Abstract
Plants produce proximal-distal growth axes with two types of growth potential: they can be indeterminate, in which case growth continues indefinitely, or they can be determinate, in which case growth is limited to the production of a single organ or a discrete set of organs. The indeterminate shoot axes of Arabidopsis pinhead/zwille mutants frequently are transformed to a determinate state. PINHEAD (PNH) is expressed in the central domain of the developing plant: the provascular tissue, the shoot apical meristem, and the adaxial (upper) sides of lateral organ primordia. Here, we show that ectopic expression of PNH on the abaxial (lower) sides of lateral organs results in upward curling of leaf blades. This phenotype correlates with a loss of cell number coordination between the two surfaces of the blade, indicating that ectopic PNH can cause changes in cell division rates. More strikingly, moving PNH expression from the central to the peripheral domain of the embryo causes transformation of the determinate cotyledon axis to an indeterminate state. We propose that growth axes are specified as determinate versus indeterminate in a PNH-mediated step. Our results add to a growing body of evidence that radial positional information is important in meristem formation. These results also indicate that genes regulating cell division and axis determinacy are likely to be among PNH targets.
INTRODUCTION
Numerous proximal-distal growth axes are found in shoot systems of angiosperm plants (Figure 1A). These include that of the main shoot, each lateral branch, each cotyledon, leaf, floral organ, and so on. Perpendicular to each proximal-distal axis is the radial axis, which runs in the central to peripheral direction (for cylindrical structures) or in the adaxial to abaxial direction (for planar structures; Figure 1B). Proximal-distal axes of plants can exhibit either indeterminate growth, resulting in the formation of an unlimited number of organs, or determinate growth, resulting in the formation of a single organ or a discrete set of organs. In flowering plants, the terms “determinate” and “indeterminate” frequently are applied to inflorescence architectural features characterized by terminal or nonterminal flowers, respectively. Here, we use the terms in their broader developmental sense to indicate growth potential of any proximal-distal axis (not just the inflorescence).
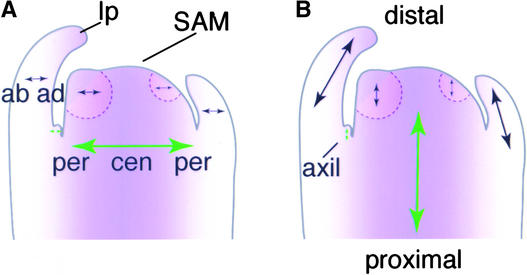
Figure 1.
Axes of the Arabidopsis Shoot.
In these cartoon representations of a longitudinal section through a vegetative shoot apex, purple represents central/adaxial positional information and arrows represent axes.
(A) Central-peripheral and adaxial-abaxial organization. The SAM is organized into central and peripheral zones. It produces new leaf primordia sequentially from its periphery. These leaf primordia have adaxial and abaxial zones that correspond to positions in the central-peripheral dimension of the meristem. ab, abaxial; ad, adaxial; cen, central; lp, leaf primordium; per, peripheral.
(B) Proximal-distal axes of the growing shoot. Arrows represent axes of growth in the proximal-distal (to the root-shoot junction) dimension. Through the action of the SAM, the main shoot axis (large green arrow), which is indeterminate, gives rise to the new growth axes of the leaf primordia (black arrows), which are determinate. Note that although the leaf will eventually be oriented nearly perpendicular to the long axis of the plant, its orientation is parallel during its primordial stages. New indeterminate axes (small green arrow) arise in the axils of the leaves with the production of an axillary meristem.
A key requirement of indeterminate growth potential for a shoot axis is the presence of a shoot apical meristem (SAM) at the distal end. The SAM is equipped specifically to give rise to organ primordia on its lateral flanks while maintaining a reservoir of self-replacing totipotent cells at its center. Several mutants have been isolated or engineered that lack or have a nonfunctional SAM. These include shoot meristemless (stm; Barton and Poethig, 1993), cup-shaped cotyledon1 and -2 double mutants (cuc; Aida et al., 1997), wuschel (wus; Laux et al., 1996), pinhead/zwille (pnh; McConnell and Barton, 1995; Moussian et al., 1998), and plants overexpressing YABBY3, FILAMENTOUS FLOWER (FIL; Siegfried et al., 1999), or KANADI (KAN; Eshed et al., 2001; Kerstetter et al., 2001). The corresponding genes act at several levels in the hierarchy that controls SAM development and function.
At the highest level, genes are responsible for establishing pattern. For instance, the SAM arises from the central, apical region of the globular embryo. Therefore, failure to elaborate central and/or apical identities would be expected to result in failure to form a SAM. Perhaps the best evidence for this is the fact that when the KAN gene (a gene normally expressed in a peripheral, or abaxial, domain of the embryo) is expressed throughout the entire embryo, the result is a lack of central, or adaxial, fates, including central vascular cylinder cells and a SAM (Eshed et al., 2001; Kerstetter et al., 2001). Alterations in organ polarity also play a role in postembryonic meristem formation. As evidence of this, adaxialized phabulosa dominant mutants form extra axillary meristems on the undersides of their leaves (McConnell and Barton, 1998), whereas the likely abaxialized revoluta mutants fail to form axillary meristems (Talbert et al., 1995; Otsuga et al., 2001).
The CUC gene also seems to act at the level of pattern formation, specifically in directing the transition from radial symmetry to bilateral symmetry. cuc mutants are largely radially symmetrical, with a single, cup-shaped cotyledon and no SAM (Aida et al., 1997).
Downstream of such pattern formation functions are genes that dictate cell fates within the embryo or the SAM. For instance, the STM gene product (a homeodomain-containing protein) is required for cells to act as meristem cells. In the absence of STM, these cells adopt fully differentiated fates according to their positions (Barton and Poethig, 1993; Long et al., 1996).
Finally, genes required for cell division and its regulation are essential for SAM function. Such genes may act globally (Cockcroft et al., 2000; De Veylder et al., 2001), whereas the action of others may be more specific to the meristem. The CLAVATA (CLV) and WUS genes define a pathway that has as its principal role the regulation of growth of stem cells within the meristem. In the absence of the CLV1 receptor or its ligand, the CLV3 gene product, these stem cells grow excessively (Clark et al., 1993, 1995, 1997; Fletcher et al., 1999), whereas in the absence of WUS, a homeodomain-containing protein, they cease dividing prematurely (Laux et al., 1996; Mayer et al., 1998).
Similar to KAN overexpressors and cuc, stm, and wus mutants, pnh mutants frequently lack a functional SAM. Although a SAM is formed during embryogenesis in pnh mutants, the indeterminate shoot axis acts like a determinate axis; these mutants produce only one organ from the first SAM. pnh mutants also exhibit a defect in axillary meristem formation, which is the mechanism by which plants create new indeterminate growth axes after embryogenesis. In addition, pnh mutants make shorter siliques and narrower leaves and petals than wild-type plants and show abnormalities in floral organ number (McConnell and Barton, 1995; Moussian et al., 1998; Lynn et al., 1999). (pnh mutants almost always go on to flower and set seed as a result of the formation of buds at the cotyledon bases.)
Collectively, the PNH expression domain comprises the central and adaxial cells of developing tissues. PNH mRNA is found at moderate levels throughout the SAM and in the adaxial domains of organ primordia (Lynn et al., 1999). This is consistent with the defects in meristem and organ development. PNH mRNA is found at higher levels in the precursors to the vasculature, especially the phloem precursors (Moussian et al., 1998; Lynn et al., 1999). Despite this fact, defects in vascular development have not been seen in pnh mutants (Moussian et al., 1998; Lynn et al., 1999).
Recently, Nishimura et al. (2002) described transgenic rice plants expressing an RNA antisense to a rice PNH homolog (OsPNH1). Similar to pnh mutants, these plants had abnormal meristems with reduced KNOX gene expression. In addition, the plants showed significant abnormalities in leaf development with altered vascular arrangement, suggesting that OsPNH1 may play a more significant role in the rice leaf than PNH does in the Arabidopsis leaf.
PNH is a member of a small gene family in Arabidopsis whose founding member is the ARGONAUTE (AGO) gene (Bohmert et al., 1998). Genetic analysis has shown that PNH and AGO, which, in contrast to PNH, is expressed ubiquitously, are partially redundant genes (Lynn et al., 1999). Mutations in AGO confer a meristem determinacy defect similar to that of PNH mutations (Lynn et al., 1999). However, ago mutants have many additional defects, including an inability to silence genes post-transcriptionally (Fagard et al., 2000). In other species, several genes similar to AGO/PNH have been described. These genes are involved in processes such as post-transcriptional gene silencing (Tabara et al., 1999; Catalanotto et al., 2000), translational regulation (Wilson et al., 1996; Zou et al., 1998; Grishok et al., 2001), and germline stem cell development (Cox et al., 1998; Reinke et al., 2000), implicating this gene family in the regulation of developmental processes, perhaps through the post-transcriptional regulation of target genes.
The Caenorhabditis elegans agl-1 and agl-2 genes encode AGO/PNH-like products required for the accumulation of short RNAs that negatively regulate the translation of genes involved in developmental timing (Grishok et al., 2001). A physical association between such short RNAs and PNH/AGO-like gene products has been seen in rabbit reticulocytes, in which the AGO/PNH-like gene product eIF2C2 has been shown to exist in 15S ribonucleoproteins that also include a helicase and micro-RNAs (Mourelatos et al., 2002).
The loss-of-function pnh phenotype is consistent with a role for the PNH gene at several levels in the hierarchy of developmental events in the shoot. It may be involved in pattern formation and/or in the more limited roles of cell division control or the promotion of the undifferentiated state. To better understand the level at which PNH functions in the shoot, we investigated PNH using gain-of-function analysis. Our results support a role for PNH at several levels in the developmental hierarchy of shoot development. Expanding the expression of PNH outside of its normal domain resulted in a cell division control defect that manifested as upward curling of leaf margins. A more dramatic result was found when PNH was moved from its normal domain to a more peripheral domain: such mutants show a conversion of the determinate cotyledon axis to an indeterminate state. Together with the loss-of-function phenotype, this result suggests that PNH regulates the indeterminacy of the proximal-distal shoot axes. Furthermore, this finding highlights the importance of radial positional information in the overall architectural development of the plant body and shows that PNH acts largely cell nonautonomously.
RESULTS
Placing PNH under the Control of the 5′ Region of the FIL Gene Causes Its Abaxial Expression
In wild-type plants, high levels of PNH mRNA are observed in developing vascular tissue (Moussian et al., 1998; Lynn et al., 1999), and low levels are seen throughout the SAM and on the adaxial, but not abaxial, sides of lateral organ primordia (Figures 2A to 2D) (Lynn et al., 1999). In effect, PNH is expressed throughout a central domain of the developing regions of the plant body. The expression of PNH was expanded by constructing a FIL::PNH transgene in which the 5′ sequences of the FIL gene were fused to a PNH cDNA (Figure 3). The FIL gene is expressed on the abaxial sides of developing lateral organ primordia (Sawa et al., 1999; Siegfried et al., 1999), in an apparently complementary fashion to PNH. FIL expression in leaves is rapidly limited to marginal domains and disappears as the leaf differentiates (Siegfried et al., 1999).
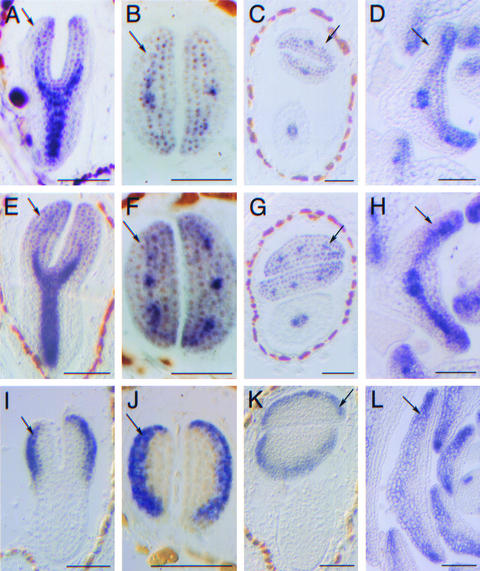
Figure 2.
Expression Analysis of Transgenic Plants.
(A) to (D) PNH antisense RNA probe on wild-type plants. PNH RNA is undetectable in abaxial cells of cotyledon ([A] to [C]) and leaf primordia (D) (arrows).
(E) to (H) PNH antisense RNA probe on PNH; FIL::PNH transgenic plants. PNH; FIL::PNH plants express PNH mRNA in the wild-type domain and in abaxial cotyledon primordia cells ([E] to [G]) and in abaxial cells near the margin in leaf primordia (H) (arrows).
(I) to (L) GUS antisense probe on FIL::GUS transgenic plants. The activity of the FIL 5′ regulatory sequences used in this analysis is limited to the abaxial cotyledon cells in the embryo ([I] to [K]) and to the abaxial cells in the leaf primordia (L) (arrows).
Bars = 50 μm.
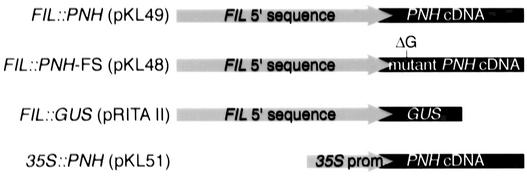
Figure 3.
Transgenes.
The 5′ regulatory sequences of the FIL gene were fused to a PNH cDNA, a mutant PNH cDNA, or the marker gene GUS. The 35S promoter of Cauliflower mosaic virus was fused to a PNH cDNA. See Methods for construct details.
To verify the new expression domain, in situ hybridization experiments were performed on PNH; FIL::PNH transgenic plants using a PNH antisense probe. In addition to the wild-type expression domain, transformants expressed PNH on the abaxial sides of cotyledons and near the margins on the abaxial sides of true leaves at levels resembling those on the adaxial sides (Figures 2E to 2H, arrows). To assess which part of the expanded PNH domain was derived from the FIL::PNH transgene, in situ hybridization using an antisense β-glucuronidase (GUS) probe was performed on FIL::GUS transgenic plants (Figure 3). These experiments confirmed that the FIL sequences used in this study drive expression in the abaxial cells of lateral organs (Figures 2I to 2L), which are the same cells in which FIL mRNA was detected under our conditions (data not shown). Note that GUS RNA persisted in medial abaxial portions of the leaf primordia longer than either endogenous FIL RNA (Siegfried et al., 1999) or PNH RNA expressed from the FIL promoter. This is most likely the result of the increased stability of the GUS RNA relative to the PNH and FIL RNAs.
Abaxial Expression of PNH Causes Upward Curling of Lateral Organs
Of 21 independent PNH; FIL::PNH lines examined, 2 gave rise to plants resembling the wild type, 3 gave rise to plants resembling pnh mutants, and 16 gave rise to plants with curled leaves, described below. Lines appearing wild type likely resulted from transgene silencing, whereas those resembling pnh mutants likely resulted from cosuppression of the endogenous PNH gene.
As a negative control, 130 independently derived lines expressing the FIL::PNH-FRAMESHIFT (FS) transgene were examined (Figure 3). This transgene is identical to FIL::PNH except for a single nucleotide deletion in the coding sequence that creates a premature stop codon. When this sequence was expressed in Escherichia coli, a truncated protein was produced (data not shown). Thirty-three lines resembled pnh mutants and likely resulted from the cosuppression of PNH. The remaining 97 lines resembled the wild type. Thus, ectopic expression of the wild-type PNH gene product is required for the curling phenotype.
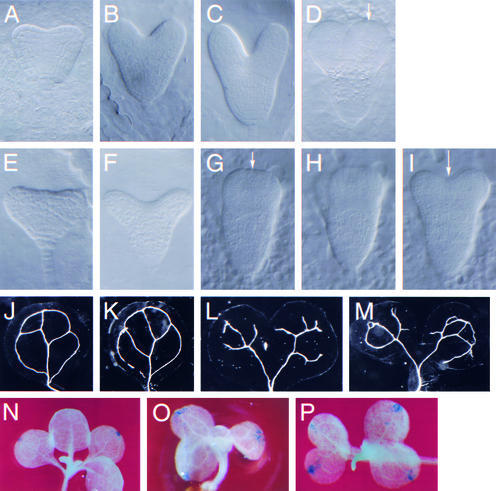
The PNH; FIL::PNH plants exhibited upwardly curled blades in rosette leaves (Figures 4A to 4C), cauline leaves (Figures 4I to 4L), petals, and sepals (Figures 4M and 4N), giving them an involute appearance compared with the wild type. Other lateral organs, such as cotyledons, stamens, and carpels, were normal.
Figure 4.
Phenotype of PNH; FIL::PNH Transformants.
(A) to (C) Rosettes from wild-type (A) and mildly (B) and severely (C) affected PNH; FIL::PNH transgenic plants. PNH; FIL::PNH rosette leaves are involute, whereas wild-type rosette leaves are slightly revolute. c, cotyledon.
(D) FIL::GUS plant stained for GUS activity. Young leaves express GUS on their abaxial side, but expanding leaves lose GUS expression in a basipetal fashion.
(E) and (G) Toluidine blue–stained paraffin sections of wild-type (E) and PNH; FIL::PNH (G) leaves. The surface at top is adaxial. Lines show the locations of blade/midrib and adaxial/abaxial cutoffs. Boxes show regions magnified in (F) and (H). Bars = 50 μm.
(F) and (H) Higher magnification of leaves in (E) and (G), respectively. The epidermal cell sizes are approximately the same in both leaves.
(I) to (L) Cauline leaves from wild-type (I) and PNH; FIL::PNH transgenic (J) to (L) plants. The leaves in (J), (K), and (L) are the youngest, middle, and oldest cauline leaves, respectively, from the same plant. Young leaves are curled up at the margins but uncurl as they mature.
(M) and (N) Flowers from wild-type (M) and PNH; FIL::PNH transgenic (N) plants. Sepals and petals of transgenic plants are involute; however, stamens and carpels are unaffected. Overall floral organ morphology and number are as in the wild type.
(O) 35S::PNH rosette with ruffled leaves.
Blade curling results from a differential between the amount of growth on the adaxial and abaxial sides of the leaf blade. This growth can be caused by cell expansion, cell division, or both. To investigate the mechanism of blade curling in PNH; FIL::PNH plants, the ratios of adaxial to abaxial epidermal cell numbers were compared in curled and wild-type rosette leaves. We expected that if abnormal cell expansion caused curling, the ratio of adaxial to abaxial cells would be the same in PNH; FIL::PNH and wild-type blades; if abnormal cell division rates caused curling, we expected this ratio to change. Cells from the youngest curled PNH; FIL::PNH leaves (plastochron 8 [P8] to P10) and similarly staged wild-type leaves were counted using transverse sections of shoot apices (Figures 4E to 4H). The ratio of adaxial to abaxial epidermal blade cells changed from an average of 1.01 ± 0.04 (se) with a range of 0.9 to 1.11 in wild-type blades to 0.63 ± 0.0095 (se) with a range of 0.60 to 0.66 in curled PNH; FIL::PNH blades (n = 5 blades for each genotype). Thus, ectopically expressing PNH in the abaxial leaf domain either causes an excess of cell divisions in this domain or represses cell division on the opposite, adaxial, side of the leaf.
The latter possibility seems less likely because plants expressing PNH in both leaf domains from the constitutive 35S promoter of Cauliflower mosaic virus did not show a similar degree of leaf curling. Plants carrying the 35S::PNH transgene had round leaf blades with gently ruffled surfaces (Figure 4O). Occasionally, the margins of the leaves curled up, but not to the same extent seen with FIL::PNH. In addition, the leaves frequently were held upright, rather than lying flat against the soil. Sepals and petals resembled those of PNH; FIL::PNH flowers, and carpels often were bumpy in appearance (data not shown). Finally, the leaves of PNH; FIL::PNH plants seemed to be the same width as wild-type leaves rather than narrower, as might have been expected if the transgene repressed cell division.
The involute morphology affected leaves from the early stages of blade outgrowth, at approximately the P8 or P9 stage, with the P1 leaf being defined as the youngest primordium that is morphologically distinguishable on the flanks of the SAM. The curling phenotype was seen until just before full expansion of the leaf. At this point, the blades corrected the curling until they nearly resembled wild-type blades (Figures 4I to 4L). This correction likely is permitted by a cessation of ectopic PNH expression in maturing leaves, because plants expressing a FIL::GUS transgene showed GUS activity in young but not maturing leaves (Figure 4D).
pnh; FIL::PNH Plants Make Double Cotyledons
We next investigated the effects of moving PNH expression from its natural domain to a new domain. To achieve this, the FIL::PNH transgene was introduced into pnh-2 mutant plants. The resulting pnh; FIL::PNH plants should have a domain of wild-type PNH expression that mimics the pattern of GUS expression in FIL::GUS plants (Figures 2I to 2L). However, it was not possible to directly test the wild-type PNH expression domain because pnh-2 mutants produced stable mutant transcripts. Therefore, mutant transcripts detected in in situ hybridization were indistinguishable from wild-type transcripts. However, we tested FIL expression in pnh; FIL::PNH plants and found that it was expressed normally (data not shown).
When the FIL::PNH transgene was expressed in a pnh mutant background, none of the pnh defects (SAM and axillary meristem formation, floral organ morphology, and number) was rescued. The inability of abaxial PNH to rescue the mutant phenotype shows that normal localization of the PNH transcript is critical. Surprisingly, unlike PNH; FIL::PNH leaves, pnh; FIL::PNH leaves did not curl. Thus, expression of PNH in its normal domain is essential for the transgene to cause leaf curling.
pnh; FIL::PNH individuals germinated and grew much more slowly than wild-type individuals. In addition, a novel phenotype was seen in pnh; FIL::PNH cotyledons. The novel phenotype consisted of cotyledon primordia splitting at various points in development to form fused or unfused “double” cotyledons (Figure 5). This phenotype segregated in four of four independent lines transformed with the FIL::PNH transgene (many other T1 individuals died before setting seed). This phenotype also was seen when the transgene was transformed into wild-type plants and crossed into a pnh background. When the FIL::PNH-FS negative control construct was transformed into homozygous pnh-2 plants, none of the 96 T1 generation plants had the novel phenotype. Furthermore, none of the hygromycin-resistant T2 progeny from 11 of 11 independent lines examined showed the novel cotyledon phenotype (Table 1). These data verify that the novel phenotype is conferred by the ectopic expression of PNH.
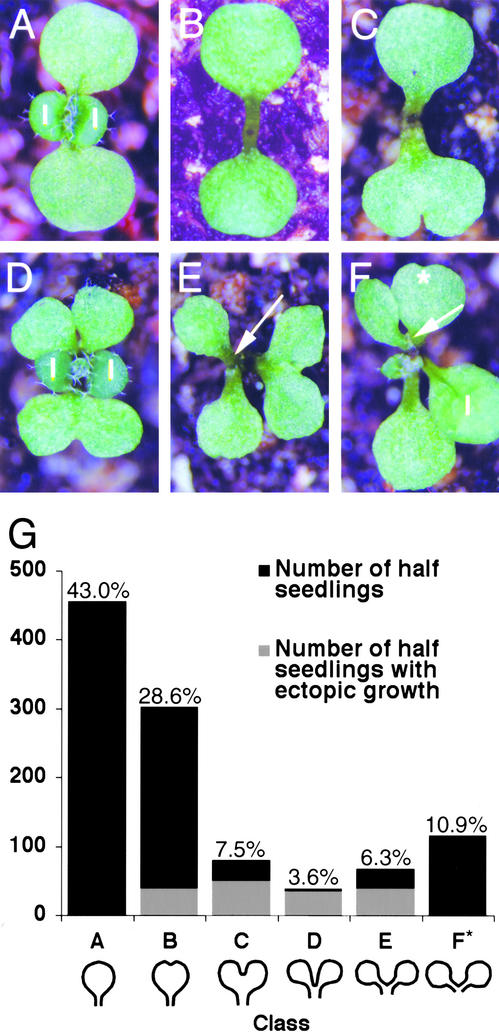
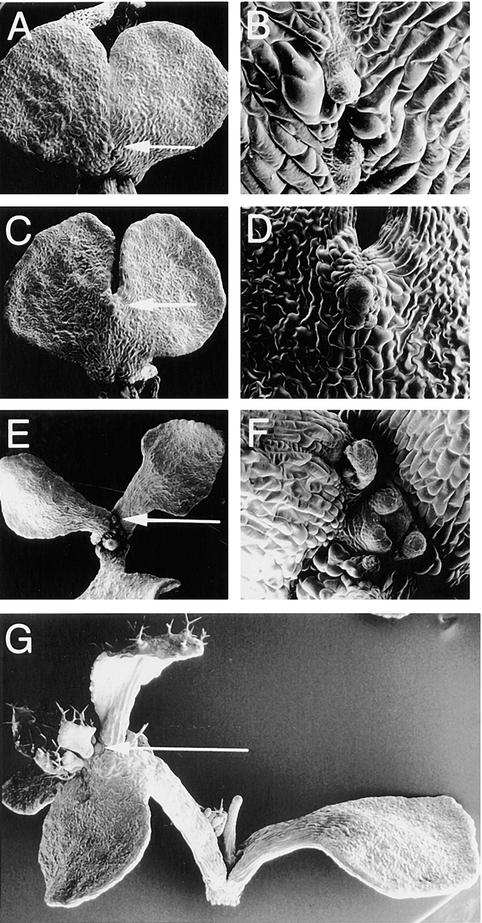
Figure 5.
Phenotypes of pnh-2; FIL::PNH Transformants.
(A) Wild-type seedling showing normal cotyledon shape. l, leaf.
(B) pnh-2 seedling with normal cotyledon morphology.
(C) to (F) pnh-2; FIL::PNH seedlings displaying the variable cotyledon phenotypes. Note the ectopic meristem formation (arrows). The double cotyledon in (F) has indentations in the two new cotyledons, as if a second round of doubling was initiated (star).
(G) Frequency of pnh-2; FIL::PNH cotyledon phenotypes and ectopic growth formation. Two- or 3-week-old seedlings from three independent transformed lines were examined, with similar results for each experiment except as discussed in the text. Cotyledon phenotypes were divided into six classes as follows: A, normal; B, indentation with most of blade fused (bottom cotyledon in [C]); C, notched with most of blade separate (cotyledon at right in [E]); D, blades completely separate but petiole fused (top cotyledon in [D]); E, petiole partially separate (top cotyledon in [F]); F, completely separate petiole and blade (cotyledon at left in [E]). Percentages refer to percent of total half-seedlings examined with that phenotype. * Cotyledons in class F could not be scored for ectopic growth formation (see text).
Table 1.
Cotyledon Phenotypes in Control Plants
| Phenotype | Number of pnh-2 Plants |
Number of pnh-2; FIL::PNH-FS Plants |
|---|---|---|
| Two cotyledons | 617 | 965 |
| Fused cotyledons | 3 (class B or C)a | 0 |
| Three cotyledons | 6 | 9 |
| Four cotyledons | 0 | 1 |
| Total | 626 | 975 |
Classes are described in Figure 6.
We classified double cotyledons according to degree of separation. For example, cotyledons in class A were completely normal, and those in class F were separated completely into two cotyledons (Figure 5G). Eighty-seven percent of hygromycin-resistant seedlings (n = 530 seedlings) showed full or partial doubling of at least one cotyledon (classes B to F). Of the double cotyledons, 80.8% were fused (classes B to E); the remainder were completely separate (class F) (n = 604 half-seedlings).
The cotyledons of each seedling were arranged in two opposite groups, just as a wild-type seedling arranges its two cotyledons. For example, individuals with three separate cotyledons (one class-A and one class-F cotyledon) usually held two smaller cotyledons in close proximity, opposite to a larger third cotyledon. We observed no cotyledons that split into unequal parts, nor did we observe multiple sinuses.
To observe the early stages of cotyledon doubling, pnh; FIL::PNH embryos were examined. Early and late heart-stage pnh; FIL::PNH embryos were very wide at the top, and their cotyledons were abnormally splayed compared with the wild type (Figures 6A, 6B, 6E, and 6F). By the early torpedo stage, evidence of cotyledon doubling emerged as a flattening (Figure 6G) or indentation (Figure 6I) of the distal tip of the cotyledon primordium compared with the wild type (Figure 6D).
Figure 6.
Morphological Analysis of pnh-2; FIL::PNH Cotyledons.
(A) to (I) Cleared embryos from wild type ([A] to [D]) or pnh-2; FIL::PNH ([E] to [I]) plants at the early heart stage ([A] and [E]), late heart stage ([B] and [F]), and early torpedo stage ([C], [D], and [G] to [I]). Torpedo-stage embryos are depicted in serial optical sections of the same embryo. Note the rounded distal tip of the wild-type cotyledon (arrow in [D]) compared with the flattened (arrow in [G]) or indented (arrow in [I]) shape of the pnh; FIL::PNH cotyledons.
(J) to (M) Cleared cotyledons from wild-type (J), pnh (K), or pnh-2; FIL::PNH ([L] and [M]) seedlings.
(N) to (P) GUS-stained seedlings expressing the hydathode::GUS marker in the wild-type (N) or pnh-2; FIL::PNH ([O] and [P]) genetic background.
Together, these observations suggest that each pnh; FIL::PNH embryo originally initiated two cotyledon primordia, as does the wild type, but many of these primordia proceeded to initiate two additional cotyledon primordia. This is consistent with our further observations of two morphological features of cotyledons: vascular patterning and hydathode formation. Wild-type cotyledons exhibited a stereotypical pattern of vascularization, which was similar in pnh mutants (Figures 6J and 6K). Although the patterning of the vasculature was aberrant in continuity and the placement of branch points with respect to the wild type, the overall pattern was duplicated in double cotyledons rather than widened to service both sides (Figures 6L and 6M). The hydathode is a specialized structure for guttation found at the margins of leaves and cotyledons (Esau, 1977). In both wild-type and pnh plants, there was only one hydathode per cotyledon, as demonstrated by staining plants carrying a hydathode:: GUS marker gene (Figure 6N and data not shown). Double cotyledons had two foci of stain, indicating that this structure had been duplicated (Figures 6O and 6P). We conclude that double cotyledons result from duplication of the cotyledon initiation process before vascular and hydathode patterning.
pnh; FIL::PNH Double Cotyledons Produce Ectopic Meristems
In the wild type, the SAM develops between the two cotyledon axes. Similarly, ectopic growths formed between the two halves of 33.0% of fused double cotyledons (classes B to E; n = 488 half-seedlings) (Figure 5G). Ectopic growths were found only along the fusion zone, at the distal tip, petiole, blade, or a combination of these sites (Figures 7A to 7D). A greater incidence of ectopic growth formation was seen in cotyledons that had two distinct lobes (classes C to E) than in those with only an indentation (class B). On separate (class F) cotyledons, the fusion zone was in virtually the same location as the normal site of adventitious SAM formation in pnh seedlings (Figures 7E and 7F); thus, these two types of SAMs could not be distinguished, and ectopic growth formation was not scored for class-F cotyledons. No ectopic growths were observed on normal (class A) cotyledons.
Figure 7.
Ectopic Meristem Formation in pnh-2; FIL::PNH cotyledons.
(A) to (F) Scanning electron micrographs of 18-day-old pnh-2; FIL::PNH seedlings forming ectopic meristems on cotyledons on the blade ([A] and [B]) or distal tip of the fusion zone ([C] to [F]). Arrows in (A), (C), and (E) indicate areas magnified in (B), (D), and (F), respectively.
(G) A typical pnh; FIL::PNH seedling at a later stage (∼23 days old). The arrow shows an ectopic meristem developing at the end of one of the cotyledons.
Ectopic growths fell into two groups: differentiated outgrowths, which were composed of large differentiated cells, and SAMs, which were defined by the production of at least one leaf-like structure such as a regular pnh SAM would produce. In 2-week-old seedlings, 56.2% of ectopic growths were SAMs (n = 73 half-seedlings), whereas in 3-week-old seedlings, 90.7% of ectopic growths were SAMs (n = 108 half-seedlings). Combined with the observation that SAMs often emerged from differentiated outgrowths, these data suggest that differentiated outgrowths typically lead to SAM formation. Ectopic SAMs occasionally were observed to produce full rosettes and inflorescences; ectopic root growth never was observed.
Consistent with the formation of ectopic SAMs on double cotyledons, the activity of the STM promoter was seen in double cotyledons. An STM::GUS transgene that confers GUS activity in the SAM of wild-type seedlings (McConnell and Barton, 1998) drove the expression of GUS activity in the double cotyledon fusion zones of pnh; FIL::PNH plants (data not shown). These data indicate that ectopic meristem formation likely occurs through the ectopic activation of meristem genes such as STM.
pnh FIL::PNH Cotyledon Primordia Ectopically Express Genes Required for SAM Initiation in the Embryo
Because the CUC1 and CUC2 genes are required for SAM formation and organ separation and are expressed in the region where separation occurs (Aida et al., 1997, 1999), we reasoned that these genes might be involved in the separation of pnh; FIL::PNH cotyledon primordia into double cotyledons. If this were true, we would expect CUC1 and CUC2 to be expressed at the distal tip of pnh; FIL::PNH cotyledon primordia before doubling. Early in development, the expression of CUC2 in pnh; FIL::PNH embryos was confined to the same region as in the wild type (Figure 8A). However, by the late heart stage, when cotyledon primordia are easily distinguishable, expression of this gene also was evident at the distal tip of some cotyledon primordia (Figure 8B). At later stages of embryogenesis, staining was seen in a higher proportion of cotyledon primordia (data not shown).
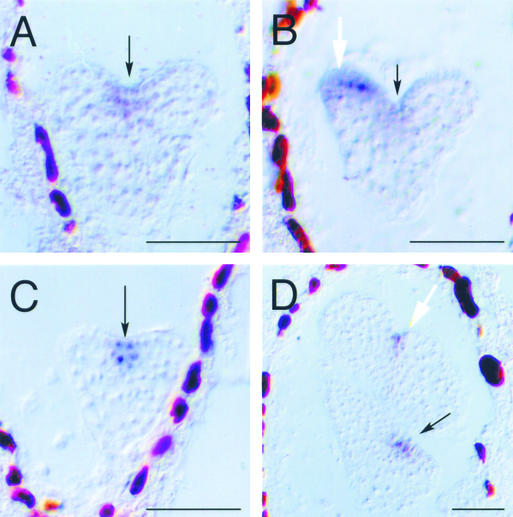
Figure 8.
Gene Expression in pnh; FIL::PNH Embryos.
(A) and (B) In situ hybridization using a CUC2 antisense probe on pnh; FIL::PNH embryos expressing CUC2 in the normal domain (A) or ectopically in the cotyledon primordium (B). Cotyledon stain was visible in multiple sections.
(C) and (D) In situ hybridization using an antisense STM probe on pnh; FIL::PNH embryos. STM is expressed in the normal domain early (C) but also is expressed in the cotyledon primordium by the bent-cotyledon stage, when cotyledon doubling is clearly distinguishable (D).
Black arrows indicate the presumptive SAM, and white arrows indicate the presumed location of ectopic meristem formation. Bars = 50 μm.
Similarly, STM was expressed only in its normal domain at the early and middle stages of embryogenesis, but it was expressed in some cotyledon primordia by the bent-cotyledon stage (Figures 8C and 8D). Unlike CUC2 expression, STM expression in cotyledon primordia was detected only after they were morphologically distinguishable as double cotyledons. The CUC2 and STM genes normally are expressed only at the shoot apex. That they were expressed in the cotyledon primordia in our experiments reveals that these cotyledon primordia had activated at least part of the program normally reserved for the indeterminate axis that forms during embryogenesis.
DISCUSSION
Through the analysis of ectopic expression phenotypes, we have gained a broader understanding of the role that PNH plays in plant development. Ectopic PNH activity is able to influence development at what appears to be a late stage in the developmental hierarchy—cell division—and also at a very early stage in the developmental hierarchy—the specification of a growth axis as indeterminate versus determinate. This finding is consistent with the highly pleiotropic nature of the pnh loss-of-function phenotype (Lynn et al., 1999).
The ability of ectopic PNH to create such substantial changes in the development of the plant when expressed ectopically suggests a regulatory role for PNH. The PNH gene is involved in the transformation of a normally determinate growth axis into an indeterminate growth axis.
Role of PNH in Cell Division Regulation and Leaf Polarity
In the wild type, the ectopic expression of PNH in the abaxial domains simply adds a new region to the normal PNH expression pattern. These new abaxial regions of PNH expression grow more than the corresponding adaxial regions, resulting in upward curling of the leaf margins. The change in growth appears to be attributable to cell division, rather than expansion. Abaxial overgrowth could be caused by an increase in abaxial cell division or by a decrease in adaxial cell division. We favor a model in which PNH promotes abaxial cell division in PNH; FIL::PNH leaves for two reasons. First, in plants that express PNH constitutively from the viral 35S promoter, both surfaces of the leaf appear to grow excessively, resulting in large ruffled leaf blades that are held upright. Second, the repression of adaxial cell division by PNH in PNH; FIL::PNH plants seems less likely because it would entail a cell-nonautonomous effect of abaxial PNH on adaxial cells, which already express PNH from the natural promoter.
The adaxial nature of PNH expression in lateral organs coupled with the abaxialized phenotype seen in some pnh organs (Lynn et al., 1999) might indicate that PNH plays a role in the polar development of the leaf. Indeed, there is some evidence that the axillary bud is an adaxial character (McConnell and Barton, 1998) that fails to develop in pnh plants (Lynn et al., 1999). Also, pnh ago/+ mutant petals show some indication of alterations in polarity. However, PNH; FIL::PNH plants did not show any obvious alterations in polar leaf development: axillary meristems, trichomes, and other polar leaf characters were normal, and PHABULOSA and FIL were expressed in the adaxial and abaxial cells, respectively, just as in the wild type (data not shown). Thus, in the leaf, ectopic PNH function is sufficient to promote only a specific subset of adaxial traits: growth and meristem formation.
PNH Is Necessary and Sufficient for the Indeterminacy of Shoot Axes
In addition to expanding the PNH expression domain, we also moved it to a new location. This was done by expressing the FIL::PNH transgene in pnh mutants. In this situation, pnh transcript was not expressed in its normal domain—a central region of the embryo—but rather was expressed in the cotyledon primordia. The result of altering the pattern of PNH expression in this way is that the cotyledons split into two new cotyledons and, in the most extreme cases, were transformed from determinate to indeterminate growth axes. In these extreme cases, each cotyledon primordium seems to recapitulate the development of the shoot pole of the globular embryo.
The development of the indeterminate pnh; FIL::PNH cotyledon axis is similar, both molecularly and morphologically, to the development of the indeterminate wild-type embryonic axis (Figure 9). In the wild-type embryo, the formation of the embryonic axis occurs as growth slows at the distal tip at the late globular stage, allowing two outgrowths to form the two cotyledon primordia on either side. This process is mediated by the CUC genes, which are expressed at the site where growth will slow at the globular stage of embryogenesis (Aida et al., 1997, 1999; Takada et al., 2001). In this region of slowed growth, a SAM forms in a process that requires the STM gene (Barton and Poethig, 1993; Long et al., 1996). This axis is indeterminate; growth will continue from this SAM, and a continuous stream of new organs will arise from this axis.
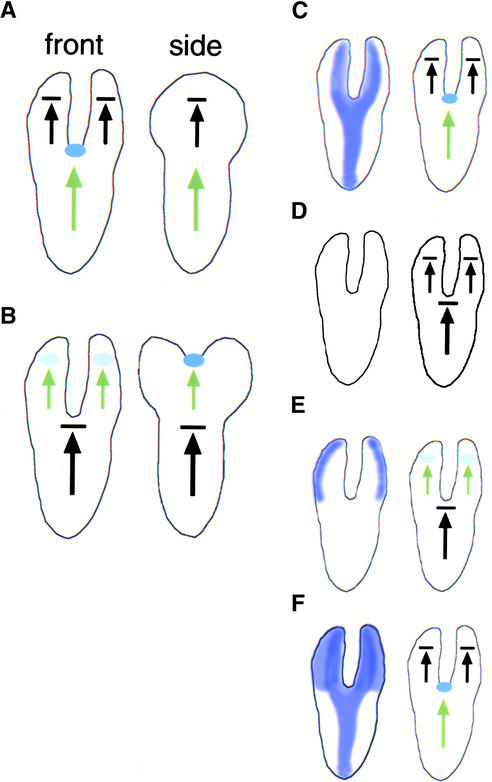
Figure 9.
Models.
(A) and (B) Cartoons of the front and side views of wild-type (A) and pnh; FIL::PNH (B) embryos. Black arrows indicate determinate axes, and green arrows indicate indeterminate axes. SAM precursors are indicated with blue ovals. Compare the side view of the cotyledon in (B) with the front view of the embryo in (A).
(C) to (F) Cartoons of embryos depicting the PNH expression domain in purple (left) and the behavior of the growth axes (right). Genotypes depicted are wild type (C), pnh (D), pnh; FIL::PNH (E), and PNH; FIL::PNH (F).
In a similar manner, growth slows at the distal end of pnh; FIL::PNH cotyledon primordia, allowing the outgrowth of two lobes. Expression of the CUC2 gene occurs at this site before the formation of an indentation. Then, a SAM is formed in the region of slowed growth. The STM gene is expressed here as it is in the indeterminate axis of the wild type. This axis also is capable of indeterminate growth, as shown by rosettes and inflorescences that can develop from pnh; FIL::PNH cotyledons.
The following model explains the conversion of the determinate cotyledon axis to an indeterminate axis in pnh; FIL::PNH plants. In the wild type (Figure 9C), PNH expression marks the central domain of the main indeterminate axis of the embryo. At the distal end of this axis is the SAM. Two new axes are specified in the cotyledon primordia; these are determinate organ axes and do not have a SAM at the distal end. In addition, the expression domain of PNH in these axes includes central and adaxial regions. In pnh embryos (Figure 9D), PNH activity is absent or reduced and the indeterminate axis behaves like a determinate axis, with only one organ at the shoot apex. In pnh; FIL::PNH embryos (Figure 9E), PNH activity is decreased or absent in its normal domain, again causing the indeterminate axis to behave like a determinate axis. However, PNH activity has been added back to the cotyledon cells, and the cotyledon primordia take on the role of the indeterminate axis in these individuals. Thus, PNH appears to be necessary and sufficient for the indeterminacy of proximal-distal shoot axes. Although the FIL promoter directs expression in the abaxial cotyledon domain throughout most of cotyledon development, this promoter may be active throughout the cotyledon primordia in their earliest stages of development (Siegfried et al., 1999). Thus, we do not know whether axis indeterminacy is caused by the expression of PNH throughout the early cotyledon primordia or by PNH expression limited to the abaxial cotyledon domain.
It is interesting that the PNH; FIL::PNH plants do not show the double cotyledon and ectopic SAM phenotypes (Figure 9F). Perhaps the main indeterminate axis, which functions normally in PNH; FIL::PNH plants, has an inhibitory effect on other axes, preventing them from becoming indeterminate. A somewhat related possibility is that a contiguous block of PNH-expressing cells can contribute to the formation of only one indeterminate axis.
These experiments underscore a growing body of evidence that indicates that radial pattern in the embryo and in the shoot is important in promoting SAM formation. Gene expression patterns show that the apical half of the embryo is divided into a central and a peripheral portion before a meristem is developed (Long and Barton, 1998). Mutants that lack a SAM, such as the stm and wus mutants, have normal adaxial/abaxial organ polarity (Barton and Poethig, 1993; Laux et al., 1996), indicating that the development of a meristem is not necessary for the proper development of organ polarity. On the other hand, alterations in polarity have profound effects on the ability of either the main SAM or axillary meristems to form (McConnell and Barton, 1998; Eshed et al., 1999, 2001; Kerstetter et al., 2001; McConnell et al., 2001). Together, these data indicate that radial polarity—that is, central (or adaxial) and peripheral (or abaxial) domains of the embryo (or leaf axil)—are established first and that the SAM is dependent on spatial cues in these distinct domains for its formation.
PNH Acts Cell Nonautonomously
Our results show that expressing PNH ectopically has consequences for both leaf and cotyledon primordia. Thus, PNH acts either cell nonautonomously or at least over a short range, emphasizing the importance of the spatially restricted PNH domain.
The new regions of PNH expression are regions in which AGO is expressed: PNH is expressed in a central domain of the developing embryo and shoot and in the presumptive vasculature, whereas the redundantly acting AGO gene is expressed throughout the embryo and the shoot apex. From this finding, we conclude either that the PNH and AGO gene products have overlapping but distinct functions or that the absolute levels of PNH/AGO function are important in development. These are testable hypotheses that have implications for the sets of genes targeted for regulation by the PNH/AGO genes: PNH and AGO may associate with overlapping sets of micro-RNAs, and this causes them to have different but overlapping sets of target genes. Alternatively, the two genes may have identical targets, but high levels of PNH/AGO activity are important for adaxially expressed targets, whereas lower levels are required for abaxially expressed targets.
The notion of distinct roles for PNH and AGO is consistent with the finding that AGO, but not PNH, is required for post-transcriptional gene silencing in Arabidopsis (Morel et al., 2002). However, it also is possible that post-transcriptional gene silencing simply requires a certain level of PNH/AGO activity and that this level is decreased more in ago mutants (because AGO is expressed ubiquitously and PNH is not) than in pnh mutants. To determine the extent of overlap between the functions of these gene products will require additional experiments.
Implications for the Molecular Function of PNH
In other systems and in plants, PNH/AGO activity is associated with the regulation of translation and/or the degradation of mRNAs (Fagard et al., 2000; Grishok et al., 2001). Therefore, it is very likely that PNH/AGO will mediate their effects at a post-transcriptional level with the specificity of their targets determined by the exact micro-RNAs the PNH and AGO gene products interact with. This study predicts that some of these targets will be mRNAs whose products regulate the cell cycle and mRNAs whose products regulate indeterminate axis formation. Identifying these targets and the mechanism by which they are regulated by PNH and AGO activity should tell us a great deal about these very basic aspects of plant development.
METHODS
Growth Conditions
Arabidopsis thaliana plants were grown at 24°C under continuous cool-white fluorescent light in Metromix 200 (Sierra, Grace, Milpitas, CA) unless noted otherwise. Sterile medium was Murashige and Skoog (1962) medium (Sigma, St. Louis, MO) with 2% Suc. Where indicated, hygromycin was added to a concentration of 40 μg/mL and carbenicillin to 100 μg/mL.
Histology
For in situ hybridization, specimens were processed as described at http://www.wisc.edu/genetics/CATG/barton/protocols.html, with color substrate incubations of 4 to 18 h. Riboprobes were synthesized from pKL3 for PNH (Lynn et al., 1999), pMACUC2 for CUC2 (Aida et al., 1999), pmeriHB1 for STM (Long et al., 1996), pJM1 for PHABULOSA (McConnell et al., 2001), pY1-Y for FIL (Siegfried et al., 1999), and pGUS for β-glucuronidase (GUS) (J. Long, unpublished data). For embryo visualization, developing seeds were cleared in Hoyer's solution (100 g of chloral hydrate and 5 mL of glycerol in 30 mL of water). For leaf cell analysis, 6-μm wax sections were stained for 15 min in 0.025% toluidine blue and then deparaffinized. For scanning electron microscopy, specimens were fixed overnight in 2% glutaraldehyde, dehydrated through a graded ethanol series, critical point dried in liquid CO2, sputter coated with gold, and visualized in a Hitachi S570 scanning electron microscope at 5 kV (Tokyo, Japan).
GUS Staining
For β-glucuronidase activity detection, samples were incubated overnight at 37°C in GUS assay buffer (100 mM phosphate buffer, pH 7, 0.1% Triton X-100, 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 5 mM FeCN, and 10 mM EDTA) and then cleared in 70% ethanol. The hydathode::GUS marker line was a fortuitous consequence of an altered STM promoter::GUS transformation. The transgene does not show expression in the STM domain.
Constructs and Plant Transformation
FIL 5′ sequences were 3971 bp of genomic DNA from 4036 to 65 bp upstream of the ATG of the FIL gene. PNH coding sequences were 3014 bp of a PNH cDNA, including 65 bp of the 5′ untranslated region. PNH-FS coding sequences have a single nucleotide deletion in codon 302, which causes a frameshift and introduces a premature stop codon in the PNH coding sequence. pKL49, pKL48, and pKL51 were constructed in pCAMBIA 1300 (www.cambia.org.au/main/r_et_quickpick.htm), which confers hygromycin resistance, sequenced, and transformed into Agrobacterium tumefaciens strain GV3101. Landsberg erecta or pnh-2 flowering plants were transformed by floral dip (Clough and Bent, 1998). Seeds resulting from self-pollination of transformed plants were sterilized in bleach with 1% Tween-20 and plated to sterile medium with hygromycin and carbenicillin to select transformants (T1 generation). pRITA II transgenic seeds were a gift from John Bowman (University of California, Davis, CA).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank John Bowman, John Emery, Yuval Eshed, and Rita Khodosh for sharing the FIL promoter as well as seeds from pRITA II transgenic lines. We are grateful for helpful comments on the manuscript from Pablo Jenik. This work was funded by National Science Foundation Grant IBN-0080853 to M.K.B. and a National Science Foundation predoctoral fellowship to K.L.N.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005132.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. [DOI] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., Azzalin, G., Macino, G., and Cogoni, C. (2000). Gene silencing in worms and fungi. Nature 404, 245. [DOI] [PubMed] [Google Scholar]
- Clark, S., Running, M., and Meyerowitz, E. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. (2000). Cyclin D control of growth rate in plants. Nature 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Cox, D.N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beemster, G.T., Beeckman, T., and Inze, D. (2001). CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 25, 617–626. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1977). Anatomy of Seed Plants. (New York: John Wiley & Sons).
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Lil, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Long, J., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J., Moan, E., Medford, J., and Barton, M. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1995). Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of shoot apical meristems. Dev. Genet. 16, 358–366. [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J.L., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. (2002). miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nishimura, A., Ito, M., Kamiya, N., Sato, Y., and Matsuoka, M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30, 189–201. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. [DOI] [PubMed] [Google Scholar]
- Reinke, V., Smith, H.E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S.J., Davis, E.B., Scherer, S., Ward, S., and Kim, S.K. (2000). A global profile of germline gene expression in C. elegans. Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. Erratum. Genes Dev. 13, 2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Alder, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Connell, J.E., and Macdonald, P.M. (1996). aubergine enhances oskar translation in the Drosophila ovary. Development 122, 1631–1639. [DOI] [PubMed] [Google Scholar]
- Zou, C., Zhang, Z., Wu, S., and Osterman, J.C. (1998). Molecular cloning and characterization of a rabbit eIF2C protein. Gene 211, 187–194. [DOI] [PubMed] [Google Scholar]