Abstract
Oscillations of cytosolic free calcium levels have been shown to influence gene regulation and cell differentiation in a variety of model systems. Intercellular calcium waves thus present a plausible mechanism for coordinating cellular processes during embryogenesis. Herein we report use of aequorin and a photon imaging microscope to directly observe a rhythmic series of intercellular calcium waves that circumnavigate zebrafish embryos over a 10-h period during gastrulation and axial segmentation. These waves first appeared at about 65% epiboly and continued to arise every 5–10 min up to at least the 16-somite stage. The waves originated from loci of high calcium activity bordering the blastoderm margin. Several initiating loci were active early in the wave series, whereas later a dorsal marginal midline locus predominated. On completion of epiboly, the dorsal locus was incorporated into the developing tail bud and continued to generate calcium waves. The locations and timing at which calcium dynamics are most active appear to correspond closely to embryonic cellular and syncytial sites of known morphogenetic importance. The observations suggest that a panembryonic calcium signaling system operating in a clock-like fashion might play a role during vertebrate axial patterning.
The vertebrate body plan emerges during gastrulation through patterns of inductive interactions, cellular rearrangements, and gene expression, some of which may be coordinated across large distances with considerable temporal precision (1–3). Morphogen gradients and propagating second messenger waves, especially those involving calcium and inositol 1,4,5-trisphosphate, have been proposed as carriers of such putative long-range coordinating messages (4–7). Intercellular calcium waves are of particular interest because oscillations of intracellular calcium levels have been shown to directly influence expression of numerous developmentally regulated genes in cultured cells (8, 9). To search for evidence of possible large-scale embryonic signaling mechanisms, we imaged whole-embryo calcium dynamics in an optically suitable vertebrate, the zebrafish (Danio rerio), by using a bioluminescent calcium reporter. An imaging system using a photon-counting spatial detector allowed for long-term imaging of calcium-triggered luminescence with high temporal and moderate spatial resolution (16-kHz sampling rate; spatial coordinate units = 12 μm2 with a ×10 objective). Because at this spatial scale, light arising from cells well above and below the nominal object plane still gave useful imaging information, an effective imaging field depth of 100–200 μm was achieved. A surprising array of spatially and temporally complex calcium transients was imaged with this technique. Herein we report initial observations on the most unusual of these patterns, a rhythmic series of intercellular calcium waves that traverse the blastoderm margin and main body axis during gastrulation and axial segmentation.
Zebrafish eggs were injected 10 min after fertilization with 10−13 mol of recombinant semisynthetic f-aequorin, a calcium reporter whose luminescence increases at the second power of the free calcium concentration (10). f-Aequorin loaded at the single cell stage spread throughout the cytoplasm and was partitioned into all cleavage stage blastomeres and the later-forming yolk syncytial layer. Individual injected embryos, selected for normal morphology and uniform aequorin loading, were transferred at the 30–40% epiboly stage to a photon imaging microscope (11, 12). Aequorin luminescence was imaged during 12 h of development up to the 16-somite stage. Luminescent imaging was interrupted periodically to capture bright-field images thus allowing correlation of luminescence and morphological features (e.g., Fig. 1 A–F). Calcium signals associated with ooplasmic segregation (12), early blastomere cleavage (13), and a variety of later morphogenetic events (14), have been studied in zebrafish using the aequorin reporter. Early zebrafish development, through mid-blastula stages, has also been studied by using fluorescent calcium reporters and confocal microscopy (15, 16). This report addresses developmental stages from 30% epiboly up to 16 somites (17), a period that covers “primary” as well as much of “secondary” gastrulation (18).
Figure 1.
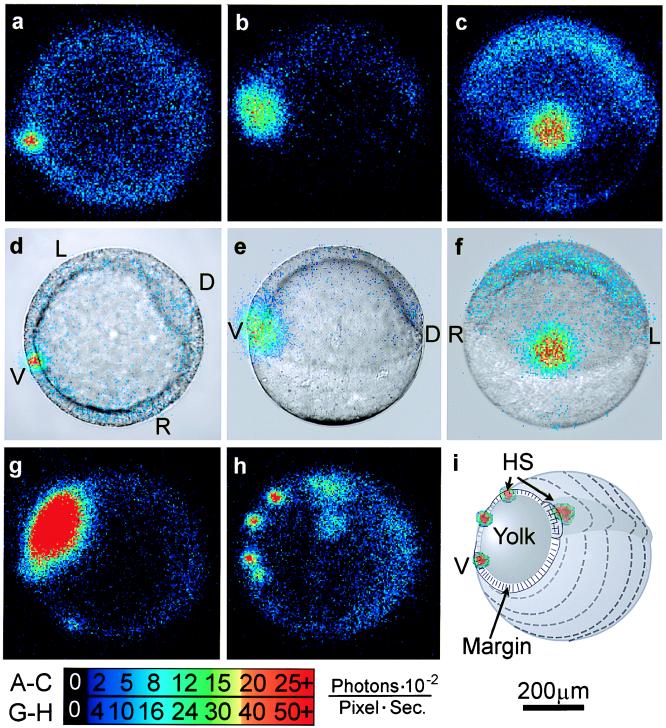
Three patterns of calcium transients that appear between 50% and 75% epiboly are ventral marginal signal (A–F), yolk flash (G), and marginal hot spots (H). (A–F) The persistent ventral signal is shown in f-aequorin-loaded zebrafish embryos at approximately 50% epiboly. (A–C) Sixty seconds of accumulated luminescence. (D–F) Same data superimposed on corresponding bright-field images. In A and D, the embryo is viewed from the vegetal pole, in B and C it is viewed from the left side, and in E and F it is viewed from the ventral side. (G) A rapid increase in calcium in the exposed portion of the yolk cell appears as a brief “yolk flash,” shown in a 30-sec integration at the late shield stage. This image corresponds to the large spike at 100 min in Fig. 2A. A time-lapse imaging sequence of the yolk flash is shown in the supplementary material on the PNAS web site. (H) Multiple loci of persistent elevated calcium levels, or marginal hot spots, are shown in a 30-sec integration of photons from minute 135 in Fig. 2B. (I) Schematic showing orientation of G and H. D, dorsal; V, ventral; L, left; R, right; HS, hot spots. The color code indicates luminescent flux in the same units as in Figs. 2 and 3. The scale bar applies to all frames.
MATERIALS AND METHODS
The methods used in this study are described in detail elsewhere (11, 13). Briefly, fertilized zebrafish eggs were collected within 5 min of spawning, enzymatically dechorionated, and injected with approximately 0.9 nl of a 1% solution of recombinant f-aequorin in 100 mM KCl, 5 mM Mops, and 50 μM EDTA. During imaging the embryos were maintained at 28°C in 30% Danieau’s medium containing penicillin (0.5 mg/ml), streptomycin (5,000 units/ml; Sigma), and 0.5% methylcellulose. Imaging was performed on a Photon Imaging Microscope (Science Wares, Falmouth, MA) that used a photon-counting spatial detector with a resistive anode output (Photek, St. Albens-on-Sea, U.K.). Digitized detector output in the form of a stream of time-labeled eight-bit x--y coordinates (256 × 256 pixels) was used to construct time-lapse imaging sequences. The imaging system software allowed the original photon data stream to be analyzed according to any chosen integration time, with the resulting image frames maintaining accurate photon quantitation up to 256 photons per pixel. Image sequences constructed with data integration times between 0.5 and 240 sec were used for the current study. The photon image frames were exported to nih image and metamorph 2.75 (Universal Imaging) for further quantitative analysis. Time- lapse digital sequences are published as supplemental material on the PNAS web site (www.pnas.org).
RESULTS
In the 1-h period after 30% epiboly, the embryos exhibited nearly uniform low calcium levels (Fig. 2A, quiet period). Over the subsequent hour (germ ring and shield stages), a small region on the ventral side of the blastoderm margin displayed free calcium levels up to 6- to 8-fold greater than the rest of the embryo for periods of 5–45 min (n = 9; Figs. 1 A–F, and 2A, ventral signal).
Figure 2.
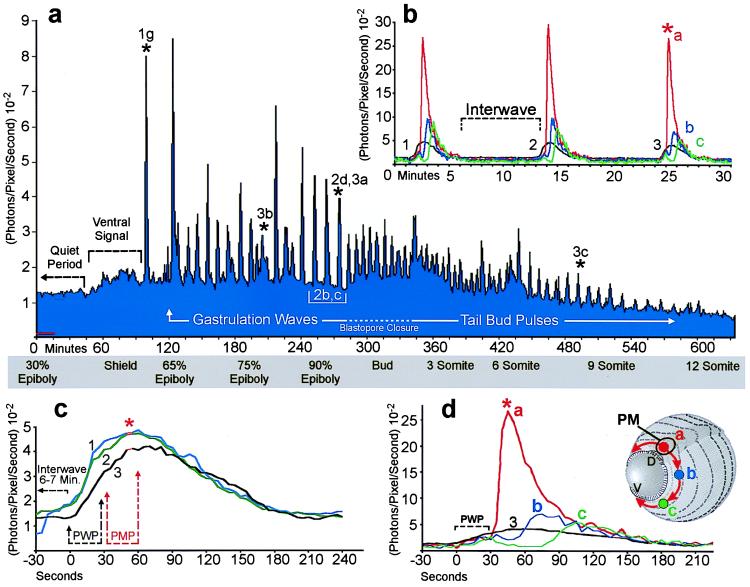
Periodic increases of intracellular calcium recur throughout gastrulation as waves that propagate around the blastoderm margin. (A) Regularity of the wave pulses is shown over 10 h of development from 30% epiboly to 12 somites. Stages are indicated in the gray box beneath the graph. The data stream was binned in 60-sec intervals for quantification. The abscissa indicates photon flux as the mean value of photons (10−2) per pixel per sec for an imaging field covering the entire embryo (approximately 14,500 pixels). Time is in min with t = 0 set at about 5 h after fertilization. The average detector background level (noise) of the imaging system is shown by the red line near the origin, representing approximately 10 “noise” photons per sec for the area covered by the embryo. Each calcium wave appears as a spike in the graph. Waves are arbitrarily divided into gastrulation waves before blastopore closure and tail bud pulses afterwards. The rise in luminescence between 60 and 100 min indicates the appearance of a prolonged locus of increased calcium on the ventral side of the blastoderm margin (see Fig. 1 A–F). Asterisks above the graph indicate the yolk flash shown in Fig. 1G and the waves in Figs. 2D and 3 A–C. (B and C) Three waves from the series in A are plotted at higher temporal resolution to demonstrate wave durations, interwave intervals, and temporal components. (B) The black trace represents whole-embryo luminescence, as in A, but quantified in 5-sec intervals. Red, blue, and green traces indicate spatial subsamples from locations indicated by the schematic embryo in D. (C) The whole-embryo luminescence data for the three waves shown in B (black trace) are aligned at the time of maximal pacemaker luminescence (red asterisk). Prewave pulse (PWP) and pacemaker pulse (PMP) time intervals are indicated. (D) Whole-embryo luminescence (black trace) and spatial subsamples (red, blue, and green) for the third wave shown in B and C are graphed at a higher temporal resolution. The imaging sequence for this wave is shown in Fig. 3A. The schematic inset shows the locations of the 400-pixel sampling regions and the pathway of wave propagation from the dorsal pacemaker hot spot (PM; arrows). Time-lapse imaging sequences for the data in a, b, and d are shown in the supplementary material on the PNAS web site.
At approximately 65% epiboly, with the ventral signal generally still persisting, a period of dynamic calcium activity ensued. This activity was often preceded by one or more brief 45- to 90-sec calcium transients covering the exposed surface of the yolk cell (Fig. 1G). Additional foci of elevated calcium also generally appeared at other locations around the blastoderm margin (Figs. 1H and 3 A and B). These marginal “hot spots” persisted for various times and served as initiating sites and sinks for a series of periodic, long-range propagating calcium waves. Fig. 2A shows the average luminescent flux (photons per pixel per sec) for a whole embryo over a 10-h period during which 64 calcium waves arose. Each wave appears as a spike in the luminescence data. The first two spikes, at 95 and 125 min along the time line, represent a yolk flash and a burst from the original ventral hot spot, respectively. Continuing immediately after the ventral burst, a series of “gastrulation waves” occurred with an average frequency of about 7 waves per h up to the time of blastopore closure (Fig. 2A). The wave frequency increased after blastopore closure, generally up to 11–12 waves per h, and then decreased during later stages of somitogenesis (Fig. 2A, tail bud pulses). Although the wave frequency varied between embryos, and at different developmental stages within individual embryos, successive waves generally originated in sequences with clear periodicity over 1–12 h.
Figure 3.
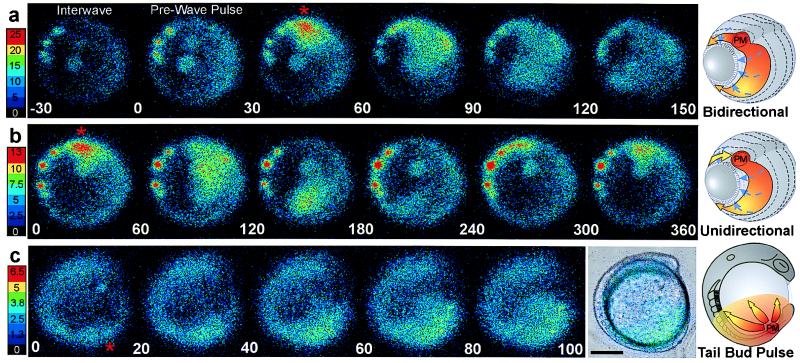
Imaging sequences and schematics showing the basic spatial wave types. The color scales indicate luminescent flux in [(photons per pixel per sec) × 10−2]. (Bar = 200 μm.) The red asterisks and PM in the schematics indicates the dorsal midline pacemaker or wave initiating site. (A) Sequence of 30-sec data integrations showing a bidirectional propagating calcium wave occurring at about 90% epiboly (Fig. 2A, asterisk). Data from this wave are plotted in matching time intervals in Fig. 2D. The end of the interwave interval is shown in the first frame, the prewave pulse is in the second frame, and the dorsal pacemaker pulse and propagating wave are in the next four frames. The schematic shows a generalized bidirectional wave oriented as in the imaging frames. Blue dashed arrows indicate epibolic movements. (B) A unidirectional wave that required about 5 min to circumnavigate the blastoderm margin is shown in sequential 60-sec integrations starting with the pacemaker pulse. (C) During somitogenesis, periodic radial calcium wave fronts spread from the tail bud region. The images in C represent a 120-sec integration window moving along the data stream in 20-sec steps at the 10-somite stage. Time-lapse movies of these imaging sequences are shown in the supplemental material on the PNAS web site.
The time course of individual waves also showed striking regularity. Three waves from the series shown in Fig. 2A are illustrated at higher temporal resolution in Fig. 2 B (black data trace) and C. The intervals between wave onset were 12.5 and 11 min, the duration of each wave was 4 min, and the interwave intervals was about 7 min (Fig. 2B). Plotting the waves with their times of peak luminescence set to coincide demonstrates that the overall time course, including rise time and wave duration, are very similar for a given spatial wave type (Fig. 2C).
The origination and termination sites and the propagation route of each wave could be observed and quantified by analysis of the time-lapse imaging sequences (see movies in supplementary materials). Waves propagating around the circumference of the blastoderm margin predominated in all experiments. These marginal waves propagated either uni- or bidirectionally from their initiation site, often a persistent hot spot (Fig. 3 A and B). In some wave series, hot spots were not evident, likely due to the position of the spots relative to the imaging focal zone. Bidirectional waves spread in two opposite directions from their point of origin. Both components of bidirectional waves traversed the blastoderm margin with roughly equal velocity, appearing to annihilate each other opposite their point of origin (Fig. 3A and movies). In contrast, unidirectional waves were imaged as single wave fronts that circumnavigated the entire blastoderm margin and generally terminated at their point of origin (Fig. 3B). The average velocity for these wave types was about 4–5 μm/sec. Thus, unidirectional waves required twice as long as bidirectional waves to traverse their routes at a given stage of epiboly (Fig. 3 A and B). In most experiments, some of the waves emanating from the dorsal midline of the blastoderm margin traveled up the main embryonic axis toward the animal pole rather than around the blastoderm margin (see movies). These “axial” waves initiated within the expected time windows for marginal waves, and thus appeared to be members of the same wave series traveling along alternative routes. Fig. 2 B and D shows luminescent flux from small spatial subsamples (400 pixels) covering a dorsal hot spot and two areas along the right arm of the bidirectional wave shown in the correlated image sequence in Fig. 3A. These data show that calcium levels at the initiating site (red asterisk) were 50–100% higher than at other parts of the wave and, therefore, at least 5-fold greater than the levels during the interwave interval. Smaller spatial samples (20 pixels) from the center of the wave initiation sites (data not shown) indicated that calcium levels reached local maxima of 10-fold greater than the resting level. If the basal intracellular calcium levels are 50–100 nM (4, 16), the peak levels during wave initiation appear to be in the range of 500–1,000 nM.
Between 65% and 85% epiboly, wave initiation was sometimes distributed among three to four loci, with a given locus generating two to six waves and then another taking the lead. In the case shown in Fig. 2A, the seven waves arising between minutes 165 and 200 originated from hot spots on the left side of the embryo (see movie). After about 85% epiboly the periodic waves almost exclusively originated from the dorsal midline, or node region, of the gastrulation margin, this site thus appearing as a stable pacemaker region (Fig. 3) over the subsequent 5–7 h. After completion of epiboly at the bud stage, the dorsal wave initiation zone became incorporated into the developing tail bud just rostral to the closing blastopore (Fig. 3C). Calcium waves continued to spread from the tail bud zone throughout the caudal half of the embryo. Some of these “tail bud pulses” continued up to the head, along either dorsal or ventral routes. The tail bud pulses declined in intensity and were difficult to visualize after 14- to 16-somite stages, likely due to the reduced concentration of unspent aequorin.
The localized peak calcium transient at the onset of each wave was generally preceded by a single brief and much more widespread calcium transient that often encompassed the entire embryo within a 15- to 30-sec period (Figs. 2 C and D and 3A). These “prewave pulses” accounted for the first 30 sec of the rise in whole-embryo luminescence preceding the steep onset of localized hot spot transients (Figs. 2D and 3A). Although the prewave calcium rise often seemed to appear earliest toward the animal pole, the time resolution of these observations was not sufficient to clearly resolve sites of origin or to allow confident estimation of the velocities of the pre-wave pulses. The more rapid propagation of the prewave calcium rise suggests a different mechanism than that responsible for the slower gastrulation waves. It is possible that the timing mechanism underlying the periodicity of the gastrulation waves involves these prewave pulses.
DISCUSSION
These findings document a level of patterned long-distance calcium-mediated cellular interaction that could provide a mechanism for coordinating the spatial and temporal regulation of highly localized processes across large cellular domains. Intercellular transmission of calcium waves is generally considered to require diffusion of either inositol 1,4,5-trisphosphate or calcium ions through gap junctions (4, 5, 19–21). Although gap junction communication compartments have not been mapped in detail in gastrulating zebrafish, the expression patterns of connexins are consistent with widespread intercellular coupling at these stages (22). Studies in closely related species have shown extensive dye coupling between yolk cell and blastomere domains, as well as significant junctional modulation during gastrulation (23). In addition, propagation of calcium signals involving gap junction-mediated membrane depolarization and entry of extracellular calcium through voltage-gated calcium channels cannot be excluded as possible mechanisms, especially for the rapid prewave pulses. Candidate calcium-responsive targets include systems regulating morphogenetic protein secretion, cytoskeletal activities, and gene expression. Calcium oscillations have recently been demonstrated to directly effect the efficiency and specificity of gene expression in lymphocytes (9), and elements of calcium signaling pathways have been shown to play roles in vertebrate axial specification, neural induction, and neuronal differentiation (24–28). Since the precise configuration of calcium-responsive elements probably differs across an embryo, the observed periodic calcium pulses might allow multiple independent calcium-responsive developmental events to be synchronized within a temporal window of a few minutes.
Some aspects of the calcium dynamics described herein appear to correspond to observations made in other embryonic systems. The early ventral marginal calcium signal might represent an aspect of the similarly localized phosphatidylinositol cycle up-regulation shown to modulate mesoderm induction during dorsoventral specification in Xenopus (24). Although the blastoderm in developing zebrafish does not exhibit significant contraction waves, the initial ventral pulse of calcium and the later emergence of propagating calcium waves could also be related to the ventral marginal pacemaker region and calcium waves that accompany blastoderm contractions in medaka (Oryzias latipes) (29, 30). In addition, the period during which the rhythmic waves occur in zebrafish overlaps with comparable stages of Xenopus in which repetitive calcium pulses, and possibly waves, with 5- to 10-min periods were reported for dissociated neural plate cells and intact spinal cords (31). Localized pacemaker foci for Ca2+ wave initiation in relation to axis determination have also been reported, albeit at the single cell stage (32, 33).
Because localized calcium hot spots often appeared to act as organizing centers for the gastrulation waves, their structural identity and the basis of their rhythmic activity pose many questions. The limited spatial resolution of the experiments did not allow precise morphological identification of the hot spot domains. However, the long-term persistence of the most stable wave initiation site at the dorsal midline of the blastoderm margin and the later apparent incorporation of this site into the rostral tail-bud region suggests close proximity or identity with a population of noninvoluting highly endocytic marginal cells described at these stages and locations (34). Other more variable hot spot activities may reside in the yolk syncytial cytoplasm directly beneath the blastoderm or, more likely, include both yolk syncytial and blastoderm components. The steady rhythmicity of wave initiation could result from intrinsic oscillatory properties of small clusters of coupled cells. However, the distribution of wave initiation among multiple hot spots during a steady periodic series suggests involvement either of higher level dynamics operating between hot spots or some timing mechanism not directly dependent upon hot spot activity. The brief and widespread calcium transients observed to occur just preceding wave initiation possibly point to a more global explanation. The cellular identity of the wave generating sites, the existence of a pacemaker-like mechanism underlying wave periodicity, and the possible roles played by these waves in development can now be studied by using pharmacological and genetic perturbations.
Supplementary Material
Acknowledgments
We thank Drs. O. Shimomura, Y. Kishi, and S. Inouye for supplying us with f-aequorin and Dr. R. Hanlon and the Marine Resources Center staff for their assistance. This work was supported by RGC HKUST 650/96m, Hong Kong JC, and National Institutes of Health Grants RR10291 and EY02007.
References
- 1.Driever W. Curr Opin Genet Dev. 1995;5:610–618. doi: 10.1016/0959-437x(95)80030-1. [DOI] [PubMed] [Google Scholar]
- 2.Stennard F, Ryan K, Gurdon J B. Curr Opin Genet Dev. 1997;7:620–627. doi: 10.1016/s0959-437x(97)80009-3. [DOI] [PubMed] [Google Scholar]
- 3.Sasai Y, De Robertis E M. Dev Biol. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- 4.Berridge M J. Nature (London) 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe L F. Cell Calcium. 1993;14:736–745. doi: 10.1016/0143-4160(93)90099-r. [DOI] [PubMed] [Google Scholar]
- 6.Nellen D, Burke R, Struhl G, Basler K. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 7.Neumann C, Cohen S. Bioessays. 1997;19:721–729. doi: 10.1002/bies.950190813. [DOI] [PubMed] [Google Scholar]
- 8.Li W-H, Lopias J, Whitney M, Zlokarnik G, Tsien R Y. Nature (London) 1998;392:936–941. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 9.Dolmetsch R E, Xu K, Lewis R S. Nature (London) 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 10.Shimomura O, Inouye S. Biochem Biophys Res Commun. 1996;221:77–81. doi: 10.1006/bbrc.1996.0548. [DOI] [PubMed] [Google Scholar]
- 11.Miller A L, Karplus E, Jaffe L F. Methods Cell Biol. 1994;40:305–338. doi: 10.1016/s0091-679x(08)61120-6. [DOI] [PubMed] [Google Scholar]
- 12.Leung C F, Webb S E, Miller A L. Dev Growth Diff. 1998;40:313–326. doi: 10.1046/j.1440-169x.1998.t01-1-00007.x. [DOI] [PubMed] [Google Scholar]
- 13.Webb S E, Lee K W, Karplus E, Miller A L. Dev Biol. 1997;192:78–92. doi: 10.1006/dbio.1997.8724. [DOI] [PubMed] [Google Scholar]
- 14.Creton R, Speksnijder J E, Jaffe L F. J Cell Sci. 1998;111:1613–1622. doi: 10.1242/jcs.111.12.1613. [DOI] [PubMed] [Google Scholar]
- 15.Chang D C, Meng C. J Cell Biol. 1995;131:1539–1543. doi: 10.1083/jcb.131.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhard E, Yokoe H, Niebling K R, Allbritton N L, Kuhn M A, Meyer T. Dev Biol. 1995;170:50–61. doi: 10.1006/dbio.1995.1194. [DOI] [PubMed] [Google Scholar]
- 17.Kimmel C B, Ballard W W, Kimmel S R, Ullmann B, Schilling T F. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 18.Kanki J P, Ho R K. Development. 1997;124:881–893. doi: 10.1242/dev.124.4.881. [DOI] [PubMed] [Google Scholar]
- 19.Boitano S, Dirksen E R, Sanderson M J. Science. 1992;258:292–295. doi: 10.1126/science.1411526. [DOI] [PubMed] [Google Scholar]
- 20.Allbritton N L, Meyer T. Cell Calcium. 1993;14:691–697. doi: 10.1016/0143-4160(93)90095-n. [DOI] [PubMed] [Google Scholar]
- 21.Sneyd J, Charles A C, Sanderson M J. Am J Physiol. 1994;266:C293–C302. doi: 10.1152/ajpcell.1994.266.1.C293. [DOI] [PubMed] [Google Scholar]
- 22.Essner J J, Laing J G, Beyer E C, Johnson R G, Hackett P B. Dev Biol. 1996;177:449–462. doi: 10.1006/dbio.1996.0177. [DOI] [PubMed] [Google Scholar]
- 23.Bozhkova V P. Membr Cell Biol. 1998;11:803–815. [PubMed] [Google Scholar]
- 24.Ault K T, Durmowicz G, Galione A, Harger P L, Busa W B. Development. 1996;122:2033–2041. doi: 10.1242/dev.122.7.2033. [DOI] [PubMed] [Google Scholar]
- 25.Slusarski D C, Corces V G, Moon R T. Nature (London) 1997;390:410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 26.Slusarski D C, Yang-Snyder J, Busa W B, Moon R T. Dev Biol. 1997;182:114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- 27.Leclerc C, Daguzan C, Nicolas M T, Chabret C, Duprat A M, Moreau M. Mech Dev. 1997;64:105–110. doi: 10.1016/s0925-4773(97)00054-3. [DOI] [PubMed] [Google Scholar]
- 28.Gu X, Spitzer N C. Dev Neurosci. 1997;19:33–41. doi: 10.1159/000111183. [DOI] [PubMed] [Google Scholar]
- 29.Barber B, da Cruz M J, DeLeon J, Fluck R A, Hasenfeld M P, Unis L A. J Exp Zool. 1987;242:35–42. doi: 10.1002/jez.1402420106. [DOI] [PubMed] [Google Scholar]
- 30.Simon J Z, Cooper M S. J Exp Zool. 1995;273:118–129. [Google Scholar]
- 31.Gu X, Olson E C, Spitzer N C. J Neurosci. 1994;14:6325–6335. doi: 10.1523/JNEUROSCI.14-11-06325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckberg W R, Miller A L. Dev Biol. 1995;172:654–664. doi: 10.1006/dbio.1995.8043. [DOI] [PubMed] [Google Scholar]
- 33.Speksnijder J E. Dev Biol. 1992;153:259–271. doi: 10.1016/0012-1606(92)90111-s. [DOI] [PubMed] [Google Scholar]
- 34.D’Amico L A, Cooper M S. Biochem Cell Biol. 1997;75:563–577. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.