Short abstract
Analysis of conservation in eight genomic regions (apterous, even-skipped, fushi tarazu, twist, and Rhodopsins 1, 2, 3 and 4) from four Drosophila species (D. erecta, D. pseudoobscura, D. willistoni, and D. littoralis) covering more than 500 kb of the D. melanogaster genome. All D. melanogaster genes (and 78-82% of coding exons) identified in divergent species such as D. pseudoobscura show evidence of functional constraint. Addition of a third species can reveal functional constraint in otherwise non-significant pairwise exon comparisons.
Abstract
Background
It is widely accepted that comparative sequence data can aid the functional annotation of genome sequences; however, the most informative species and features of genome evolution for comparison remain to be determined.
Results
We analyzed conservation in eight genomic regions (apterous, even-skipped, fushi tarazu, twist, and Rhodopsins 1, 2, 3 and 4) from four Drosophila species (D. erecta, D. pseudoobscura, D. willistoni, and D. littoralis) covering more than 500 kb of the D. melanogaster genome. All D. melanogaster genes (and 78-82% of coding exons) identified in divergent species such as D. pseudoobscura show evidence of functional constraint. Addition of a third species can reveal functional constraint in otherwise non-significant pairwise exon comparisons. Microsynteny is largely conserved, with rearrangement breakpoints, novel transposable element insertions, and gene transpositions occurring in similar numbers. Rates of amino-acid substitution are higher in uncharacterized genes relative to genes that have previously been studied. Conserved non-coding sequences (CNCSs) tend to be spatially clustered with conserved spacing between CNCSs, and clusters of CNCSs can be used to predict enhancer sequences.
Conclusions
Our results provide the basis for choosing species whose genome sequences would be most useful in aiding the functional annotation of coding and cis-regulatory sequences in Drosophila. Furthermore, this work shows how decoding the spatial organization of conserved sequences, such as the clustering of CNCSs, can complement efforts to annotate eukaryotic genomes on the basis of sequence conservation alone.
Background
The functional annotation of metazoan genome sequences represents one of the greatest challenges in modern biological research. For example, even with structural constraints imposed by the genetic code to guide algorithm design, the identification of all protein-coding genes in a metazoan genome remains an unsolved computational problem. The identification of functional non-coding sequences, such as untranslated regions (UTRs), genes for non-protein-coding RNAs, and cis-regulatory elements, poses an even more difficult problem for comprehensive genome annotation, as the rules governing their structure and function remain more elusive. Despite these difficulties, it is increasingly clear that comparative genomic approaches will substantially aid efforts to annotate these and other important sequence features. With whole-genome sequence data quickly becoming available for several organisms, it is important to determine which species comparisons and features of genome evolution will be most useful for comparative genome annotation.
The genus Drosophila offers a well-characterized evolutionary genetic system for developing and testing methods for comparative genome annotation. From the seminal population-genetic and phylogenetic studies of Dobzhansky and co-workers [1], and the classification of taxonomic relationships in the genus by Patterson, Stone and others [2], Drosophila has long served as a model system for developing and testing evolutionary principles at the morphological and cytological levels. The genus Drosophila has also served as a proving ground for developing and testing evolutionary principles at the protein [3] and DNA sequence levels [4]. In addition, for over a decade and a half, comparative sequence analysis has had an important role in the functional analysis of genes and cis-regulatory sequences in Drosophila (see, for example, [5,6]). This history of research has culminated in a rich understanding of the pattern and process of molecular evolution in the genus Drosophila [7]. With the complete sequencing of the euchromatic portion of the Drosophila melanogaster genome [8,9], this prior knowledge can be applied to the task of comparative genome annotation.
We have undertaken a pilot study to assess the contribution of large-scale comparative genomic sequence data on the functional annotation of the Drosophila genome. Our goals are to identify the species whose genome sequences would be most useful in annotating the D. melanogaster genome, and to identify features of genome evolution that can assist the annotation of protein-coding genes and the non-coding cis-regulatory sequences controlling their transcription. The lessons learned from this study have implications for efforts to annotate the entire D. melanogaster genome using comparative sequence data from the forthcoming D. pseudoobscura genome [10] as well as the recently completed Anopheles gambiae genome [11]. Beyond the initial analyses presented here, these data also serve as materials for the further study of molecular evolutionary processes in Drosophila and the calibration of comparative sequence analysis tools.
Here, we report the isolation and analysis of genomic sequences from eight candidate regions representing both gene-rich and gene-poor regions of the Drosophila genome, totaling over 1.25 megabases (Mb) of DNA sequence. These regions were isolated from fosmid libraries of four divergent Drosophila species (D. erecta, D. pseudoobscura, D. willistoni, and D. littoralis) chosen to cover a range of divergence times (6-15, 46, 53 and 61-65 million years, respectively) from the reference species, D. melanogaster [7]. Using the annotation pipeline and curation tools described in accompanying papers [12,13,14], we predicted the coding sequence content of these sequences for subsequent comparative analyses. Our results indicate that the majority of coding sequences predicted in D. melanogaster can be identified in divergent Drosophila species and show evidence of functional constraint. Microsynteny is generally maintained at the scale of individual fosmid clones, and the few rearrangement breakpoints, transposable elements and gene transpositions can readily be identified. Analysis of coding sequence evolution suggests that uncharacterized genes, which we will refer to as 'predicted' genes, tend to have a higher rate of protein evolution than 'known' genes - those genes that have been selected for experimental study and thus are more likely to have easily discerned functions. Analysis of non-coding sequence evolution reveals that levels of conservation vary with divergence time, and that conserved non-coding sequences (CNCSs) exhibit a striking pattern of spatial clustering in Drosophila. Using transgenic reporter assays we show that CNCS clusters can be used to accurately predict a developmentally regulated enhancer in the apterous (ap) region. We discuss the implications of our results for comparative approaches to protein-coding and cis-regulatory sequence prediction in the genus Drosophila.
Results
Isolation and sequencing of genomic regions from divergent Drosophila species
On the basis of genome size considerations and the desire to investigate a range of divergence times in the genus Drosophila, we constructed fosmid libraries (approximately 40-kb inserts) for D. erecta, D. pseudoobscura, D. willistoni and D. littoralis (Figure 1). D. littoralis is closely related to the well-studied species, D. virilis, but has been reported to have less dispersed repetitive DNA than D. virilis (Kevin White, personal communication). We designed degenerate PCR primers for a set of eight well-characterized genes (apterous (ap), even-skipped (eve), fushi-tarazu (ftz), twist (twi), and Rhodopsins 1, 2, 3 and 4 (Rh1, Rh2, Rh3 and Rh4)) to obtain species-specific sequence-tagged sites (STSs) that were subsequently used for hybridization to gridded fosmid filters (see Materials and methods). Positive clones from the library screen were verified by PCR and restriction mapped to choose the longest clone containing the candidate gene and its regulatory regions.
Figure 1.
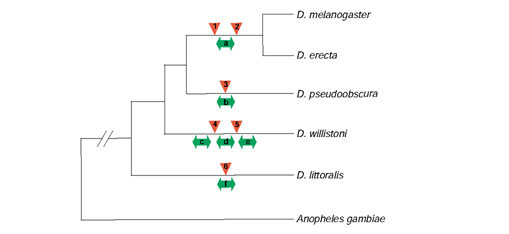
Phylogenetic relationships of the five Drosophila species studied in this paper and the outgroup species, the mosquito Anopheles gambiae. The topology of this tree is based on the accepted relationship of these six species; the divergence times from D. melanogaster are approximately 6-15, 46, 53, 61-65, and 250 million years for D. erecta, D. pseudoobscura, D. willistoni, D. littoralis and A. gambiae, respectively [7,84]. D. melanogaster, D. erecta, D. pseudoobscura and D. willistoni belong to the subgenus Sophophora and D. littoralis belongs to the subgenus Drosophila. Rearrangements are indicated by double-headed arrows below each branch and gene transpositions are indicated by triangles above each branch. Rearrangements are inferred to occur on the lineages leading to (a) the ancestor of the D. melanogaster/D. erecta eve region, (b) the D. pseudoobscura Rh1 region, the D. willistoni (c) eve, (d) Rh1, and (e) Rh3 regions, and (f) the D. littoralis ftz region. Gene transpositions are inferred to occur for the (1) CG13029 and (2) CG12133 genes in the ancestor of the D. melanogaster/D. erecta lineage, (3) the CG5245-like gene in the D. pseudoobscura lineage, (4) the CG8319-like gene in the D. willistoni lineage, (5) the CG2222-like gene in the D. willistoni lineage, and (6) the Rh4 gene in the D. littoralis lineage. We note that the event classified as a rearrangement involving the D. pseudoobscura CG31155 gene at the end of the Rh1 clone may be a gene transposition as this gene is a partial gene spanning the edge of the clone. In addition, we note that rearrangement involving the D. littoralis ftz gene may have occurred on the branch leading to the ancestor of the Sophophoran species since, although the orientation of ftz with respect to Antp is ambiguous in A. gambiae ([85,86] and data not shown), it shares a similar configuration to D. littoralis in the outgroup, Tribolium castaneum [87].
In the initial design of this project, comparative sequence data was to be collected from a D. virilis P1 library [15]. Using a PCR-based plate-pool screening strategy, we isolated a P1 clone from this library containing an 83.2-kb insert from the ap region of D. virilis. Sequencing of this clone revealed long stretches of repetitive DNA, which complicated both assembly and comparative analyses. In addition, the insert size of the D. virilis P1 library (approximately 60-80 kb) was greater than necessary for comparative analysis of single gene regions. This clone was used to guide transgenic reporter analysis (see below), but has not been included in the other analyses reported here.
In total, 30 fosmid clones were isolated and sequenced using methods described in [9] which sum to 1,257,069 bp. All clones were finished to an estimated error rate of fewer than 0.17 errors per 10 kb, with an average estimated error rate of 0.03 errors per 10 kb. The lengths of fosmids sequenced for the eight candidate regions are shown in Table 1. Though we were able to obtain species-specific STSs for the D. willistoni twi gene, we were not able to obtain clones for this region from the D. willistoni fosmid library. We were also not able to obtain a species-specific probe for D. willistoni ftz, nor could we obtain any D. willistoni ftz clones using probes from other non-melanogaster species. Also shown in Table 1 are the lengths and locations of D. melanogaster genomic regions corresponding to the union of the Release 3 sequences homologous to all four non-melanogaster species. The union of sequences from all non-melanogaster species for the eight candidate regions covers 494.6 kb of the D. melanogaster genome; an additional 65.3 kb of D. melanogaster genomic sequence was sampled owing to rearrangements in non-melanogaster species. Thus the 1.25 Mb of comparative data presented here span over 0.5 Mb of coding and non-coding sequences of the D. melanogaster genome.
Table 1.
Summary of candidate gene regions and lengths of sequences analyzed in this study
| Region | Arm | Cytological location | D. melanogaster | D. erecta | D. pseudoobscura | D. willistoni | D. littoralis |
| Rh1 | 3R | 92B3-6 | 54,450 | 38,418 | 45,873 | 43,804 | 35,983 |
| Rh2 | 3R | 91D3-5 | 58,172 | 43,599 | 42,336 | 35,954 | 43,945 |
| Rh3 | 3R | 92C3-D1 | 83,394 | 43,180 | 42,117 | 41,651 | 45,428 |
| Rh4 | 3L | 73D1-6 | 53,470 | 41,352 | 44,117 | 36,325 | 44,255 |
| ap | 2R | 41F8 | 50,314 | 37,077 | 38,050 | 40,487 | 39,016 |
| eve | 2R | 47C6-D4 | 46,587 | 45,909 | 44,139 | 38,059 | 43,320 |
| ftz | 3R | 84A5-B2 | 66,214 | 44,340 | 42,627 | NA | 43,155 |
| twi | 2R | 59C1-3 | 82,029 | 43,101 | 43,025 | NA | 46,427 |
| Total | 494,630* | 336,976 | 342,284 | 236,280 | 341,529 |
Cytological locations are for sequences in D. melanogaster. The D. willistoni ftz and twi region (NA) were not isolated in our library screen. All fosmid clones sequenced have estimated error rates of fewer than 0.17 errors/10 kb. *An additional 65.3 kb of sequence was surveyed from other regions of the D. melanogaster genome as a result of rearrangements (see text for details).
Comparative annotation of coding sequences
The 30 non-melanogaster fosmid (and the D. virilis ap P1) sequences were computationally processed using the pipeline used to re-annotate the D. melanogaster genome [12]. The only major modification to this pipeline was to add an additional tier of evidence containing the results of TBLASTN searches of all Release 3 D. melanogaster peptides [14] against non-melanogaster sequences. Predicted coding sequences were manually verified and refined using the Apollo annotation tool [13]. As no expressed sequence tag (EST) information exists to annotate transcribed non-coding sequences (such as UTRs) for the four non-melanogaster species, we annotated only protein-coding gene and exon models. Thus, in keeping with other gene-prediction studies (for example [16]), we use the terms gene and exon to refer to the translated components of genes and exons.
In the 30 fosmids, we predict a total of 164 protein-coding genes in non-melanogaster species (53 in D. erecta, 41 in D. pseudoobscura, 39 in D. willistoni, 31 in D. littoralis) that form orthologous clusters with 81 D. melanogaster genes. Of the 81 genes, 30 are 'known' genes that have been functionally characterized in some way by the community of Drosophila researchers; the remaining 51 genes are 'predicted' genes based only on the evidence in the Release 3 annotations ([14] and see Supplementary Table 1 in the Additional data files section). Of the 164 genes predicted in non-melanogaster species, 133 (81%) are full length; the remaining 31 (19%) are partial coding sequences that span the edge of the sequenced genomic clone. In non-melanogaster species, we predict 495 coding exons (148 in D. erecta, 133 in D. pseudoobscura, 111 in D. willistoni, 103 in D. littoralis) that form orthologous clusters with 264 D. melanogaster coding exons. On average, there are approximately two non-melanogaster species sampled per orthologous gene and coding exon cluster. Fifteen genes (10 complete) and 39 coding exons were sequenced in all four non-melanogaster species.
Qualitatively, our data reveal that the majority of D. melanogaster Release 3 gene models are highly conserved in divergent Drosophila species. This made it possible to automatically identify orthologous genes in non-melanogaster species using TBLASTN results in conjunction with Genie [17] and/or GENSCAN [18] predictions to improve intron-exon boundaries and identify small/divergent exons in Apollo. In the few discrepant cases where no clear ortholog could be unambiguously identified (such as the four closely related members of the Rhodopsin gene family), we used the conserved microsyntenic gene orders maintained in these species to resolve orthologs (see below). With the exception of the retrotransposition events discussed below, the intron-exon structure of gene models is highly conserved as well: only one case of intron gain was observed in the D. littoralis Rh2, as has been reported previously for Rh2 in the closely related species, D. virilis [19]. For a small class of genes (BcDNA:LD21213, Gr59a, Gr59b, CG9895, CG10887, CG17186, CG4733), orthologs could be identified in divergent species, but amino-acid sequences could not be reliably aligned with the D. melanogaster gene model. In addition, orthologs of four genes (CG13029, CG14294, CG12133, CG12378) could not be identified in non-melanogaster species except in D. erecta, the species most closely related to D. melanogaster. The absence of these genes is not simply due to insufficient sampling, since in these cases both 5' and 3' neighboring genes could be identified in more divergent species (see Figure 2, for example). These may represent genes overpredicted in both D. erecta and D. melanogaster, lineage-specific genes which evolved before the divergence of D. melanogaster from D. erecta, or genes which have transposed from (or to) other locations in the genomes of the more divergent species.
Figure 2.
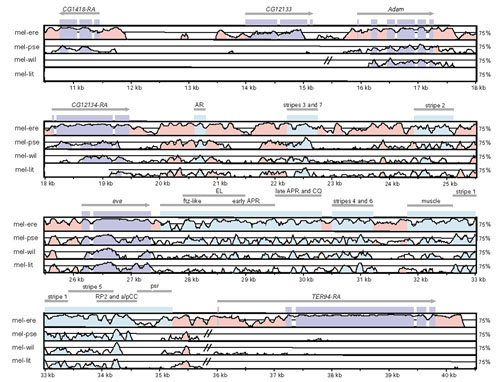
VISTA plot of genome organization and sequence conservation in the Drosophila eve region. Sequences were aligned using AVID, and conserved sequences were visualized using default parameters of VISTA. From top to bottom are pairwise comparisons between D. melanogaster and D. erecta (mel-ere), D. pseudoobscura (mel-pse), D. willistoni (mel-wil) and D. littoralis (mel-lit), respectively. In each panel, conserved segments from 50-100% are plotted, with the midline indicating 75% identity; regions with no midline represent sequences not sampled in a pairwise comparison. Double bars crossing a midline represent rearrangement breakpoints. The location and orientation of coding sequences are indicated by arrows; purple boxes represent coding exons and light-blue boxes represent functionally characterized cis-regulatory sequences [50,88,89,90]; pink regions represent uncharacterized CNCSs. Suffixes on gene names (for example, TER94-RA) indicate the particular transcript displayed for genes with multiple transcripts. Note that the predicted gene CG12133 is restricted to the D. melanogaster/D. erecta lineage but is absent in D. pseudoobscura, although both flanking genes are present.
We used an evolutionary genetic approach, the Ka/Ks test, to assess the accuracy of these gene and exon predictions [20]. This test relies on the assumption that functionally constrained protein-coding sequences should exhibit significantly lower rates of evolution in amino-acid-encoding nucleotide sites (typically first and second positions in a codon) relative to silent sites (typically third positions in a codon). Quantitatively, this leads to the prediction that the ratio of the average rate of amino-acid substitution per site (Ka) relative to the average rate of silent substitution per site (Ks) for functionally constrained coding sequences should be significantly less than 1 [21]. Genes or exons which have a Ka/Ks ≈ 1 are inferred to evolve in the absence of functional constraint; genes or exons which have a Ka/Ks > 1 are inferred to evolve under the influence of positive selection. The significance of a Ka/Ks ratio can be determined by a likelihood ratio test of the probabilities of the data under the alternative hypotheses of functional constraint relative to no constraint [22]. Genes or coding exons with a Ka/Ks ratio significantly less than 1 'pass' the Ka/Ks test; genes or coding exons with a Ka/Ks ratio not significantly less than 1 'fail' the Ka/Ks test. The power of this test to detect functional constraint is influenced both by evolutionary distance and sequence length [20]; thus we analyzed both genes and coding exons in pairwise comparison with all four non-melanogaster species.
All pairwise gene-level comparisons studied here exhibited Ka/Ks ratios less than one (see Supplementary Table 1 in the Additional data files section). One hundred and fifty-five of 164 (94.5%) of these Ka/Ks ratios were significantly less than 1, indicating that the vast majority of genes in our sample show evidence of functional constraint. All nine pairwise comparisons that fail the Ka/Ks test at the gene level were D. melanogaster-D. erecta comparisons, and eight out of nine involved predicted genes (Supplementary Table 1). Genomic sequences for six of the nine genes which fail the Ka/Ks test at the gene level were sampled in more divergent species: four of these six genes could be identified in more divergent species (Lmpt, CG10887, CG14292, and CG4468), whereas two could not (CG12378 and CG14294), indicating that genes conserved in divergent species can fail gene-level Ka/Ks tests in comparisons among closely related species like D. erecta. Of the four genes identified only in D. melanogaster and D. erecta and not in more distantly related species, two pass CG12133 and CG13029) and two fail (CG12378 and CG14294) the gene-level Ka/Ks test. We note that of these four genes, the two genes that pass (CG12133 and CG13029) have multiple exons, whereas the two genes that fail (CG12378 and CG14294) have only a single exon. This result indicates that at least some of the genes found only in D. melanogaster and D. erecta are likely to be real genes under functional constraint.
Though the majority of pairwise exon level comparisons have Ka/Ks ratios less than one (Figure 3), a much lower proportion of pairwise comparisons at the exon level pass the Ka/Ks test. In total, 71.9% (356/495) of pairwise comparisons at the exon level pass the Ka/Ks test: 54.0% (80/148) for D. erecta; 78.9% (105/133) for D. pseudoobscura; 81.1% (90/111) for D. willistoni; and 79.6% (82/103) for D. littoralis. Coding exons from known and predicted genes pass the Ka/Ks test at similar rates: overall, (72.2% known versus 71.1% predicted), D. erecta (56.6% known versus 50.0% predicted), D. pseudoobscura (80.4% known versus 75.6% predicted), D. willistoni (81.5% known versus 82.1% predicted), D. littoralis (78.0% known versus 80.6% predicted). The majority of exons that fail the Ka/Ks test still have Ka/Ks ratios less than 1; only six non-significant pairwise exon comparisons (one in D. erecta, one in D. pseudoobscura, two in D. willistoni, and two in D. littoralis) have Ka/Ks ratios greater than 1 (Figure 3). As with gene-level comparisons, the most closely related species, D. erecta, fails the highest proportion of exon-level Ka/Ks tests. In contrast to gene-level comparisons, there is no tendency for exons from predicted genes to fail Ka/Ks tests relative to exons from known genes.
Figure 3.
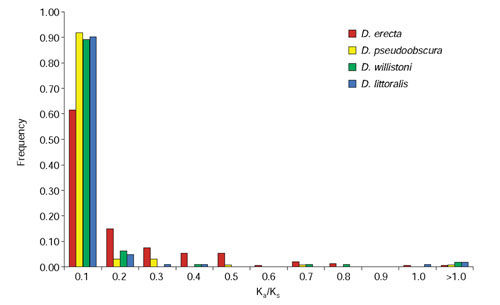
Frequency distribution of Ka/Ks ratios for pairwise exon-level comparisons between D. melanogaster and either D. erecta, D. pseudoobscura, D. willistoni, or D. littoralis. Ka/Ks ratios were estimated using the codeml program of PAML 3.12 using runmode = -2.
Pairwise comparisons that do not pass the Ka/Ks test could result from misannotated exons or an insufficient amount of divergence to resolve differential rates of amino acid and silent site evolution. Failure to pass exon-level Ka/Ks tests because of insufficient divergence is a function of divergence time and exon length [20]. Our results suggest that both factors contribute to non-significant exon-level Ka/Ks tests between species in the genus Drosophila. The fact that the most closely related species, D. erecta, fails the highest proportion of gene-and exon-level Ka/Ks tests indicates that insufficient divergence time contributes to non-significant comparisons. Exon length is also a factor, as there is a tendency for shorter exons to fail Ka/Ks tests in our data. For example, the average length of all exons failing the Ka/Ks test in comparisons between D. melanogaster and D. pseudoobscura is 22.1 codons, and the average length of all exons passing the Ka/Ks test is 152.1 codons. Similar results are obtained for pairwise comparisons involving D. melanogaster and D. erecta, D. willistoni or D. littoralis, and for both known and predicted genes (data not shown).
To determine if insufficient divergence time is the major cause of non-significant exon-level Ka/Ks tests, we performed multi-species exon-level Ka/Ks tests that capitalize on a greater total amount of divergence for a given exon [20]. The question addressed by this analysis is: does the addition of a third species to D. melanogaster-D. pseudoobscura pairwise comparisons increase the proportion of exons that pass exon-level Ka/Ks tests? For this analysis, we analyzed exons that failed pairwise tests between D. melanogaster and D. pseudoobscura using triplets involving D. melanogaster, D. pseudoobscura and one other non-melanogaster species. Using the same cutoffs for the pairwise exon-level analyses and a guide tree based on Figure 1, we tested 16, 14 and 13 exons which did not show evidence of functional constraint between D. melanogaster and D. pseudoobscura, for which we have sequence data available in D. erecta, D. willistoni and D. littoralis, respectively. Only 2 of the 16 (12.5%) non-significant exon-level D. melanogaster-D. pseudoobscura comparisons pass the Ka/Ks test when D. erecta is included as a third species, whereas 6 of 14 (42.8%) and 6 of 13 (46.1%) pass when D. willistoni and D. littoralis are included as a third species, respectively. These results demonstrate that multiple comparisons among divergent species can reveal functional constraint acting on coding exons that cannot otherwise be detected in pairwise comparisons.
Finally, as a preliminary assessment of the relative utility of A. gambiae genome sequences for comparative gene prediction in Drosophila, we attempted to identify homologs in A. gambiae of the 81 genes for which we have comparative sequence data in Drosophila. For 21 of the 81 genes in our study (25.9%) we were not able to obtain a clear homolog (defined as a high-scoring pair (HSP) with an expected (E) value less than 10-20 and greater than or equal to 30% identity over 100 amino acids using default parameters of TBLASTN) in the A. gambiae mapped scaffold sequences; 11 of these 21 genes did not yield any HSPs at all. These results are compatible with a recent whole-genome analysis showing that 18.6% of D. melanogaster genes have no clear homolog in A. gambiae [23]. No clear homolog could be identified in the A. gambiae genome sequences for three of 30 (10.0%) known genes in our dataset, whereas a greater than three times higher proportion of predicted genes, 18 of 51 (35.3%), had no clear homolog in A. gambiae. Five D. melanogaster genes - the four members of the Rhodopsin gene family and CG5245 - have multiple HSPs in the A. gambiae genome sequences. We were able to resolve orthology for Rh4 only, as the sina gene in A. gambiae is contained within the Rh4 gene as in D. melanogaster and other species in the subgenus Sophophora.
Rearrangement and transposition of genomic sequences
Using the gene predictions discussed above as orthologous markers, we addressed the question of whether the microsyntenic relationships in the D. melanogaster genomic sequence surveyed are conserved in non-melanogaster species. In general, our data indicate that the microsyntenic order of coding and non-coding sequences is highly conserved in the genus Drosophila at the scale of individual fosmids (approximately 40 kb). Our data provide evidence for only six genomic rearrangements in these sequences occurring in the phylogeny of these five species, one each in the lineages leading to the D. littoralis ftz, D. pseudoobscura Rh1, D. willistoni eve, Rh1, and Rh3 regions, as well as in the ancestor of the D. melanogaster/D. erecta eve region (see Figure 1). All of these unique events occurred in non-coding intergenic regions and none of the rearrangement breakpoints is associated with detectable transposable element sequences (see also [24]). Although it is difficult to estimate the length distribution of microsyntenic regions in Drosophila from our data, it is clear that very small microsyntenic regions can be delimited in the Drosophila genome through multiple species comparisons. For example, the two independent rearrangements in the vicinity of the eve locus reduce this microsyntenic region to a approximately 20-kb interval of the D. melanogaster genome containing only three neighboring genes (Adam, CG12134 and eve) and their flanking non-coding sequences (Figure 2).
We can directly confirm the nature of one rearrangement (D. littoralis ftz) as a paracentric micro-inversion since both breakpoints are contained within a single fosmid clone. In this case, a small (approximately 14 kb) region containing the ftz coding sequence and flanking non-coding DNA is inverted between the Antp and Scr genes relative to D. melanogaster. Maier et al. [25] provide hybrization data for a similar rearrangement in the ftz locus of D. hydei, another member of the subgenus Drosophila. It is likely that the other rearrangement breakpoints we observe also result from paracentric inversions, the predominant form of genome rearrangement in Drosophila [26]. Consistent with this is the fact that rearranged sequences can be inferred to come from the same chromosome arm. At least two other breakpoints (in the D. willistoni Rh1 and Rh3 regions) also have probably arisen from micro-inversions, as in both cases only two genes are inferred to have switched order locally on the chromosome.
We also identified eight examples of novel genetic elements in non-melanogaster species, seven of which occur in intergenic regions (Figure 1). Four of these cases involve the insertion of novel transposable element sequences: full length Bari-1-like elements in both the D. pseudoobscura Rh1 region and the D. willistoni Rh3 regions, a partial I-like element in the D. willistoni Rh4 region, and a partial blastopia-like element in the D. littoralis Rh3 region. Identification of Bari-1-like transposon sequences in D. pseudoobscura and D. willistoni is consistent with previous observations [27]; I-like elements have been shown to exist in the melanogaster and obscura species [28], but this is the first report of I-like elements in the willistoni group. The other four cases arise from gene transposition including: a homolog of the D. melanogaster X-chromosome gene CG2222 in the D. willistoni eve region; a homolog of the D. melanogaster 3R-chromosome gene CG5245 in the D. pseudoobscura Rh1 region; a homolog of the D. melanogaster 3R-chromosome gene CG8319 in the D. willistoni Rh1 region, and the Rh4 gene in D. littoralis (see below). The CG5245-like gene in D. pseudoobscura and the CG8319-like gene in D. willistoni both are located in the same intergenic region between the Arc42 and PK92B genes, but result from independent events since they involve different ancestral sequences and occur on opposite strands in this intergenic region. This result suggests the possibility of hotspots for gene transposition in the Drosophila genome.
At least one novel gene, the CG2222-like gene D. willistoni, is likely to have originated from a retrotransposition event as this gene lacks introns while its closest homolog, found on a different chromosome arm in the D. melanogaster genome, has two introns. Another striking example of retrotransposition involves the D. littoralis Rh4 gene and illustrates the fact that functionally important genes can undergo dramatic changes in location and gene structure during genome evolution [29]. This gene maintains its microsyntenic relationship with neighboring genes in the 72D2-3 region of the D. melanogaster genome in Sophophoran species, but has retrotransposed into the intron of another gene, CG10967, in a region of the D. littoralis genome that corresponds to the 69E1-2 region of the D. melanogaster genome. As a result, genes contained in the intron of the Sophophoran Rh4 (sina, CG13030 and CG13029) have been lost in the process. Cytological evidence for transposition of Rh4 exists for the closely related species D. virilis and the more distantly related species D. repleta [29,30].
In contrast to the stability of microsyntenic gene order in the genus Drosophila, we found that the sample of genes studied here are scattered widely throughout the Anopheles genome. For example, of the 55 Drosophila genes that had a single clear homolog in Anopheles, 27 are located on D. melanogaster chromosome arm 2R. Of these 27 genes, ten, five, six and six are located on A. gambiae chromosome arms 2L, 2R, 3L and 3R. These results are consistent with previous reports comparing the locations of genes in D. melanogaster with A. gambiae, which indicate that extensive genome rearrangement has occurred since the divergence of these two lineages [23,31,32]. Some D. melanogaster genes in our sample do maintain microsyntenic relationships in A. gambiae, such as the Rh4 and sina genes. In this case, conservation of microsynteny is most probably maintained because of the nested relationship of these genes, and this configuration in the outgroup Anopheles supports the scenario that transposition of Rh4 occurred at some point in the lineage leading to the Drosophila subgenus (see above, and [29,30]).
Patterns of coding sequence evolution
In addition to providing a useful resource for studying comparative gene prediction and genome rearrangements, our data confirm and extend emerging trends in Drosophila coding sequence evolution. Table 2 summarizes the average rates of amino-acid and silent site substitution for all, known and predicted genes in our dataset. These data show that predicted genes tend to have a higher rate of amino-acid substitution than known genes in the genus Drosophila. This trend is significant for the three most closely related pairwise comparisons (D. melanogaster versus D. erecta, D. pseudoobscura or D. willistoni) but non-significant in the comparison involving the most distantly related species (D. melanogaster versus D. littoralis). No significant differences were detected in the rates of silent site substitution between known and predicted genes in any pairwise comparison, although predicted genes in D. pseudoobscura, D. willistoni and D. littoralis tend to show elevated rates of silent site substitution.
Table 2.
Rates of amino-acid (Ka) and silent (Ks) substitution in Drosophila genes
| Species | All genes | Known genes | Predicted genes | p-value | |||||||
| Ka | Ks | N | Ka | Ks | N | Ka | Ks | N | Ka | Ks | |
| D. erecta | 0.057 | 0.357 | 53 | 0.042 | 0.366 | 25 | 0.071 | 0.349 | 28 | 0.0001 | 0.1299 |
| D. pseudoobscura | 0.146 | 2.313 | 41 | 0.071 | 1.830 | 17 | 0.199 | 2.655 | 24 | 0.0009 | 0.0262 |
| D. willistoni | 0.220 | 2.627 | 39 | 0.089 | 2.225 | 15 | 0.302 | 2.878 | 24 | 0.0001 | 0.0735 |
| D. littoralis | 0.170 | 2.166 | 31 | 0.126 | 1.923 | 14 | 0.206 | 2.366 | 17 | 0.1315 | 0.6058 |
Rates of substitution per site between D. melanogaster and D. erecta, D. pseudooobscura, D. willistoni, or D. littoralis are estimated using the method of Yang and Nielsen [81]. Shown are the average rates of substitution per site (and sample sizes) of all, known or predicted genes. p-values are the results of Mann-Whitney U-tests for differences in the distribution of Ka and Ks values between known and predicted genes for a given pairwise comparison. Values in bold represent significant differences in rates of evolution between known and predicted genes at the 0.006 (= 0.05/8) level.
In contrast to expectation, average rates of amino-acid substitution are highest in comparisons between D. melanogaster and D. willistoni, not D. melanogaster and D. littoralis (Table 2, Figure 1). The overall increased rate of amino-acid substitution for genes in the D. willistoni lineage is caused by an increased rate of amino-acid substitution in predicted genes. For known genes, average rates of amino-acid substitution are consistent with the accepted phylogenetic relationships of these species: D. erecta is most closely related to D. melanogaster, followed by D. pseudoobscura, D. willistoni and D. littoralis, respectively. Average rates of silent site substitution also do not show a pattern consistent with the accepted phylogeny of these species (Table 2, Figure 1). This is a consequence of the fact that, for comparisons between D. melanogaster and either D. pseudoobscura, D. willistoni or D. littoralis, average rates of silent site substitution exceed an expectation of one substitution per site, indicating that silent sites are 'saturated' in these comparisons. Even so, it is apparent that there may be an increased rate of silent site substitution as well in the D. willistoni lineage. It is unlikely that these results are simply a consequence of an incorrect phylogeny, since the phylogenetic relationships of these species are well established [7].
Our estimate of the average rate of amino-acid substitution per site in known genes between D. melanogaster and D. pseudoobscura (0.071) is nearly the same as previous estimates (0.076) using a different sample of known genes and estimation procedure [33]. In addition, our estimate of the average rate of amino-acid substitution for predicted genes between D. melanogaster and D. erecta (0.071) is similar to that estimated using different methods for a sample of rapidly evolving genes between D. melanogaster and D. yakuba (0.067) [34], a species approximately as divergent from D. melanogaster as D. erecta [35]. Thus the categorical and lineage effects we detect are unlikely to be artifacts of our data or methods. The cause(s) of the increased rate of amino-acid substitution in predicted genes in the D. willistoni lineage remain to be clarified, but are most probably related to increased rates of protein evolution detected previously in the D. saltans lineage [36], which have been explained by a shift in base composition in the common ancestor of the D. saltans and D. willistoni groups (see below, and [37]).
In D. melanogaster, it is well established that coding sequences have a higher GC content, relative to genomic averages, due to the preferential use of codons ending in C or G [38,39]. This pattern holds in the closely related species D. erecta, as well as in the more distantly related species D. pseudoobscura and D. littoralis (Table 3). In contrast, our data show that D. willistoni coding sequences have a higher frequency of AT (53%) base-pairs than GC (47%) base-pairs. This shift in base usage in D. willistoni coding sequences is apparent at the dinucelotide level as well, predominantly affecting those dinucleotides that exclusively contain AT or GC. Non-coding sequences of all non-melanogaster species are AT-rich, as in D. melanogaster [40]; slight shifts towards higher AT frequency are observed in the non-coding sequences of the D. willistoni lineage (Table 3).
Table 3.
Mono- and dinucleotide frequencies of coding and non-coding sequences in Drosophila species
| Mononucleotide | D. melanogaster | D. erecta | D. pseudoobscura | D. willistoni | D. littoralis |
| Coding | |||||
| A = T | 0.231 | 0.222 | 0.220 | 0.265 | 0.224 |
| G = C | 0.269 | 0.278 | 0.280 | 0.235 | 0.276 |
| Non-coding | |||||
| A = T | 0.300 | 0.295 | 0.281 | 0.324 | 0.305 |
| G = C | 0.200 | 0.205 | 0.219 | 0.176 | 0.195 |
| Dinucleotide | D. melanogaster | D. erecta | D. pseudoobscura | D. willistoni | D. littoralis |
| Coding | |||||
| TA | 0.032 | 0.028 | 0.027 | 0.046 | 0.030 |
| AT | 0.057 | 0.053 | 0.056 | 0.080 | 0.058 |
| AA = TT | 0.057 | 0.052 | 0.049 | 0.076 | 0.056 |
| AC = GT | 0.054 | 0.053 | 0.051 | 0.051 | 0.051 |
| AG = CT | 0.063 | 0.064 | 0.064 | 0.057 | 0.059 |
| GA = TC | 0.067 | 0.068 | 0.067 | 0.063 | 0.055 |
| CA = TG | 0.075 | 0.074 | 0.077 | 0.078 | 0.082 |
| CG | 0.064 | 0.068 | 0.068 | 0.046 | 0.073 |
| GC | 0.081 | 0.085 | 0.091 | 0.067 | 0.109 |
| CC = GG | 0.067 | 0.072 | 0.071 | 0.054 | 0.061 |
| Non-coding | |||||
| TA | 0.069 | 0.068 | 0.058 | 0.080 | 0.075 |
| AT | 0.086 | 0.084 | 0.077 | 0.094 | 0.089 |
| AA = TT | 0.110 | 0.106 | 0.094 | 0.126 | 0.111 |
| AC = GT | 0.052 | 0.052 | 0.052 | 0.053 | 0.053 |
| AG = CT | 0.052 | 0.053 | 0.058 | 0.051 | 0.052 |
| GA = TC | 0.053 | 0.053 | 0.059 | 0.052 | 0.048 |
| CA = TG | 0.068 | 0.068 | 0.070 | 0.066 | 0.071 |
| CG | 0.037 | 0.040 | 0.039 | 0.025 | 0.037 |
| GC | 0.052 | 0.056 | 0.057 | 0.038 | 0.058 |
| CC = GG | 0.043 | 0.044 | 0.052 | 0.033 | 0.035 |
Values for D. melanogaster are genome-wide averages based on Release 3 sequences/annotations [9,14] and include unmapped scaffolds derived from heterochromatic regions (see [83]). Values in bold indicate the most frequently used mono- or dinucleotide. Frequencies of complementary mono- and dinucleotides were averaged to account for the double-stranded nature of DNA.
The shift in base usage in the D. willistoni lineage is also detected in the pattern of synonymous codon usage (see Supplementary Table 2 in the additional data files). Previous analyses of a limited number of coding sequences revealed a shift away from preferred C-ending codons used in the D. melanogaster lineage, towards T-ending codons in the D. willistoni lineage [7,41]. Our data indicate that this trend holds for a much larger sample of genes (see Supplementary Table 2 in additional data files). For 10 of the 18 amino acids with more than one codon (Arg, Asn, His, Ile, Leu, Lys, Phe, Pro, Thr, Tyr), the most frequently used codon in D. willistoni differs from that in D. melanogaster. All 10 of these changes in synonymous codon usage involve D. willistoni most frequently using an A- or T-ending (or beginning, for example, Leu) codon with D. melanogaster using a G- or C-ending (or beginning) codon, supporting a trend identified originally using only the Adh coding sequence [41]. The most frequently used codon differs between D. melanogaster and D. erecta, D. pseudoobscura and D. littoralis for only two (Asp, Ser), one (Asn) and four (Asn, Ile, Pro, Thr) amino acids, respectively.
Patterns of non-coding sequence evolution
Our data also provide an opportunity to study basic features of non-coding conservation in Drosophila. which remain largely unexplored. As shown in Figures 2 and 4, a substantial proportion of non-coding sequences are conserved in Drosophila, especially in pairwise comparisons between D. melanogaster and D. erecta. Levels of conservation appear to plateau in more divergent comparisons, with a tendency for D. pseudoobscura to show higher levels of non-coding conservation relative to D. willistoni or D. littoralis in pairwise comparisons with D. melanogaster. Few, if any, non-coding sequences are conserved between D. melanogaster and A. gambiae (Figure 4, see also [23]). There is also regional variation in levels of non-coding conservation in the Drosophila genome, as illustrated by contrasting conservation between D. melanogaster and D. erecta, for example, in the eve (Figure 2) and ap (Figure 4) regions.
Figure 4.
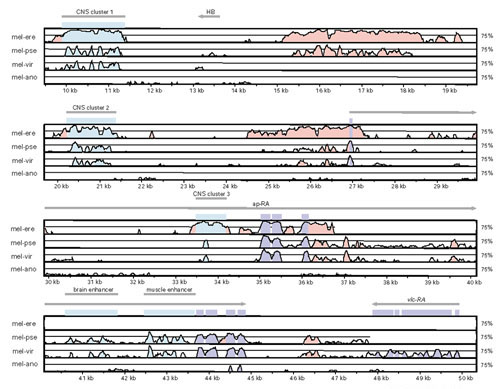
VISTA plot of genome organization and sequence conservation in the Drosophila ap region. From top to bottom are pairwise comparisons between D. melanogaster and D. erecta (mel-ere), D. pseudoobscura (mel-pse), D. virilis (mel-vir) and A. gambiae (mel-ano), respectively. Features of this plot are as in Figure 3. Shown are five CNCS clusters corresponding to the muscle enhancer [91], the brain-specific enhancer empirically verified in this study (Figure 8), and three predicted enhancers labeled CNCS clusters 1, 2 and 3. Note that the HB transposable element in the region 5' to ap is located between CNCS clusters and is not conserved between species.
To estimate levels of sequence conservation in non-coding regions and to contrast patterns of coding with non-coding conservation, we aligned genomic sequences using the AVID alignment tool [42]. AVID is a global alignment tool that works by recursively finding co-linear 'anchors' of maximal sequence identity; therefore, locally inverted or transposed sequences that might be conserved will not be included in our analysis. Conserved non-coding sequences (CNCSs), defined as windows of 10 bp or greater with 90% or greater nucleotide identity, were identified in pairwise alignments using the VISTA program [43]. These parameters were chosen to identify short, highly conserved sequences found in Drosophila non-coding regions [44]. We used D. melanogaster as the reference species in pairwise comparisons with non-melanogaster species, and Release 3 annotations [14] exported from Gadfly in VISTA format to classify conserved segments as either coding or non-coding. Transcribed and nontranscribed non-coding sequences were analyzed together, since previous results showed similar patterns of conservation for intergenic and intronic sequences in Drosophila [44].
The results of this analysis are shown in Table 4, which contrasts features of conservation in both coding and non-coding sequences by species. For all species analyzed, coding regions have a higher proportion of sequences that meet our definition of conservation relative to non-coding sequences. In addition, the median segment length surpassing our criterion for conservation is longer for coding sequences relative to non-coding sequences for all species analyzed. These results are expected, as coding sequences are on average thought to experience more intense purifying selection than non-coding sequences [21]. In contrast, the average percent identity of conserved segments is higher for non-coding sequences than coding sequences. This is probably a result of silent site substitution in otherwise functionally constrained coding sequences.
Table 4.
Estimates of pairwise sequence conservation in coding and non-coding regions between D. melanogaster and D. erecta, D. pseudoobscura, D. willistoni or D. littoralis
| Species | Number of bp surveyed | Number of bp conserved | % conserved (bp) | Median length of conserved segment | Average % identity of conserved segment |
| Coding | |||||
| D. erecta | 63,655 | 60,327 | 94% | 39 | 93% |
| D. pseudoobscura | 46,626 | 26,978 | 61% | 20 | 91% |
| D. willistoni | 42,224 | 18,774 | 45% | 17 | 91% |
| D. littoralis | 19,717 | 10,997 | 63% | 17 | 92% |
| Non-coding | |||||
| D. erecta | 272,366 | 186,895 | 69% | 24 | 94% |
| D. pseudoobscura | 276,731 | 77,391 | 28% | 17 | 95% |
| D. willistoni | 174,421 | 19,700 | 13% | 14 | 95% |
| D. littoralis | 138,866 | 24,633 | 18% | 15 | 95% |
| D. virilis | 114,015 | 30,564 | 27% | 16 | 95% |
| D. virilis [44] | 114,015 | 29,915 | 26% | 19 | 93% |
Microsyntenic regions were globally aligned using AVID and conserved sequences greater than or equal to 10 bp and 90% identity were identified using VISTA. Sequences were classified as coding or non-coding using Release 3 annotations [14] exported from GadFly in VISTA format. Shown for comparison are a re-analysis of conservation between D. melanogaster and D. virilis using the current methods, as well as previous results, for a sample of non-coding regions published in [44].
Analysis of levels of conservation by species shows that the increased rate of amino-acid sequence evolution in the D. willistoni lineage detected above may reflect a more widespread phenomenon in the genome of this species. As shown in Table 4, D. willistoni shows unexpectedly low levels of both non-coding and coding conservation, given the accepted phylogeny of the species. These data show that the increased rate of evolution in the D. willistoni lineage is not restricted to coding sequences, rendering coding-sequence-based interpretations of the unusual patterns of molecular evolution in this lineage less tenable (see, for example [7,41]). Together with the changes in base composition in both coding and non-coding sequences noted above, the increased rate of evolution in both coding and non-coding sequences detected in the D. willistoni suggests a genome-wide effect, possibly resulting from a change in mutation pressure or a change in population size at some time during the history of this lineage (see also [37]).
Despite the lineage effect in levels of conservation in the D. willistoni genome, the median length of conserved coding or non-coding segments generally decreases with increasing divergence time as expected (Table 4). However, the average percent identity of conserved coding or non-coding segments identified does not decrease with increasing divergence time. Finally, the ratio of conserved sequences that are coding relative to non-coding increases with increasing divergence time. The ratio of conserved sequences that are coding relative to non-coding is 1.36 for comparisons with D. erecta, but increases to 2.21 for comparisons involving D. pseudoobscura and approximately 3.5 for comparisons involving D. willistoni or D. littoralis.
Changes in the median CNCS length reflect changes in the overall distribution of CNCS lengths in pairwise comparisons between D. melanogaster and either D. erecta, D. pseudoobscura, D. willistoni, or D. littoralis (Figure 5). These data quantitatively describe the pattern of non-coding conservation shown in Figures 2 and 4: CNCS lengths become shorter with increasing divergence but plateau to approximately the same length in the most distant comparisons. The stability of this distribution at more extreme evolutionary distances is apparently insensitive to changes in the proportion of non-coding DNA that is conserved (compare D. willistoni and D. littoralis). Shown for comparison is the distribution of CNCS lengths between D. melanogaster and D. virilis from [44], as well as a reanalysis of this data using the current methods. Differences between the present and previous results for the D. virilis data show the effect of different methods for detecting CNCSs. The differences observed in the distribution of CNCS lengths between the closely related species D. virilis and D. littoralis using the AVID-VISTA method reflect the fact that the D. virilis data were obtained from non-coding regions with known or suspected cis-regulatory function, whereas the data here represent a more random sampling of non-coding regions in the Drosophila genome.
Figure 5.
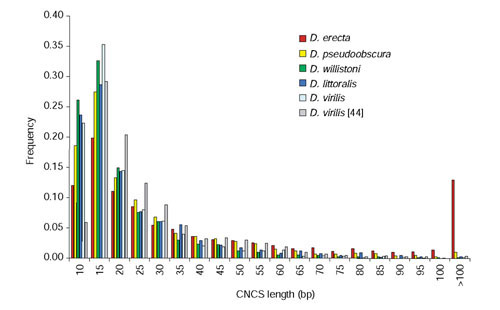
Frequency distribution of CNCS lengths in Drosophila species. The distributions of CNCS lengths are shown for comparisons between D. melanogaster and either D. erecta, D. pseudoobscura, D. willistoni or D. littoralis. CNCSs of 10 bp or greater with 90% or greater nucleotide identity were identified using VISTA. Also shown for comparison is a re-analysis of the length distribution of CNCSs between D. melanogaster and D. virilis using the current methods, as well as previous results for a sample of noncoding regions published in [44].
Conservation of non-coding sequences is typically interpreted as evidence of functional constraint and this assumption underlies most phylogenetic footprinting methods. This assumption was questioned by Clark [45], who proposed an alternative hypothesis for non-coding conservation based on heterogeneity in mutation rates (that is, mutational cold spots). To resolve these alternatives we studied the spatial distribution and conservation of spacing between CNCSs in the Drosophila genome. Under a simple mutational cold-spot hypothesis, CNCSs should occur randomly in non-coding DNA and the lengths of 'spacer intervals' between CNCSs should be exponentially distributed [46,47]. In addition, there should be no tendency for the spacing between mutational cold spots to remain conserved between divergent Drosophila species, given the rapid rate of DNA loss in unconstrained sequences in the Drosophila genome [48,49].
As shown in Figure 6 for non-coding comparisons between D. melanogaster and D. pseudoobscura, the frequency spacer interval lengths between CNCSs in the D. melanogaster genome differ significantly from the exponential distribution. The deviation from expected results from an excess of short and long spacer intervals, indicating that CNCSs are clustered in the Drosophila genome. Non-random spacing of CNCSs is also observed in other pairwise species comparisons in the genus Drosophila ([46] and data not shown). In addition, the lengths of homologous spacer intervals are highly correlated across species (Figure 7). This correlation is unlikely to be an artifact of alignment, as the AVID method first aligns regions of local similarity before generating a global alignment. Moreover, similar results have been obtained using non-global alignment methods [46]. These results suggest that spacer interval sequences between CNCSs (and therefore CNCSs themselves) are functionally constrained, and provide evidence against the hypothesis that CNCSs are simply mutational cold spots.
Figure 6.
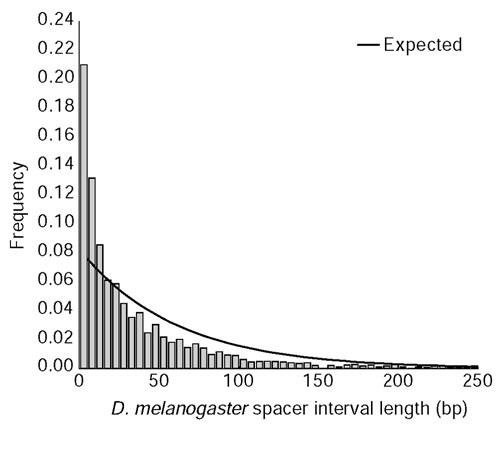
Frequency distribution of spacer interval lengths separating CNCSs between D. melanogaster and D. pseudoobscura. Plotted is a histogram of the length in D. melanogaster of 'nonconserved' spacer interval sequences between CNCSs identified using VISTA (10-bp window, 90% identity). Spacer intervals separating a CNCS and a conserved coding segment, or between two conserved coding segments were omitted from this analysis. Note that only spacer interval lengths less than 250 bp are displayed for clarity. Solid lines represent the expectation under an exponential distribution using an estimate of the rate parameter λ based on the inverse of the mean spacer interval length to be 0.0165. The null hypothesis that spacer interval lengths are exponentially distributed can be rejected (χ2 = 2,040.1, df = 30, p < 10-6), indicating that Drosophila CNCSs are non-randomly spaced.
Figure 7.
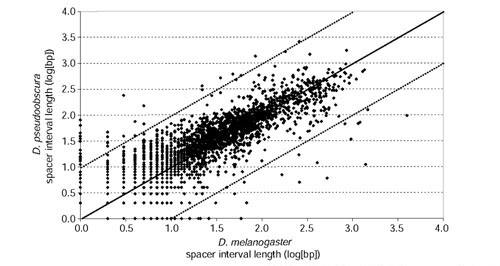
Correlation of spacer interval lengths separating CNCSs between D. melanogaster and D. pseudoobscura. Each point represents the log10-transformed lengths for a homologous pair of spacer intervals. Spacer intervals separating a CNCS and a conserved coding segment, or between two conserved coding segments were omitted from this analysis. The solid line represents perfect spacer interval length conservation; the dashed lines represent order of magnitude size changes in spacer interval length between these two species. The correlation coefficient for homologous spacer interval lengths is r = 0.85 (p < 0.01).
Clusters of CNCSs are readily apparent in VISTA plots of complex gene regions with known cis-regulatory function (Figures 2 and 4). In addition, there is a strong tendency for known cis-regulatory elements to overlap clusters of CNCSs. For example, discrete enhancers that control embryonic expression of eve are contained within discrete CNCS clusters in the region 5' to eve (Figure 2). In contrast, discrete CNCSs clusters are not observed in the region 3' to eve where enhancers overlap one another [50,51]. The correspondence of CNCS clusters and functional enhancers is observed in other regions of the Drosophila genome, such as the discrete muscle-specific enhancer in the fourth intron of ap (Figure 4). The inexact correspondence between enhancer sequences and CNCS clusters is perhaps not unexpected as enhancers are typically defined as the minimal sequence sufficient to recapitulate native expression in a reporter gene assay. Nevertheless, this pattern suggests a functional relationship between cis-regulatory elements and discrete CNCS clusters.
To test the hypothesis that CNCS clusters can predict the location of cis-regulatory elements in the Drosophila genome, we carried out P-element-mediated reporter gene analysis of genomic sequences corresponding to a CNCS cluster in the fourth intron of ap. This CNCS cluster is apparent in pairwise comparisons between D. melanogaster and D. pseudoobscura as well as between D. melanogaster and D. virilis (Figure 4). ap is a LIM-homeobox transcription factor expressed in many tissues in Drosophila, including embryonic expression in the developing brain [52,53]. As shown in Figure 8, the D. melanogaster genomic sequences corresponding to the CNCS cluster in the ap intron 4 drives reporter gene expression in the Drosophila embryo in a specific pattern that recapitulates native ap expression in the developing brain. In addition, when introduced into the genome of D. melanogaster, the homologous fragment from the D. virilis genome also drives reporter gene expression in the same pattern, indicating that the expression pattern resulting from this enhancer has been conserved since the divergence of these two species. Experiments to test the function of CNCS clusters 1, 2, and 3 in the ap region are currently underway.
Figure 8.
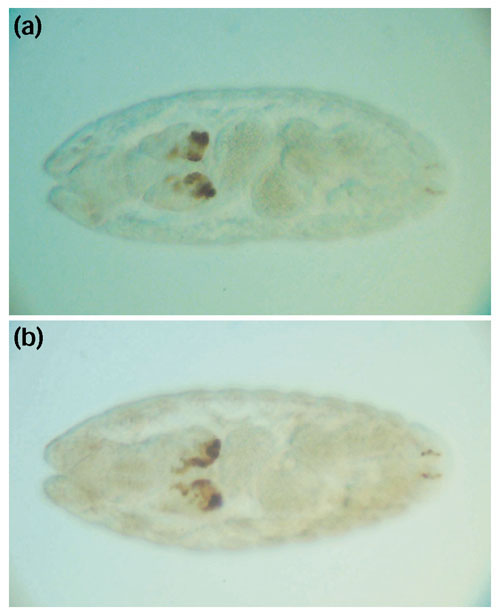
Reporter gene expression driven by genomic sequences corresponding to the CNCS cluster in ap intron 4. Specific expression in the embryonic brain is driven by both (a) D. melanogaster and (b) D. virilis sequences, indicating that the function of this enhancer has been conserved in these two species.
Discussion
Prospects for comparative gene prediction in Drosophila
Although great progress has been made towards understanding the protein-coding component of eukaryotic genome sequences [54,55,56,57], comprehensive genome annotation is far from complete in any metazoan. State-of-the-art statistical and remote-homology gene-prediction methods are successful at identifying the location of exons in unannotated genomic DNA, but are often quite poor at predicting the details of gene structure, necessitating human curation [14]. One of the most useful sources of information for accurately predicting complex gene structures is EST/cDNA data [58]. Predicting the structure of genes for which no EST/cDNA data exists will require alternative approaches, such as comparative gene modeling among divergent species with conserved proteomes in the same group of organisms.
The results of our Ka/Ks analyses presented here give preliminary insight into the prospects of comparative gene modeling using large-scale sequence data in the genus Drosophila. From our findings, we expect that structural details of most Release 3 coding sequences can be verified and improved using pairwise sequence data between divergent species like D. melanogaster and D. pseudoobscura. Our results also indicate that, although it may not be necessary for many genes in the D. melanogaster genome, de novo comparative gene prediction between these species will find the vast majority of as-yet unidentified genes lacking EST/cDNA data. It is important to note that we do not expect to detect all coding exons (especially short exons [20]) in pairwise comparisons, highlighting the added value of multiple species data for comparative exon prediction. In addition, important details of gene models will prove difficult to predict using only comparative data, as amino-acid divergence (especially insertions or deletions) can obscure intron-exon boundaries and other details of gene structure. Moreover, there is inherent uncertainty in the 'correct' gene structure developed from comparative data alone, since two divergent sequences are simultaneously being modeled. Finally, the comparative annotation of UTR sequences awaits the development of methods that accurately predict the non-coding components of gene models.
The patterns of protein-coding sequence evolution detected in our data have important implications for comparative gene prediction. Most notably, the trend we detect for predicted genes to show an increased rate of amino-acid substitution relative to known genes is important, as it may reflect differences in functional constraint or quality of gene models between the two classes of genes. For at least three reasons, we believe that the elevated rate of amino-acid substitution in predicted genes is not a result of poor-quality gene models in this class of genes. First, many of the genes in the predicted class have EST/cDNA data (see Supplementary Table 1), so the details of these gene models are likely to be correct. Second, estimates of Ka (and Ks) are based on aligned sequences; thus gross inaccuracies in gene models that would create gaps in the alignment are excluded from estimates of evolutionary rates. Third, differential rates in these two classes of genes maybe expected, as a high proportion of known genes were selected for study because their mutational inactivation resulted in an obvious phenotype. Thus we favor the interpretation that increased rates of amino-acid substitution in predicted genes results from lower levels of functional constraint.
If this interpretation is correct, our results confirm those of Schmid and co-workers [34,59] who have shown that a large fraction of randomly sampled coding sequences and orphan genes are rapidly evolving in the genus Drosophila. Our results are also consistent with those of Ashburner et al. [60] who show that genes with known mutant phenotypes in D. melanogaster are more likely to have a conserved homolog in GenBank relative to predicted genes with no known phenotype. Similarly, Zdobnov et al. [23] show that D. melanogaster orphan genes tend to exhibit lower levels of conservation in pairwise comparison with A. gambiae [23]. Finally, the interpretation that known and predicted genes differ in their levels of functional constraint is supported by the fact that increased rates of protein evolution in D. willistoni affect predicted genes more strongly than known genes (Table 2). Together these results suggest that there is a large class of functional protein-coding sequences evolving under weak selective constraint in the Drosophila genome [34]. Rates of evolution for this class of genes may be too fast to allow the identification of homologs from extremely divergent species (such as Anopheles) for comparative gene prediction, but slow enough to use comparative data within the genus Drosophila.
Rearrangement, transposition and genome annotation
Genome rearrangement in Drosophila typically occurs through paracentric inversion, allowing the homology of chromosome arms to be maintained over millions of years. Homologous chromosome arms in the genus Drosophila are referred to as Muller's elements, and represent a clearly established level of synteny between species [7]. The homology of Muller's elements can be extended to A. gambiae [23,31]; however it is clear that a substantial fraction of rearrangements between these species must occur by other mechanisms than paracentric inversion. Levels of synteny below the chromosome arm are more difficult to establish, and the description of these levels of conserved gene order is currently arbitrary [23]. Thus it is important to point out that the strictly co-linear microsynteny we detect between Drosophila species differs from the 'hyphenated' microsynteny with multiple gene losses and gains between Drosophila and Anopheles [32].
Rates of genome rearrangement between species in the genus Drosophila based on cytological evidence are thought to be among the highest for any metazoan [61]. Before this study only a single fixed inversion breakpoint had been characterized at the sequence level in Drosophila [24]. Unfortunately, the limited number of events in our data makes it difficult to estimate the absolute rate of genome rearrangement at the sequence level in Drosophila. More extensive analysis of genome rearrangement at the sequence level in Caenorhabditis by Coghlan and Wolfe [62], suggests that rates of genome rearrangement are fourfold higher in nematodes than in flies. As micro-inversions (such as that seen in the D. littoralis ftz region) are not expected to be included in cytological estimates, this claim will have to be re-evaluated using large-scale comparative sequence data in Drosophila.
There is mounting evidence for the role of transposable elements in the origin of inversion breakpoints in D. melanogaster and other species [63,64,65]. In contrast, our data provide no evidence that rearrangement breakpoints fixed between Drosophila species are associated with transposable element sequences (see also [24]). Together, these observations suggest a 'hit-and-run' scenario in which transposable elements may play a part in the origin of chromosome rearrangements, but are lost (either through deletion or transposition) by the time rearrangements reach high frequency or fixation between species [66]. Consistent with this scenario is the fact that transposable element sequences are not conserved between species within unrearranged microsyntenic regions (for example, the HB element in the ap region, Figure 4).
The maintenance of microsyntenic regions we observe at the scale of multi-gene fosmid-sized regions (approximately 40 kb) should improve identification of orthologs for comparative gene modeling. On the other hand, de novo comparative gene identification can be complicated if, for example, nested genes maintain microsyntenic relationships [67]. In addition, as the few rearrangement breakpoints we observed occur exclusively in intergenic regions (see also [24]), rearrangement breakpoints may also help define the boundaries of complex genetic loci. Conservation of microsynteny between genes and flanking intergenic regions may reveal structural and functional connections in the genome, and thus it may be possible to associate functional non-coding sequences with the appropriate flanking gene through genome rearrangements (see also [30]). Proof of this principle can be seen for the intergenic region 3' to eve and 5' to TER94, which maintains association with eve, but not TER94, in non-melanogaster group species (Figure 2). This conservation of microsynteny between the eve coding and 3' CNCSs is consistent with the fact that this region is known to contain multiple enhancer sequences that regulate embryonic eve expression [50,51].
Prospects for comparative cis-regulatory prediction in Drosophila
Conservation of non-coding sequences is rapidly becoming one of the most powerful methods of predicting individual cis-regulatory elements in genomic sequences [68]. Most computational methods designed to identify CNCSs rely on the assumption that non-coding conservation implies functional constraint, rather than heterogeneity in mutation rates [45]. Our results provide two pieces of evidence that support this assumption and argue against a simple mutational cold-spot interpretation of non-coding conservation. First, we demonstrate that the lengths of spacer intervals separating CNCSs are non-randomly distributed in the Drosophila genome, indicating a tendency for CNCSs to be clustered in non-coding DNA. Second, we show that the spatial relationships of neighboring CNCSs are generally conserved as revealed by the strong correlation of spacer interval lengths in divergent species. Clustering of CNCSs and conservation of CNCS spacing are not expected under a mutational cold-spot hypothesis, and suggest that the spacer intervals between CNCSs, and therefore CNCSs themselves, are under functional constraint. Given the rapid rate of DNA loss for unconstrained sequences in Drosophila [48,49], it is difficult to understand the mere existence of spacer intervals, as well as their conservation of length, without invoking some form of functional constraint acting on these sequences.
Clustering of CNCSs has also been recently reported in the worm genome [47], and future research will determine if CNCS clustering is a general feature of non-coding conservation in metazoan genomes. As yet, there has been no report of a general tendency for CNCSs to be clustered in mammalian genomes, although certain regions of the mammalian genome (such as the H19 region [69]) show a strong pattern of CNCS clustering. The general functional significance of CNCS clusters remains to be explored, but it is clear that some CNCS clusters correspond to functional enhancer sequences. We show here proof of the principle that CNCS clusters can be used to identify functional enhancer sequences in the Drosophila genome (Figure 7, 8), as Ishihara et al. [69] have shown for CNCS clusters in the mammalian genome.
Using CNCS clusters to identify enhancers represents the comparative genomic analogue of efforts to predict cis-regulatory sequences by exploiting the clustering of predicted transcription factor binding sites [70,71]. However, unlike binding-site clusters, CNCS clusters can be rapidly identified in the absence of any a priori information about transcription factor specificity, and may therefore provide a more general approach to genome-wide cis-regulatory prediction. In fact, CNCS clusters may be a powerful source of data for inferring transcription factor specificity, as specific binding-site motifs are likely to be locally over-represented in CNCS clusters with demonstrable enhancer function. Using the intrinsic clustering of CNCSs provides a new way of circumventing the reliance on expression data implicit in current approaches to CNCS-based motif discovery [72]. Finally, successfully linking CNCS clusters with enhancer function will provide an alternative means of defining enhancer structure based on evolutionary rather than operational criteria.
Conclusions
The patterns of divergence in both coding and cis-regulatory sequences described here indicate that D. pseudoobscura will greatly aid efforts to functionally annotate the D. melanogaster genome, and justify the choice of D. pseudoobscura as the second Drosophila species for complete genome sequencing. The divergence between these species is sufficient such that there does not appear to be any need to go to more distantly related species to obtain a high level of signal to noise to detect functionally constrained sequences. This observation suggests that the search for additional Drosophila species whose genome sequence would help interpret the D. melanogaster sequence should focus on species at a similar evolutionary distance as D. pseudoobscura.
Of the species studied here, D. erecta is too closely related to D. melanogaster for comprehensive gene and cis-regulatory prediction. Neither coding nor non-coding sequences have experienced sufficient divergence to differentiate whether sequences 'conserved' between D. erecta and D. melanogaster result from functional constraint or shared ancestry (Figures 2,4). At the other extreme, using A. gambiae genome sequences may not substantially aid comprehensive genome annotation, as a large proportion of Drosophila genes may not be present in the Anopheles genome and the lack of non-coding conservation between these groups (Figure 7 and [23]) means that this powerful source of data is unavailable for cis-regulatory annotation. In contrast, divergent species in the genus Drosophila (such as D. littoralis and D. willistoni) show similar properties to each other and to D. pseudoobscura in terms of their utility for identifying functionally constrained coding or non-coding sequences.
D. willistoni and D. melanogaster are both members of the subgenus Sophophora; thus these two species are expected to share more aspects of their biology in common than either will with D. littoralis. Therefore, we propose that of the species studied here, D. willistoni is the most suitable candidate for the third Drosophila species for complete genome sequencing. We make this suggestion despite the increased rate of evolution and changes in base composition and codon usage observed in this lineage. In fact, it may be possible to take advantage of these unusual patterns of molecular evolution in the D. willistoni lineage to dissect regions of the Drosophila genome under different levels of functional constraint. Given a more thorough understanding of these phenomena and their cause(s), we believe that a D. willistoni genome sequence would complement efforts to annotate the Drosophila genome based on whole genome comparisons between D. melanogaster and D. pseudoobscura.
Materials and methods
Construction of Drosophila species fosmid libraries
Four Drosophila species spanning a range of divergence times in the genus were selected for study: D. erecta (strain 14021-0224.0), D. pseudoobscura (strain 14011-0121.4), D. willistoni (strain 14030-0814.10) and D. littoralis (strain 15010-1001.10). All strains are available from the Tucson Drosophila species stock center [73]. To construct fosmid libraries, genomic DNA from adult flies was prepared by partial digestion with MboI and size-selecting fragments using pulsed-field gel electrophoresis, then cloned in the BamHI site of the fosmid vector pFOS1 [74] and transformed into Escherichia coli strain XL1-Blue MR (Stratagene). Detailed information about the Drosophila stocks, construction of the genomic fosmid libraries and their availability can be found at the Children's Hospital of Oakland Research Institute BACPAC Resources website [75].
Probe design and library screening
To amplify gene-specific STSs, D. erecta, D. pseudoobscura, D. willistoni and D. littoralis genomic DNA was isolated from adult flies and used as template for PCR with degenerate primers. Primer sequences are available upon request. Double-stranded 40-nucleotide oligomers designed from the gene and species specific STSs were radioactively labeled with 32P and hybridized to genomic colony filters [76]. Positive clones were subjected to PCR to remove false positives and restriction mapped using SalI-NotI, EcoRI-NotI double digestions. The largest clone overlapping all positive hits was sheared and subcloned into 3-kb plasmids, and sequenced using methods described in [9]. From the results of this screen, we estimated the average number of unique hits per library per species to be 4.1 for D. erecta, 3.2 for D. pseudoobscura, 2.2 for D. willistoni and 2.9 for D. littoralis. These figures indicate that these libraries have fewer unique hits than the expected approximately 5× coverage. Sequences for these clones have been submitted to GenBank under the accession numbers AY186999 and AY190934-AY190963.
Comparative gene prediction
Coding sequences in these fosmid clones were predicted using the computational pipeline used to re-annotate the D. melanogaster genome [12]. The only major modification to this pipeline was to add an additional tier of evidence containing the results of TBLASTN searches of all Release 3 D. melanogaster peptides [14] against non-melanogaster sequences. Gene predictions were manually verified and refined using the annotation platform Apollo [13]. As no EST information exists to annotate transcribed non-coding sequences (such as UTRs) for non-melanogaster species, we annotated only protein-coding gene and exon models. For similar reasons, and to minimize the insertion/deletion of amino acids at intron/exon boundaries, we did not require non-melanogaster models to obey consensus splice site rules. Annotated sequences were stored and queried in Gadcompara, a cloned version of the D. melanogaster annotation database, Gadfly.
To identify orthologous genes in Anopheles gambiae, 8,987 scaffolds from project accession number AAAB00000000 were downloaded from GenBank and searched using default parameters of TBLASTN 2.0 [77] with the D. melanogaster peptides listed in Supplementary Table 1 (see additional data files) as queries. Homologs were identified as HSPs with an expected value < 10-20 and ≥ 30% identity over 100 amino acids.
Alignment and estimation of sequence conservation
Orthologous regions of the D. melanogaster genome corresponding to sequenced fosmids were identified using default parameters of BLAT [78] against a database made up of the Release 3 sequences [9]. The union of sequences spanning hits to each candidate region was extracted and is listed in Table 1. Orthologous coding and peptide sequences from D. melanogaster were identified by unique gene symbols in Gadfly using Release 3 annotations [14]. For genes with alternative transcripts, the transcript leading to the longest translation product was chosen in most, but not all, cases. Coding and translated amino-acid sequences from non-melanogaster species were extracted from Gadcompara using MySQL queries.
Multiple alignment of orthologous amino-acid sequences was carried out using the default parameters of DIALIGN 2.1 [79]. Amino-acid alignments were used to align coding sequences using the gap_cds utility of the SEALS package [80] to ensure that nucleotide alignment gaps were inserted between codons. Pairwise Ka/Ks tests were carried out using PAML 3.12 [22] with runmode = -2, as outlined in [20]. Evidence of functional constraint was inferred when twice the difference in likelihoods between a model with Ka/Ks ratio fixed at 1 versus one with Ka/Ks as a free parameter exceeded a cutoff such that the p-value was less than 0.05 per number of tests. Estimates of Ka and Ks in Table 2 were obtained using the method of Yang and Nielson [81], which accounts for differences in nucleotide and codon frequencies as well as transition:transversion rate bias, implemented in PAML 3.12 [22]. Multiple amino-acid and coding sequence alignments are available on request.
Annotations of candidate regions in Table 1 were exported from Gadfly in VISTA format [43], and used to evaluate conservation in non-coding sequences. Nontranscribed (intergenic) and transcribed (UTR and intron) non-coding sequences were analyzed together [44]. Pairwise alignment of homologous genomic regions between D. melanogaster and individual non-melanogaster species was performed using default parameters of the AVID global alignment tool [42]. CNCSs were identified in VISTA using a window size of 10 bp with an identity of 90% with manual post-processing to remove spurious matches at the beginning and end of the alignments. Repeat masking was performed before coding sequence annotation to identify transposable elements; however, it is possible that some CNCSs are simple repetitive sequences. Only sequences from candidate regions that clearly maintain microsynteny in non-melanogaster species were included in estimates of non-coding conservation. Thus, the entire D. littoralis ftz, Rh3 and Rh4 clones as well as rearranged segments of the D. willistoni, D. pseudoobscura and D. littoralis eve clones, D. willistoni and D. pseudoobscura Rh1 clones, and D. willistoni Rh3 clones were omitted from analyses of non-coding conservation. In addition, we also reanalyzed non-coding conservation between D. melanogaster and D. virilis using this same approach for the dataset of [44]. Pairwise genomic alignments and VISTA regions files are available on request.
Generation of transgenic flies and expression analysis
The CNCS cluster carrying the ap brain enhancer was amplified from D. melanogaster (BAC) and D. virilis (P1) clones bearing the ap gene, respectively. The following pairs of oligonucleotides were used for PCR amplification:
DmelBR-F:5'-AAACCATCTCACTCGCATGA-3'
DmelBR-R: 5'-TGCTTCCAGACAACGACAAA-3'
DvirBR-F: 5'-TTCGCTGGTCAATGGTTCAA-3'
DvirBR-R: 5'-ATGGGCTGGCAACATACAACA-3'
The resulting PCR products from D. melanogaster (1,235 bp) and D. virilis (1,658 bp) were cloned into pCaSpeRhs43β-gal to generate pChabMelBR and pChabVirBR, respectively. These constructs were injected into D. melanogaster y;w embryos by standard P-element transformation [82]. Two independent lines were generated with pChabMelBR and three with pChabVirBR, all of which showed similar expression patterns. Transgenic embryos were immunostained with monoclonal anti-β-gal antibody (1:4,000; Promega) as described in [53].
Additional data files
Supplementary Table 1, describes the number of coding exons, amino acids, ESTs, and Ka/Ks ratio for D. melanogaster genes in this study. Supplementary Table 2, details the codon usage in D. melanogaster compared with D. erecta, D. pseudoobscura, D. willistoni, and D. littoralis.
Supplementary Material
Supplementary table 1
Supplementary table 2
Acknowledgments
Acknowledgements
We thank J. Powell and K. White for providing fly strains and D. Grimaldi for confirming the identity of the strains used in this study; J. Kaminker, S. Misra and S. Prochnik for advice on using Apollo; D. Bensasson and D. Pollard for providing scripts to split multiple alignments by exon, and calculate base composition, respectively; I. Dubchak and A. Poliakov for advice on VISTA analyses; S. Prochnik and J. Carlson for assistance with GenBank submissions; and E. Frise, E. Smith and J. Carlson for technical support. We also thank M. Eisen, J. Fay, A. Nekrutenko, D. Pollard, K. Schmid, M. Yandell, and three anonymous reviewers for comments on the manuscript. This work was supported by NIH grant HG00750 to G.M.R., by NIH Grant HG00739 to FlyBase (W.M. Gelbart), and by HHMI (C.J.M. and G.M.R.). C.M.B is supported by NIH training grant T32 HL07279 to E. Rubin. Research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC0376SF00098, University of California.
This article is part of a series of refereed research articles from Berkeley Drosophila Genome Project, FlyBase and colleagues, describing Release 3 of the Drosophila genome, which are freely available at http://genomebiology.com/drosophila/.
References
- Lewontin RC, Moore JA, Provine WB, Wallace B, Eds . Dobzhansky's Genetics of Natural Populations I-XLIII. New York: Columbia University Press; 1981. [Google Scholar]
- Patterson JT, Stone WS. Evolution in the Genus Drosophila. New York: Macmillan; 1952. [Google Scholar]
- Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural population of D. pseudoobscura. Genetics. 1966;54:595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Blackman RK, Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986;188:499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Bray SJ, Hirsh J. The Drosophila virilis dopa decarboxylase gene is developmentally regulated when integrated into Drosophila melanogaster. EMBO J. 1986;5:2305–2311. doi: 10.1002/j.1460-2075.1986.tb04498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford: Oxford University Press; 1997. [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: Release 3 of the Drosophila euchromatic genome sequence. Genome Biol. 2002;3:research0079.1–0079.14. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human genome sequencing center at Baylor College of Medicine: Drosophila genome project http://www.hgsc.bcm.tmc.edu/projects/drosophila/update.html
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Mungall CJ, Misra S, Berman BP, Carlson J, Frise E, Harris NL, Marshall B, Shu S, Kaminker JS, Prochnik SE, et al. An integrated computational pipeline and database to support whole-genome sequence annotation. Genome Biol. 2002;3:research0081.1–0081.11. doi: 10.1186/gb-2002-3-12-research0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE, Searle SMJ, Harris N, Gibson ML, Iyer VR, Richter J, Wiel C, Bayraktaroglu L, Birney E, Crosby MA, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3:research0082.1–0082.14. doi: 10.1186/gb-2002-3-12-research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila euchromatic genome: a systematic review. Genome Biol. 2002;3:research0083.1–0083.22. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovskaya ER, Petrov DA, Hartl DL. A combined molecular and cytogenetic approach to genome evolution in Drosophila using large-fragment DNA cloning. Chromosoma. 1993;102:253–266. doi: 10.1007/BF00352399. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, Gelfand MS, Mironov AA. Gene recognition in eukaryotic DNA by comparison of genomic sequences. Bioinformatics. 2001;17:1011–1018. doi: 10.1093/bioinformatics/17.11.1011. [DOI] [PubMed] [Google Scholar]
- Reese MG, Kulp D, Tammana H, Haussler D. Genie - gene finding in Drosophila melanogaster. Genome Res. 2000;10:529–538. doi: 10.1101/gr.10.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Carulli JP, Chen DM, Stark WS, Hartl DL. Phylogeny and physiology of Drosophila opsins. J Mol Evol. 1994;38:250–262. doi: 10.1007/BF00176087. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A, Makova KD, Li WH. The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 2002;12:198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- Yang Z. Phylogenetic Analysis by Maximum Likelihood (PAML) 3.0. London, University College London; 2000. [Google Scholar]
- Zdobnov EM, Von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Cirera S, Martin-Campos JM, Segarra C, Aguade M. Molecular characterization of the breakpoints of an inversion fixed between Drosophila melanogaster and D. subobscura. Genetics. 1995;139:321–326. doi: 10.1093/genetics/139.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D, Preiss A, Powell JR. Regulation of the segmentation gene fushi tarazu has been functionally conserved in Drosophila. EMBO J. 1990;9:3957–3966. doi: 10.1002/j.1460-2075.1990.tb07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR, Eds . Drosophila Inversion Polymorphism. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- Moschetti R, Caggese C, Barsanti P, Caizzi R. Intra- and interspecies variation among Bari-1 elements of the melanogaster species group. Genetics. 1998;150:239–250. doi: 10.1093/genetics/150.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Frutos R, Peterson KR, Kidwell MG. Distribution of Drosophila melanogaster transposable element sequences in species of the obscura group. Chromosoma. 1992;101:293–300. doi: 10.1007/BF00346008. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Carthew RW, Rubin GM. Evolution of gene position: chromosomal arrangement and sequence comparison of the Drosophila melanogaster and Drosophila virilis sina and Rh4 genes. Proc Natl Acad Sci USA. 1991;88:10203–10207. doi: 10.1073/pnas.88.22.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Ranz JM, Ruiz A. Chromosomal elements evolve at different rates in the Drosophila genome. Genetics. 2002;161:1137–1154. doi: 10.1093/genetics/161.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshakov VN, Topalis P, Blass C, Kokoza E, della Torre A, Kafatos FC, Louis C. A comparative genomic analysis of two distant diptera, the fruit fly, Drosophila melanogaster, and the malaria mosquito, Anopheles gambiae. Genome Res. 2002;12:57–66. doi: 10.1101/gr.196101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasova D, Ton LQ, Copley RR, Zdobnov EM, Wang X, Hong YS, Sim C, Bork P, Kafatos FC, Collins FH. Comparative genomic analysis in the region of a major Plasmodium-refractoriness locus of Anopheles gambiae. Proc Natl Acad Sci USA. 2002;99:8179–8184. doi: 10.1073/pnas.082235599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LW, Comeron JM, Chen B, Kreitman M. The molecular clock revisited: the rate of synonymous versus replacement change in Drosophila. Genetica. 1998;102-103:369–382. [PubMed] [Google Scholar]
- Schmid KJ, Tautz D. A screen for fast evolving genes from Drosophila. Proc Natl Acad Sci USA. 1997;94:9746–9750. doi: 10.1073/pnas.94.18.9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs PS, Holmes EC, Ashburner M. The molecular evolution of the alcohol dehydrogenase and alcohol dehydrogenase-related genes in the Drosophila melanogaster species subgroup. Mol Biol Evol. 1994;11:287–304. doi: 10.1093/oxfordjournals.molbev.a040110. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. Switch in codon bias and increased rates of amino acid substitution in the Drosophila saltans species group. Genetics. 1999;153:339–350. doi: 10.1093/genetics/153.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. Fluctuating mutation bias and the evolution of base composition in Drosophila. J Mol Evol. 2000;50:1–10. doi: 10.1007/s002399910001. [DOI] [PubMed] [Google Scholar]
- Shields DC, Sharp PM, Higgins DG, Wright F. 'Silent' sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- Laird CD. DNA of Drosophila chromosomes. Annu Rev Genet. 1973;7:177–204. doi: 10.1146/annurev.ge.07.120173.001141. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Hartl DL. Codon usage bias and base composition of nuclear genes in Drosophila. Genetics. 1993;134:847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CL, Carew EA, Powell JR. Evolution of the Adh locus in the Drosophila willistoni group: the loss of an intron, and shift in codon usage. Mol Biol Evol. 1993;10:605–618. doi: 10.1093/oxfordjournals.molbev.a040027. [DOI] [PubMed] [Google Scholar]
- Visualization tools for alignments http://www-gsd.lbl.gov/vista/details_avid.htm
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Kreitman M. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 2001;11:1335–1345. doi: 10.1101/gr.178701. [DOI] [PubMed] [Google Scholar]
- Clark AG. The search for meaning in noncoding DNA. Genome Res. 2001;11:1319–1320. doi: 10.1101/gr.201601. [DOI] [PubMed] [Google Scholar]
- Bergman CM. Evolutionary Analyses of Transcriptional Control Sequences in Drosophila. Chicago: University of Chicago; 2001. [Google Scholar]
- Webb CT, Shabalina SA, Ogurtsov AY, Kondrashov AS. Analysis of similarity within 142 pairs of orthologous intergenic regions of Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acids Res. 2002;30:1233–1239. doi: 10.1093/nar/30.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol. 1998;15:293–302. doi: 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackerson C, Fujioka M, Goto T. The even-skipped locus is contained in a 16-kb chromatin domain. Dev Biol. 1999;211:39–52. doi: 10.1006/dbio.1999.9301. [DOI] [PubMed] [Google Scholar]
- Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- Rincon-Limas DE, Lu CH, Canal I, Calleja M, Rodriguez-Esteban C, Izpisua-Belmonte JC, Botas J. Conservation of the expression and function of apterous orthologs in Drosophila and mammals. Proc Natl Acad Sci USA. 1999;96:2165–2170. doi: 10.1073/pnas.96.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Aravind L, Sherlock G, Ball CA, Koonin EV, Dwight SS, Harris MA, Dolinski K, Mohr S, Smith T, et al. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey DA. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Intiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- International Human Genome Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL. Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 2002;3:research0029.1–0029.12. doi: 10.1186/gb-2002-3-6-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Aquadro CF. The evolutionary analysis of 'orphans' from the Drosophila genome identifies rapidly diverging and incorrectly annotated genes. Genetics. 2001;159:589–598. doi: 10.1093/genetics/159.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Casals F, Ruiz A. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 2001;11:230–239. doi: 10.1101/gr.162901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A, Wolfe KH. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle TW, Haymer DS. The role of the transposable element hobo in the origin of endemic inversions in wild populations of Drosophila melanogaster. Genetica. 1992;86:113–126. doi: 10.1007/BF00133715. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Wall JD, Kreitman M. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics. 1999;153:1297–1311. doi: 10.1093/genetics/153.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres M, Puig M, Ruiz A. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 2001;11:1353–1364. doi: 10.1101/gr.174001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiopoulos KD, della Torre A, Predazzi V, Petrarca V, Coluzzi M. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc Natl Acad Sci USA. 1998;95:12444–12449. doi: 10.1073/pnas.95.21.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachter L, Alexandersson M, Cawley S. Applications of generalized pair hidden Markov models to alignment and gene finding problems. J Comput Biol. 2002;9:389–399. doi: 10.1089/10665270252935520. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Hatano N, Furuumi H, Kato R, Iwaki T, Miura K, Jinno Y, Sasaki H. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in Igf2/H19 imprinting. Genome Res. 2000;10:664–671. doi: 10.1101/gr.10.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley EM, Roeder K, Bina M. A statistical model for locating regulatory regions in genomic DNA. J Mol Biol. 1997;268:8–14. doi: 10.1006/jmbi.1997.0965. [DOI] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence C. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Tucson Drosophila Species Stock Center http://stockcenter.arl.arizona.edu
- Kim UJ, Shizuya H, de Jong PJ, Birren B, Simon MI. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 1992;20:1083–1085. doi: 10.1093/nar/20.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BACPAC Resources Center http://www.chori.org/bacpac
- Ross MT, LaBrie J, McPherson VP, Stanton P., Jr . Screening large insert libraries by hybridization. In: Dracopoli NC, Haines JL, Korf BR, Moir DT, Morton CC, Seidman CE, Seidman JG, Smith DR, editor. In Current Protocols in Human Genetics. Vol. 1. New York: Wiley and Sons; 1999. pp. 5.6.1–5.6.52. [Google Scholar]
- Washington University BLAST archives http://blast.wustl.edu
- Kent WJ. BLAT - the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Walker DR, Koonin EV. SEALS: a system for easy analysis of lots of sequences. Proc Int Conf Intell Syst Mol Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, Kaminker JS, Kennedy C, Mungall CJ, Sullivan BA, Sutton GG, et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002;3:research0085.1–0085.16. doi: 10.1186/gb-2002-3-12-research0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Powers TP, Hogan J, Ke Z, Dymbrowski K, Wang X, Collins FH, Kaufman TC. Characterization of the Hox cluster from the mosquito Anopheles gambiae (Diptera: Culicidae). Evol Dev. 2000;2:311–325. doi: 10.1046/j.1525-142x.2000.00072.x. [DOI] [PubMed] [Google Scholar]
- Devenport MP, Blass C, Eggleston P. Characterization of the Hox gene cluster in the malaria vector mosquito, Anopheles gambiae. Evol Dev. 2000;2:326–339. doi: 10.1046/j.1525-142x.2000.00074.x. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Fellers JP, Shippy TD, Richardson EA, Maxwell M, Stuart JJ, Denell RE. Sequence of the Tribolium castaneum homeotic complex: the region corresponding to the Drosophila melanogaster antennapedia complex. Genetics. 2002;160:1067–1074. doi: 10.1093/genetics/160.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Hoey T, Levine M. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 1991;5:265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- Capovilla M, Kambris Z, Botas J. Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development. 2001;128:1221–1230. doi: 10.1242/dev.128.8.1221. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, Guarin H, Kronmiller B, Pacleb J, Park S, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3:research0080.1–0080.8. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1
Supplementary table 2


