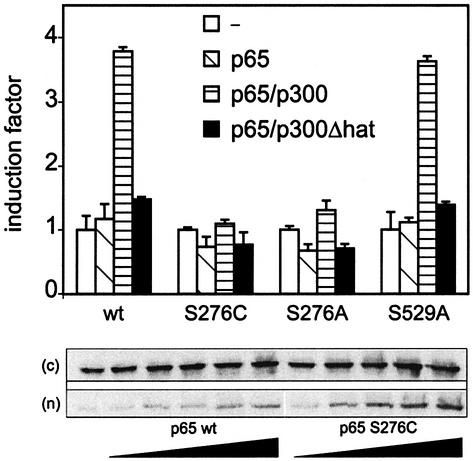
As TNF induces both p38 and ERK MAPKs (Boone et al., 1998), MSK1 should also become activated by these two MAPK pathways in response to TNF. Therefore, MSK1 was immunoprecipitated from L929sA cells either left untreated or treated with TNF for various times. The corresponding MSK1 activity was measured by an in vitro kinase assay. As shown in Figure 4A, MSK1 is rapidly, but transiently activated upon TNF stimulation, the maximum activity being between 10 and 15 min. Notably, the nuclear appearance of NF-κB p65 coincides well with this time point (Figure 4C). Pre-treatment of L929sA cells with PD98059 or SB203580 partially inhibited MSK1 activity, whereas inhibition of p38 and ERK MAPK together completely reversed that activity to the basal level (Figure 4B). These results are consistent with previous observations that, when one pathway is blocked, the other can take over (Deak et al., 1998). Therefore, complete inhibition of MSK1 might only occur when both p38 and ERK are blocked simultaneously. The incubation of activated MSK1 with 10 µM H89 also completely blocked phosphorylation of CREBtide (or p65-tide), in agreement with earlier results identifying H89 as a potent MSK1 inhibitor (Thomson et al., 1999).
Fig. 4. MSK1 is activated by TNF in vivo and phosphorylates Ser276 of p65 in vitro. (A) L929sA cells were starved for 48 h in serum-free medium and stimulated with 2000 IU/ml TNF. Cells were lysed and endogenous MSK1 was isolated by immunoprecipitation. The activity of MSK1 was assessed by an in vitro kinase assay. (B) After 2 days of serum starvation, L929sA cells were incubated for 4 h in serum-free medium supplemented with 10 µM SB203580, 10 µM PD98059 or a combination. Cells were treated with 2000 IU/ml TNF for 15 min in the presence or absence of these inhibitors. After cell lysis, MSK1 was immunoprecipitated and assayed for its ability to phosphorylate CREBtide. Where indicated, H89 was included in the in vitro reaction. (C) L929sA cells were treated with 2000 IU/ml TNF. The presence of p65 in the nuclear extracts was revealed by western blotting. (D) MSK1 was isolated from HEK293 cells overexpressing either wt MSK1 or a kinase-dead mutant, together with the upstream activators p38 and MKK6. Immunoprecipitates were used in an in vitro kinase reaction with either wt GSTp6512–317 or the corresponding Ser276 mutant (S276C) for 20 min at 30°C, followed by SDS–PAGE. Similar experiments were performed with 15 ng of purified PKAc.
MSK1 phosphorylates Ser276 of p65 in vitro
As several kinases, such as PKA, MSK1, MSK2, RSK1, RSK2 and MAPKAPK2, have already been shown to phosphorylate CREB at Ser133 (Mayr and Montminy, 2001), we also tested some of these kinases for their ability to phosphorylate Ser276 of p65 in an in vitro kinase assay, using wt or S276C-mutated GSTp6512–317 as a substrate. Figure 4D demonstrates specific phosphorylation of Ser276 by activated MSK1, but not by the C-terminal kinase-dead mutant of MSK1. This confirms that the observed phosphorylation is specific for MSK1 and does not result from a contaminating kinase in the extract. As a positive control, specific in vitro phosphorylation of Ser276 was also demonstrated by purified PKAc. It should be noted that none of these kinases was able to phosphorylate the substrate after Ser276 had been mutated to cysteine. This specificity has been confirmed further by phospho-specific western analysis (Supplementary figure 3) and phosphopeptide mapping. Briefly, after in vitro kinase assays of GST–p65 (RHD) with recombinant purified PKA or MSK1 (10 U/ml), in-gel trypsin digestion and HPLC/MS identification of phosphorylated peptides, the phosphorylated peak was identified as the peptide RPSDR corresponding to phosphorylation of Ser276; from the results, it appeared that MSK1 is at least as potent as PKA for phosphorylation of Ser276 in vitro (data not shown).
MSK1 phosphorylates p65 in vivo
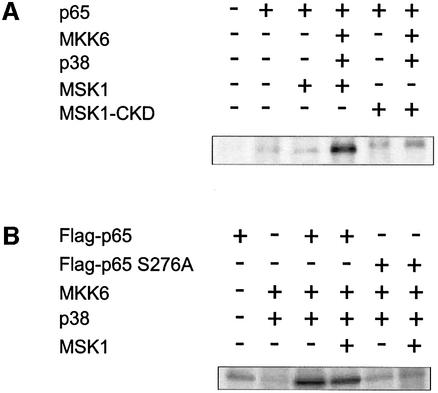
To investigate the biological relevance of MSK1 as a p65-specific kinase, we performed in vivo labeling experiments. First, HEK293 cells were transiently transfected with expression vectors for p65 and/or Flag-MSK1 (wt or C-terminal kinase-dead mutant), either alone or in combination with the activators MKK6 and p38, and were then labeled with [32P]orthophosphate. Figure 5A clearly shows phosphorylation of p65 in the case of overexpression of MSK1 and its activating proteins. On the contrary, no enhanced phosphorylation was observed when a C-terminal kinase-dead mutant was used. Alternatively, we also expressed wt Flag-p65 or the S276A mutant either alone or in combination with MSK1 and/or its activators MKK6 and p38. Figure 5B demonstrates that only wt p65 and not the Ser276 mutant is hyperphosphorylated after MSK1 activation. Moreover, overexpression of MSK1 does not seem to enhance the MKK6/p38-mediated phosphorylation of p65 further, which indicates that activation of the endogenous MSK1 is sufficient. Evidence for the active role of MSK1 itself and not the upstream activator kinases is also provided by Figure 5A in those lanes where the endogenous MSK1 in the presence of the kinase-dead mutant (acting as a dominant-negative player) is not able to phosphorylate wt p65. Earlier in vitro data are in agreement with this interpretation and have already showed p65 not to be a direct substrate for phosphorylation by p38 MAPK (Wesselborg et al., 1997). Similar results were obtained with the phospho-specific antibody (Supplementary figure 3).
Fig. 5. MSK1 phosphorylates p65 in vivo. (A) HEK293 cells, transiently transfected with the relevant expression vectors, were labeled with [32P]orthophosphate for 4 h. Cell lysates were subjected to immunoprecipitation with anti-p65 antibody. Precipitated proteins were separated by SDS–PAGE and analyzed using PhosphorImager technology. (B) Similar experiment to that in (A), but immunoprecipitation was performed with anti-Flag antibody.
MSK1 associates with p65 in vivo
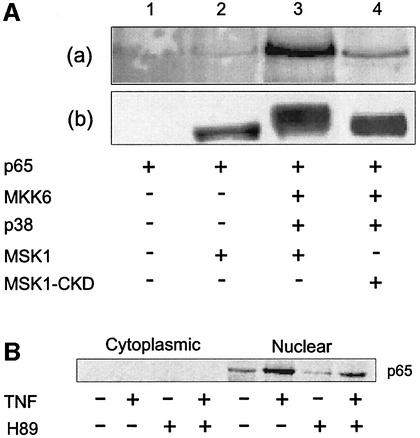
In order to demonstrate a physical interaction between p65 and MSK1, co-immunoprecipitations were performed. HEK293 cells were transiently transfected with expression vectors for p65 and/or Flag-MSK1 (wt or C-terminal kinase-dead mutant), either alone or in combination with the activators MKK6 and p38. MSK1 complexes, isolated by anti-Flag immunoprecipitation, were analyzed by western blotting for co-precipitation of p65. Figure 6A, panel a shows the presence of p65 in the immune complex in the case of activated MSK1 (lane 3). When no activating proteins were co-expressed (lane 2) or when the kinase-dead mutant was used (lane 4), no or significantly less p65 was found to be associated with MSK1. The activation status of MSK1 was monitored by the altered mobility shift upon activation. Interaction between p65 and MSK1 was also seen using GST–MSK1 fusion proteins and 35S-labeled p65 fragments (Supplementary figure 4).
Fig. 6. p65 interacts with activated MSK1 in vivo. (A) HEK293 cells were transiently transfected using expression vectors for p65, Flag-tagged wt MSK1 or C-terminal kinase-dead MSK1 (MSK1-CKD). Activation of MSK1 was achieved by co-transfecting MKK6 and p38 kinase. At 48 h post-transfection, Flag-MSK1 was isolated by immunoprecipitation. Co-precipitating p65 was detected by immunoblotting using an anti-p65 antibody (a). To monitor the activation status of MSK1, blots were stripped and reprobed using an anti-Flag antibody (b). (B) L929sA cells were pre-treated with 10 µM H89 for 2 h, either followed or not by induction with 2000 IU/ml TNF for 15 min. Cytoplasmic and nuclear cell lysates were prepared and subjected to immunoprecipitation with an MSK1 antibody. Co-precipitating p65 was detected by western analysis using an anti-p65 antibody.
In Figure 6B, the interplay between the endogenous factors p65 and MSK1 is also shown to be strictly TNF stimulus dependent, and can be significantly reduced with H89, reconfirming an activation-dependent association of MSK1 with the p65 subunit of NF-κB.
MSK1 selects and activates NF-κB-driven genes
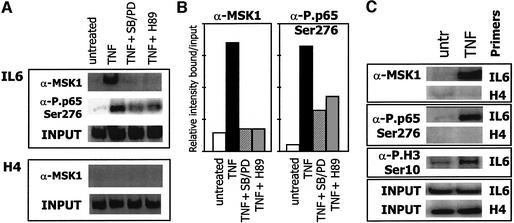
Further in vivo evidence for a specific role for MSK1 in NF-κB-dependent gene expression was obtained by chromatin immunoprecipitation (ChIP) analysis. From Figure 7A, it appears that, in response to TNF, MSK1 localizes at the endogenous NF-κB-containing IL-6 promoter, but not at the histone H4 promoter, which does not contain an NF-κB-responsive element. Consequently, it is not surprising that the phosphorylated MSK1 substrates, such as phospho-Ser276 p65 and phospho-histone H3, are also localized at the endogenous IL-6 promoter following TNF stimulation (Figure 7C). This demonstrates that activated MSK1 and its phosphorylated substrates are an essential component of the transcriptionally active IL-6 promoter enhanceosome, centered at the NF-κB site (Vanden Berghe et al., 1999). Interestingly, although MAPK inhibitors and H89 can completely block MSK1 recruitment (Figure 7A), phosphorylation of NF-κB Ser276 is only partially reduced at the IL-6 promoter, suggesting the involvement of multiple kinases in Ser276 phosphorylation.
Fig. 7. MSK1 and its phosphorylated substrates position at the endogenous IL-6 promoter upon TNF treatment. L929sA mouse fibroblasts were starved for 24–48 h in 0.5% serum. Quiescent cells were treated for 30 min with 2500 IU/ml TNF alone, or following 2 h pre-treatment with the inhibitors SB203580 + PD98059 (10 µM) or H89 (10 µM). ChIP analysis was performed against MSK1, or against phospho NF-κB p65 (Ser276) (A and C) or phospho H3 (Ser10) (C). After reversal of cross-linking, co-immunoprecipitated genomic DNA fragments were analyzed by quantitative PCR for 27 cycles with IL-6 or H4 promoter-specific primer sets. Input reflects the relative amounts of sonicated DNA fragments present before immunoprecipitation and revealed by quantitative PCR with either IL-6- or H4-specific primers. (B) Schematic representation of the results obtained in (A).
Disruption of MSK genes selectively decreases TNF-dependent p65 transactivation
To analyze further the physiological importance of MSK in p65-driven gene expression, we studied p65 transcriptional activity in MSK1/MSK2 ‘double knockout’ MEFs. Studies were performed in ‘double knockout’ cells to prevent misleading results caused by the possible functional redundancy of the two kinases (Wiggin et al., 2002).
From Figure 8, it is clear that MSK kinases are essential in the control of p65 transcriptional activity in response to TNF, although IκB degradation and nuclear translocation (Figure 8A) as well as DNA binding (Figure 8B) remain unaffected. A significant reduction in gal4-p65 transactivation can be observed on a Gal4-driven promoter in MSK1–/–/MSK2–/– as compared with wt MEFs (Figure 8C). Confocal microscopy of TNF-treated MSK knockout cells further reveals a reduction in nuclear staining intensities of phospho-p65 Ser276 as compared with wt cells (Figure 8D). Finally, and most interestingly, upon detailed analysis of expression levels of endogenous target genes of NF-κB, selective reduction in the expression of the inflammatory gene IL-6 was observed, whereas expression of another regulatory NF-κB target gene (NFKB2) remained unaffected (Figure 8E).
Fig. 8. Characterization of MSK1–/–/MSK2–/– MEFs. MSK1–/–/MSK2–/– and wt cells were treated with 2000 IU/ml TNF for the indicated times. Nuclear and total cell extracts were prepared to visualize nuclear import of p65 and IκBα degradation, respectively (A) and NF-κB DNA-binding capacity (B). (C) MSK1/MSK2 double knockout and wt MEFs were transiently transfected with Gal4-p65 expression vector together with p(gal4)2hu.IL6-luc+. At 48 h post-transfection, cells were left untreated or were treated with 2000 IU/ml TNF for 6 h. Corresponding cell lysates were analyzed for luciferase activity and normalized for transfection efficiency by quantifying β-galactosidase levels expressed upon co-transfection of pPGKβgeobpA. (D) Confocal microscope images of TNF-induced wt or MSK1–/–/MSK2–/– MEFs showing Ser276-phosphorylated p65 (green) and PI-stained (red) nuclei. (E) L929sA, wt and MSK1–/–/MSK2–/– MEFs were left untreated or were treated for 4 h with 2000 IU/ml TNF, either pre-treated or not with 10 µM SB203580 and 10 µM U0126 for 2 h. Total RNA was isolated and analyzed using GEArray technology according to the manufacturer’s instructions. Specific mRNA expression was normalized for loading differences.
Discussion
In the present study, we show that phosphorylation of the p65 NF-κB subunit is a prerequisite for gene expression in response to TNF, and point to Ser276 in the RHD of p65 as the crucial residue for regulation of the nuclear transactivation capacity. More particularly, phosphorylation of Ser276 is an essential element for engagement of nuclear cofactors and their associated HAT activity, necessary for elaborate gene expression. Whether this engagement relies on conformational changes of CBP upon interaction with NF-κB (Parker et al., 1998) and/or is the result of phosphorylations of NF-κB and CBP, e.g. by TNF induction, is not clear at present (Zhong et al., 1998, 2002). Interestingly, reconstitution of p65–/– MEFs with various serine to alanine substituted mutants of p65 recently was found to completely rescue TNF-induced IL-6 production with S529A, S536A and S529A/S536A mutants, whereas the S276A mutant had an impaired ability to rescue this response (Okazaki et al., 2003). We identified the dually regulated nuclear kinase MSK1 as a candidate for phosphorylation of Ser276. This kinase, which acts downstream of p38 and ERK MAPKs, is indeed activated by TNF, associates with p65 in a stimulus-dependent manner and specifically phosphorylates the serime residue at position 276, thus leading to its positioning at NF-κB-containing promoter sections and selective stimulation of particular NF-κB-driven genes. Subsequent to MSK1-mediated phosphorylation of p65, a conformational switch may allow engagement of CBP to create a transcriptionally competent enhanceosome. Although deacetylase inhibitors have been shown to prolong phosphorylation of p65 (Ashburner et al., 2001; Chen et al., 2001), it is not clear yet how transcription factor phosphorylation is linked further to acetylation.
Whether MSK1 is the only nuclear p65 kinase is uncertain. It should be noted that besides MSK1 and MSK2, H89 inhibits several protein kinases, such as PKA, RSK, etc. (Davies et al., 2000). Furthermore, reduction of p65 phosphorylation in MSK1–/–/MSK2–/– MEFs in response to TNF is not complete (data not shown) and the transactivation of Gal4-p65 is not entirely knocked out in MSK1–/–/MSK2–/– cells (Figure 8C). Several growth factor and stress signals have been shown to promote phosphorylation of CREB at Ser133, with comparable stoichiometry and kinetics. Coincident with this wide profile of inducibility, CREB is a substrate for various cellular kinases including pp90rsk, PKA, PKC, Akt, MSK1, MSK2, MAPKAPK-2 and Ca2+/calmodulin kinases II and IV (Mayr and Montminy, 2001). The question can be raised of whether p65 Ser276, considering its high homology with the CREB Ser133 motif, displays a similar promiscuity for different kinases and how the transcriptional system centered at p65 can distinguish between various stimuli to produce different gene responses (Deisseroth and Tsien, 2002). The ability of two stimuli to activate distinct genetic programs through a common component has now been noted in eukaryotes and yeast (Mayr and Montminy, 2001; Murphy et al., 2002).
Zhong et al. (1997) pointed to a PKAc subunit present in the cytoplasmic NF-κB–IκB complex, which is able to phosphorylate a subfraction of the cytoplasmic p65 molecules at Ser276 in response to LPS. Whether the same occurs upon stimulation with TNF has not been described. Alternatively, another kinase(s) may also contribute to specific Ser276 phosphorylation of p65. Depending on starvation conditions, cellular confluence and senescence, varying background phosphorylation intensities have been observed, which may explain conflicting results of inducible (Zhong et al., 1997) versus constitutive (Anrather et al., 1999) phosphorylated p65 Ser276 levels; persistent activation of the Ras-MAPK pathway in oncogene-transformed or embryonal cells regularly has been found to result in elevated basal phosphorylation levels of histone H3 and NF-κB p65 (Anrather et al., 1999; Bradbury, 2002; Strelkov and Davie, 2002).
Whether multiple kinases are present simultaneously at the enhanceosome (static model) (Vanden Berghe et al., 2000, 2002) or act subsequently at different stages (dynamic model) (Freeman and Yamamoto, 2002; Morimoto, 2002) and/or at different locations (different subnuclear structures) (Hager et al., 2002) to ensure transcription needs to be investigated further. So far, the lack of specific inhibitors without cross-reactivity does not allow the relative contribution of each kinase in NF-κB-driven gene expression to be untangled easily; therefore, comparison of PKA-, MSK- or RSK-defective MEFs might possibly reveal unique or overlapping signaling functions. Interestingly, whereas p65 S276A mutant mice are lethal (S.Ghosh, personal communication), MSK1–/–/MSK2–/– and RSK2–/– mice are viable (Dufresne et al., 2001; Wiggin et al., 2002). This may suggest a more selective involvement of these kinases in particular subsets of genes. To what extent the core promoter architecture, other MSK1 targets such as CREB and histone H3, and other (as yet unidentified) mechanistic factors also contribute to determine a specific gene expression pattern of NF-κB-driven genes, needs to be explored further.
Although multiple kinases may target the same NF-κB p65 Ser276 residue, a specific biological response may depend on the complete pattern of modifications (such as acetylation, ubiquitylation, methylation or SUMOylation) present in surrounding transcription factor or chromatin domains at a particular moment (Mayr and Montminy, 2001; Deisseroth and Tsien, 2002); by analogy with the proposed histone (Strahl and Allis, 2000; Jenuwein and Allis, 2001) and cofactor code (Rosenfeld and Glass, 2001; Gamble and Freedman, 2002), the complete set of p65 modifications may also determine a unique transcription factor code (Tansey, 2001). Alternatively, specificity may also originate from variable activation kinetics of the kinases (Merienne et al, 2001; Hazzalin and Mahadevan, 2002; Murphy et al., 2002; Vanden Berghe et al., 2002). Part of the specificity and kinetics may also be determined by histone modifications; differences in NF-κB responses (early versus late genes) have now been correlated with differences in H4 acetylation and H3 phosphorylation, as well as with methylation patterns at particular promoters (Saccani et al., 2001, 2002; Saccani and Natoli, 2002). Of special interest is the direct interaction of transcription factor transactivation domains with histones H3/H4, which may restrict signaling effects to promoters with selected nucleosome settings and a particular transcription factor/histone code (Agalioti et al., 2002; Cirillo et al., 2002; Lomvardas and Thanos, 2002).
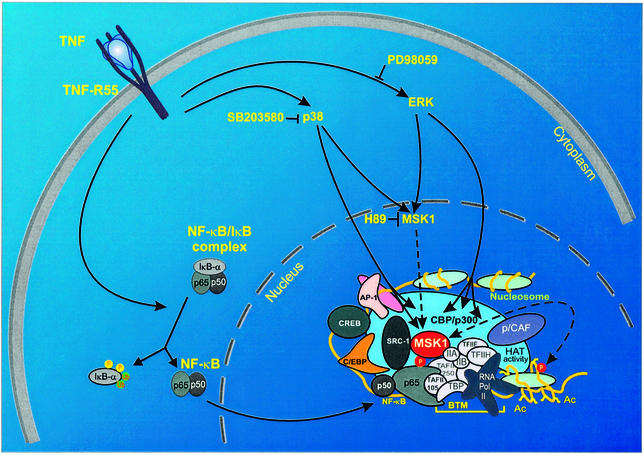
Taken together, we propose a model (Figure 9) in which cytoplasmic NF-κB activation is followed by nuclear phosphorylation of the p65 subunit at Ser276. This involves effective recruitment of the dually MAPK-activated, nuclear kinase MSK1 into the enhanceosome, followed by recruitment of CBP/p300 and accompanying phosphorylation of the chromatin environment, e.g. histone H3. This is, to our knowledge, the first evidence of a kinase phosphorylating both the driving transcription factor and the surrounding chromatin components. However, as (co)factor/histone modifications may occur selectively on a subset of target genes, it will now be interesting to investigate the spatio-temporal relationships between various kinases and their activation pattern in order to drive NF-κB-responsive genes differentially, in relation to surrounding chromatin and factor modifications. Tandem phosphorylation of transcription factor and chromatin components may therefore be an attractive potential target for therapeutic inhibition of various affections. Therefore, the search for other possible dual histone- and p65-specific kinases and their corresponding specific inhibitors will certainly be the subject of further fundamental and applied research.
Fig. 9. Overview of the phosphorylation pathways leading to activation of the IL-6 enhanceosome. Ac, acetyl; AP-1, activator protein-1; BTM, basal transcription machinery; C/EBP, CCAAT enhancer-binding protein; CREB, cAMP-responsive element binding protein; p/CAF, p300/CBP-associated factor; Pol, polymerase; SRC, steroid receptor activator.
Materials and methods
Cell culture, cytokines, inhibitors and antibodies
Maintenance of mouse fibroblast L929sA cells, HEK293 cells, primary fibroblasts from wt and MSK1–/–/MSK2–/– mice, kindly provided by Dr J.S.Arthur (University of Dundee, Dundee, UK), has been described previously (Vanden Berghe et al., 1999; Wiggin et al., 2002). MDA-MB-231 cells were cultured in L-15 medium supplemented with 10% fetal calf serum. Recombinant TNF, produced in Escherichia coli and purified to at least 99% homogeneity in our laboratory, has been described previously (Vanden Berghe et al., 1998). SB203580, PD98059, LY294002 and wortmannin were purchased from Alexis Corporation (Lausen, Switzerland); U0126 was supplied by Promega Biotec (Madison, WI), and H89 was obtained from Calbiochem-Novabiochem International (San Diego, CA). p65 antibody used for immunoprecipitation was from Biomol Research Laboratories (Plymouth Meeting, PA); for western detection of p65, SC-273 was used (Santa Cruz Biotechnology, Santa Cruz, CA). Specific MSK1 antibody and purified PKAc were obtained from Upstate Biotechnology (Lake Placid, NY). CREBtide was a kind gift from Dr M.Gaestel (Martin-Luther University, Halle, Germany). Anti-phospho-H3 antibodies were described previously (Clayton et al., 2000); anti-phospho-Ser276 NF-κB p65 was raised against the CMQLRRPS(phospho)DRELSE peptide and was characterized for specificity.
Plasmids
The reporter plasmids p1168hu.IL6P-luc+ and p(NF-κB)350hu.IL6P-luc+, as well as p(gal4)250hu.IL6P-luc+, were described previously (Vanden Berghe et al., 1998). The expression plasmids pGal4, pGal4-p65 and pGal4-VP16, containing the Gal4 DNA-binding domain, either alone or fused to the full-length p65 or VP16, were obtained from Dr M.L.Schmitz (University of Bern, Bern, Switzerland). Site-directed mutations of pGal4-p65 were performed in order to create specific Ser276→Cys, or Ser529 and Ser536→Ala mutants. Human p65 cDNA was cloned into the expression vector pRcRSV (Invitrogen, San Diego, CA). Mutation of the obtained pRcRSVp65 was achieved by exchange of the AflII–BstEII fragment of the mutated Gal4-p65 variants. pRSVp65 S276A was a kind gift from Dr G.Nabel (National Institutes of Health, Bethesda, MD) and Dr N.Perkins (University of Dundee, Dundee, UK). Expression vectors for wt Flag-tagged or S276A mutant p65 were obtained from Dr A.Baldwin (University of North Carolina, Chapel Hill, NC). pGEXp6512–317 was generously provided by Dr R.Hay (University of St Andrews, St Andrews, UK). A Ser276→Cys mutant of the last construct was prepared by exchange of a SacII–NdeI fragment from pGal4-p65 S276C. An expression vector for MSK1, C-terminally fused to an E-tag (pMSK1-E), was obtained from cloning the human expressed sequence tag (EST) cDNA (AA158571) into pcDNA1. For construction of the kinase-dead mutant (Asp565→Ala), a PCR-based mutation method was used. Expression vectors for Flag-tagged MSK1 and the respective C- and N-terminal kinase-dead mutants as well as the RSK1 expression vector were kindly provided by Dr D.Alessi (University of Dundee, Dundee, UK). The expression vector for MKK6 was kindly provided by Dr R.Davis (Howard Hughes Medical Institute, Worcester, MA) and Dr P.Cohen (University of Dundee, Dundee, UK). The plasmids pPGKβgeobpA, pELAMP-Luc+ and p1481.IL8P-luc+ have been described elsewhere (Vanden Berghe et al., 1999; and references therein). CREB- and AP1-dependent reporter plasmids were part of the PathDetect® system from Stratagene Cloning Systems (La Jolla, CA). The expression vectors containing wt p300 (pCIp300) and the corresponding HAT deletion variant (pCIp300HATΔ1472–1522) were a gift of Dr J.Boyes (MRC Clinical Sciences Centre, London, UK).
Transfections and cytoplasmic/nuclear protein extraction
L929sA and HEK293 cells were transiently or stably transfected by the DEAE–dextran method or the calcium phosphate precipitation procedure, respectively, as described previously (Vanden Berghe et al., 1998, 1999). MEFs were transfected using the fugene transfection method (Roche Molecular Biochemicals, Basel, Switzerland) according to the manufacturer’s instructions. Cytoplasmic and nuclear cell extracts were prepared as described previously (Plaisance et al., 1997).
Analysis of mRNA expression, reporter gene analysis and IL-6 production
Northern blot experiments were performed as described previously (De Bosscher et al., 1997). Alternatively, gene-specific mRNA quantification was performed using GEArray NF-κB Q series filters (Superarray, Bethesda, MD). The arrays were employed according to the manufacturer’s instructions. Quantification and normalization of the obtained hybridization signals were performed using PhosphorImager and SuperArray software. Comparison of the GEArray and northern results revealed good correlation and confirmed signal specificity.
Reporter gene assays were carried out essentially as described previously (Vanden Berghe et al., 1998), and IL-6 production was measured using mouse and human IL-6 enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN).
In vivo 32P-labeling and EMSA
Metabolic labelling of the cells was carried out essentially as described by Beyaert et al. (1996). The EMSA procedure has been described previously (De Bosscher et al., 1997).
In vitro kinase assay
GST, GSTp6512–317 and GSTp6512–317 S276C were produced and purified according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Rainham, UK). A 5 µg aliquot of the purified proteins was incubated at 30°C for 20 min in PKA kinase buffer (50 mM Tris pH 7.4, 10 mM MgCl2, 0.1 mM EGTA, 0.1% β-mercaptoethanol and 10 mM NaF), supplemented with 10 µCi [γ-32P]ATP and 15 ng of purified PKA.
For MSK1 in vitro kinase assays, activated MSK1-E was isolated by E-tag immunoprecipitation from HEK293 cells overexpressing MSK1-E, MKK6 and p38. The isolated MSK1 was added to GST, GSTp6512–317 or GSTp6512–317 S276C as a substrate. The kinase reaction was performed for 20 min at 30°C in the same reaction buffer as described above, supplemented with 2.5 µM PKI. Phosphorylated GST or fusion proteins were subjected to SDS–PAGE; phosphate incorporation was analyzed using PhosphorImager technology.
For CREBtide phosphorylation tests, endogenous MSK1 was immunoprecipitated from L929sA cells, pre-treated or not for 4 h with SB203580, PD98059 or their combination, and/or induced with TNF for 15 min. Isolated MSK1 was incubated with 30 µM CREBtide for 20 min at 30°C in the phosphorylation buffer described above. Incorporation of phosphate into peptides was determined using p81 phosphocellulose paper (Alessi et al., 1995).
Immunofluorescence staining
Immunofluorescence assay and 4′,6-diamidino-2-phenylindole (DAPI) staining of the cells were performed as previously described (Vanden Berghe et al., 1999). Non-specific recognition of unphosphorylated p65 by anti-phospho-Ser276 antibody was blocked by incubation of the antibody with the unphosphorylated CMQLRRPSDRELSE peptide for 2 h, before administration to the cells.
Chromatin immunoprecipitation assay
ChIP analysis was performed as described previously (Clayton et al., 2000), under modified cross-linking and sonication conditions (Nissen and Yamamoto, 2000). Input controls reflect relative amounts of sonicated DNA fragments present before immunoprecipitation. Quantitative PCR of co-immunoprecipitated genomic DNA fragments was performed with the following promoter-specific primers: IL-6 sense 5′-TGACTTCAGCTTTACTCTTGT-3′, IL-6 antisense 5′-CTGATTGGAAACCTTATTAAG-3′; H4 sense 5′GACACCGCATGCAAAGAATAGCTG-3′ and H4 antisense 5′-CTTTCCCAAGGCCTTTACCACC-3′.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank N.De Waele, D.Peelman and K.Van Wesemael for technical assistance, F.Jacquemotte (Institut Meurice, Brussels, Belgium) for synthesis of p65-tide, Dr P.Cohen, Dr J.S.Arthur and G.Wiggin (University of Dundee, Dundee, UK) for helpful discussions and for giving us the opportunity to use the MSK1–/–/MSK2–/– MEFs in our studies, Drs C.Armstrong and J.Leitch for production and purification of the phospho-specific Ser276 p65 antibody, and Drs L.Mahadevan and A.Clayton (Department of Biochemistry, Nuclear Signaling Laboratory, University of Oxford, Oxford, UK) for H3 immunoreagents and excellent assistance during ChIP assay performance. Research was supported by the IUAP and the IWT. L.V. and W.V.B. were granted an EMBO short-term fellowship. W.V.B. is a post-doctoral fellow with the F.W.O-Vlaanderen.
References
- Agalioti T., Chen,G. and Thanos,D. (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111, 381–392. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Cohen,P., Ashworth,A., Cowley,S., Leevers,S.J. and Marshall,C.J. (1995) Assay and expression of mitogen-activated protein kinase, MAP kinase kinase and Raf. Methods Enzymol., 255, 279–290. [DOI] [PubMed] [Google Scholar]
- Anrather J., Csizmadia,V., Soares,M.P. and Winkler,H. (1999) Regulation of NF-κB RelA phosphorylation and transcriptional activity by p21ras and protein kinase Cζ in primary endothelial cells. J. Biol. Chem., 274, 13594–13603. [DOI] [PubMed] [Google Scholar]
- Ashburner B.P., Westerheide,S.D. and Baldwin,A.S.,Jr (2001) The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol., 21, 7065–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaert R., Cuenda,A., Vanden Berghe,W., Plaisance,S., Lee,J.C., Haegeman,G., Cohen,P. and Fiers,W. (1996) The p38/RK mitogen-activated protein kinase pathway regulates interleukin-6 synthesis in response to tumour necrosis factor. EMBO J., 15, 1914–1923. [PMC free article] [PubMed] [Google Scholar]
- Boone E., Vandevoorde,V., De Wilde,G. and Haegeman,G. (1998) Activation of p42/p44 mitogen-activated protein kinases (MAPK) and p38 MAPK by tumor necrosis factor (TNF) is mediated through the death domain of the 55-kDa TNF receptor. FEBS Lett., 441, 275–280. [DOI] [PubMed] [Google Scholar]
- Bradbury E.M. (2002) Chromatin structure and dynamics: state-of-the-art. Mol. Cell, 10, 13–19. [DOI] [PubMed] [Google Scholar]
- Chen L., Fischle,W., Verdin,E. and Greene,W.C. (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science, 293, 1653–1657. [DOI] [PubMed] [Google Scholar]
- Cirillo L.A., Lin,F.R., Cuesta,I., Friedman,D., Jarnik,M. and Zaret,K.S. (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell, 9, 279–289. [DOI] [PubMed] [Google Scholar]
- Clayton A.L., Rose,S., Barratt,M.J. and Mahadevan,L.C. (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J., 19, 3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S.P., Reddy,H., Caivano,M. and Cohen,P. (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J., 351, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bosscher K., Schmitz,M.L., Vanden Berghe,W., Plaisance,S., Fiers,W. and Haegeman,G. (1997) Glucocorticoid-mediated repression of NF-κB-dependent transcription involves direct interference with transactivation. Proc. Natl Acad. Sci. USA, 94, 13504–13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak M., Clifton,A.D., Lucocq,L.M. and Alessi,D.R. (1998) Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38 and may mediate activation of CREB. EMBO J., 17, 4426–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. and Tsien,R.W. (2002) Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron, 34, 179–182. [DOI] [PubMed] [Google Scholar]
- Dong C., Davis,R.J. and Flavell,R.A. (2002) MAP kinases in the immune response. Annu. Rev. Immunol., 20, 55–72. [DOI] [PubMed] [Google Scholar]
- Dufresne S.D., Bjorbaek,C., El-Haschimi,K., Zhao,Y., Aschenbach,W.G., Moller,D.E. and Goodyear,L.J. (2001) Altered extracellular signal-regulated kinase signaling and glycogen metabolism in skeletal muscle from p90 ribosomal S6 kinase 2 knockout mice. Mol. Cell. Biol., 21, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B.C. and Yamamoto,K.R. (2002) Disassembly of transcrip tional regulatory complexes by molecular chaperones. Science, 296, 2232–2235. [DOI] [PubMed] [Google Scholar]
- Gamble M.J. and Freedman,L.P. (2002) A coactivator code for transcription. Trends Biochem. Sci., 27, 165–167. [DOI] [PubMed] [Google Scholar]
- Ghosh S. and Karin,M. (2002) Missing pieces in the NF-κB puzzle. Cell, 109Suppl., S81–S96. [DOI] [PubMed] [Google Scholar]
- Hager G.L., Elbi,C. and Becker,M. (2002) Protein dynamics in the nuclear compartment. Curr. Opin. Genet. Dev., 12, 137–141. [DOI] [PubMed] [Google Scholar]
- Hazzalin C.A. and Mahadevan,L.C. (2002) Transcription MAPK-regulated transcription: a continuously variable gene switch? Nat. Rev. Mol. Cell Biol., 3, 30–40. [DOI] [PubMed] [Google Scholar]
- Holtmann H. et al. (1999) Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol., 19, 6742–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P. (1992) Regulating transcription factor activity by phosphorylation. Trends Cell Biol., 2, 104–108. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Karin M. and Ben-Neriah,Y. (2000) Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol., 18, 621–663. [DOI] [PubMed] [Google Scholar]
- Kracht M. and Saklatvala,J. (2002) Transcriptional and post-transcrip tional control of gene expression in inflammation. Cytokine, 20, 91–106. [DOI] [PubMed] [Google Scholar]
- Krause A., Holtmann,H., Eickemeier,S., Winzen,R., Szamel,M., Resch,K., Saklatvala,J. and Kracht,M. (1998) Stress-activated protein kinase/Jun N-terminal kinase is required for interleukin (IL)-1-induced IL-6 and IL-8 gene expression in the human epidermal carcinoma cell line KB. J. Biol. Chem., 273, 23681–23689. [DOI] [PubMed] [Google Scholar]
- Kunsch C. and Rosen,C.A. (1993) NF-κB subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol., 13, 6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. and Verma,I.M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol., 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Lomvardas S. and Thanos,D. (2002) Opening chromatin. Mol. Cell, 9, 209–211. [DOI] [PubMed] [Google Scholar]
- Mayr B. and Montminy,M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol., 2, 599–609. [DOI] [PubMed] [Google Scholar]
- Merienne K., Pannetier,S., Harel-Bellan,A. and Sassone-Corsi,P. (2001) Mitogen-regulated RSK2–CBP interaction controls their kinase and acetylase activities. Mol. Cell. Biol., 21, 7089–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R. (2002) Dynamic remodeling of transcription complexes by molecular chaperones. Cell, 110, 281. [DOI] [PubMed] [Google Scholar]
- Mukaida N., Mahe,Y. and Matsushima,K. (1990) Cooperative interaction of nuclear factor-κB- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J. Biol. Chem., 265, 21128–21133. [PubMed] [Google Scholar]
- Murphy L.O., Smith,S., Chen,R.H., Fingar,D.C. and Blenis,J. (2002) Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol., 4, 556–564. [DOI] [PubMed] [Google Scholar]
- Nissen R.M. and Yamamoto,K.R. (2000) The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev., 14, 2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T., Sakon,S., Sasazuki,T., Sakurai,H., Doi,T., Yagita,H., Okumura,K. and Nakano,H. (2003) Phosphorylation of serine 276 is essential for p65 NF-κB subunit-dependent cellular responses. Biochem. Biophys. Res. Commun., 300, 807–812. [DOI] [PubMed] [Google Scholar]
- Parker D., Jhala,U.S., Radhakrishnan,I., Yaffe,M.B., Reyes,C., Shulman,A.I., Cantley,L.C., Wright,P.E. and Montminy,M. (1998) Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell, 2, 353–359. [DOI] [PubMed] [Google Scholar]
- Pearson G., Robinson,F., Beers Gibson,T., Xu,B.E., Karandikar,M., Berman,K. and Cobb,M.H. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev., 22, 153–183. [DOI] [PubMed] [Google Scholar]
- Plaisance S., Vanden Berghe,W., Boone,E., Fiers,W. and Haegeman,G. (1997) Recombinant signal sequence binding protein Jκ is constitutively bound to the NF-κB site of the interleukin-6 promotor and acts as a negative regulatory factor. Mol. Cell. Biol., 17, 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M.G. and Glass,C.K. (2001) Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem., 276, 36865–36868. [DOI] [PubMed] [Google Scholar]
- Saccani S. and Natoli,G. (2002) Dynamic changes in histone H3 Lys9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev., 16, 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S., Pantano,S. and Natoli,G. (2001) Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med., 193, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S., Pantano,S. and Natoli,G. (2002) p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol., 3, 69–75. [DOI] [PubMed] [Google Scholar]
- Sakurai H., Chiba,H., Miyoshi,H., Sugita,T. and Toriumi,W. (1999) IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem., 274, 30353–30356. [DOI] [PubMed] [Google Scholar]
- Schindler U. and Baichwal,V.R. (1994) Three NF-κB binding sites in the human E-selectin gene required for maximal tumor necrosis factor α-induced expression. Mol. Cell. Biol., 14, 5820–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M.L. and Baeuerle,P.A. (1991) The p65 subunit is responsible for the strong transcription activating potential of NF-κB. EMBO J., 10, 3805–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M.L., Bacher,S. and Kracht,M. (2001) IκB-independent control of NF-κB activity by modulatory phosphorylations. Trends Biochem. Sci., 26, 186–190. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strelkov I.S. and Davie,J.R. (2002) Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res., 62, 75–78. [PubMed] [Google Scholar]
- Tansey W.P. (2001) Transcriptional activation: risky business. Genes Dev., 15, 1045–1050. [DOI] [PubMed] [Google Scholar]
- Thomson S., Clayton,A.L., Hazzalin,C.A., Rose,S., Barratt,M.J. and Mahadevan,L.C. (1999) The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J., 18, 4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe W., Plaisance,S., Boone,E., De Bosscher,K., Schmitz,M.L., Fiers,W. and Haegeman,G. (1998) p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-κB p65 transactivation mediated by tumor necrosis factor. J. Biol. Chem., 273, 3285–3290. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W., De Bosscher,K., Boone,E., Plaisance,S. and Haegeman,G. (1999) The nuclear factor-κB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J. Biol. Chem., 274, 32091–32098. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W., Vermeulen,L., De Wilde,G., De Bosscher,K., Boone,E. and Haegeman,G. (2000) Signal transduction by tumor necrosis factor and gene regulation of the inflammatory cytokine interleukin-6. Biochem. Pharmacol., 60, 1185–1195. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W., De Bosscher,K., Vermeulen,L., De Wilde,G. and Haegeman,G. (2002) Induction and repression of NF-κB driven inflammatory genes. Ernst Schering Res. Found. Workshop, 40, 233–278. [DOI] [PubMed] [Google Scholar]
- Vermeulen L., De Wilde,G., Notebaert,S., Vanden Berghe,W. and Haegeman,G. (2002) Regulation of the transcriptional activity of the nuclear factor-κB p65 subunit. Biochem. Pharmacol., 64, 963–970. [DOI] [PubMed] [Google Scholar]
- Wang D. and Baldwin,A.S.,Jr (1998) Activation of nuclear factor-κB-dependent transcription by tumor necrosis factor-α is mediated through phosphorylation of RelA/p65 on serine 529. J. Biol. Chem., 273, 29411–29416. [DOI] [PubMed] [Google Scholar]
- Wang D., Westerheide,S.D., Hanson,J.L. and Baldwin,A.S.,Jr (2000) Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem., 275, 32592–32597. [DOI] [PubMed] [Google Scholar]
- Wesselborg S., Bauer,M.K.A., Vogt,M., Schmitz,M.L. and Schulze Osthoff,K. (1997) Activation of transcription factor NF-κB and p38 mitogen-activated protein kinase is mediated by distinct and separate stress effector pathways. J. Biol. Chem., 272, 12422–12429. [DOI] [PubMed] [Google Scholar]
- Wiggin G.R., Soloaga,A., Foster,J.M., Murray-Tait,V., Cohen,P. and Arthur,J.S. (2002) MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol., 22, 2871–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., SuYang,H., Erdjument Bromage,H., Tempst,P. and Ghosh,S. (1997) The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell, 89, 413–424. [DOI] [PubMed] [Google Scholar]
- Zhong H., Voll,R.E. and Ghosh,S. (1998) Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell, 1, 661–671. [DOI] [PubMed] [Google Scholar]
- Zhong H., May,M.J., Jimi,E. and Ghosh,S. (2002) The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell, 9, 625–636. [DOI] [PubMed] [Google Scholar]