Abstract
Cortical microtubules have long been hypothesized to regulate the oriented deposition of cellulose microfibrils. However, the molecular mechanisms of how microtubules direct the orientation of cellulose microfibril deposition are not known. We have used fibers in the inflorescence stems of Arabidopsis to study secondary wall deposition and cell wall strength and found a fragile fiber (fra1) mutant with a dramatic reduction in the mechanical strength of fibers. The fra1 mutation did not cause any defects in cell wall composition, secondary wall thickening, or cortical microtubule organization in fiber cells. An apparent alteration was found in the orientation of cellulose microfibrils in fra1 fiber walls, indicating that the reduced mechanical strength of fra1 fibers probably was attributable to altered cellulose microfibril deposition. The FRA1 gene was cloned and found to encode a kinesin-like protein with an N-terminal microtubule binding motor domain. The FRA1 protein was shown to be concentrated around the periphery of the cytoplasm but absent in the nucleus. Based on these findings, we propose that the FRA1 kinesin-like protein is involved in the microtubule control of cellulose microfibril order.
INTRODUCTION
Plant cells are enclosed in rigid cell walls composed of a load-bearing cellulose-hemicellulose network and matrix polysaccharides. The cell wall not only defines the shape of a differentiated cell but also determines the direction of cell elongation and, hence, cell morphogenesis. It is known that cellulose microfibrils in the cell walls play pivotal roles in the direction of cell elongation (Kost and Chua, 2002). Cellulose microfibrils in elongating cells typically are oriented transversely to the elongation axis. Such an orientation presumably allows the rigid cellulose microfibril and hemicellulose network to be loosened longitudinally during cell elongation (Baskin, 2000; Carpita and McCann, 2000). Understanding how the oriented deposition of cellulose microfibrils is controlled will provide important insights into the process of cell morphogenesis.
Since the first report by Ledbetter and Porter (1963) that cortical microtubules lie in parallel with cellulose microfibrils, many studies have suggested that cortical microtubules lying underneath the plasma membrane are involved in the control of the oriented deposition of cellulose microfibrils (Baskin, 2001). The first evidence came from the observations that both cortical microtubules and cellulose microfibrils are oriented transversely along the long axis of elongating cells. The second evidence came from pharmacological studies showing that treatment of cells with various drugs that affect microtubule organization also alters the orientation of cellulose microfibril deposition. Except for a few cases in which no parallel alignment of cortical microtubules and cellulose microfibrils is found (Preston, 1988; Emons et al., 1992; Wasteneys, 2000), the correlative alignment of cortical microtubules and cellulose microfibrils has been demonstrated in both elongating cells and cells undergoing secondary wall thickening, such as tracheary elements and fiber cells (Baskin, 2001). Recently, it was shown that an alteration in cortical microtubule orientation caused by mutation of the katanin-like microtubule-severing protein AtKTN1 in the fragile fiber2 (fra2) mutant accompanies the aberrant deposition of cellulose microfibrils in the primary walls of elongating cells and the secondary walls of fiber cells (Burk et al., 2001; Burk and Ye, 2002), which further supports the idea of microtubule/microfibril parallelism.
Although available evidence suggests that the cortical microtubules control the oriented deposition of cellulose microfibrils, the underlying molecular mechanisms are not known. Two hypotheses have been proposed to explain the microtubule control of microfibril order at the molecular level. The first hypothesis states that cortical microtubules control cellulose microfibril deposition by acting as rails on which cellulose synthase complexes move—the so-called monorail model first proposed by Heath (1974). In this model, cortical microtubules are proposed to interact with cellulose synthase complexes through linker proteins such as motor proteins. Motor proteins might act as locomotives to pull or guide cellulose synthase complexes along cortical microtubules.
The second hypothesis states that cortical microtubules act as rails between which cellulose synthase complexes move—the so-called guard rail model (Herth, 1980; Giddings and Staehelin, 1991). In this model, cortical microtubules are proposed not to interact with cellulose synthase complexes. Instead, they may delimit the path of cellulose synthase complex movement by aligning putative barrier proteins. It was proposed that the putative barrier proteins align closely along cortical microtubules so that the cellulose synthase complexes can move only between the microtubule/barrier-aligned rails. The driving force for the movement of cellulose synthase complexes is considered to be generated from the polymerization and crystallization of cellulose microfibrils during cellulose synthesis (Giddings and Staehelin, 1991).
Ultrastructural studies have provided evidence to support both hypotheses. It has been shown that in taxol-treated tobacco BY2 cells, newly synthesized cellulose microfibrils are located directly atop cortical microtubules, which is in favor of the monorail model (Hasezawa and Nozaki, 1999). Evidence in support of the guard rail model came from freeze-fracture studies of the plasma membrane of the alga Closterium, in which rows of cellulose synthase complexes were found to be located between individual cortical microtubules (Giddings and Staehelin, 1988). It is not known whether these distinct differences in cellulose microfibril localization reflect differences in the molecular mechanisms of microtubule control of cellulose microfibril deposition between algae and land plants. A critical issue in these two models is how microtubules interact with either cellulose synthase complexes in the monorail model or putative barrier proteins in the guard rail model. Ultrastructural studies have shown that some types of physical linkers are present between cortical microtubules and the plasma membrane (Hardham and Gunning, 1978; Lancelle et al., 1986, 1987; Giddings and Staehelin, 1988), which suggests that cortical microtubules might bind certain types of microtubule binding proteins, which in turn interact with cellulose synthase complexes or barrier proteins located in the plasma membrane (Giddings and Staehelin, 1991).
Kinesins are motor proteins that move along microtubules. It has been suggested that kinesins are likely one of the microtubule binding proteins involved in the microtubule control of cellulose microfibril order (Asada et al., 1997). Although a number of kinesin-like proteins have been characterized in plants (Lee et al., 2001; Matsui et al., 2001; Reddy, 2001) and two of them have been shown to localize in the cortical region of cells (Asada et al., 1997; Barroso et al., 2000), none has been demonstrated to play a role in the deposition of cellulose microfibrils. In addition, four Arabidopsis mutants with mutations in kinesin-like genes have been reported (Oppenheimer et al., 1997; Chen et al., 2002; Nishihama et al., 2002; Strompen et al., 2002), and they were not shown to affect cellulose microfibril deposition.
Arabidopsis inflorescence stems develop three to four layers of interfascicular fibers next to the endodermis to provide mechanical support to the stems (Zhong et al., 1997, 2001). Fibers in Arabidopsis have been shown to be an excellent model in which to study cell differentiation, cell elongation, and cell wall formation. Mutants that affect various aspects of fiber cell formation have been described, and some of their corresponding genes have been isolated (Zhong et al., 2001). Because the thick secondary walls of fiber cells consist primarily of orderly deposited cellulose microfibrils (Harada and Coté, 1985), fiber cells also are an ideal system in which to study the molecular mechanisms that regulate the oriented deposition of cellulose microfibrils.
In this study, we describe the isolation and characterization of an Arabidopsis mutant, fra1, and the cloning of the FRA1 gene. We show that the fra1 mutation causes a dramatic reduction in fiber mechanical strength without an apparent alteration in cell wall composition. We provide evidence that the reduced mechanical strength of fra1 fibers is correlated with an alteration in the oriented deposition of cellulose microfibrils in fiber cell walls. We show that the gene responsible for the fra1 mutation encodes a kinesin-like protein. Our findings provide direct evidence that FRA1, a kinesin-like motor protein, is involved in the oriented deposition of cellulose microfibrils.
RESULTS
Isolation of fra1 Mutants Defective in Fiber Mechanical Strength
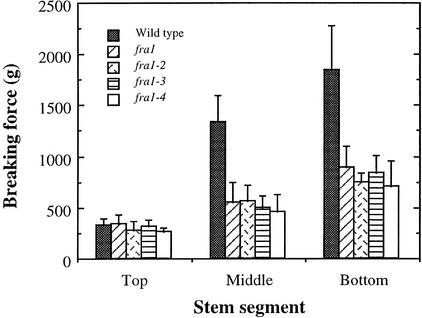
Arabidopsis inflorescence stems develop interfascicular fiber cells to provide mechanical support for the plant body (Zhong et al., 2001). The formation of interfascicular fibers correlates directly with an increase in the mechanical strength of mature stems. This has been demonstrated in the ifl1 mutant, in which disruption in the formation of interfascicular fibers in mature stems dramatically reduces their mechanical strength (Zhong et al., 1997). This finding indicates that the high mechanical strength of mature stems is conferred mainly by the presence of interfascicular fibers. To investigate the mechanisms that regulate the mechanical strength of fibers, we have screened ethyl methanesulfonate–mutagenized populations of Arabidopsis for mutants with reductions in fiber strength. We found four allelic mutants, fra1, fra1-2, fra1-3, and fra1-4, which showed dramatic reductions in stem strength. These four alleles were isolated from different batches of ethyl methanesulfonate–mutagenized populations of Arabidopsis. The mechanical strength of mature internodes of stems in these mutants was reduced to ∼45% of that of the wild type (Figure 1). Because stems from all four mutant alleles showed similar mechanical strength, one of them (fra1) was chosen for further analysis.
Figure 1.
Breaking Strengths of Stems of the Wild Type and the fra1 Mutants.
The main inflorescence stems of 8-week-old plants were divided into three equal segments, and each segment was used to measure breaking forces. Breaking strength was measured as the force needed to break stem segments apart. Data are means ± se of 15 plants.
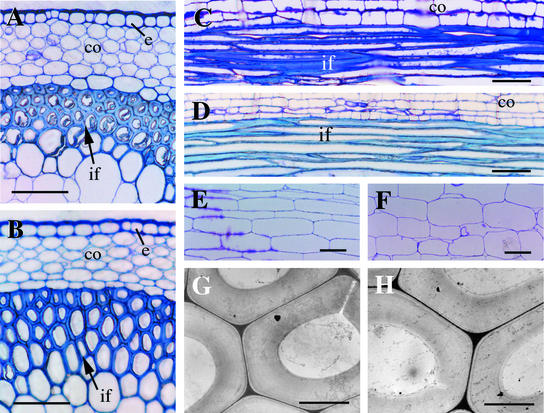
To determine whether the reduced mechanical strength of fra1 mutant stems was caused by an alteration in fiber cell formation, we examined the anatomical structure of interfascicular fiber cells. Cross-sections and longitudinal sections of the mutant stems showed the presence of interfascicular fiber cells (Figure 2B), as in the wild type (Figure 2A), and their lengths (Figure 2D) were similar to those in the wild type (Figure 2C). Transmission electron microscopy showed that the thickness of fiber cell walls in the mutant (Figure 2H) was not noticeably different from that of the wild type (Figure 2G). Examination of the surface anatomy of the broken ends of the stems by scanning electron microscopy showed that the breaking points occurred randomly within fiber cell walls (data not shown), thus excluding the possibility that the defects occurred in the adhesion between fiber cells. These results indicated that the reduced mechanical strength in fra1 mutant stems was not accompanied by abnormal formation or reduced wall thickness of interfascicular fibers.
Figure 2.
Anatomy of Interfascicular Fibers and Pith Cells in Mature Stems of the Wild Type and the fra1 Mutant.
(A) and (B) Cross-sections of interfascicular regions of stems of the wild type (A) and fra1 (B) showing the presence of fiber cells.
(C) and (D) Longitudinal sections of interfascicular regions of stems of the wild type (C) and fra1 (D) showing the long fiber cells.
(E) and (F) Longitudinal sections of stems showing pith cells of the wild type (E) and fra1 (F).
(G) and (H) Transmission electron micrographs of fiber cells of the wild type (G) and fra1 (H) showing thick walls.
co, cortex; e, endodermis; if, interfascicular fiber. Bars = 75 μm in (A) to (F) and 5 μm in (G) and (H).
Cell Wall Composition in the fra1 Mutant
To determine whether the reduced mechanical strength of fra1 stems was caused by alterations in cell wall composition, we analyzed the amount of cell wall polysaccharides and lignin in mature stems. Because thick fiber walls constitute a major fraction of total wall materials in mature stems, it was expected that any significant change in fiber wall composition should be detected by analyzing the cell wall composition of stems, as demonstrated in irx and fra2 mutants (Turner and Somerville, 1997; Burk et al., 2001). Quantitative analysis of crystalline cellulose in mature stems of fra1 did not show any reduction in cellulose amount compared with the wild type. Both the wild type and the fra1 mutant had cellulose constituting 23% of total cell wall residues (Table 1). This finding was confirmed by cell wall sugar analysis, which showed that the cell walls of both wild-type and fra1 mutant stems had ∼25% Glc (Table 1). In addition, no significant alterations in the amount of other sugars or total Klason lignin were detected in the fra1 mutant compared with the wild type (Table 1).
Table 1.
Cell Wall Composition of Stems of the Wild Type and the fra1 Mutant (mg/g)
| Sample | Rhamnose | Arabinose | Xylose | Mannose | Galactose | Glucose | Fucose | Lignin | Cellulose |
|---|---|---|---|---|---|---|---|---|---|
| Wild type | 6.9 ± 0.2 | 11.2 ± 0.7 | 124 ± 3 | 16.8 ± 1.9 | 14.4 ± 0.1 | 256 ± 15 | 1.8 ± 0.1 | 158 ± 4 | 230 ± 13 |
| fra1 | 6.4 ± 0.1 | 10.5 ± 3.1 | 125 ± 12 | 13.2 ± 1.1 | 12.6 ± 1.2 | 252 ± 35 | 1.4 ± 0.2 | 166 ± 3 | 231 ± 9 |
Cell wall residues prepared from mature stems of 8-week-old plants were used for compositional analysis. Each wall component was calculated as milligram per gram of total cell wall residues. Values shown are means ± se of three independent assays.
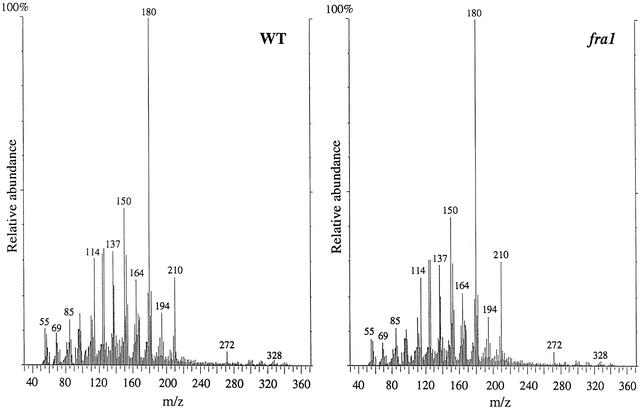
We further applied in-source pyrolysis mass spectrometry to analyze the relative abundances and degree of cross-linking of cell wall components (van der Hage et al., 1993). No apparent changes in the mass peak abundances of polysaccharides or lignin were observed between the wild type and the fra1 mutant (Figure 3). Both monomeric (in a mass-to-charge ratio range of 50 to 200) and dimeric (in a mass-to-charge ratio range of 270 to 360) wall components released from pyrolysis were similar in their mass peak abundances. These results indicated that the reduced mechanical strength in fibers of the fra1 mutant was not accompanied by any alterations in cell wall composition.
Figure 3.
In-Source Pyrolysis Mass Spectrometry of Cell Walls of Mature Stems of the Wild Type and the fra1 Mutant.
In-source pyrolysis mass spectrometry was performed using a Finnigan GCQ mass spectrometer equipped with a direct exposure probe (rhenium loop). Mass peaks of guaiacyl lignin had mass-to-charge ratio (m/z) values of 124, 137, 138, 150, 152, 164, 166, 178, and 180. Mass peaks of syringyl lignin had m/z values of 154, 167, 168, 180, 182, 194, 196, 208, and 210. Mass peaks of cellulose and amylose had m/z values of 57, 60, 73, 85, 86, 96, 98, 100, 102, 110, 112, 126, and 144. Mass peaks of hemicellulose had m/z values of 58, 85, 86, and 114. Note that there are no apparent differences in the relative intensities of mass peaks for cell wall polysaccharides and lignin between the wild type (WT) and the fra1 mutant.
Visualization of Cellulose Microfibrils in Cell Walls of the fra1 Mutant
Because the reduced mechanical strength of fra1 fibers was not associated with any alterations in fiber anatomy or cell wall composition, we reasoned that it was likely caused by an alteration in cell wall architecture, such as the deposition or assembly of cell wall components. Because the main component contributing to the mechanical strength of cell walls is cellulose microfibrils, we examined the pattern of cellulose microfibril deposition in walls of fra1 fibers. It has been shown that in the secondary walls of fibers, cellulose microfibrils are deposited in three distinct layers (S1, S2, and S3). Cellulose microfibrils in the S1 and S3 layers are oriented in a flat helix, and those in the S2 layer are oriented in a steep helix (Harada and Coté, 1985).
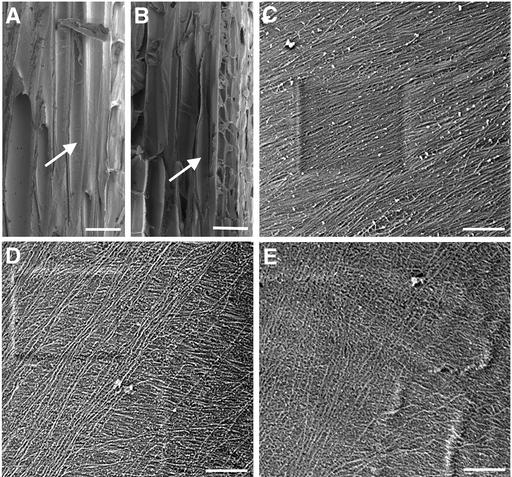
Using field emission scanning electron microscopy, we visualized cellulose microfibrils in the innermost layer of secondary walls of mature fibers (Figure 4). In wild-type fiber walls, cellulose microfibrils were densely packed and ran in parallel in a flat helix along the long axis (Figures 4A and 4C). By contrast, cellulose microfibrils in fra1 fiber walls were not as densely packed or arranged in the same orderly manner as in the wild type, and they appeared to be oriented in different directions (Figures 4B, 4D, and 4E). These results indicated that the fra1 mutation caused an aberrant deposition of cellulose microfibrils in the secondary walls of fibers, which might be the cause of the reduced mechanical strength of fibers. Examination of cellulose microfibrils in the innermost layer of walls of elongating pith cells in stems and elongating cortical cells in hypocotyls and roots did not reveal any apparent differences in their orientation compared with the wild type (data not shown).
Figure 4.
Visualization of Cellulose Microfibrils in the Innermost Layer of Fiber Cell Walls.
Mature stems were sectioned longitudinally through interfascicular fiber cells, and the innermost layer of microfibrils in fiber cell walls was visualized using a field emission scanning electron microscope. The vertical direction of the cellulose microfibril images corresponds to the long axis of the fiber cells. The square marks in (C) and (D) are the result of beam focusing.
(A) and (B) Longitudinal sections of interfascicular regions of stems of the wild type (A) and fra1 (B) showing representative fiber cells (arrows) that were used for cellulose microfibril visualization.
(C) Cellulose microfibrils in the middle part of a wild-type fiber cell showing their parallel alignment in a small angle relative to the transverse orientation.
(D) and (E) Cellulose microfibrils in the middle parts of fra1 fiber cells showing their aberrantly oriented pattern with various directions.
Bars = 25 μm in (A) and (B) and 0.5 μm in (C) to (E).
Cortical Microtubules Were Not Affected in the fra1 Mutant
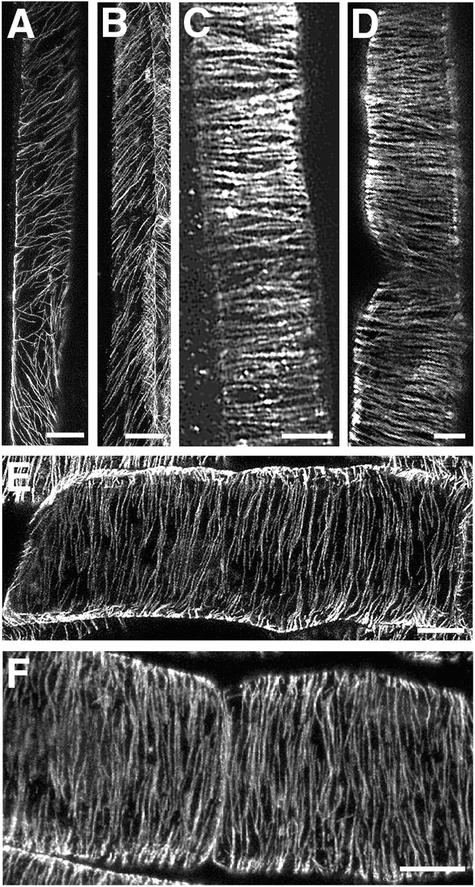
Because it is known that an alteration in the orientation of cortical microtubules could cause the aberrant deposition of cellulose microfibrils (Baskin, 2001), we next examined whether the altered cellulose microfibril deposition in the fra1 mutant was caused by an alteration in cortical microtubule organization. We applied immunocytochemistry to visualize cortical microtubules in fibers of both the wild type and the fra1 mutant. In developing wild-type fiber cells, cortical microtubules were oriented in a flat helix along the long axis (Figure 5A). Cortical microtubules in developing fra1 fiber cells (Figure 5B) appeared to be arranged in a pattern similar to that of wild-type fibers. This finding indicated that the fra1 mutation did not affect the organization of cortical microtubules; thus, the aberrantly oriented deposition of cellulose microfibrils in fra1 fibers was not the result of an altered arrangement of cortical microtubules. In addition, examination of cortical microtubules in elongating epidermal cells of roots (Figures 5C and 5D) and elongating pith cells of stems (Figures 5E and 5F) did not show any noticeable differences between the wild type and the fra1 mutant.
Figure 5.
Visualization of Cortical Microtubules in Cells of the Wild Type and the fra1 Mutant.
Cortical microtubules were immunolabeled with monoclonal antibody against α-tubulin and fluorescein isothiocyanate–conjugated secondary antibody. Fluorescence-labeled microtubules were visualized using a confocal laser microscope. The vertical ([A] to [D]) or horizontal ([E] and [F]) direction of the images corresponds to the long axis of the cells.
(A) and (B) Developing fiber cells in stems of the wild type (A) and fra1 (B) showing cortical microtubules oriented in a flat helix.
(C) and (D) Elongating root epidermal cells from 3-day-old seedlings of the wild type (C) and fra1 (D) showing cortical microtubules oriented transversely along the long axis.
(E) and (F) Elongating pith cells in stems of the wild type (E) and fra1 (F) showing cortical microtubules oriented transversely along the long axis.
Bars = 10 μm in (A) and (B), 5 μm in (C), 7 μm in (D), and 10 μm in (E) and (F).
Effects of the fra1 Mutation on Plant Morphology
To determine whether the fra1 mutation had any effects on plant growth and development, we examined the morphology of fra1 adult plants and seedlings. The inflorescence stems of fra1 mutant plants appeared to stand more upright than those of the wild type (Figure 6A). This result indicated that the altered deposition of cellulose microfibrils in fra1 fibers might cause an increase in cell wall rigidity, leading to the appearance of more erect stem morphology. This possibility does not contradict the fragile fiber phenotype. The increase in cell wall rigidity might cause a reduction in the extensibility of cell walls so that fibers become easily broken by sheering. In addition, the fra1 mutation caused a reduction in the height of inflorescence stems (Figure 6A). The height of the fra1 mutant stems was reduced to 65% of that of the wild type, whereas the diameter of the stems was not altered (Table 2). Anatomical examination of pith cells in fra1 stems showed a slight reduction in cell length compared with the wild type (Figures 2E and 2F), indicating that the reduced stem length in the fra1 mutant probably was caused by a reduction in cell length.
Figure 6.
Morphology of the Wild Type and the fra1 Mutant.
(A) Eight-week-old plants showing shorter but more erect stems in fra1 (right) compared with the wild type (left).
(B) and (C) Four-day-old dark-grown seedlings showing shorter hypocotyls in fra1 (C) compared with the wild type (B).
(D) and (E) Three-day-old light-grown seedlings showing shorter roots in fra1 (E) compared with the wild type (D).
Bars = 3 cm in (A) and 1.55 mm in (B) to (E).
Table 2.
Length and Diameter of Different Organs of the Wild Type and the fra1 Mutant
| Organs | Wild Type | fra1 |
|---|---|---|
| Light-grown seedlinga | ||
| Root length (mm) | 6.0 ± 0.7 | 3.5 ± 1.0 |
| Root diameter (mm) | 0.13 ± 0.02 | 0.14 ± 0.01 |
| Dark-grown seedlingb | ||
| Hypocotyl length (mm) | 14.6 ± 0.8 | 11.0 ± 1.2 |
| Hypocotyl diameter (mm) | 0.24 ± 0.03 | 0.23 ± 0.03 |
| Main inflorescence stemc | ||
| Height (cm) | 23.5 ± 3.1 | 15.2 ± 2.8 |
| Diameter (mm) | 1.6 ± 0.2 | 1.5 ± 0.1 |
Values shown are means ± se from 20 plants.
Seeds were germinated and grown on agar for 3 days before measurement.
Seeds were germinated and grown on agar for 4 days before measurement.
Eight-week-old plants grown on soil were used for measurement.
Examination of fra1 seedlings showed that the fra1 mutation also caused a slight reduction in the lengths of hypocotyls of dark-grown seedlings (Figures 6B and 6C) and roots of light-grown seedlings (Figures 6D and 6E). Quantitative analysis showed that although the diameters of these organs remained unchanged, the lengths of roots and hypocotyls were reduced to 58 and 79%, respectively, of those of the wild type (Table 2). The reduced lengths of these organs was accompanied by a decrease in cell length (data not shown). These results indicated that, in addition to the dramatic effect on fiber cell wall strength, the fra1 mutation also affected overall plant growth. The reduced organ lengths in the fra1 mutant were not caused by defects in cortical microtubule organization, because the fra1 mutation did not affect cortical microtubule pattern (Figure 5). Although we did not observe any apparent differences in the cellulose microfibril orientation in primary walls of elongating cells between the wild type and the fra1 mutant (data not shown), we could not exclude the possibility of a subtle change in cellulose microfibril deposition in fra1 elongating cells, which could result in the reduced cell and organ lengths. Alternatively, the mild fra1 seedling phenotype might be caused by changes in the biosynthesis or deposition of other cell wall components.
Map-Based Cloning of the FRA1 Gene
To investigate the molecular nature of the fra1 mutation, we cloned the FRA1 gene using a map-based approach. Genetic analysis showed that the fra1 mutation was recessive and occurred in a single locus. To map the chromosomal location of the fra1 locus, we used 2120 F2 homozygous mapping plants generated from crossing the fra1 mutant to the wild-type Landsberg erecta for linkage analysis with cleaved codominant amplified polymorphic sequence markers (Konieczny and Ausubel, 1993). The fra1 locus was shown to be located between the DFR and m588 markers on chromosome 5. Fine mapping with more cleaved codominant amplified polymorphic sequence markers placed the fra1 locus in a small region with a genetic distance of 0.2 centimorgan between markers PME2 and MCA1. According to Arabidopsis genome sequencing information, the physical distance between these two markers is 127 kb, indicating a low recombination rate in this region.
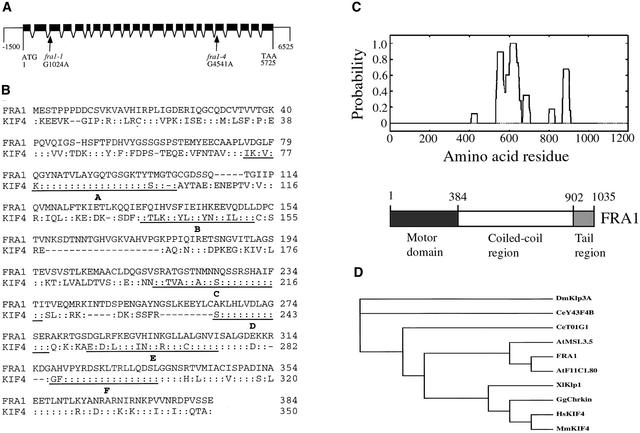
Sequencing analysis of putative genes in the 127-kb region revealed that in the fra1 mutant, there was a G-to-A mutation in a putative gene MCA23.16 (Figure 7A). Sequencing of the other fra1 alleles showed that fra1-2 and fra1-3 harbored the same mutation as fra1 and that fra1-4 had a G-to-A mutation in a different region of the same gene (Figure 7A). To confirm that the gene harboring the mutations was responsible for the fra1 mutant phenotypes, an 8-kb wild-type genomic DNA containing the MCA23.16 gene was ligated into a binary vector and introduced into the fra1 mutant plants by Agrobacterium-mediated transformation. Transgenic fra1 plants containing the wild-type gene completely restored the mechanical strength of stems to the wild-type level, and the orien-tation of cellulose microfibrils in fiber walls in these plants was indistinguishable from that in the wild type (data not shown). In addition, these plants showed wild-type morphology in seedlings and adult plants (data not shown). These results demonstrated unequivocally that mutations in the MCA23.16 gene were responsible for the fra1 mutant phenotypes and that the MCA23.16 gene represents the FRA1 gene.
Figure 7.
Structure of the FRA1 Gene and Analysis of the FRA1 Protein.
(A) The FRA1 gene is composed of 23 exons and 22 introns. Point mutations were found in the acceptor sites of the 2nd intron (fra1-1) and the 18th intron (fra1-4). Black boxes indicate exons, and lines between exons indicate introns.
(B) Amino acid sequence alignment of the motor domains of FRA1 and human KIF4. Identical amino acid residues are indicated by colons. Dashed lines indicate gaps introduced to maximize the identity. The motifs conserved among kinesin motor domains (http://mc11.mcri.ac.uk/khome/logos.html) are underlined. Motif A is known to be the ATP binding site. The fra1 mutation causes a deletion of part of the B motif (amino acid residues 142 to169).
(C) Location of the coiled coils in the FRA1 amino acid sequence (top) and diagram of the functional domains of FRA1 (bottom). The FRA1 sequence was analyzed for coiled-coil formation using the Paircoil program (Berger et al., 1995), and the cutoff for scoring a coiled coil as positive was 0.5. The fra1-4 mutation causes an insertion of 33 amino acids in the coiled-coil region between amino acid residues 756 and 757.
(D) Phylogenetic tree of FRA1 and other kinesin-like proteins in the KIF4 family. The phylogenetic relationship of the KIF4 kinesin-like proteins was analyzed based on the amino acid sequences of the motor domains. The sequences were aligned using the CLUSTAL W program (http://ebi.ac.uk/clustalw), and the resulting alignment was used to generate a phylogenetic tree using TREEVIEW (Page, 1996). The kinesin-like protein sequences are from Arabidopsis (FRA1; AtF11C1.80 and AtMSL3.5), human (HsKIF4), mouse (MmKIF4), chicken (GgChrkin), Xenopus laevis (Xlklp1), Caenorhabditis elegans (CeT01G1 and CeY43F4B), and Drosophila melanogaster (Dmklp3A).
Nature of the fra1 Mutations
To analyze the exon and intron organization of the FRA1 gene, we isolated and sequenced the full-length FRA1 cDNA from an Arabidopsis cDNA library. Comparison of FRA1 and its cDNA sequence showed that the gene is 5725 bp long from the start codon to the stop codon and contains 23 exons and 22 introns (Figure 7A). The longest open reading frame in the FRA1 cDNA is 3108 bp; it encodes a polypeptide of 1035 amino acids with a predicted molecular mass of 116,734 D and a predicted pI of 8.93.
The mutation of FRA1 in the fra1, fra1-2, and fra1-3 alleles occurred in the acceptor site of the second intron, which changed AG to AA (Figure 7A). Sequencing of the fra1 mutant cDNA showed that the intron acceptor site was shifted to an AG located 84 bp downstream of the authentic acceptor site. This shift resulted in a deletion of 84 nucleotides in the third exon, which led to a deletion of 28 amino acids of the predicted protein.
The mutation of FRA1 in the fra1-4 allele occurred in the acceptor site of the 18th intron, which changed AG to AA (Figure 7A). Sequencing of the fra1-4 mutant cDNA showed that the 18th intron was not spliced out in the mutant. This resulted in an insertion of 99 nucleotides in the mutant cDNA and an insertion of 33 amino acids in the predicted protein. It is interesting that neither fra1 nor fra1-4 causes a frameshift of the coding sequence or creates a premature stop codon; thus, the predicted mutant proteins retain the full-length wild-type sequence except for the partial internal deletion or insertion.
Sequence Analysis of the FRA1 Protein
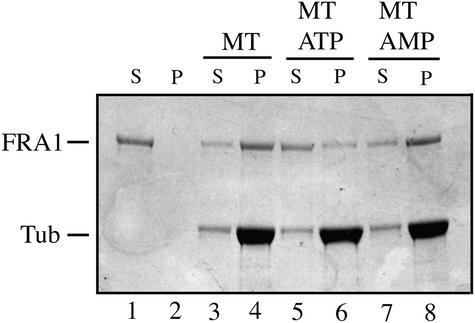
Comparison of the deduced amino acid sequence of FRA1 with those of proteins deposited in the GenBank database revealed that FRA1 exhibits high similarity to a group of animal kinesin-like proteins in the KIF4 family (Figures 7B and 7D). Some members of this family have been proposed to be involved in vesicle transport or mitotic division (Sekine et al., 1994; Wang and Adler, 1995; Hirokawa, 1996). Like KIF4 proteins, FRA1 has an N-terminal motor domain that presumably is involved in binding to and movement along microtubules in the expense of ATP hydrolysis and a C-terminal tail region, which are separated by a long coiled-coil region (Figure 7C). The FRA1 motor domain (amino acid residues 1 to 384) shares 51% amino acid sequence identity and 63% similarity with the motor domains of human KIF4 (Figure 7B) (Oh et al., 2000). It contains all of the conserved motifs pres-ent in the motor domains of typical kinesins. The mutation in the fra1 allele results in the deletion of part of the second conserved motif in the motor domain (Figure 7B). Cosedimentation experiments showed that, like typical kinesins, recombinant FRA1 motor domain has the ability to bind microtubules in an ATP-dependent manner (Figure 8).
Figure 8.
In Vitro Binding of Microtubules by the FRA1 Motor Domain.
The FRA1 motor domain fused with maltose binding protein was assayed for its ability to bind microtubules in the microtubule cosedimentation assay. After sedimentation, the fusion protein remained in the supernatant fraction in the absence of microtubules (lanes 1 and 2), whereas a large fraction of the fusion protein cosedimented with microtubules in the presence of microtubules (lanes 3 and 4). The addition of ATP significantly reduced the cosedimentation of the fusion protein with microtubules (lanes 5 and 6), and the nonhydrolyzable ATP analog adenylylimidodiphosphate (AMP) had no effect on the cosedimentation of the fusion protein with microtubules (lanes 7 and 8). MT, microtubule; P, pellet; S, supernatant; Tub, tubulin.
Following the motor domain is the putative coiled-coil region (amino acid residues 385 to 902), which likely is involved in dimerization. The amino acid sequences in this region were predicted by the Paircoil program (Berger et al., 1995) to form coiled-coil structures (Figure 7C). The FRA1 coiled-coil region shares 25% amino acid sequence identity and 44% similarity with that of human KIF4 protein. The mutation in the fra1-4 allele leads to an insertion of 33 amino acids in the coiled-coil region.
The C-terminal tail region of FRA1 (amino acid residues 903 to 1035) showed no sequence similarity with any known proteins, including members in the KIF4 family. Because the tail regions of KIF4 kinesin-like proteins have been suggested to be involved in binding cargos, the apparent differences in the tail regions of FRA1 and animal KIF4 proteins indicate that FRA1 might bind cargos that are different from those of animal KIF4 proteins.
Sequence comparison of FRA1 with all of the predicted proteins in the Arabidopsis genome revealed two proteins, AtF11C1.80 and AtMSL3.5, that show high sequence similarity and the same domain organization with FRA1 (Figure 7D) (Reddy and Day, 2001). The FRA1 motor domain shares 70 and 66% sequence identity with those of AtF11C1.80 and AtMSL3.5, respectively. The coiled-coil region shares 54 and 38% sequence identity with those of AtF11C1.80 and AtMSL3.5, respectively. Although the tail region of AtF11C1.80 exhibits 41% sequence identity with that of FRA1, the tail region of AtMSL3.5 does not show any sequence similarity with that of FRA1. Because AtF11C1.80 and FRA1 share high similarity throughout their sequences, it is tempting to propose that they are involved in the regulation of similar plant cellular processes.
FRA1 Protein Localization
It has been shown that some members of the animal KIF4 family were present in the nucleus and some were able to bind certain DNA sequences (Wang and Adler, 1995). This is consistent with the fact that these kinesin-like proteins have several nuclear localization signals (Hirokawa, 1996). Despite the high sequence similarity between FRA1 and animal KIF4 family members, FRA1 does not possess any nuclear localization signals in its entire sequence, as predicted by the PSORT program (http://psort.nibb.ac.jp) (data not shown). To determine the subcellular localization of FRA1, we used the green fluorescent protein (GFP)–tagging approach. The GFP-tagging approach has been used widely to study the subcellular locations of various proteins (Zuo et al., 2000; Hall and Cannon, 2002). In addition, it is known that the nuclear targeting of a protein is dictated by nuclear localization signals that are rich in basic amino acids and that are position independent (Smith and Raikhel, 1999).
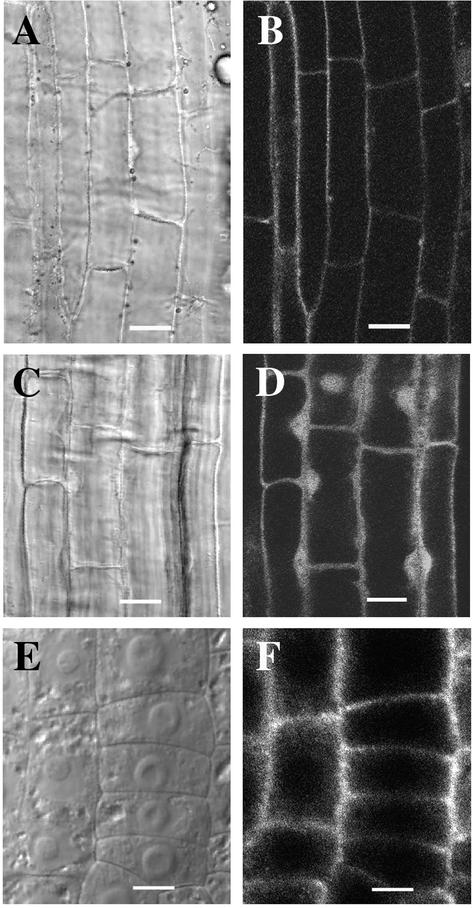
Full-length FRA1 tagged with GFP driven by the 35S promoter of Cauliflower mosaic virus was expressed in Arabidopsis plants. Analysis of 12 transgenic lines expressing various levels of FRA1-GFP fusion protein showed that the green fluorescence signals were located around the periphery of elongating root cells (Figures 9A and 9B), indicating that the FRA1 protein was present only in the cytoplasm. FRA1-GFP expressed under the FRA1 gene promoter showed the same localization pattern (data not shown). FRA1-GFP was not easily detectable in newly divided cells at the root tips. Transgenic plants expressing GFP alone showed green fluorescence signals in the cytoplasm and the nucleus (Figures 9C and 9D). Immunolocalization using affinity-purified polyclonal antibody against recombinant FRA1 confirmed that FRA1 was concentrated around the periphery of the cytoplasm but absent in the nucleus in early-elongating root cells (Figures 9E and 9F). Root cells incubated with the preimmune serum and fluorescein isothiocyanate–labeled secondary antibodies did not reveal any fluorescent signals (data not shown). These results indicate that, unlike animal KIF4 proteins, FRA1 is not targeted to the nucleus and it may play a role in cellular activities different from that of animal KIF4 proteins.
Figure 9.
Localization of FRA1 in Root Cells.
Roots of 3-day-old seedlings expressing the FRA1-GFP fusion protein ([A] and [B]) or GFP alone ([C] and [D]) were used to visualize GFP signals. Roots of 3-day-old wild-type seedlings were used for the immunolocalization of FRA1 ([E] and [F]).
(A) and (B) Differential interference contrast (DIC) image (A) of root cells and corresponding fluorescence signals of FRA1-GFP (B). The signal was present in the cytoplasm but absent in the nucleus.
(C) and (D) DIC image (C) of root cells and corresponding fluorescence signals of GFP (D). The signal was present in both the cytoplasm and the nucleus.
(E) and (F) DIC image (E) of root cells and corresponding FRA1 protein localization (F). FRA1 was present in the cytoplasm but absent in the nucleus. Note that the DIC image (E) shows the prominent nuclei in the centers of the cells.
Bars = 12 μm in (A) to (D) and 6 μm in (E) and (F).
Expression Pattern of the FRA1 Gene
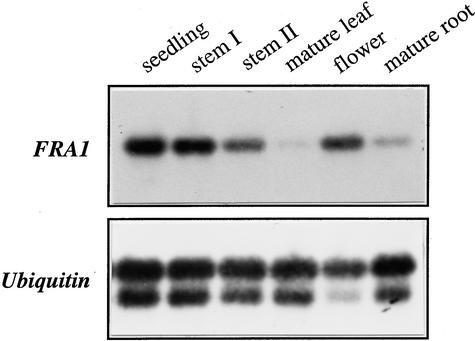
Examination of the FRA1 expression pattern showed that FRA1 was expressed in all organs (Figure 10). A higher level of FRA1 mRNA was seen in seedlings, floral buds, and elongating and nonelongating internodes of stems than in mature leaves and mature roots (Figure 10). FRA1 mRNA also was present in roots of 3-day-old seedlings (data not shown). The ubiquitous expression of FRA1 is consistent with the phenotypes of the fra1 mutant, showing defects in roots and hypocotyls of seedlings and in inflorescence stems of adult plants.
Figure 10.
FRA1 Gene Expression in Arabidopsis Organs.
RNA from different organs was used for reverse transcription–PCR. A ubiquitin gene was used as an internal control. The FRA1 mRNA was detected in all organs, with a lower level of expression in mature leaves and mature roots. The seedlings were 2 weeks old. Mature leaves were from 6-week-old plants. Flowers and mature roots were from 8-week-old plants. Stem I and stem II were from 4- and 8-week- old plants, respectively.
DISCUSSION
Cortical microtubules have long been proposed to influence the oriented deposition of cellulose microfibrils (Heath, 1974). Although there is considerable evidence to support this hypothesis, the molecular mechanisms of how cortical microtubules direct the oriented deposition of cellulose microfibrils are not clear (Baskin, 2001). Here, we have shown that mutation of a kinesin-like protein in the Arabidopsis fra1 mutant causes a dramatic alteration in the oriented deposition of cellulose microfibrils in fibers and a significant reduction in the mechanical strength of stems. This finding suggests that FRA1, a kinesin-like protein, is essential for the oriented deposition of cellulose microfibrils. Because the aberrant cellulose microfibril deposition in the fra1 mutant was not caused by an alteration in the organization of cortical microtubules, the FRA1 kinesin-like protein most likely plays an essential role in a step between cortical microtubules and cellulose microfibrils. Our finding marks an important step in understanding the mechanisms that underlie the microtubule control of cellulose microfibril order.
FRA1 Encodes a Kinesin-Like Motor Protein
The FRA1 protein shows high sequence similarity with members of the animal KIF4 kinesin-like protein family. The high sequence identity between FRA1 and KIF4 members is seen in both the microtubule binding motor domain and the coiled-coil region involved in dimerization. Based on sequence analysis (Figure 7) and microtubule binding assay (Figure 8), it is reasonable to suggest that FRA1 is a member of the KIF4 kinesin-like protein family. KIF4 proteins were first characterized in mouse, and they were proposed to be involved in vesicle transport (Sekine et al., 1994). Several KIF4 members contain nucleus-targeting sequences (Hirokawa, 1996) and have been shown to be present in the nucleus; thus, they are proposed to function as mitotic motors (Wang and Adler, 1995). Therefore, it was concluded that proteins in the KIF4 family play diverse roles in animal cells (Hirokawa, 1996). No mutants have been obtained to ascertain the functions of KIF4 kinesin-like proteins in cellular activities.
Although FRA1 shares overall sequence similarity with animal KIF4 proteins, it has several distinct features. First, FRA1 does not have any apparent nuclear localization sequences, and subcellular localization experiments did not detect its presence in the nucleus (Figure 9). Second, the tail region of FRA1 does not share any sequence similarity with the tail region of any animal KIF4 proteins. Because the tail region of KIF4 kinesin-like proteins is thought to be involved in binding cargos, this feature suggests that FRA1 may bind cargos that are different from those of animal KIF4s. Thus, it may play a role in plant-specific cellular activities.
The fra1 Mutation Causes an Alteration in the Oriented Deposition of Cellulose Microfibrils in Fibers
It was shown clearly that mutation of the FRA1 kinesin-like protein affects the oriented deposition of cellulose microfibrils in secondary walls of fibers. The prominent difference between the wild type and the fra1 mutant is that cellulose microfibrils in fiber walls are less organized in the fra1 mutant than in the wild type. It is interesting that although the orientation of cellulose microfibrils in fra1 fiber walls obviously is altered, the microfibrils are not oriented randomly (Figures 4D and 4E). This finding may be attributable to the nature of the mutations. Because the mutations in the fra1 mutant alleles did not cause truncation of the encoded protein, these mutant alleles are not null alleles of the FRA1 gene. It is possible as well that other mechanisms regulate the oriented deposition of cellulose microfibrils in fiber walls. It has been suggested that the oriented deposition of cellulose microfibrils might be controlled by cellular geometry (Emons and Mulder, 1998). The geometric path has been proposed to dictate the regular movement pattern of cellulose synthase complexes even without microtubule control.
Another possibility is that FRA1 may be one of several kinesin-like proteins involved in the control of cellulose microfibril deposition. Thus, the loss of FRA1 function could be compensated for partially by other kinesin-like proteins. This possibility is supported by the existence in the Arabidopsis genome of another kinesin-like protein (AtF11C1.80) that exhibits high overall sequence identity with FRA1. This also could explain the fact that although the FRA1 gene is expressed in young seedlings and in all organs of adult plants, an alteration in cellulose microfibril deposition was apparent only in the secondary walls of fibers. It is likely that FRA1 plays a dominant role in the oriented deposition of cellulose microfibrils during secondary wall thickening of fiber cells and that other proteins play dominant roles in other cell types.
Possible Roles of FRA1 in the Microtubule Control of Microfibril Orientation
It is not known how FRA1 affects the oriented deposition of cellulose microfibrils. Based on sequence analysis and microtubule binding assays, it is clear that FRA1 is a kinesin-like microtubule binding protein. Because mutation of the FRA1 gene caused altered deposition of cellulose microfibrils without affecting the organization of cortical microtubules, it is unlikely that FRA1 influences cellulose microfibril deposition by altering the pattern of cortical microtubules.
It is important to note that FRA1 does not have domains involved in formation of tetramers, a characteristic of kinesin-like proteins in the BimC family (Kashina et al., 1997). A kinesin-like protein in this family has been shown to be localized in the cortical region of carrot cells, and it was proposed to be involved in the sliding of cortical microtubules (Barroso et al., 2000). The fact that FRA1 does not show sequence similarity with the BimC family of kinesin-like proteins indicates that FRA1 is unlikely to be involved in the sliding of microtubules.
It has been suggested that some members of the KIF4 family are involved in vesicle transport (Hirokawa, 1996). However, to date, there is no evidence that microtubules in interphase cells are involved in vesicle transport in plants; in fact, the elimination of microtubules has been shown not to affect vesicle transport in interphase cells (Steinborn et al., 2002). Therefore, it appears unlikely that FRA1 influences cellulose microfibril deposition by affecting vesicle transport, although we could not exclude this possibility completely. This is consistent with the fact that there were no detectable changes in the cell wall composition in the fra1 mutant.
Available evidence suggests that the FRA1 kinesin-like protein may play a role in the microtubule control of cellulose microfibril orientation. Two models have been proposed to explain the microtubule control of cellulose microfibril order (Giddings and Staehelin, 1991). The FRA1 kinesin-like protein appears to fit well into the monorail model proposed by Heath (1974). In this model, motor proteins such as kinesins and dyneins were proposed to drag or guide cellulose synthase complexes along cortical microtubules, thus resulting in the coalignment of microtubules and cellulose microfibrils. If this is the case, the FRA1 kinesin-like protein would be one of the motor proteins involved in guiding the movement of cellulose synthase complexes along cortical microtubules.
The FRA1 kinesin-like protein also appears to fit into the guard rail model. In this model, the movement of cellulose synthase complexes was proposed to be constricted physically by guard rails that were composed of cortical microtubules and putative barrier proteins located in the plasma membrane, resulting in the coalignment of microtubules and cellulose microfibrils. In this case, motor proteins such as FRA1 might be important in aligning putative barrier proteins along cortical microtubules to form guard rails.
It is not known which of these two models operates in plant cells. Nevertheless, mutation of the FRA1 gene most likely causes a loss in the regulation of the directional movement of cellulose synthase complexes, which leads to aberrant deposition of cellulose microfibrils. It is important to note that FRA1 did not interact with the cytoplasmic domains of the putative cellulose synthase catalytic subunit AtCesA7 in the yeast two-hybrid system (data not shown). AtCesA7 has been shown to be involved in cellulose biosynthesis in secondary walls of xylem cells and fibers (Taylor et al., 1999). This result suggests that FRA1 may not bind directly to cellulose synthase catalytic subunits. Identification of the putative cargo proteins that the FRA1 motor protein binds is critical to our understanding of how FRA1 influences the oriented deposition of cellulose microfibrils.
The fra1 Mutation Dramatically Reduces the Mechanical Strength of Fibers
The fra1 mutant was isolated based on its dramatically reduced breaking strength in mature stems. Because the high mechanical strength of mature stems is conferred largely by the presence of interfascicular fibers (Zhong et al., 1997), we expected the mechanical strength of fibers to be reduced dramatically in the fra1 mutant. Because no changes in fiber cell morphology and cell wall composition were seen in the fra1 mutant, the reduced mechanical strength of these fibers most likely is caused by the altered deposition of cellulose microfibrils in the secondary walls of fibers. It is intriguing that the alteration of microfibril orientation in fiber walls without apparent changes in cell wall composition could lead to such a striking effect on fiber strength. This finding supports the early studies showing the importance of cellulose microfibril angles in the mechanical strength of wood (Chaffey, 2000).
In conclusion, we have demonstrated that FRA1, a kinesin-like protein, influences the oriented deposition of cellulose microfibrils in the secondary walls of fiber cells without affecting the organization of cortical microtubules. We propose that the FRA1 kinesin-like protein is involved in the microtubule control of the oriented deposition of cellulose microfibrils. Further investigation of how FRA1 performs its functions is expected to provide insights into the molecular mechanisms that underlie the microtubule control of cellulose microfibril deposition.
METHODS
Materials
M2 Arabidopsis thaliana (ecotype Columbia) plants generated from ethyl methanesulfonate mutagenization were grown in a greenhouse. Mature inflorescence stems of plants were measured for their mechanical strength. Plants showing a reduction in stem strength were selected as putative mutants. Mutant lines were backcrossed with wild-type Columbia three times before analysis.
Breaking Strength Measurement
The main inflorescence stems of 8-week-old plants were divided into three equal segments, and each segment was measured for their breaking forces with a digital force/length tester (model DHT4-50; Larson System, Minneapolis, MN). The ends of a stem segment were held, and a force was applied manually to the segment (Zhong et al., 1997). The breaking force was calculated as the force needed to break the stem segments.
Histology
Basal internodes of mature inflorescence stems were fixed in 2% glutaraldehyde in PBS (33 mM Na2HPO4, 1.8 mM NaH2PO4, and 140 mM NaCl, pH 7.2). After fixation, tissues were dehydrated through a gradient of ethanol, cleared in propylene oxide, and embedded in Araldite resin (Electron Microscopy Sciences, Fort Washington, PA). One-micrometer-thick sections were cut with a microtome and stained with toluidine blue.
Transmission Electron Microscopy
Stem segments embedded in Araldite resin as described above were cut into 90-nm-thick sections with a Reichert Jung ultrathin microtome (C. Reichert Optische Werke AG, Vienna, Austria). Sections were mounted on formvar-coated gold slot grids, poststained with uranyl acetate and lead citrate, and visualized with a Zeiss EM 902A electron microscope (Jena, Germany).
Field Emission Scanning Electron Microscopy
Cellulose microfibrils in the innermost layer of cell walls were visualized using field emission scanning electron microscopy according to Sugimoto et al. (2000). After fixation and dehydration, tissues were dried in a semidry critical point drier (Tousimis, Rockville, MD) and mounted on stubs with carbon paste. After being coated with platinum using an Edwards 306 vacuum evaporator (Edwards High Vacuum International, Wilmington, MA), samples were examined for cellulose microfibrils with a LEO 982 FE scanning electron microscope (LEO, Thornwood, NY).
Cell Wall Analysis
Mature inflorescence stems were ground into fine power in liquid nitrogen. The powder was washed in cold phosphate buffer (50 mM, pH 7.2) five times and extracted twice with 70% ethanol at 70°C for 1 h. After vacuum drying, cell wall materials were assayed for cellulose content with the anthrone reagent according to Updegraff (1969). Whatman 3MM paper (Clifton, NJ) was used as standard cellulose for quantitation. Cell wall sugars (as alditol acetates) were measured according to Hoebler et al. (1989) with the initial digestion time increased from 30 to 90 min. Klason lignin content in cell wall materials was measured according to Kirk and Obst (1988).
In Source Pyrolysis Mass Spectrometry
Cell wall materials were analyzed for their composition with a Finnigan GCQ mass spectrometer equipped with a direct exposure probe (rhenium loop) (Thermoquest, San Jose, CA) according to Morrison and Archibald (1998). Analysis conditions were as follows: ionization energy of 20 electron volts; mass-to-charge ratio of 50 to 500; scan time of 1 s; temperature increase of ∼10°C/s to 700°C; and ion source temperature of 175°C. All samples were run in triplicate.
Immunolocalization of Microtubules
Microtubules in epidermal cells, pith cells, and fiber cells were visualized by immunolabeling α-tubulins according to Sugimoto et al. (2000). Samples were probed with mouse monoclonal antibody against chicken α-tubulin (Sigma, St. Louis, MO) and then with fluorescein isothiocyanate–conjugated goat antibody against mouse IgG (Sigma). The fluorescence-labeled microtubules were visualized using a Leica TCs SP2 spectral confocal microscope (Leica Microsystems, Heidelberg, Germany). Images were saved and processed with Adobe Photoshop version 5.0 (Adobe Systems, San Jose, CA).
Map-Based Cloning
The fra1 mutant was crossed to the wild-type Arabidopsis ecotype Landsberg erecta to generate F1 seeds. F1 plants were self-pollinated to generate F2 seeds, which were germinated and grown to maturity for mapping study. F2 plants showing the fra1 mutant phenotype were selected and used to determine the chromosomal location of the fra1 locus with cleaved codominant amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993). For fine mapping of the fra1 locus, 2120 F2 mapping plants were analyzed to delimit the boundaries of the locus. The information on CAPS markers DFR, m558, and RPS4CT came from the Arabidopsis database. CAPS markers MCA1, MIF2, MNJ1, MQD2, and PME2 were developed by PCR amplification of genomic DNA fragments from ecotypes Columbia and Landsberg and digestion of the DNA fragments with various restriction enzymes.
For complementation analysis, the wild-type FRA1 gene was amplified by PCR (primers 5′-GATGGCTTCTGTTAATGAATCCA-3′ and 5′-TACAGCAGATGCTCATGGCATATC-3′), confirmed by sequencing, and then ligated into the binary vector pBI101 (Clontech, Palo Alto, CA). The construct was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation and introduced into the fra1 mutant plants using the vacuum infiltration–mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants containing the wild-type FRA1 gene were selected and analyzed for the mechanical strengths of inflorescence stems.
The FRA1 cDNA was isolated from an Arabidopsis cDNA library constructed with stem mRNA (Zhong and Ye, 1999). FRA1 genomic DNA and cDNA were sequenced using a dye-based cycle sequencing kit (Applied Biosystems, Foster City, CA).
Gene Expression Analysis
Total RNA was isolated from Arabidopsis seedlings, mature roots, mature leaves, and inflorescence stems using an RNA isolation kit (Qiagen, Valencia, CA). After being treated with DNase, mRNA was reverse transcribed to synthesize first-stand cDNA that was further used for PCR amplification of FRA1 cDNA (primers 5′-AAGACAAGT-AGATGAGTTTGCAGTG-3′ and 5′-TACGGCTGATAGTTTTCTCAT-CGA-3′) and ubiquitin cDNA. The amplified cDNA fragments were separated on an agarose gel and transferred onto a nylon membrane. The membrane was probed with digoxigenin-labeled DNA probes, and the hybridized DNA signal was detected with alkaline phosphatase–conjugated antibody against digoxigenin in conjunction with a chemiluminescence detection kit (Roche Molecular Biochemicals, Indianapolis, IN).
Microtubule Cosedimentation Assay
A 1.2-kb partial FRA1 cDNA encoding the FRA1 motor domain (amino acid residues 1 to 400) was fused in frame with the maltose binding protein (MBP) cDNA in the pMAL-C2 expression vector (New England Biolabs, Beverly, MA). The fusion protein was expressed in Escherichia coli and purified on an amylose resin affinity column. The purified fusion protein was used in the microtubule cosedimentation assay.
Microtubules were prepared by incubation of 5 mg/mL tubulin (Cytoskeleton, Denver, CO) in PEM buffer (80 mM Na-Pipes, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 1 mM GTP and 10% glycerol at 37°C for 50 min. Microtubules were stabilized by adding an equal volume of PEM buffer containing 40 μM taxol and incubated at 37°C for 10 min. The microtubule binding assay was performed in a 100-μL reaction volume containing 25 μg of taxol-stabilized microtubules and 10 μg of MBP-FRA1 fusion protein in PEM buffer containing 20 μM taxol. The reaction mixture was incubated at room temperature for 30 min. The MBP was used as a control, and it did not show any ability to bind microtubules in the assay. To test the ATP dependence of FRA1 binding to microtubules, ATP (5 mM) or a nonhydrolyzable ATP analog, adenylylimidodiphosphate (5 mM), was included in some of the reactions. After incubation, the reaction mixture was centrifuged at 14,000g for 25 min to sediment microtubules (Gardiner et al., 2001). The supernatant and pellet fractions were mixed with SDS sample buffer and analyzed by SDS-PAGE. Tubulin subunits and the fusion protein were visualized by staining the gel with Coomassie Brilliant Blue R 250 (Sigma).
Localization of the FRA1–Green Fluorescent Protein Fusion Protein
The FRA1 cDNA was fused in frame to the green fluorescent protein (GFP) cDNA (ABRC, Columbus, OH; developed by S.J. Davis and R.D. Vierstra) at the N terminus, and the fusion DNA was ligated downstream of the 35S promoter of Cauliflower mosaic virus or the FRA1 gene promoter in the pBI121 vector (Clontech). The FRA1-GFP constructs were transformed into Agrobacterium strain GV3101, which then was introduced into Arabidopsis plants by the vacuum infiltration–mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants were selected on Murashige and Skoog (1962) medium containing 50 μg/mL kanamycin, and T2 progeny were used for analysis.
Roots of 3-day-old seedlings expressing FRA1-GFP were visualized for GFP signals with a Leica TCs SP2 spectral confocal microscope. The same cells were imaged simultaneously for cellular morphology by differential interference contrast microscopy. Images were saved and processed with Adobe Photoshop version 5.0.
Immunolocalization of FRA1
A partial FRA1 cDNA encoding the tail region (amino acid residues 903 to 1035) of FRA1 was ligated in frame with the MBP cDNA in the pMAL-C2 expression vector (New England Biolabs). The recombinant FRA1 protein was purified on an amylose resin affinity column and used for polyclonal antibody production in rabbits. The antibodies were purified by affinity chromatography (Harlow and Lane, 1988) with a different recombinant FRA1 fusion protein, GST-FRA1, that was created by fusing the FRA1 tail region with glutathione S-transferase in the pGEX-2T expression vector (Amersham Pharmacia Biotech, Piscataway, NJ). The purified FRA1 antibody reacted specifically with the recombinant GST-FRA1 protein but not with the recombinant GST-F11C1.80 protein on immunoblots (data not shown). Further immunoblot analysis of protein extracts from Arabidopsis stems and 3-day-old roots showed that the purified FRA1 antibody detected a single protein band with an apparent molecular mass of ∼120 kD that corresponds to FRA1 (data not shown).
Roots of 3-day-old wild-type Arabidopsis seedlings were used for the immunodetection of FRA1 according to Sugimoto et al. (2000). Roots were incubated with the purified FRA1 antibody or with the preimmune serum as a control and then with fluorescein isothiocyanate–conjugated goat antibodies against rabbit IgG (Sigma). The fluorescent signals were visualized with a Leica TCs SP2 spectral confocal microscope. The same cells were imaged simultaneously for cellular morphology by differential interference contrast microscopy. Images were saved and processed with Adobe Photoshop version 5.0.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences described in this article and shown in Figure 7 are AY158083 and AY158084 (FRA1), AL132976 (AtF11C1.80), AB008269 (AtMSL3.5), NM_080314 (DmKlp3A), NM_067341 (CeY43F4B), Z92811 (CeT01G1), X82012 (XlKlp1), U18309 (GgChrkin), AF071592 (HsKIF4), and NM_008446 (MmKIF4).
Acknowledgments
We thank Nick Carpita, Alan Darvill, L. Andrew Staehelin, and William York for helpful discussions during the early stage of this work, John Shields for his assistance with confocal microscopy, and the ABRC for providing the GFP cDNA. This work was supported by the Cooperative State Research, Education, and Extension Service of the U.S. Department of Agriculture.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005801.
References
- Asada, T., Kuriyama, R., and Shibaoka, H. (1997). TKRP125, a kinesin-related protein involved in the centrosome-independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J. Cell Sci. 110, 179–189. [DOI] [PubMed] [Google Scholar]
- Barroso, C., Chan, J., Allan, V., Doonan, J., Hussey, P., and Lloyd, C. (2000). Two kinesin-related proteins associated with the cold-stable cytoskeleton of carrot cells: Characterization of a novel kinesin, DcKRP120-2. Plant J. 24, 859–868. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2000). The cytoskeleton. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 202–258.
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Berger, B., Wilson, D.B., Wolf, E., Tonchev, T., Milla, M., and Kim, P.S. (1995). Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.-H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Chaffey, N. (2000). Microfibril orientation in wood cells: New angles on an old topic. Trends Plant Sci. 5, 360–362. [DOI] [PubMed] [Google Scholar]
- Chen, C., Marcus, A., Li, W., Hu, Y., Calzada, J.-P.V., Grossniklaus, U., Cyr, R.J., and Ma, H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129, 2401–2409. [DOI] [PubMed] [Google Scholar]
- Emons, A.M.C., Derksen, J., and Sassen, M.M.A. (1992). Do microtubules orient plant cell wall microtubules? Physiol. Plant. 84, 486–493. [Google Scholar]
- Emons, A.M.C., and Mulder, B.M. (1998). The making of the architecture of the plant cell wall: How cells exploit geometry. Proc. Natl. Acad. Sci. USA 95, 7215–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J.C., Harper, J.D.I., Weerakoon, N.D., Collings, D.A., Ritchie, S., Gilroy, S., Cyr, R.J., and Marc, J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13, 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1988). Spatial relationship between microtubules and plasma-membrane rosettes during the deposition of primary wall microfibrils in Closterium sp. Planta 173, 22–30. [DOI] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (San Diego, CA: Academic Press), pp. 85–99.
- Hall, Q., and Cannon, M.C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, T., and Coté, W.A. (1985). Structure of wood. In Biosynthesis and Biodegradation of Wood Components, T. Higuchi, ed (San Diego, CA: Academic Press), pp. 1–42.
- Hardham, A.R., and Gunning, B.E.S. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77, 14–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hasezawa, S., and Nozaki, H. (1999). Role of cortical microtubules in the orientation of cellulose microfibril deposition in higher-plant cells. Protoplasma 209, 98–104. [DOI] [PubMed] [Google Scholar]
- Heath, I.B. (1974). A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J. Theor. Biol. 48, 445–449. [DOI] [PubMed] [Google Scholar]
- Herth, W. (1980). Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: Evidence for a gap between polymerization and microfibril formation. J. Cell Biol. 87, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N. (1996). Organelle transport along microtubules: The role of KIFs. Trends Cell Biol. 6, 135–141. [DOI] [PubMed] [Google Scholar]
- Hoebler, C., Barry, L.D., and Delort-Laval, J. (1989). Rapid hydrolysis of plant cell wall polysaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Kashina, A.S., Rogers, G.C., and Scholey, J.M. (1997). The bimC family of kinesins: Essential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta 1357, 257–271. [DOI] [PubMed] [Google Scholar]
- Kirk, T.K., and Obst, J.R. (1988). Lignin determination. Methods Enzymol. 161, 87–101. [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kost, B., and Chua, N.-H. (2002). The plant cytoskeleton: Vacuoles and cell walls make the difference. Cell 108, 9–12. [DOI] [PubMed] [Google Scholar]
- Lancelle, S.A., Callaham, D.A., and Hepler, P.K. (1986). A method for rapid freeze-fixation of plant cells. Protoplasma 131, 153–165. [Google Scholar]
- Lancelle, S.A., Cresti, M., and Hepler, P.K. (1987). Ultrastructure of the cytoskeleton in freeze-substituted pollen tubes of Nicotiana alata. Protoplasma 140, 141–150. [Google Scholar]
- Ledbetter, M.C., and Porter, K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.-R.J., Giang, H.M., and Liu, B. (2001). A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13, 2427–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, K., Collings, D., and Asada, T. (2001). Identification of a novel plant-specific kinesin-like protein that is highly expressed in interphase tobacco BY-2 cells. Protoplasma 215, 105–115. [DOI] [PubMed] [Google Scholar]
- Morrison, W.H., and Archibald, D.D. (1998). Analysis of graded flax fiber and yarn by pyrolysis mass spectrometry and pyrolysis gas chromatography mass spectrometry. J. Agric. Food Chem. 46, 1870–1876. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nishihama, R., Soyano, T., Ishikawa, M., Araki, S., Tanaka, H., Asada, T., Irie, K., Ito, M., Terada, M., Banno, H., Yamazaki, Y., and Machida, Y. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109, 87–99. [DOI] [PubMed] [Google Scholar]
- Oh, S., Hahn, H., Torrey, T.A., Shin, H., Choi, W., Lee, Y.M., Morse, H.C., and Kim, W. (2000). Identification of the human homologue of mouse KIF4, a kinesin superfamily motor protein. Biochim. Biophys. Acta 1493, 219–224. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, M.D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Preston, R.D. (1988). Cellulose-microfibril-orienting mechanisms in plant cell walls. Planta 174, 67–74. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001). Molecular motors and their functions in plants. Int. Rev. Cytol. 204, 97–177. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N., and Day, I.S. (2001). Kinesins in the Arabidopsis ge-nome: A comparative analysis among eukaryotes. BMC Genomics 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, Y., Okada, Y., Noda, Y., Kondo, S., Aizawa, H., Takemura, R., and Hirokawa, N. (1994). A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J. Cell Biol. 1, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., and Raikhel, N.V. (1999). Protein targeting to the nuclear pore: What can we learn from plants? Plant Physiol. 119, 1157–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn, K., Maulbetsch, C., Priester, B., Trautmann, S., Pacher, T., Geiges, B., Küttner, F., Lepiniec, L., Stierhof, Y.-D., Schwarz, H., Jürgens, G., and Mayer, U. (2002). The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 16, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen, G., Kasmi, F.E., Richter, S., Lukowitz, W., Assaad, F.F., Jürgens, G., and Mayer, U. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12, 153–158. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.-R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff, D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- van der Hage, E.R.E., Mulder, M.M., and Boon, J.J. (1993). Structural characterization of lignin polymers by temperature-resolved in-source pyrolysis-mass spectrometry and Curie-point pyrolysis-gas chromatography/mass spectrometry. J. Anal. Appl. Pyrolysis 25, 149–183. [Google Scholar]
- Wang, S.-Z., and Adler, R. (1995). Chromokinesin: A DNA-binding, kinesin-like nuclear protein. J. Cell Biol. 128, 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2000). The cytoskeleton and growth polarity. Curr. Opin. Plant Biol. 3, 503–511. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., and Ye, Z.-H. (2001). Fibers: A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.-H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.-W., Nishizawa, N., Wu, Y., Kost, B., and Chua, N.-H. (2000). KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12, 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]