Abstract
We have examined the expression of three alternative oxidase (aox) genes in two types of maize mitochondrial mutants. Nonchromosomal stripe (NCS) mutants carry mitochondrial DNA deletions that affect subunits of respiratory complexes and show constitutively defective growth. Cytoplasmic male-sterile (CMS) mutants have mitochondrial DNA rearrangements, but they are impaired for mitochondrial function only during anther development. In contrast to normal plants, which have very low levels of AOX, NCS mutants exhibit high expression of aox genes in all nonphotosynthetic tissues tested. The expression pattern is specific for each type of mitochondrial lesion: the NADH dehydrogenase–defective NCS2 mutant has high expression of aox2, whereas the cytochrome oxidase–defective NCS6 mutant predominantly expresses aox3. Similarly, aox2 and aox3 can be induced differentially in normal maize seedlings by specific inhibitors of these two respiratory complexes. Translation-defective NCS4 plants show induction of both aox2 and aox3. AOX2 and AOX3 proteins differ in their ability to be regulated by reversible dimerization. CMS mutants show relatively high levels of aox2 mRNAs in young tassels but none in ear shoots. Significant expression of aox1 is detected only in NCS and CMS tassels. The induction pattern of maize aox genes could serve as a selective marker for diverse mitochondrial defects.
INTRODUCTION
Several maternally inherited mitochondrial DNA deletion mutants (nonchromosomal stripe; NCS) have been characterized in maize. NCS2 plants have a truncated nad4 gene and lack a fully assembled respiratory complex I (CI) in the mutant mitochondria (Marienfeld and Newton, 1994; Karpova and Newton, 1999). NCS5 and NCS6 plants carry different deletions of the 5′ end of the cox2 gene, which encodes a subunit of respiratory complex IV (CIV; cytochrome oxidase) (Lauer et al., 1990; Newton et al., 1990). NCS3 and NCS4 are two different deletions of the mitochondrial gene encoding the RPS3 ribosomal protein and are associated with very reduced levels of mitochondrial protein synthesis (Hunt and Newton, 1991; Newton et al., 1996). Another type of plant mitochondrial defect, cytoplasmic male sterility (CMS), causes respiratory failure specifically during pollen development (reviewed by Conley and Hanson, 1995; Schnable and Wise, 1998). CMS is usually associated with the expression of chimeric mitochondrial proteins that become toxic during microsporogenesis. In contrast to CMS, homoplasmic NCS mu-tations are lethal during kernel development (with very rare exceptions) (Yamato and Newton, 1999). Thus, the NCS mutations are propagated in heteroplasmic NCS plants that carry a mixture of mutant and normal mitochondria (Newton and Coe, 1986; Gu et al., 1993; Marienfeld and Newton, 1994). During development, somatic segregation of mutant from normal mitochondria leads to clonal sectors of defective growth.
Because NCS mutations have blocks in the normal cytochrome pathway of mitochondrial electron transfer, mutant mitochondria could be expected to show an increase in the alternative respiratory pathway that is characterized by the KCN-insensitive terminal oxidase, alternative oxidase (AOX). AOX transfers electrons directly from the ubiquinone pool, bypassing two of the three sites at which the cytochrome pathway is coupled to ATP synthesis (Moore and Siedow, 1991). Although the alternative pathway is energetically wasteful, it could be used to help maintain normal levels of metabolites and to reduce levels of reactive oxygen species (ROS) in mitochondria when electron flow through the cytochrome pathway is limited (Wagner and Moore, 1997). Also, in addition to the rotenone-sensitive CI, plants contain up to four NAD(P)H dehydrogenases that can introduce electrons into the ubiquinone pool (Soole and Menz, 1995; Bhattramakki and Elthon, 1997; Møller, 2001).
The combined actions of multiple NAD(P)H dehydrogenases and AOX should make plant mitochondria more tolerant of respiratory defects than are animal mitochondria, which lack these additional enzymes. Indeed, homoplasmic nad7 (CI-defective) mutants of Nicotiana sylvestris show increased alternative respiration and activities of the additional NAD(P)H dehydrogenases (Gutierres et al., 1997; Sabar et al., 2000). However, the pathways to cope with respiratory arrest vary, because no increase in external NAD(P)H dehydrogenase activities was detected in the nad4 (CI-deficient) mutant of maize (Marienfeld and Newton, 1994; Karpova and Newton, 1999).
AOX has been shown to be encoded by a small family (three to four members) of nuclear genes in a number of plant species and appears to be subject to complex regulation during development (McCabe et al., 1998; Considine et al., 2001) and in different tissues (Finnegan et al., 1997; Saika et al., 2002). Two mechanisms are known to regulate AOX at the post-translational level: activation by pyruvate and reversible inactivation by redox dimerization (Millar et al., 1993; Umbach and Siedow, 1993, 1996; Vanlerberghe and McIntosh, 1997).
AOX has been shown to be induced in response to stress or inhibition of the respiratory chain (Vanlerberghe and McIntosh, 1997). Increasing evidence suggests that stressed plant mitochondria signal the nucleus to induce the transcription of genes whose products are needed to cope with altered metabolic conditions (Maxwell et al., 2002). Signaling from plastids to activate nuclear genes also is known to occur (reviewed by Surpin et al., 2002).
Here, we examined three members of the aox gene family in maize and used respiratory-deficient mutants to determine whether the expression of different AOX isoforms varies depending on which part of the electron transfer chain (ETC) is blocked. Each NCS mutant provides a metabolically stable model for a molecularly defined mitochondrial defect. Interestingly, CI- and CIV-deficient mutants were found to have different aox genes expressed to high levels, encoding a putative redox-regulated, Cys-containing isoform (AOX2) and a less commonly studied Cys-minus isoform (AOX3), respectively. Although these results have been corroborated by experiments using specific ETC inhibitors on seedlings, the use of mutants allows one to analyze the expression of the aox genes as the plants develop and without the loss of viability that occurs when seedlings are treated with inhibitors of respiration. In maize CMS plants, high levels of aox2 and increased aox1 mRNAs have been found in the developing male florets, which is the site of mitochondrial dysfunction. The constitutive expression of AOX in the NCS mutants under normal growing conditions could reflect an adaptation of NCS mutants to continuous mitochondrial dysfunction. Furthermore, evidence from mutant analysis suggests that defective maize mitochondria not only signal the nucleus that mitochondrial function is compromised but also convey information about where the lesion is located, resulting in a differential induction of aox genes.
RESULTS
Constitutive Expression of AOX in the NCS Mutants
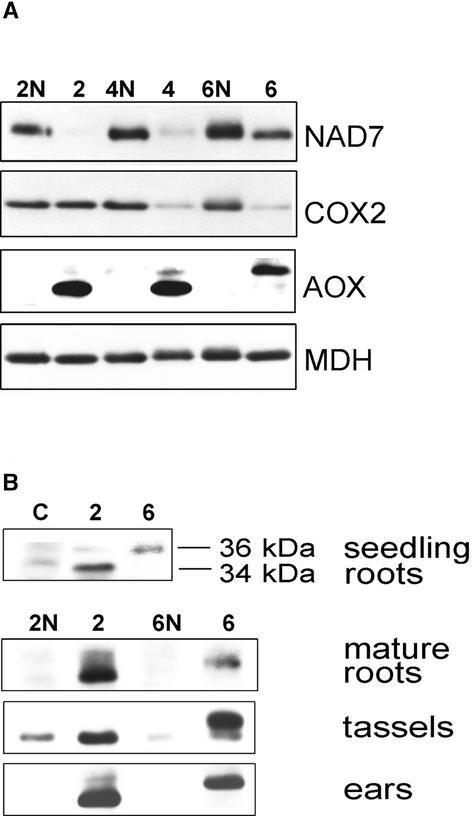
The expression of several proteins was analyzed initially in extracts of mitochondria isolated from young, unpollinated ear shoots. As a result of heteroplasmy, it is crucial to work with NCS plants that are phenotypically very affected and to monitor the reduction of mitochondrial proteins from the affected complexes (Karpova and Newton, 1999). In Figure 1A, immunoblot analysis shows that mitochondrially encoded COX2 was reduced dramatically in heteroplasmic NCS6 (cox2-deficient) and NCS4 (mitochondrial translation–deficient) ear shoots. The CI subunit, NAD7, was reduced in NCS2 (nad4-deficient) and NCS4 plants.
Figure 1.
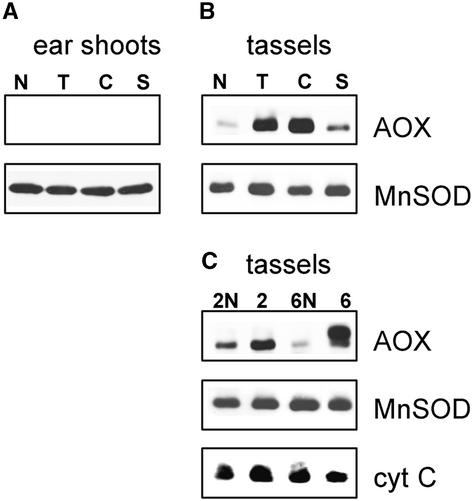
NCS Mutants Express High Levels of AOX Protein.
(A) Immunoblot analysis of mitochondrial proteins and AOX isolated from ear shoots. Mitochondrial proteins (50 μg/lane) from ear shoots of NCS plants (lanes 2, 4, and 6, corresponding to NCS2, NCS4, and NCS6 samples, respectively) and their normal relatives (lanes 2N, 4N, and 6N) were loaded pairwise for comparison. Immunoblots were reacted with antisera to the NAD7 subunit of CI, the COX2 subunit of CIV, and AOX. Reprobing the blot with antibody to mitochondrial malate dehydrogenase (MDH) was used for a loading control.
(B) Expression of AOX in nonphotosynthetic tissues of NCS mutants. Immunoblot analysis of mitochondrial proteins from different tissues of NCS2 (lane 2) and NCS6 (lane 6) mutants and their normal relatives (lanes 2N and 6N, respectively). Mitochondria from normal A619 seedling roots were used as a control (lane C). Results for roots are shown at a longer exposure because of lower loading (30 μg/lane instead of 50 μg/lane for other samples).
Although many plant tissues have been reported to have AOX, there is no evidence for AOX protein in unpollinated ear shoots of nonmutant maize plants, suggesting that AOX is not normally a constituent of the mitochondrial ETC in this organ (Figure 1A, lanes 2N, 4N, and 6N). However, in ear shoots from all of the NCS mutants, AOX was present at high levels (Figure 1A, lanes 2, 4, and 6). Careful inspection reveals that the migrations of the major AOX polypeptides in NCS2 and NCS6 are different from each other and that NCS4 has both forms of the protein.
To further characterize the expression of AOX proteins in NCS mutants, immunoblot analysis of AOX in three other nonphotosynthetic tissues was performed. Figure 1B shows the immunodetection of AOX in mitochondrial protein extracts isolated from seedling roots and from mature roots and tassels of NCS2 and NCS6 plants. Roots of a normal seedling showed low levels of both AOX isoforms, with apparent molecular masses of 34 and 36 kD. However, it is clear that tissues of NCS mutants carrying specific lesions in respiratory complexes always contained more AOX than the corresponding normal tissues. Moreover, for each mutant, the AOX pattern remained the same: NCS2 had mostly the 34-kD AOX induced, whereas in NCS6, a 36-kD AOX was the major isoform induced in all tissues tested (Figure 1B, lanes 2 and 6). These results demonstrate that a mutant in CI (NCS2) and one in CIV (NCS6) had high expression of different AOX isoforms in multiple tissues.
Differential Expression of Three aox Genes in NCS Mutants
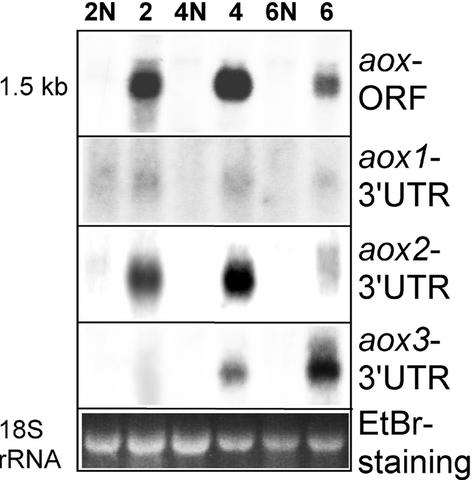
To determine whether the induction of AOX protein in NCS mutants correlates with increased transcript levels, RNA gel blot analysis was performed using total RNA isolated from ear shoots of NCS plants and their normal relatives. When a fragment corresponding to the open reading frame of a maize aox (Figure 2, aox-ORF) was used as a probe, no traces of the transcripts were detected in normal ear shoots. This finding parallels the protein results and suggests that aox is not expressed routinely in ear shoots (Figure 2, lanes 2N, 4N, and 6N). However, in all of the NCS mutants, aox transcripts were highly induced (Figure 2, lanes 2, 4, and 6).
Figure 2.
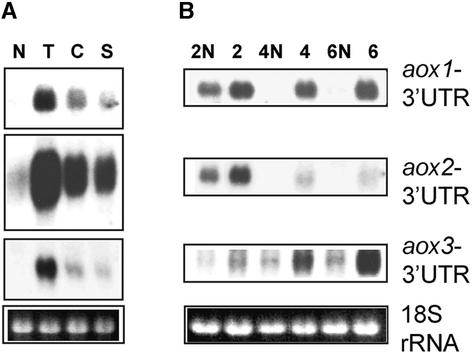
RNA Gel Blot Analysis Shows Increased Expression of Specific aox Genes in Different NCS Mutants.
Total RNAs were isolated from ear shoots of very affected NCS mutants (lanes 2, 4, and 6) and their corresponding normal relatives (lanes 2N, 4N, and 6N). The gel at bottom shows ethidium bromide (EtBr) staining of the 18S RNA as a loading control. Hybridization was with the riboprobes indicated at right. UTR, untranslated region.
AOX has been shown to be encoded by a small multigene family in soybean (Whelan et al., 1996), Arabidopsis (Saisho et al., 1997), mango (Considine et al., 2001), and rice (Ito et al., 1997; Saika et al., 2002). When we started this study, there were no published data concerning the number and organization of aox genes in maize. From Pioneer Hi-Bred International's maize cDNA libraries, we obtained full-length aox1, aox2, and partial aox3 cDNA clones that showed homology with a partial sequence of a maize aox gene in GenBank. After completion of the aox3 cDNA to full length (see Methods) and sequencing of all three aox cDNAs, the clones were used to analyze the aox gene–specific transcripts in NCS mutants.
Hybridization with aox 3′ untranslated region gene-specific riboprobes showed that the ratios of aox2 and aox3 transcripts differed dramatically in NCS mutants (Figure 2). The amounts of aox1 transcripts appeared to be extremely low in all of the ear shoot samples. aox2 transcripts were seen at high levels in NCS2 and NCS4 mutants but were detected at low levels in the NCS6 mutant. aox3 transcripts were abundant in NCS6 samples, present at lower levels in NCS4, and undetectable in NCS2. Thus, the RNA gel blot data demonstrated that different aox genes were expressed to different levels in each of the NCS mutants.
Differential Expression of aox Genes in Seedlings Treated with ETC Inhibitors
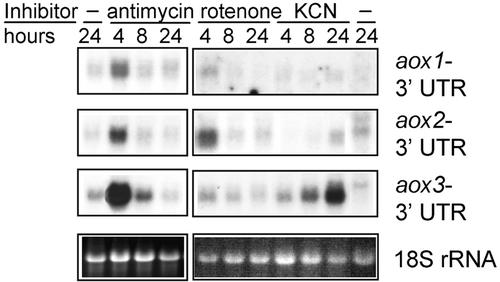
Because there was a clear difference in which aox genes were expressed predominantly in the NCS mutants affecting CI and CIV, it was of interest to determine whether a differential induction of aox genes could be produced in nonmutant maize plants by treatment with specific inhibitors of the ETC. Antimycin A, an inhibitor of respiratory complex III, and cyanide, an inhibitor of CIV, had been shown previously to induce aox transcription in some fungi and plants (Sakajo et al., 1991; Vanlerberghe and McIntosh, 1994, 1997). Five-day-old etiolated maize seedlings were treated for up to 24 h with inhibitors specific to respiratory CI (rotenone), CIV (KCN), and CIII (antimycin A). At 4, 8, and 24 h after the addition of each inhibitor, samples of total RNA were isolated from seedling roots. Figure 3 shows aox transcripts in normal seedlings upon treatment with rotenone, KCN, and antimycin A. Neither rotenone nor KCN induced the aox1 transcripts to an appreciable level. During incubation with rotenone, a transient induction of aox2 transcripts was detected within 4 h. In KCN-treated seedlings, aox3 transcripts accumulated more slowly but increased throughout the 24 h. Notably, only one major aox transcript was detected after each treatment: aox2 in rotenone-treated seedlings and aox3 in KCN-treated seedlings. Antimycin A treatment caused an induction of all three aox genes, although aox3 transcripts were the most abundant. For all aox genes, the highest expression level was seen within 4 h of incubation with antimycin A. These experiments demonstrate directly that maize aox genes can be upregulated individually by inhibition of the ETC at specific sites and that the results mimic what is seen in the NCS mutants.
Figure 3.
RNA Gel Blot Analysis of aox Gene Expression in Normal Maize Seedlings after Specific Inhibition of the Mitochondrial ETC.
Maize seedlings were incubated with the CI inhibitor rotenone (40 μM), the CIV inhibitor KCN (10 mM), the CIII inhibitor antimycin A (20 μM), or with no addition (−). Shown are RNA gel blot hybridization results of total RNA samples isolated from seedling roots after the treatment times indicated. Riboprobes are listed at right. The gel at bottom shows ethidium bromide staining of the 18S RNA as a loading control. UTR, untranslated region.
NCS Mutants Express Distinctive AOX Proteins
Deduced amino acid sequences of maize aox genes compared with those from other plants are shown in Figure 4A. The predicted maize AOX proteins cluster with AOXs from other monocots, particularly those from rice (Figure 4B).
Figure 4.
Deduced Protein Alignment of Maize aox Genes with Those from Other Plants.
(A) The deduced amino acid sequences for maize (Zm) AOX1, AOX2, and AOX3, Arabidopsis AOX1a (Ath AOX1a), and rice AOX1b (Osa AOX1b) were aligned using the CLUSTAL V method in Megalign (DNASTAR, Madison, WI). The positions of three introns in the Sauromatum guttatum aox1 gene are designated i1, i2, and i3 (cited by Vanlerberghe and McIntosh, 1997). Nonconsensus amino acid residues are shown on a black background. Conserved Cys residues and the target epitope for the AOX antibody are boxed. The regulatory Cys is marked with a star. The position of the N-terminal Ala of the mature maize AOX proteins is predicted by homology with the S. guttatum mature AOX protein (Rhoads and McIntosh, 1991) and is marked with a triangle. The deduced molecular masses for precursor (mature) maize AOX proteins are as follows: AOX1, 38.7 kD (32.6 kD); AOX2, 36.9 kD (31.5 kD); and AOX3, 36.9 kD (31.7 kD).
(B) Phylogenetic tree showing the relationship of maize AOX1, AOX2, and AOX3 to AOX proteins from rice (AOX1a, AOX1b, and AOX1c), wheat (AOX1a and AOX1c), and Arabidopsis (AOX1a, AOX1b, AOX1c, and AOX2). The maize gene names are consistent with the standard maize nomenclature. Alternative names for the maize aox genes (according to the suggestion of Considine et al., 2002) are shown in parentheses. The number of differences per 100 amino acid residues is derived from the CLUSTAL V alignment.
The predicted AOX2 and AOX3 proteins contain a consensus sequence, 5′-RADEAHHRDVNH-3′, that was shown to be the target epitope for the monoclonal antibody (AOA) used in our AOX analyses (Finnegan et al., 1999). The Ala in position 2 of this sequence is changed to a Ser residue in the AOX1 predicted protein (Figure 4A). Substituting Val for Ala at position 2 of the consensus sequence was shown to cause a loss of recognition by the AOX antibody (Finnegan et al., 1999), which raises the possibility that AOX1 might not be detected in our immunoblot experiments (Figure 1). However, AOX1 does not appear to be expressed in ear shoots of normal or NCS plants, because few, if any, aox1 transcripts were detected (Figure 2).
To clarify which gene product corresponds to each of the major AOX proteins seen in NCS mitochondria, analysis of maize AOX isoforms was performed. AOX1 and AOX2 are predicted to belong to the main class of plant AOXs that interact with α-keto acids (pyruvate) and that can be inactivated reversibly by dimerization via the oxidation of sulfhydryl groups (Umbach and Siedow, 1993, 1996; Vanlerberghe and McIntosh, 1997). Both regulatory mechanisms are associated with the presence of a highly conserved Cys residue at the N terminus of the AOX protein (Cys-127 in Arabidopsis AOX1a) (Rhoads et al., 1998). As seen in Figure 4A, AOX1 and AOX2 do contain this essential Cys, as well as a second Cys that is conserved in plant AOXs (Vanlerberghe and McIntosh, 1997). AOX3 does not contain either Cys and represents a naturally occurring AOX protein with Cys-to-Ser replacements. When the regulatory Cys was converted to Ser by site-directed mutagenesis of a soybean aox gene, the resulting protein did not form dimers through reversible covalent disulfide bonds (Djajanegara et al., 1999). However, it should be noted that Cys-substituted AOX proteins have been found to exist within membranes as dimers (Umbach et al., 2002), formed apparently via noncovalent associations.
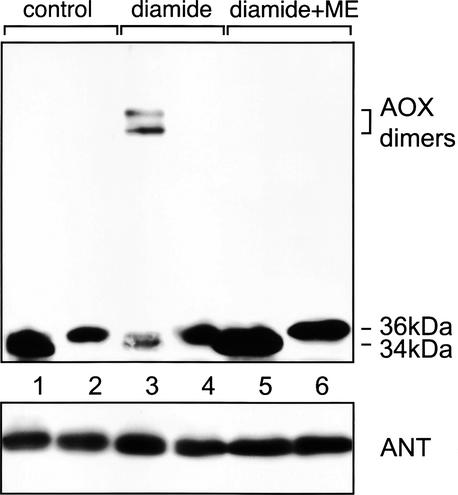
We used the predicted differences in dimerization through reversible disulfide bonds to distinguish between AOX2 and AOX3 expression in NCS mutants. Figure 5 shows immunodetection of AOX in protein extracts from NCS2 and NCS6 mitochondria after treatment with the sulfhydryl-oxidizing reagent diamide. In untreated NCS2 and NCS6 mitochondria, AOX proteins are present mostly as monomers with apparent molecular masses of 34 and 36 kD, respectively (Figure 5, lanes 1 and 2). However, a striking difference between the AOX proteins from NCS2 and NCS6 mitochondria was observed after sulfhydryl bond oxidation with diamide. NCS2 AOX was converted to dimers (with apparent molecular masses of 71 and 77 kD), leaving traces of monomers (Figure 5, lane 3). The dimers could be reduced to monomers by mercaptoethanol (Figure 5, lane 5). Thus, in NCS2 mitochondria, there are Cys-containing AOX isoforms that give dimers upon oxidation. Because this result correlates with the induction of mostly aox2 mRNA in the NCS2 mutants, the major AOX in the CI mutant now can be identified as AOX2. The doublet of oxidized forms of NCS2 AOX could reflect variation in the processing or modification of AOX, as was suggested previously (Hiser et al., 1996). Multiple dimers for a single type of AOX have been reported (Rhoads et al., 1998) and may be attributable to alternative conformations of the dimer. In contrast to the results with NCS2, diamide treatment had no effect on NCS6 AOX (Figure 5, lanes 2 and 4). This experiment confirms that the 36-kD AOX seen in NCS6 mitochondria does not contain the regulatory Cys necessary for dimerization and most likely corresponds to the aox3 gene product.
Figure 5.
Sulfhydryl Oxidation of AOX from NCS2 and NCS6 Mitochondria.
Mitochondria from ear shoots of NCS2 (lanes 1, 3, and 5) and NCS6 (lanes 2, 4, and 6) mutants were treated with diamide (lanes 3 and 4) or with diamide plus mercaptoethanol (ME; lanes 5 and 6), and protein extracts (50 μg/lane) were subjected to immunoblot analysis and probed with antibodies to AOX. The gel at bottom shows probing with antiserum against adenine nucleotide translocator (ANT) as a loading control.
Analysis of AOX Expression in Maize CMS Mutants
To compare the impact of different types of mitochondrial deficiencies on AOX expression, we determined the AOX profiles in the three types of CMS (CMS-T, -C, and -S) known for maize. Unlike the NCS mutations, maize CMS mutations are tissue specific, affecting pollen development (reviewed by Schnable and Wise, 1998). Pollen abortion is caused by mitochondrial failure, the biochemical basis of which is well characterized for CMS-T (Rhoads et al., 1995) but is unknown for CMS-C and CMS-S. In CMS mutants, all tissues contain functional mitochondria except for the anthers.
The parallel analysis of aox expression in CMS-T plants represents an important control for the NCS2 mutation because it arose in the CMS-T genotype and the NCS2 plants are fully male sterile. The NCS4 and NCS6 plants used in this study arose in two different spontaneous revertants to fertility of CMS-S plants.
Our previous results indicated that increased levels of AOX in nonphotosynthetic maize tissues correlate with respiratory defects. Thus, we hypothesized that AOX would be highly expressed in male florets only when mitochondrial function was defective (i.e., in both CMS and NCS plants). However, because the rest of the plant is normal, a CMS plant would not be expected to have increased levels of AOX in ear shoots.
Figure 6 shows the steady state levels of AOX and additional stress-related marker proteins in ear shoots and tassels of NCS and CMS mutants. In contrast to the NCS mutants, CMS mutants did not express AOX in ear shoots (Figure 6A). However, in preemergent tassels of each of the CMS types, increased AOX levels were found (C > T >> S). Only low amounts of AOX were detected in tassels of fertile plants, which had the same nuclear background as the CMS plants tested (Figure 6B, lane N). Also, AOX was detected only at low levels in tassels of the normal relatives of NCS6 mutants (Figure 6C, lane 6N). Male florets of NCS mutants appeared to accumulate the same AOX proteins as their respective ear shoots: 34 kD in NCS2 and 36 kD in NCS6 (Figure 6C, lanes 2 and 6). However, significant levels of 34-kD AOX also were present in male florets of NCS2 normal relatives that have CMS-T cytoplasm.
Figure 6.
Comparative Analysis of AOX and Other Stress-Related Proteins in CMS and NCS Mutants.
Immunoblots are shown from mitochondrial proteins isolated from ear shoots of CMS mutants, types T, C, and S, and a fertile control, N (A), and mitochondrial proteins isolated from preemergent tassels of CMS (B) and NCS (C) mutants. The antisera used are indicated at right. Gels were loaded uniformly with 50 μg protein/lane.
Results of RNA gel blot analyses demonstrated that different CMS mutants contained mainly aox2 mRNA in their tassels (Figure 7A). By contrast, the pattern of aox transcripts found in tassels of the NCS mutants depended on the specific type of defect. In the CMS-T derivative, NCS2, there was an additional increase in the expression of aox2 in tassels over that of its non-NCS but male-sterile relative (Figure 7B, lanes 2N and 2). This “superinduction” of aox at the mRNA level correlated with the significant increase in the level of AOX protein in NCS2 tassels. The major transcripts in the anthers of the NCS6 mutant corresponded to those from the aox3 gene (Figure 7B, lane 6), which correlated with the observed induction of the 36-kD AOX protein. Considering the low level of aox1 transcripts in ear shoots, it is interesting that aox1 expression was found to be induced in tassels of NCS4 and NCS6 mutants and to be superinduced in the NCS2 mutant (Figure 7B).
Figure 7.
RNA Gel Blot Analysis of aox Gene Expression in Preemergent Tassels of CMS and NCS Mutants.
Total RNAs were isolated from tassels of CMS (A) and NCS (B) mutants and their normal relatives. Hybridization was with the aox1-, aox2-, and aox3-specific riboprobes indicated at right. The exposure of the aox2 hybridization shown in (B) is short relative to that of the other probes, so that the induction of aox2 transcripts in NCS2 can be seen clearly. This figure depicts the relative levels of specific aox transcripts in the different NCS mutants. The gel at bottom shows ethidium bromide staining of the 18S RNA as a loading control. UTR, untranslated region.
Analysis of Oxidative Stress in NCS Mutants
In nonphotosynthetic plant tissues, mitochondria are considered to be a major source of ROS, superoxide, and H2O2 (Maxwell et al., 1999). ROS production is known to increase dramatically when oxidative phosphorylation is damaged, which can cause oxidative stress (reviewed by Møller, 2001). Thus, respiratory deficiencies associated with mitochondrial mutations might be expected to induce antioxidant defense systems in addition to AOX. ROS-scavenging enzymes such as mitochondrial Mn-dependent superoxide dismutase (MnSOD) and cytosolic CuZnSOD might be involved in enhancing the survival of NCS plants.
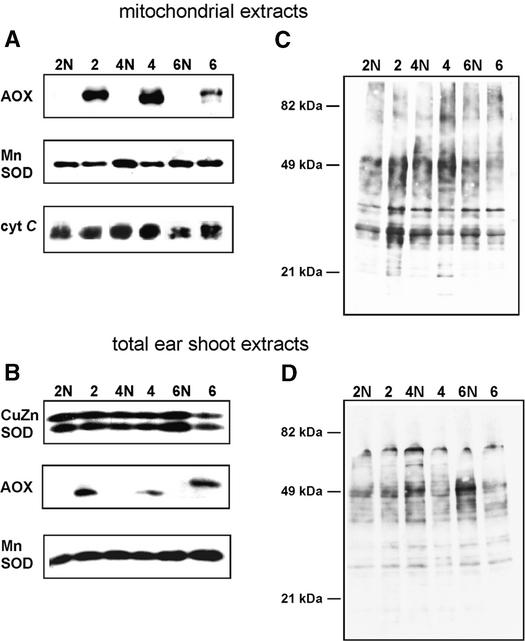
Unlike AOX, mitochondrial MnSOD showed no significant difference between mutant (NCS or CMS) and normal mitochondria in ear shoots and tassels (Figures 6, 8A, and 8B). Cytosolic CuZnSOD, which is induced by oxidative stress in plants (Kliebenstein et al., 1998), was analyzed in total protein extracts from maize ear shoots (Figure 8B) and did not appear to be increased in NCS mutants. Cytochrome c, which, when lost from mitochondria, serves as an indicator of impending apoptosis (Balk et al., 1999), was found at normal levels in mitochondria of all NCS mutants (Figures 6 and 8A).
Figure 8.
Analysis of Stress-Related Proteins and Protein Oxidation in Ear Shoots of NCS Mutants.
(A) Immunoblots of mitochondrial proteins (50 μg/lane) from each NCS mutant (lanes 2, 4, and 6) and their corresponding normal relatives (lanes 2N, 4N, and 6N) reacted with antisera as indicated.
(B) Immunoblot analysis of the cytosolic CuZnSOD and mitochondrial MnSOD enzymes in total protein extracts from ear shoots (30 μg/lane). The antiserum against Arabidopsis CuZnSOD stained 16- and 15-kD proteins, corresponding to the cytosolic SOD2 and SOD4 isozymes described previously for maize (Baum and Scandalios, 1981). Antisera against AOX used in a subsequent probing of the same immunoblot showed that the NCS samples used had high expression of AOX.
(C) and (D) Oxidation of mitochondrial and total proteins in ear shoots of NCS mutants. Immunoblots of mitochondrial ([C]; 25 μg/lane) and total tissue ([D]; 10 μg/lane) protein extracts after dinitrophenyl derivatization were reacted with anti-DNP antibodies to visualize oxidatively modified proteins.
Mitochondrial and total proteins of NCS mutants were tested for ROS-mediated oxidation, which is a reliable indicator of ROS accumulation (Berlett and Stadtman, 1997). Immunoblot analyses of oxidized proteins of mitochondrial and total extracts from ear shoots of NCS mutants did not detect increases in the level of protein oxidation, which would have been expected if there were excessive production of ROS within the mitochondria or cells (Figures 8C and 8D).
To determine if NCS plants have ROS levels high enough to trigger the induction of oxidative stress–responsive transcripts, the levels of three known H2O2-inducible transcripts were examined in the ear shoots of NCS mutants. Maize orthologs of cytosolic glutathione S-transferases type I (gstI and gstIIIb; McGonigle et al., 2000) and ascorbate peroxidase (apx; Shigeoka et al., 2002) were chosen for analysis (Table 1). The maize gstIIIb and apx genes are related closely to Arabidopsis and tobacco genes that serve as markers for the oxidative burst response (Alvarez et al., 1998; Mittler et al., 1999). Induction by H2O2 was confirmed for all three genes in maize seedlings (Figure 9A, gels at right). Levels of these transcripts were similar between NCS4 and NCS6 mutants and their related normal plants (Figure 9A, lanes 4 and 6), whereas all three transcripts were increased in the NCS2 mutant (Figure 9A, lane 2). These results suggest that H2O2 levels are not increased in NCS4 and NCS6 ear shoot tissues and that H2O2 is not the primary inducer of aox expression in these mutants. Although all three mRNAs were increased in NCS2 samples, suggesting that H2O2 overproduction might occur, the lack of extensive protein oxidation in NCS2 does not support severe oxidative stress conditions in this mutant.
Table 1.
Maize Clones and PCR Primers for the Preparation of Hybridization Probes
| Maize Clone NCBI Accession No. |
Amino Acid Sequence Accession No. |
Protein Homolog NCBI Accession No. |
Amino Acid Identity (%) |
Nucleotide Sequence Accession No. |
PCR Primers |
|---|---|---|---|---|---|
| Cytosolic GSTI M16901 |
AAA33470 | Forward, 5′-CACCATGGCTCCGATGAAGCTG-3′; reverse, 5′-GTTTCAAGCAGATGGCTTCATCAGG-3′ |
|||
| Cytosolic GSTIIIb X04455 |
CAB38119 | Arabidopsis cytosolic GST |
44 | Z26426 | Forward, 5′-CCATGGCGCCTCTGAAGCTGTACG-3′; reverse, 5′-GCAACGCAAGGCGAGGTCAAGC-3′ |
| Cytosolic APX Z34934 |
CAA84406 | Tobacco cytosolic APX AAA86689 |
81 | U15933 | Forward, 5′-ACCATGAAGAACCCCGTCGAG-3′; reverse, 5′-GCTTCATATCAAACCTTCTCCG-3′ |
| StUCP homolog AI745875 |
Potato UCP CAB60277 |
84 | Y11220 | Forward, 5′-GAATGATGGGTGAYTCARCSTACA-3′; reverse, 5′-CAGGGATCGTTACCGATCAGAG-3′ |
|
| TaUCP homolog AW216323 |
Wheat UCP1a BAB16384 |
89 | AB042428 | Forward, 5′-GAATGATGGGTGAYTCARCSTACA-3′; reverse, 5′-AGACGTAGGTGGCAGTATGGCA-3′ |
Figure 9.
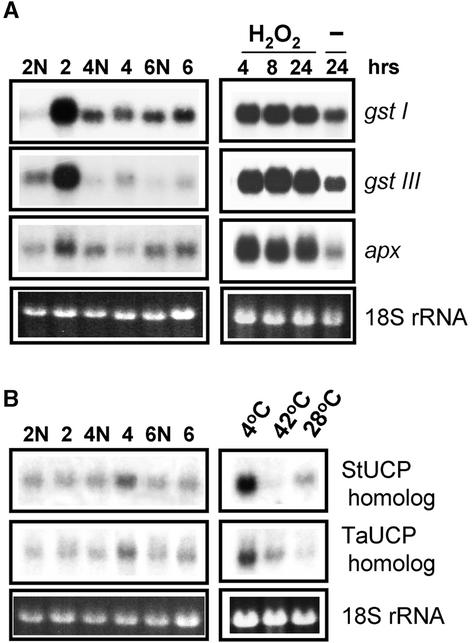
RNA Gel Blot Analysis of the Expression of H2O2-Responsive Genes and Genes That Encode UCPs in Ear Shoots of NCS Mutants.
(A) Total RNA from ear shoots of NCS mutants (lanes 2, 4, and 6) and normal relatives (lanes 2N, 4N, and 6N) were hybridized with probes specific to gstI, gstIIIb, and apx (described in Table 1). The H2O2 inducibility of the genes assayed is shown at right after hybridization with RNA samples from normal maize seedlings treated with 10 mM H2O2 for the times indicated or after no treatment (−). The gel at bottom shows ethidium bromide staining of the 18S RNA as a loading control.
(B) Total NCS RNAs similar to those described above were hybridized with the riboprobes specific to StUCP and TaUCP. The cold inducibility of the ucp gene probes used is shown at right by hybridization to RNAs from normal maize seedlings treated as follows: 4°C cold shock for 48 h; 42°C heat shock for 24 h; and 28°C control for 24 h. Riboprobes used (Table 1) are indicated at right. The gel at bottom shows ethidium bromide staining of the 18S RNA as a loading control.
The uncoupling proteins (UCPs) constitute another energy-dissipating pathway in mitochondria. UCPs facilitate fatty acid–dependent proton reuptake through the inner mitochondrial membrane, causing dissipation of the electrochemical gradient as heat (Garlid et al., 1996). Like mammalian UCPs, plant UCP homologs can be regulated by cold, and like AOX, they are postulated to decrease ROS production (Laloi et al., 1997; Kowaltowski et al., 1998; Sluse and Jarmuszkiewicz, 2000). Figure 9B shows the expression of two maize ucp genes that were identified by their amino acid similarities to cold-inducible potato UCP (StUCP; Nantes et al., 1999) and cold-insensitive wheat UCP1a (TaUCP; Murayama and Handa, 2000). Interestingly, unlike wheat ucp, the maize TaUCP homolog could be upregulated by cold (Figure 9B, gels at right). In maize ear shoots, transcripts of both genes were found to be present at very low levels, particularly the TaUcp homolog. (Long exposures of the films were necessary to achieve the results seen in Figure 9B.) Slight increases of both types of ucp mRNAs were detected only in NCS4, a translation-defective mitochondrial mutant. Thus, the ucp transcripts did not show significant changes compared with the aox transcripts in the same mutants.
DISCUSSION
In this study, we characterized the RNA and protein products of three maize aox genes that encode AOX and examined their expression in plants carrying mitochondrial mutations. Unexpectedly, our data showed that the induction of individual aox genes in the mutants correlated with the location of the mitochondrial defect. The CI-deficient NCS2 and CIV-deficient NCS6 mutants had distinctive protein isoforms of AOX, corresponding to aox2 and aox3 gene products, respectively, whereas the translation-defective NCS4 mutant had both types of AOX, with a strong bias toward AOX2. This differential induction of aox genes was seen in all tested nonphotosynthetic tissues of NCS mutants, although the overall levels of AOX protein varied (ear shoots > tassels > roots > seedling roots). In contrast to the differential expression of aox2 and aox3, aox1 appeared to be tissue specific, with no detectable transcripts in ear shoots but relatively high levels in young tassels of all of the NCS mutants. aox1 also was found to be induced in the developing anthers of CMS-T mutants, although aox2 was the predominant form in all of the CMS mutants.
In plants, AOX is detected typically as a set of isoforms (Elthon and McIntosh, 1987; Millenaar et al., 2001), and in soybean, there is strong evidence that AOX isoforms correspond to products of different genes that can be expressed differentially during cotyledon development (Finnegan et al., 1997; McCabe et al., 1998). The data presented here show the induction of specific AOXs in different mitochondrial mutants.
The results with the NCS mutants were corroborated strongly by studies using specific inhibitors of mitochondrial respiratory complexes. Treatment of normal maize seedlings with inhibitors of CI (rotenone) or CIV (KCN) also led to the predominant expression of aox2 or aox3 genes, respectively. Inhibition of CIII by antimycin A resulted in the induction of all three aox transcripts, but the aox3 increase was significantly higher. Thus, aox3 expression appears to be especially responsive to the over-reduced state of the ubiquinone pool that may be achieved by KCN or antimycin A treatment. These data confirm that the differential expression of aox genes is sensitive to the respiratory status of the ETC.
The maize AOX1 and AOX2 proteins are similar to the majority of plant AOX proteins, which contain a conserved Cys. The product of the aox3 gene, which is the major AOX that accumulates in the NCS6 mutant, represents an example of AOX with Ser substituted for the conserved Cys. Although naturally occurring plant aox genes with predicted Cys-to-Ser substitutions have been reported to exist previously (rice AOX1b [Ito et al., 1997] and a tomato AOX AOX [Djajanegara et al., 1999]), no data on their expression have been published. Our finding that the AOX3 isoform is accumulated selectively upon permanent CIV deficiency in maize indicates that such genes are functional in plants. When the conserved regulatory Cys in soybean or Arabidopsis aox genes was converted to Ser by site-directed mutagenesis, the resulting protein was found to be activated by succinate instead of pyruvate (Djajanegara et al., 1999). None of the known fungal or algal AOXs are activated by pyruvate, which is consistent with the absence of the corresponding Cys in their N-terminal domains (Joseph-Horne et al., 2000; Dinant et al., 2001). The enzymatic properties and regulation of the maize Cys-less AOX3 remain to be determined. Transgenic plants that overproduce AOX3 would be useful for the characterization of this isoform in vivo.
Recent expression studies of two energy-dissipating pathways in plant mitochondria, represented by AOX and UCP, gave conflicting results regarding whether they function together or sequentially (Sluse and Jarmuszkiewicz, 2000; Considine et al., 2001). We observed only low levels of ucp transcripts in the NCS mutants, with some increase in NCS4, suggesting that AOX is used preferentially in maize when the ETC is blocked constitutively.
AOX was first discovered as the activity responsible for noncoupled respiration during thermogenesis associated with the attraction of insect pollinators in aroid plants (Meeuse, 1975). However, it is not known what role this enzyme serves in other plants, including maize. One compelling idea is that AOX can control the production of free radicals by stabilizing the ratio of reduced ubiquinone to total ubiquinone (Purvis and Shewfelt, 1993; Wagner, 1995; Wagner and Moore, 1997; Millenaar et al., 1998). Indeed, isolated mitochondria were found to produce more H2O2 when the alternative pathway was inhibited (Popov et al., 1997). Experiments with transgenic tobacco confirmed an inverse relationship between AOX expression and ROS levels in plant cells (McIntosh et al., 1998; Maxwell et al., 1999).
In this study, we found no significant difference in the levels of mitochondrial (MnSOD) and cytosolic (CuZnSOD) ROS-scavenging enzymes, either between NCS mutants and normals or between different NCS mutants, suggesting that the mutants are not being exposed to endogenous oxidative stress. Correspondingly, there was no increase in the oxidation of mitochondrial and cytosolic proteins. Also, there was no leakage of cytochrome c from mitochondria that might indicate apoptotic events (Balk et al., 1999; Sun et al., 1999) in any NCS mutant. To detect changes in ROS that could be physiologically significant, we tested the expression of marker genes that are known to be highly responsive to H2O2. Transcripts of all three genes tested (gstI, gstIII, and apx) were found to be induced only in NCS2 and not in NCS4 or NCS6. Although we observed an increased expression of H2O2-responsive transcripts in the NCS2 mutants, this does not necessarily mean that the induction of aox2 is activated by H2O2 or ROS. When tobacco cell cultures were incubated with antimycin A, the induction of aox transcripts occurred faster than when H2O2 was applied and faster than ROS accumulation (Maxwell et al., 2002). In NCS mutants with permanent respiratory defects, potential mitochondrial signals could be emitted continuously, leading to the constitutive expression of aox genes in the nucleus. The high level of AOX apparently prevents oxidative stress in the NCS plants during their life cycle. Conversely, tobacco cell cultures with antisense suppression of AOX (AS8) were found to have excessive ROS levels in mitochondria (Maxwell et al., 1999), as well as altered growth under phosphate limitation (Parsons et al., 1999; Yip and Vanlerberghe, 2001). Thus, AOX appears to be vital for the survival and development of plant cells with high levels of mutant mitochondria.
Insights into the general role of the AOX in maize may be provided by comparative analyses of aox expression in NCS and CMS mutants. A large increase in the amount of AOX2 protein and its mRNA has been found in developing anthers of CMS-T, in which the expression of a chimeric mitochondrial protein, T-URF13, is associated with the depolarization of mitochondrial membranes in the tapetum during microsporogenesis (Rhoads et al., 1995). However, in the CMS-T derivative NCS2 mutants, introduction of the additional nad4 mutation caused a further increase in aox2 expression. The additive effect of CMS-T and nad4 mutations on aox2 induction indicates that there might be distinct mechanisms of aox2 regulation, one associated with alterations in membrane integrity and another associated with the inhibition of respiratory CI.
Although the actual molecular basis of male sterility in CMS-C of maize is not understood, it is known that CMS-C sterility is sporophytic and that it acts relatively early during pollen maturation (reviewed by Newton and Gabay-Laughnan, 1998). The large induction of aox in the young anthers reflects this early event. CMS-S acts later in pollen development and is gametophytic in its mode of action—that is, the mitochondrial defect leading to pollen abortion is expressed within the developing pollen grain itself. Although AOX induction is seen in the young tassels, it is less than in the CMS-T and CMS-C mutants.
Sterile tassels of all CMS mutants contain variable amounts of AOX2, whereas their ear shoots, which have functionally normal mitochondria, have no detectable AOX. These data also indicate that expression of AOX to high levels occurs under conditions of respiratory deficiency or membrane depolarization in maize. Thus, as a reliable marker of mitochondrial dysfunction, AOX could be used to identify new respiration-impaired mutants in maize.
Signals from defective mitochondria must be transmitted to the nucleus, resulting in the induction of stress-responsive AOX proteins. Our studies indicate that this signaling can be sensitive to the actual site of disruption within the ETC, resulting in the induction of different AOX isoforms encoded by separate genes. The NCS and CMS mutants of maize could serve as model systems for the analysis of mitochondrial–nuclear signaling pathways.
METHODS
Plant Material and Mitochondrial Isolation
The NCS2 mutation arose spontaneously in maize (Zea mays) plants carrying the cytoplasmic male sterility (CMS) T cytoplasm in a WF9-derived nuclear background (Coe, 1983; Newton and Coe, 1986). The NCS4 and NCS6 mutations were found in the WF9 nuclear genotype and a fertile revertant maize mitochondrial genotype of the CMS-S group (Lauer et al., 1990; Newton et al., 1996). Although plants carrying the mitochondrial deletions are heteroplasmic, the mutations themselves are exceptionally stable, with no alterations detectable over many years of propagation. Normal plants that no longer give rise to defective progeny are found occasionally within mutant families and are the sources of the normal relatives used in these studies. Thus, the nuclear backgrounds of all of the NCS mutants and their normal controls are very similar, and the male parent in the final crosses was A619. The normal and CMS-T, -C, and -S cytoplasms all were present in the B37 inbred nuclear background. The following plant materials were used as sources of mitochondria: seedling and mature plant roots, unpollinated ear shoots, and preemergent tassels (4 to 7 days before pollen shedding, when tassels are still within the whorl of leaves at the top of the plant). All samples were from field-grown plants in Missouri, with the exception of mature roots, which were from potted greenhouse plants. Mitochondria were isolated and purified on Suc step gradients as described previously (Cooper and Newton, 1989).
Inhibitor Studies
Five-day-old etiolated maize seedlings with normal cytoplasm (type NB in the Mo17 and A619 lines) were grown in paper rolls and then transferred to trays in which the roots were submerged in the following solutions: rotenone (40 μM), KCN (10 mM), antimycin A (20 μM), or H2O2 (10 mM; changed twice at 4 and 8 h). Rotenone and antimycin A were dissolved in DMSO and subsequently diluted with water (to a final concentration of 0.1% DMSO). After 4, 8, and 24 h of incubation at 28°C in the dark, samples of seedling roots were processed for the isolation of total RNA. Control seedlings were incubated under the same conditions in water (to parallel experiments with H2O2) or in 0.1% DMSO (controls for respiratory inhibitors).
Immunoblot Analysis of Mitochondrial and Total Cellular Proteins
Total protein extracts of mitochondria were prepared according to Karpova and Newton (1999). Protein concentrations were determined by the Lowry method (Ausubel et al., 1992). For total protein extracts from ear shoots, tissue was powdered in liquid N2, homogenized in 6 mM Tris-HCl, pH 8.0, 10 mM NaCl, 0.1 mM EDTA, 0.25 M Suc, 0.5 mM aminocaproic acid, and 1 mM phenylmethylsulfonyl fluoride, sonicated, treated with SDS (final concentration of 2% [w/v]) at 100°C for 5 min, and centrifuged (16,000g for 30 min), and the supernatants were used for analysis. SDS-PAGE using Tris-Gly buffers was performed according to Laemmli (1970). For separation, 12 to 18% (w/v) gradient or 15% (w/v) polyacrylamide gels were used; stacking gels were uniformly 6% (w/v). Prestained protein standards (6.5 to 200 kD; Bio-Rad) were used for molecular mass estimation. Proteins were transferred to polyvinylidene difluoride membranes (Schleicher & Schull [Keene, NH] or Millipore [Bedford, MA]) by electroblotting as described previously (Karpova and Newton, 1999). The following antisera were used in this study: polyclonal antisera raised in rabbits against the 49-kD subunit of Neurospora crassa complex I (NAD7; provided by T. Friedrich and U. Schulte, Institut fur Biochemie, Düsseldorf, Germany); maize COX2 (provided by C.S. Levings III, North Carolina State University, Raleigh), mitochondrial malate dehydrogenase (Newton, 1980), and ADP/ATP translocator (provided by C. Leaver, Oxford University, UK); and Arabidopsis mitochondrial Mn-dependent superoxide dismutase and cytosolic CuZn-dependent superoxide dismutase (provided by D. Kliebenstein, Cornell University, Ithaca, NY) (Kliebenstein et al., 1998). Monoclonal antibodies included antibodies to Sauromatum guttatum AOX (Elthon et al., 1989) and cytochrome c (clone 7H8.2C12; PharMingen, San Diego, CA). For detection, anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibodies and the enhanced chemiluminescence kit (DuPont–New England Nuclear) were used.
Analysis of Protein Oxidation
Sulfhydryl oxidation of mitochondria was conducted essentially as described by Rhoads et al. (1998). Aliquots of mitochondria in PBS buffer were treated with 3 mM diamide at 4°C for 1 h. Sulfhydryl-oxidized mitochondria and control (untreated) samples were extracted for proteins with the buffer containing either no reductant or 3% (v/v) mercaptoethanol (final concentration of 75 mM Tris-HCl, pH 6.8, and 3% [w/v] SDS). Analysis of protein oxidation was performed using the OxyBlot kit (Oncor, Gaithersburg, MD). Protein extracts of mitochondria or total tissue (ear shoots) were treated with dinitrophenylhydrazine to convert the carbonyl groups of oxidized proteins to dinitrophenylhydrazone. The dinitrophenylhydrazone-containing proteins were detected by protein gel blot hybridization with anti-DNP primary antibodies. Each experiment was repeated three times with independent samples of NCS mitochondria.
Sequencing of Maize aox cDNA Clones
Three cDNA clones in pSport I that showed homology with a fragment of the maize aox gene were provided by Pioneer Hi-Bred International (Johnston, IA). All clones were sequenced using the ABI dRhodamine terminator sequencing kit and the ABI 3700 automated DNA sequencer (Perkin-Elmer Applied Biosystems). Sequence analysis demonstrated that clones CSSTN34R and CHSSG24R represent full-length cDNAs (ZmAOX1 and ZmAOX2, respectively). Clone CTUTY62R (ZmAOX3) did not contain the 5′ untranslated region (UTR) and ∼25% of the open reading frame (ORF). To obtain the 5′ UTR and the 5′ end of the aox3 ORF, 5′ rapid amplification of cDNA ends (GeneRacer kit; Invitrogen, Carlsbad, CA) was performed with total RNA (2.5 μg) from the NCS6 mutant as a template. The gene-specific primer from the partial aox3 cDNA sequence was 5′-GTC-GGCGCGTGGTGCTTCCCTAT-3′. The PCR fragment (∼550 bp) was cloned directly into the pGEMT-Easy vector. A DraIII fragment containing the 5′ RACE insert and a DraIII fragment of the original ZmAOX3 were ligated together to complete the aox3 cDNA. The genes were named according to the standard nomenclature used by maize geneticists (www.agron.missouri.edu).
RNA Gel Blot Analysis
Total RNAs were isolated from unpollinated ear shoots and preemergent tassels and seedling roots using the guanidine isothiocyanate method (Chomczynski and Sacchi, 1987), separated by formaldehyde-denaturing electrophoresis in 1.4% (w/v) agarose, and transferred to a Hybond+ membrane (Amersham). Loading (∼5 μg) was controlled by staining gels with ethidium bromide. For determination of transcript size, RNA standards (0.28 to 6.58 kb; Promega) were used. Hybridization was performed with 32P-UTP–labeled antisense riboprobes in a phosphate and 50% formamide mixture at 58°C. Blots were washed twice in 1 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.5% SDS at 65°C and once in 0.1 × SSC and 0.5% SDS at 70°C. For these experiments, three different sets of total RNAs isolated from NCS mutants were used.
Hybridization Probes
aox-ORF Probe
A 420-bp fragment of the maize aox gene was obtained by Taq-polymerase–directed PCR with B37 maize total DNA as a template. Conditions were as follows: 96°C for 5 min; 35 cycles of 94°C for 40 s, 52°C for 40 s, and 72°C for 1 min; and 72°C for 5 min. Primer sequences were as follows: AOX probe forward, 5′-TGATGCTGG-AGACGGTGGCT-3′; AOX probe reverse, 5′ -GACGGTGACTAC-GTCCTTGA-3′. The PCR fragment was cloned into the pGEMT-Easy vector (Promega) and sequenced. It showed 98% identity to the previously published maize aox partial sequence.
aox Gene–Specific Probes
To generate the antisense riboprobes, the aox cDNA clones were digested with PvuII (ZmAOX1) or HincII (ZmAOX2 and AOX3) and were labeled with 32P-UTP (3000 Ci/mmol) using SP6 RNA polymerase. The aox1-specific 484-bp probe consisted of the 3′ UTR and the C-terminal part of the ORF, including the RADEAHHRDVNH region (epitope). The aox2-specific 435-bp probe and the aox3-specific 316-bp probe included the 3′ UTR and the C terminus of the ORF immediately downstream of the epitope.
Other Hybridization Probes
Maize cDNA clones homologous with Arabidopsis gst (clone ATTS1553), tobacco apx, potato ucp, and wheat ucp1a were identified in the EST library of the ZmDataBase (www.zmdb.iastate.edu) or in GenBank by amino acid homology with the corresponding proteins (Table 1). Fragments of the ORFs (gstI and gstIIIb), or fragments containing 3′ UTRs joined to the 3′ ends of the ORF (apx, StUCP, and TaUCP), were obtained by reverse transcriptase–mediated PCR with total RNA from normal maize seedlings as a template and were cloned into pGEMT-Easy. Although GSTI and GSTIIIb proteins are homologous (50% identity) and belong to the same family of maize GSTs (type I; McGonigle et al., 2000), there was no nucleotide homology detected for the gstI and gstIII PCR fragments used for analysis.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the complete sequences of the maize aox cDNAs described in this article are AY059646 (aox1), AY059647 (aox2), and AY059648 (aox3). The accession number for the maize aox gene fragment is AF040566. The accession numbers for the complete sequences of AOX proteins that were used in the protein alignments are as follows: Q39219 (Arabidopsis AOX1a), O23913 (Arabidopsis AOX1b), O22048 (Arabidopsis AOX1c), O22049 (Arabidopsis AOX2), BAA28773 (rice AOX1a), BAA28774 (rice AOX1b), BAB71944 (rice AOX1c), BAB88645 (wheat AOX1a), and BAB88646 (wheat AOX1c).
Acknowledgments
We thank T. Friedrich and U. Schulte, C.S. Levings III, C. Leaver, and D. Kliebenstein for generous gifts of antisera. We are very grateful to M. Guo (Pioneer Hi-Bred International) for providing the maize AOX cDNA clones and D. Bergstrøm (University of Missouri, Columbia) for DNA automated sequencing. This research was supported by grants from the National Science Foundation and the Illinois-Missouri Biotechnology Alliance (to K.J.N.) and from Pioneer Hi-Bred International and the National Science Foundation Experimental Program to Stimulate Competitive Research (to T.E.E.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005603.
References
- Alvarez, M.E., Pennell, R.I., Meijer, P.-J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1992). Short Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Balk, J., Leaver, C.J., and McCabe, P.F. (1999). Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463, 151–154. [DOI] [PubMed] [Google Scholar]
- Baum, J.A., and Scandalios, J.G. (1981). Isolation and characterization of the cytosolic and mitochondrial superoxide dismutases of maize. Arch. Biochem. Biophys. 206, 249–264. [DOI] [PubMed] [Google Scholar]
- Berlett, B.S., and Stadtman, E.R. (1997). Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 272, 20313–20316. [DOI] [PubMed] [Google Scholar]
- Bhattramakki, D., and Elthon, T.E. (1997). Exogenous NAD(P)H dehydrogenases of plant mitochondria. Recent Res. Dev. Plant Physiol. 1, 93–104. [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 161, 156–159. [DOI] [PubMed] [Google Scholar]
- Coe, E. (1983). Maternally inherited abnormal plant types in maize. Maydica 28, 151–167. [Google Scholar]
- Conley, C.S., and Hanson, M.R. (1995). How do alterations in plant mitochondrial genomes disrupt pollen development? J. Bioenerg. Biomembr. 27, 447–457. [DOI] [PubMed] [Google Scholar]
- Considine, M.J., Daley, D.O., and Whelan, J. (2001). The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiol. 126, 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine, M.J., Holtzapffel, R.C., Day, D.A., Whelan, J., and Millar, A.H. (2002). Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 129, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, P., and Newton, K.J. (1989). Maize nuclear background regulates the synthesis of a 22-kDa polypeptide in Zea luxurians mitochondria. Proc. Natl. Acad. Sci. USA 86, 7423–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant, M., Baurain, D., Coosemans, N., Joris, B., and Matagne, R.F. (2001). Characterization of two genes encoding the mitochondrial alternative oxidase in Chlamydomonas reinhardtii. Curr. Genet. 39, 101–108. [DOI] [PubMed] [Google Scholar]
- Djajanegara, I., Holtzapffel, R., Finnegan, P.M., Hoefnagel, M.H., Berthold, D.A., Wiskich, J.T., and Day, D.A. (1999). A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett. 454, 220–224. [DOI] [PubMed] [Google Scholar]
- Elthon, T.E., and McIntosh, L. (1987). Identification of the alternative terminal oxidase of higher plant mitochondria. Proc. Natl. Acad. Sci. USA 84, 8399–8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon, T.E., Nickels, R.L., and McIntosh, L. (1989). Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89, 1311–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, P.M., Whelan, J., Millar, A.H., Zhang, Q., Smith, M.K., Wiskich, J.T., and Day, D.A. (1997). Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol. 114, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, P.M., Wooding, A.R., and Day, D.A. (1999). An alternative oxidase monoclonal antibody recognises a highly conserved se-quence among alternative oxidase subunits. FEBS Lett. 447, 21–24. [DOI] [PubMed] [Google Scholar]
- Garlid, K.D., Orosz, D.E., Modriansky, M., Vassanelli, S., and Jezek, P. (1996). On the mechanism of fatty acid-induced proton transport by mitochondrial uncoupling protein. J. Biol. Chem. 269, 2615–2620. [DOI] [PubMed] [Google Scholar]
- Gu, J., Miles, D., and Newton, K.J. (1993). Analysis of leaf sectors in the NCS6 mitochondrial mutant of maize. Plant Cell 5, 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres, S., Sabar, M., Lelandais, C., Chetrit, P., Diolez, P., Degand, H., Boutry, M., Vedel, F., de Kouchkovsky, Y., and De Paepe, R. (1997). Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc. Natl. Acad. Sci. USA 94, 3436–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiser, C., Kapranov, P., and McIntosh, L. (1996). Genetic modification of respiratory capacity in potato. Plant Physiol. 110, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M.D., and Newton, K.J. (1991). The NCS3 mutation: Genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y., Saisho, D., Nakazono, M., Tsutsumi, N., and Hirai, A. (1997). Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 203, 121–129. [DOI] [PubMed] [Google Scholar]
- Joseph-Horne, T., Babij, J., Wood, P.M., Hollomon, D., and Sessions, R.B. (2000). New sequence data enable modelling of the fungal alternative oxidase and explain an absence of regulation by pyruvate. FEBS Lett. 481, 141–146. [DOI] [PubMed] [Google Scholar]
- Karpova, O.V., and Newton, K.J. (1999). A partially assembled complex I in NAD4-deficient mitochondria of maize. Plant J. 17, 511–521. [Google Scholar]
- Kliebenstein, D.J., Monde, R.A., and Last, R.L. (1998). Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaltowski, A.J., Costa, A.D.T., and Verseci, A.E. (1998). Activation of the potato plant uncoupling mitochondrial protein inhibits reactive oxygen species generation by the respiratory chain. FEBS Lett. 425, 213–216. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Laloi, M., Klein, M., Riesmeier, J.W., Muller-Rober, B., Fleury, C., Bouillaud, F., and Ricquier, D. (1997). A plant cold-induced uncoupling protein. Nature 389, 135–136. [DOI] [PubMed] [Google Scholar]
- Lauer, M., Knudsen, C., Newton, K.J., Gabay-Laughnan, S., and Laughnan, J.R. (1990). A partially deleted mitochondrial cytochrome oxidase gene in the NCS6 abnormal growth mutant of maize. New Biol. 2, 179–186. [PubMed] [Google Scholar]
- Marienfeld, J.R., and Newton, K.J. (1994). The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138, 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, D., Nickels, R.L., and McIntosh, L. (2002). Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence. Plant J. 29, 263–279. [DOI] [PubMed] [Google Scholar]
- Maxwell, D.P., Wang, Y., and McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96, 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe, T.C., Finnegan, P.M., Millar, A.H., Day, D.A., and Whelan, J. (1998). Differential expression of alternative oxidase genes in soybean cotyledons during postgerminative development. Plant Physiol. 118, 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle, B., Keeler, S.J., Lau, S.-M.C., Koeppe, M.K., and O'Keefe, D.P. (2000). A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 124, 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, L., Eichler, T., Gray, G., Maxwell, D., Nickels, R.N., and Wong, Y. (1998). Biochemical and genetic controls exerted by plant mitochondria. Biochim. Biophys. Acta 1365, 278–284. [Google Scholar]
- Meeuse, B.J.D. (1975). Thermogenic respiration in aroids. Annu. Rev. Plant Physiol. 26, 117–126. [Google Scholar]
- Millar, A.H., Wiskich, J.T., Whelan, J., and Day, D.A. (1993). Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett. 329, 259–262. [DOI] [PubMed] [Google Scholar]
- Millenaar, F.F., Benschop, J.J., Wagner, A.M., and Lambers, H. (1998). The role of the alternative oxidase in stabilizing the in vivo reduction state of the ubiquinone pool and the activation state of the alternative oxidase. Plant Physiol. 118, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar, F.F., Gonzalez-Meler, M.A., Fiorani, F., Welschen, R., Ribas-Carbo, M., Siedow, J.N., Wagner, A.M., and Lambers, H. (2001). Regulation of alternative oxidase activity in six wild monocotyledonous species: An in vivo study at the whole root level. Plant Physiol. 126, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Lam, E., Shulaev, V., and Cohen, M. (1999). Signals controlling the expression of cytosolic ascorbate peroxidase during pathogen-induced programmed cell death in tobacco. Plant Mol. Biol. 39, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Møller, I.M. (2001). Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591. [DOI] [PubMed] [Google Scholar]
- Moore, A.L., and Siedow, J.N. (1991). The regulation and nature of the cyanide-resistant alternative oxidase of plant mitochondria. Biochim. Biophys. Acta 1059, 121–140. [DOI] [PubMed] [Google Scholar]
- Murayama, S., and Handa, H. (2000). Isolation and characterization of cDNAs encoding mitochondrial uncoupling proteins in wheat: Wheat UCP genes are not regulated by low temperature. Mol. Gen. Genet. 264, 112–118. [DOI] [PubMed] [Google Scholar]
- Nantes, I.L., Fagian, M.M., Catisti, R., Arruda, P., Maia, I.G., and Vercesi, A.E. (1999). Low temperature and aging-promoted expression of PUMP in potato tuber mitochondria. FEBS Lett. 457, 103–106. [DOI] [PubMed] [Google Scholar]
- Newton, K., and Coe, E.J. (1986). Mitochondrial DNA changes in abnormal growth mutants of maize. Proc. Natl. Acad. Sci. USA 83, 7363–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K.J. (1980). Studies of the Malate Dehydrogenase Isozymes in Zea mays. PhD dissertation (Bloomington, IN: Indiana University).
- Newton, K.J., and Gabay-Laughnan, S. (1998). Abnormal growth and male sterility associated with mitochondrial DNA rearrangements in plants. In Mitochondrial DNA Mutations in Aging, Disease and Cancer, K.K. Singh, ed (Berlin: Springer-Verlag), pp. 365–381.
- Newton, K.J., Knudsen, C., Gabay-Laughnan, S., and Laughnan, J.R. (1990). An abnormal growth mutant in maize has a defective mitochondrial cytochrome oxidase gene. Plant Cell 2, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K.J., Mariano, J.M., Gibson, C.M., Kuzmin, E., and Gabay- Laughnan, S. (1996). Involvement of S2 episomal sequences in the generation of NCS4 deletion mutation in maize mitochondria. Dev. Genet. 19, 277–286. [DOI] [PubMed] [Google Scholar]
- Parsons, H.L., Yip, J.Y.H., and Vanlerberghe, G.C. (1999). Increased respiratory restriction during phosphate-limited growth in transgenic tobacco cells lacking alternative oxidase. Plant Physiol. 121,1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov, V.N., Simonian, R.A., Skulachev, V.P., and Starkov, A.A. (1997). Inhibition of the alternative oxidase stimulates H2O2 production in plant mitochondria. FEBS Lett. 415, 87–90. [DOI] [PubMed] [Google Scholar]
- Purvis, A.C., and Shewfelt, R.L. (1993). Does the alternative pathway ameliorate chilling injury in sensitive plant tissues? Physiol. Plant. 88, 712–718. [DOI] [PubMed] [Google Scholar]
- Rhoads, D.M., Levings, C.S., III, and Siedow, J.N. (1995). URF13, a ligand-gated, pore-forming receptor for T-toxin in the inner membrane of cms-T mitochondria. J. Bioenerg. Biomembr. 27, 437–445. [DOI] [PubMed] [Google Scholar]
- Rhoads, D.M., and McIntosh, L. (1991). Isolation and characterization of a cDNA clone encoding an alternative oxidase protein of Sauromatum guttatum (Schott). Proc. Natl. Acad. Sci. USA 88, 2122–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads, D.M., Umbach, A.L., Sweet, C.R., Lennon, A.M., Rauch, G.S., and Siedow, J.N. (1998). Regulation of the cyanide-resistant alternative oxidase of plant mitochondria: Identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. J. Biol. Chem. 273, 30750–30756. [DOI] [PubMed] [Google Scholar]
- Sabar, M., De Paepe, R., and de Kouchkovsky, Y. (2000). Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 124, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika, H., Ohtsu, K., Hamanaka, S., Nakazono, M., Tsutsumi, N., and Hirai, A. (2002). AOX1c, a novel rice gene for alternative oxidase: Comparison with rice AOX1a and AOX1b. Genes Genet. Syst. 77, 31–38. [DOI] [PubMed] [Google Scholar]
- Saisho, D., Nambara, E., Naito, S., Tsutsumi, N., Hirai, A., and Nakazono, M. (1997). Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol. Biol. 35, 585–596. [DOI] [PubMed] [Google Scholar]
- Sakajo, S., Minagawa, N., Komiyama, T., and Yoshimoto, A. (1991). Molecular cloning of cDNA for antimycin A-inducible mRNA and its role in cyanide-resistant respiration in Hansenula anomala. Biochim. Biophys. Acta 1090, 102–108. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., and Wise, R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. [Google Scholar]
- Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., and Yoshimura, K. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. [PubMed] [Google Scholar]
- Sluse, F.E., and Jarmuszkiewicz, W. (2000). Activity and functional interaction of alternative oxidase and uncoupling protein in mitochondria from tomato fruit. Braz. J. Med. Biol. Res. 33, 259–268. [DOI] [PubMed] [Google Scholar]
- Soole, K.L., and Menz, R.I. (1995). Functional molecular aspects of the NADH dehydrogenases of plant mitochondria. J. Bioenerg. Biomembr. 27, 397–406. [DOI] [PubMed] [Google Scholar]
- Sun, Y.-L., Zhao, Y., Hong, X., and Zhai, Z.-H. (1999). Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 462, 317–321. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Larkin, R.M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14 (suppl.), S327.–S338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach, A.L., Gonzalez-Meler, M.A., Sweet, C.R., and Siedow, J.N. (2002). Activation of the plant mitochondrial alternative oxidase: Insights from site-directed mutagenesis. Biochim. Biophys. Acta 1554, 118–128. [DOI] [PubMed] [Google Scholar]
- Umbach, A.L., and Siedow, J.N. (1993). Covalent and non-covalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 103, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach, A.L., and Siedow, J.N. (1996). The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that alpha-keto acid activation involves the formation of a thioacetal. J. Biol. Chem. 271, 25019–25026. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, G. C., and McIntosh, L. (1994). Mitochondrial electron transport regulation of nuclear gene expression: Studies with the alternative oxidase gene of tobacco. Plant Physiol. 105, 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., and McIntosh, L. (1997). Alternative oxidase: From gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 703–734. [DOI] [PubMed] [Google Scholar]
- Wagner, A.M. (1995). A role for active oxygen species as second messengers in the induction of alternative oxidase gene expression in Petunia hybrida cells. FEBS Lett. 368, 339–342. [DOI] [PubMed] [Google Scholar]
- Wagner, A.M., and Moore, A.L. (1997). Structure and function of the plant alternative oxidase: Its putative role in the oxygen defence mechanism. Biosci. Rep. 17, 319–333. [DOI] [PubMed] [Google Scholar]
- Whelan, J., Millar, A.H., and Day, D.A. (1996). The alternative oxidase is encoded in a multigene family in soybean. Planta 198, 197–201. [DOI] [PubMed] [Google Scholar]
- Yamato, K.T., and Newton, K.J. (1999). Heteroplasmy and homoplasmy for maize mitochondrial mutants: A rare homoplasmic nad4 deletion mutant plant. J. Hered. 90, 369–373. [Google Scholar]
- Yip, J.Y., and Vanlerberghe, G.C. (2001). Mitochondrial alternative oxidase acts to dampen the generation of active oxygen species during a period of rapid respiration induced to support a high rate of nutrient uptake. Physiol. Plant 112, 327–333. [DOI] [PubMed] [Google Scholar]