Abstract
Microarray gene expression profiling was used to examine the role of COP1 in the light control of Arabidopsis genome expression. Qualitatively similar gene expression profiles were observed between wild-type seedlings grown in white light and multiple cop1 mutant alleles grown in the dark. Furthermore, overexpression of the dominant-negative-acting N terminus of COP1 (N282) in darkness produced a genome expression profile similar to those produced by white light and the cop1 mutations. Different cop1 mutant alleles, N282, and light treatment also resulted in distinct expression profiles in a small fraction of the genes examined. In the light, the genome expression of cop1 mutations displayed an exaggerated light response. COP1-regulated genes in the dark were estimated to account for >20% of the genome. Analysis of these COP1-regulated genes revealed that >28 cellular pathways are coordinately but antagonistically regulated by light and COP1. Interestingly, the gene expression regulation attributable to HY5 in the light is included largely within those genes regulated by COP1 in the dark. Thus, this genomic study supports the hypothesis that COP1 acts as a repressor of photomorphogenesis, possibly by controlling the degradation of transcription factors and their target gene expression. The majority of light-controlled genome expression could be accounted for by the negative regulation of COP1 activity.
INTRODUCTION
As sessile organisms, higher plants are characterized by a high degree of developmental plasticity in response to environmental cues, thereby optimizing their development in a way that maximizes their chances of survival and reproduction. Light is an important environmental factor for plant growth and development (Kendrick and Kronenberg, 1994; Deng and Quail, 1999; Neff et al., 2000). Plants undergo dramatic changes in developmental patterns depending on the presence and absence of light in the growth environment. Light is perceived by light quality–specific photoreceptors, including blue/UV-A light receptor cryptochromes (CRY1 and CRY2) and far-red/red light receptor phytochromes (PHYA to PHYE) (Quail et al., 1995; Deng and Quail, 1999; Neff et al., 2000).
For light-controlled development, the photoreceptors perceive and interpret incident light and transduce the signals to modulate light-responsive nuclear genes, which in turn direct appropriate growth and developmental responses (Kendrick and Kronenberg, 1994; Deng and Quail, 1999). Recent results suggest that plant photomorphogenesis involves a regulated change in the expression of up to 30% of the genes in the Arabidopsis genome (Ma et al., 2001), and this massive change in gene expression likely is the result of a transcriptional cascade (Tepperman et al., 2001). Therefore, the contrasting developmental patterns are mediated primarily by coordinated changes in light-regulated gene expression (Terzaghi and Cashmore, 1995; Puente et al., 1996; Ma et al., 2001; Tepperman et al., 2001). Furthermore, different light signals seem to be perceived by distinct photosensory systems and transduced by their signaling pathways to achieve the control of expression of a largely common fraction of the genome (Ma et al., 2001).
A group of 10 pleiotropic COP/DET/FUS loci have been defined by a photomorphogenic seedling phenotype in darkness by their loss-of-function mutations (Chory et al., 1989; Deng et al., 1991; Castle and Meinke, 1994; Misera et al., 1994; Wei and Deng, 1996, 1999). This constitutive photomorphogenic seedling phenotype is characterized not only by morphological changes, such as open, expanded cotyledons, suppression of hypocotyl growth, and anthocyanin accumulation, but also by corresponding changes in subcellular features, such as chloroplast differentiation and derepressed expression for some representatives of normally light-induced genes, such as RbcS, CAB, and CHS (Chory et al., 1989; Deng et al., 1991, 1992; Wei and Deng, 1992; Castle and Meinke, 1994; Misera et al., 1994; Wei et al., 1994). The 10 COP/DET/FUS loci define four biochemical entities: eight loci are required for the COP9 signalosome, and the other three, COP1, DET1, and COP10, exist as separate entities (Serino et al., 1999; Wei and Deng, 1999; Deng et al., 2000). It is assumed that the COP/DET/FUS genes define a pathway designed to repress photomorphogenic development, which acts in the dark to repress the default photomorphogenic development pattern (Wei et al., 1994). Among these four biochemical entities, COP1 has been proposed to act as a limiting step (McNellis et al., 1994a, 1994b). The light repression of COP1 involves a quantitative reduction of COP1 abundance in the nucleus according to the light intensity (von Arnim and Deng, 1994), whereas COP1 activity is regulated negatively by light perceived by multiple photoreceptors (Osterlund and Deng, 1998). In the case of cryptochromes, a direct interaction between COP1 and photoreceptors is involved in the blue light inactivation of COP1 activity (Wang et al., 2001; Yang et al., 2001).
Recent evidence suggests that COP1 may act as a putative E3 ubiquitin ligase within the nucleus, interacting directly with the transcription factor HY5 and targeting its degradation via the 26S proteasome (Osterlund et al., 2000; Schwechheimer and Deng, 2000). Therefore, it is reasonable to hypothesize that COP1 modulates genome expression by targeting key transcription factors for degradation in the nucleus. Thus, a major part of the light control of genome expression could be achieved by regulating the activity of COP1. Because different light signals activate largely common genome expression profiles (Ma et al., 2001), our hypothesis predicts that the genome expression profile attributable to COP1 would essentially overlap the common genome expression regulated by light signals. Furthermore, the COP1-regulated genome expression profile should encompass those genes controlled by COP1-regulated transcription factors.
We used a microarray with 6126 unique genes (Ma et al., 2001) to examine the gene expression profiles controlled by COP1 in the dark and in distinct light conditions during Arabidopsis photomorphogenesis. The expression profiles were compared with the light-regulated genome expression profiles of wild-type seedlings. This comparative analysis helps to define the set of genes affected by COP1 in the dark. Our data are consistent with the hypothesis that a large extent of light-controlled genome expression can be achieved by regulating COP1 activity.
RESULTS
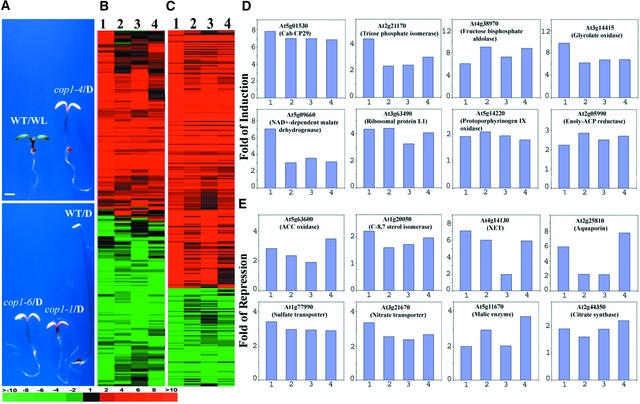
Three cop1 Mutant Alleles Share a Similar Genome Expression Profile in Darkness
To reveal the genome expression profile attributable to COP1 in dark-grown Arabidopsis seedlings, the gene expressions of three representative cop1 mutant alleles (cop1-4, cop1-6, and cop1-1) were examined. Phenotypically, these three alleles represent both weak (cop1-4 and cop1-6) and strong (cop1-1) mutations of COP1 (Figure 1A) (McNellis et al., 1994a). We used a previously reported microarray that includes 9216 EST clones defining 6126 unique expressed genes (Ma et al., 2001) to profile the genome-wide gene expression. For each experiment, we prepared RNA samples from at least two independent biological samples. Furthermore, each experiment included at least four highly reproducible microarray hybridization data sets that met our quality-control standard (correlation coefficient of ratios ≥ 0.95 [Ma et al., 2001]). Examination of the expression ratios of the genes in the microarray between dark-grown cop1 mutants and dark-grown wild-type seedlings (see Methods) revealed that COP1 regulates the expression of a large proportion of the genes (Figure 1B). A total of 1199, 1172, and 1321 of 6126 genes displayed twofold or greater differential expression in the cop1-4, cop1-6, and cop1-1 alleles, respectively, with the stronger cop1-1 mutation having the largest number of differentially expressed genes over the twofold cutoff. Among them, 696, 691, and 680 genes exhibited inducible expression by cop1-4, cop1-6, and cop1-1 mutations, whereas 503, 471, and 641 genes exhibited downregulation in dark-grown cop1-4, cop1-6, and cop1-1 seedlings (Figure 1E and data not shown).
Figure 1.
Genome Expression Profile Comparison between Dark-Grown cop1 Mutants and White Light–Grown Wild-Type Seedlings.
(A) Morphological comparison of continuous white light-grown wild-type, dark-grown cop1, and wild-type Arabidopsis seedlings. All seedlings were 6 days old and photographed at the same magnification. D, darkness; WL, white light; WT, wild-type seedlings. Bar = 1 mm.
(B) Hierarchical clustering display of expression ratios from wild-type seedlings grown under white light versus dark-grown seedlings and dark-grown cop1 mutants versus dark-grown wild-type seedlings. Only those genes that exhibited twofold or greater differential expression in at least one sample pair among the four tested were included for comparison. A total of 2261 genes were included in the cluster.
(C) Overview of the hierarchical cluster display for those genes in our microarray that represent the 28 cellular and metabolic pathways regulated by light and COP1. Only those genes that exhibited twofold or greater differential change in at least one sample pair among the four pairs tested were included for comparison. A total of 311 genes were included in the cluster (see supplementary data at http://plantgenomics.biology.yale.edu/ for details).
(D) Expression profiles of eight representative genes from some of the cellular and metabolic pathways upregulated in dark-grown cop1 mutants.
(E) Expression profiles of eight representative genes from some of the cellular and metabolic pathways downregulated in dark-grown cop1 mutants. XET, xyloglucan endotransglycosylase.
The four bars in each graph of (D) and (E) correspond to the lanes in (B) and (C). Lane 1, expression ratios of white light– and dark-grown wild-type seedlings; lane 2, expression ratios of dark-grown cop1-4 and wild-type seedlings; lane 3, expression ratios of dark-grown cop1-6 and wild-type seedlings; lane 4, expression ratios of dark-grown cop1-1 and wild-type seedlings. The color scale for (B) and (C) is shown at bottom left. See supplementary data at http://plantgenomics.biology.yale.edu/ for more information.
The gene expression profiles in different cop1 mutant alleles were compared further through cluster analysis (Eisen et al., 1998), which groups genes with similar patterns of expression. The white light–induced gene expression profile was included in this cluster analysis for comparison. Only the genes with twofold or greater differential expression in at least one experimental condition (including the white light control) were selected and analyzed. As shown in Figure 1B, there was remarkable similarity in gene expression patterns among cop1 mutant alleles (cop1-4/D versus WT/D, cop1-6/D versus WT/D, and cop1-1/D versus WT/D).
The Gene Expression Profile of Dark-Grown cop1 Mutants Is Similar to That of Light-Grown Wild-Type Seedlings
Among the 2261 genes analyzed (Figure 1B), the vast majority showed qualitatively similar expression (as defined by similar red or green colors of variable shades) by either white light or any of the three cop1 mutations. Compared with white light, only 12, 18, and 18 genes exhibited opposite expression patterns in cop1-4, cop1-6, and cop1-1, respectively, with the twofold cutoff (Figure 1B; see also supplemental data online). It is interesting that among the three cop1 mutations, 13, 8, and 13 genes seemed to exhibit contrasting expression patterns for cop1-4 versus cop1-6, cop1-4 versus cop1-1, and cop1-6 versus cop1-1, respectively. Thus, the similarly small gene numbers with opposite expression among the cop1 mutant alleles and between the cop1 mutants and white light–treated wild-type alleles imply that this low level of contrasting expressed genes may be attributable largely to individual variations.
Despite this qualitatively similar expression pattern among the dark-grown cop1 mutants and the white light–grown wild type, there was quantitatively less differential expression in the dark-grown cop1 mutants (Figure 1B). In fact, for a significant fraction of those genes exhibiting twofold or greater differential expression in the white light–grown wild type, their differential expression values in dark-grown cop1 mutants were just below the twofold cutoff. This is the major reason why the total number of genes with twofold or greater differential expression in the cop1 mutants is in the range of 1200 to 1300, less than the reported ∼1800 genes affected by white light (Figure 1B) (Ma et al., 2001). In fact, if we reduce the cutoff ratios of the cop1 mutants to 1.2, ∼85 to 90% of those genes that exhibited twofold or greater differential expression in the wild type under white light also showed a similar qualitative regulation by the cop1 mutations in darkness (see supplementary data online).
In another indication, ∼88, 88, and 83% of upregulated genes that displayed twofold or greater differential expression in cop1-4, cop1-6, and cop1-1, respectively, were included in the profile of white light–induced genes (see supplementary data online), whereas ∼73, 66, and 67% of downregulated genes that displayed twofold or greater differentiation expression in cop1-4, cop1-6, and cop1-1, respectively, were included in the profile of white light–repressed genes (see supplementary data online). With the arbitrary twofold cutoff, this degree of overlap is approximately the same as that among the cop1 mutant alleles themselves (Figure 1B; see also supplementary data online) and among the wild type grown under distinct light quality conditions (Ma et al., 2001). It should be noted that even these numbers are likely to be an underestimate of the similarity in gene expression patterns. This is because there were fractions of genes that exhibited twofold or greater differential expression in the cop1 mutant that were not included in the white light–regulated genes (with the twofold differential expression cutoff) but that exhibited similar but less than twofold differential expression in white light. In summary, there was very high overlap among the genes differentially expressed in dark-grown cop1 mutants and those regulated by white light in wild-type seedlings. Thus, the loss of COP1 activity has a similar effect on the genome-wide expression pattern triggered by light.
Functional Classification of COP1-Regulated Genes
In a previous study, we showed that a wide range of cellular and biochemical functions, including DNA replication, transcription, translation, metabolism, protein degradation, plant defense, and developmental regulation, are controlled by light (Ma et al., 2001). The genome expression profiling described above suggests that light and COP1 antagonistically regulate the expression of a common set of genes (Figure 1B). Therefore, it is likely that COP1 and light may regulate those same cellular and metabolic pathways in an antagonistic manner. As expected, light and cop1 mutations coordinately upregulated or downregulated at least 17 or 11 cellular or metabolic pathways, respectively (Table 1). Another cluster analysis of selected genes in all 28 pathways presented in our microarray suggested that they share similar differential expression in white light or different cop1 alleles in the dark (Figure 1C). A selected set of sample genes for these representative cellular or metabolic pathways is shown in Figures 1D and 1E. However, most of those pathways exhibited quantitatively variable sensitivities to the light signal and COP1 mutations. For most of the pathways and their genes, the strong white light we used seemed to have a quantitatively stronger effect on their expression than the cop1 mutations. It is worth noting that none of the three alleles we used was a null mutation, because the cop1 null mutants are too retarded in growth to be compared properly.
Table 1.
Summary of the 28 Cellular and Metabolic Pathways and the Effect of COP1 on Three Representative Genes from Each Pathway
| Pathways | Three Representative Genesa |
|---|---|
| Upregulated | |
| Photosynthetic light reactions | At1g15820 (9.2), At1g30380 (8.3), At5g66750 (6.2) |
| Photosynthetic carbon metabolism | At3g54050 (3.4), At3g55800 (8.8), At5g38430 (6.5) |
| Starch synthesis | At5g48300 (3.9), At2g36460 (2.8), At4g38970 (9.1) |
| Suc synthesis | At2g21170 (3.0), At5g46110 (8.7), At3g03250 (6.4) |
| Photorespiration | At2g26080 (9.0), At3g14420 (9.5), At1g68010 (11.4) |
| Glycolysis | At5g63620 (2.5), At3g22960 (1.7), At3g26650 (9.1) |
| Trichloroacetic acid cycle | At5g50850 (2.5), At5g58330 (3.6), At5g66760 (2.5) |
| Fatty acid synthesis | At2g05990 (2.9), At2g26250 (4.1), At3g25110 (2.6) |
| Starch degradation | At4g17090 (3.6), At5g63840 (4.0), At5g12890 (2.1) |
| Cell wall synthesis and cell wall protein | At1g71220 (4.3), At4g19200 (4.3), At4g30140 (2.1) |
| Protein synthesis in chloroplast | At1g17220 (5.0), At1g35680 (4.1), At1g79850 (3.7) |
| Protein synthesis in cytoplasm | At3g55400 (2.1), At2g09990 (3.8), At3g53020 (4.1) |
| Phenylpropanoid biosynthesis | At3g55120 (6.6), At3g51240 (1.8), At5g08640 (4.6) |
| Amino acid synthesis pathways | At2g39800 (10.4), At3g48560 (2.4), At5g04140 (3.8) |
| Chlorophyll synthesis | At1g58290 (3.2), At5g14220 (2.3), At5g63570 (1.4) |
| Transcription factors | At4g23750 (9.8), At5g10280 (6.3), At5g15850 (3.2) |
| Ubiquitin-proteasome pathway | At1g09100 (3.3), At3g17000 (3.4), At3g20060 (9.6) |
| Downregulated | |
| Ethylene biosynthesis | At1g05010 (1.5), At1g62380 (1.2), At5g63600 (3.9) |
| Brassinosteroid biosynthesis | At1g20050 (2.0), At1g20330 (1.2), At1g47290 (1.7) |
| Cell wall degradation | At1g65310 (3.6), At5g02260 (4.1), At1g70710 (7.9) |
| Water transport across tonoplast | At2g37180 (1.8), At2g25810 (7.8), At3g16240 (2.2) |
| Water transport across plasma membrane | At4g17340 (2.1), At1g01620 (2.0), At4g35100 (3.4) |
| Sulfur assimilation | At1g02920 (14.3), At1g59870 (3.7), At3g47340 (2.9) |
| Nitrogen assimilation | At2g26690 (2.2), At3g21670 (3.6), At3g47340 (5.1) |
| Fatty acid oxidation | At1g79750 (2.4), At2g18230 (2.0), At5g11670 (3.8) |
| Glyoxylate cycle | At2g44350 (2.0), At3g21720 (3.3), At5g25880 (3.1) |
| Transcription factors | At1g43160 (4.7), At2g22770 (3.3), At3g57390 (7.5) |
| Ubiquitin-proteasome pathway | At4g14800 (9.0), At5g05780 (2.1), At5g23540 (2.0) |
For each pathway, only three representative genes are listed.
The number in parentheses following each gene identification number represents induction or repression in one of three cop1 mutations examined.
Although the gene expression profiles seen in white light and the cop1 mutations were largely overlapping, there was a notable exception. This involved the gene encoding a key enzyme in the chlorophyll biosynthetic pathway, At4g27440 for protochlorophyllide reductase B, which was upregulated by white light by >26-fold but was downregulated by 6.0-, 4.9-, and 6.7-fold in cop1-4, cop1-6, and cop1-1 seedlings, respectively. This is in contrast to the three genes encoding enzymes that act upstream in the chlorophyll biosynthesis pathway (At1g58290 for glutamyl-tRNA reductase, At5g14220 for protoporphyrinogen IV oxidase, and At5g63570 for glutamate-1-semialdehyde aminotransferase), all of which were induced in white light–grown seedlings as well as in all three cop1 mutants in darkness. This difference may be related to the fact that light is required for reactions catalyzed by protochlorophyllide reductase; thus, At4g27440's expression may exhibit a large feedback regulation by light per se (Su et al., 2001) but not by the cop1 mutations.
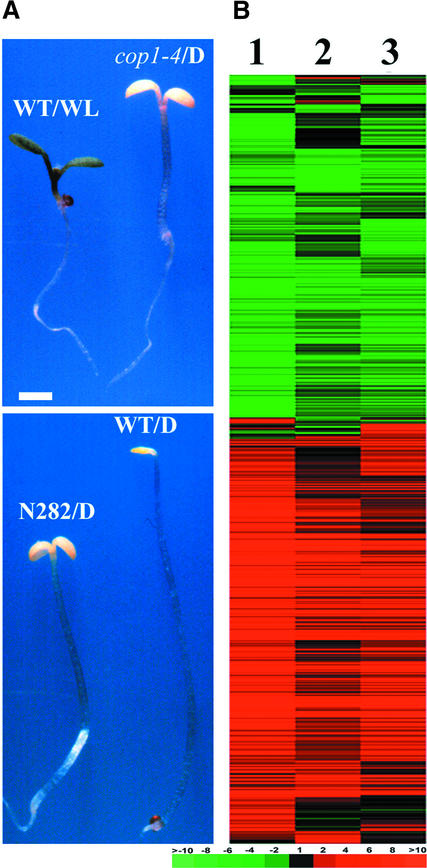
Dominant Interference of COP1 Activity by Overexpression of the N Terminus of COP1 in the Dark Results in an Expression Profile Similar to That in cop1 Mutations
The COP1 protein contains three functional domains: a RING finger domain near the N terminus, followed by a coiled-coil domain, and a domain with WD-40 repeats in the C-terminal half (Deng et al., 1992; McNellis et al., 1994a). It has been shown that overexpression of N282 (with the RING finger and coiled-coil domains) causes a dominant-negative phenotype resembling that of weak loss-of-function cop1 mutants (McNellis et al., 1996; Stoop-Myer et al., 1999) (Figure 2A). To determine whether the dominant-negative N282 also results in similar changes in genome expression as the cop1 mutations, the gene expression profile caused by the overexpression of N282 in the wild-type background in the dark was analyzed. As shown in Figure 2B, the gene expression patterns were quite similar among N282 (N282/D versus WT/D), cop1-4 (cop1-4/D versus WT/D), and white light (WT/WL versus WT/D). As in the weak cop1-4 and cop1-6 mutants, there was a large overlap for the genes expressed differentially in N282 and white light–regulated genes, although the degree of differential expression caused by N282 was quantitatively less than that caused by white light. The quantitative changes in gene expression caused by N282 were very similar to those of the three cop1 mutants examined (see supplementary data online). The finding that the same set of genes is affected in cop1 mutants and by the overexpression of N282 indicates that N282 interferes with the activity of the endogenous COP1.
Figure 2.
Comparison of Genome Expression Profiles among the Dominant-Negative COP1 Mutant Form (N282), the cop1-4 Mutation, and White Light.
(A) Morphological comparison of continuous white light–grown wild-type, dark-grown N282, cop1-4, and wild-type Arabidopsis seedlings. All seedlings were 6 days old and photographed at the same magnification. D, darkness; N282, seedling overexpressing the N-terminal 282 amino acids of COP1; WL, white light; WT, wild-type seedlings. Bar = 1 mm.
(B) Hierarchical clustering display of expression ratios from wild-type seedlings grown under white light versus darkness, dark-grown N282 seedlings versus dark-grown wild-type seedlings, and dark-grown cop1-4 seedlings versus dark-grown wild-type seedlings. Lane 1, expression ratios of white light– and dark-grown wild-type seedlings; lane 2, expression ratios of dark-grown N282 and wild-type seedlings; lane 3, expression ratios of dark-grown cop1-4 and wild-type seedlings. Only those genes that exhibited twofold or greater differential change in at least one sample pair among the three pairs examined were included for comparison. A total of 2021 genes were included in the cluster (see supplementary data at http://plantgenomics.biology.yale.edu/ for details). The color scale for (B) is shown at bottom right.
The Genome Expression Profile Is Sensitive to the Modulation of COP1 Activity in Planta
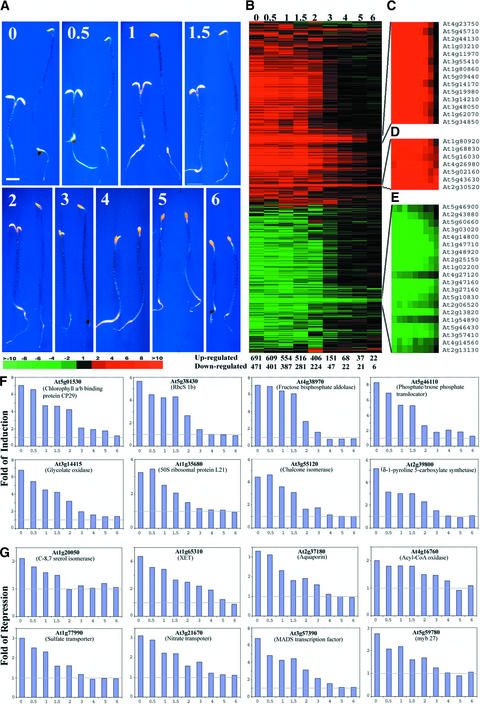
It has been reported that cop1-6 has a small in-frame six–amino acid deletion (McNellis et al., 1994a) and exhibits a temperature-dependent phenotype (Hsieh et al., 2000). Dark-grown cop1-6 shows a (weak) mutant phenotype under ambient temperature of <28°C, whereas it behaves like the wild type when grown at >28°C. Thus, the cop1-6 mutation allows us to modulate endogenous COP1 activity by shifting the temperature and to examine the genome expression profile during the developmental period when COP1 activity is critically required for seedling development. To this end, cop1-6 and wild-type seedlings were germinated at 30°C for 0 to 6 days in the dark and then transferred to a 22°C dark chamber for 6 days before examination. As reported previously and shown in Figure 3A, the cop1-6 seedlings had a dramatically different phenotype than the wild type when grown at 22°C in the dark. The cop1-6 seedlings grown at 30°C for <2 days before they were transferred to 22°C clearly showed largely photomorphogenic development. However, after growing at 30°C for 4 days, there was almost no difference in phenotype between cop1-6 and wild-type seedlings (Figure 3A).
Figure 3.
Kinetics of the Developmental Characteristics and Genome Expression Profile Changes during an in Planta Modulation of COP1 Activity.
(A) Phenotypic characteristics of 6-day-old dark-grown wild-type and cop1-6 mutant seedlings grown at 30°C for different times before transfer to 22°C in the dark. In each panel, the cop1-6 mutant seedlings are shown at left and wild-type seedlings are shown at right. Both the cop1-6 and wild-type seedlings were grown for 0, 0.5, 1, 1.5, 2, 3, 4, 5, and 6 days at 30°C before seedlings were transferred to 22°C. All seedlings were 6 days old and photographed at the same magnification. Bar = 1 mm.
(B) Overview of the hierarchical cluster display of the genome expression profiles among all treated cop1-6 and wild-type sample pairs shown in (A). Each lane represents the expression ratios of 6-day-old dark-grown cop1-6 and wild-type seedlings that were grown initially at 30°C for various times (days are listed at top). A total of 1484 genes that had twofold or greater differential expression in at least one sample pair were included in the cluster (see supplementary data at http://plantgenomics.biology.yale.edu/ for details). The number of genes exhibiting twofold or greater differential expression between dark-grown cop1-6 and wild-type seedlings changed according to their duration of growth at 30°C before being shifted to 22°C; these values are indicated at bottom.
(C) to (E) Three representative subclusters of genes exhibited delayed alteration in their genome expression profiles.
(F) Expression profiles of eight representative genes from some of the cellular and metabolic pathways upregulated in the dark-grown cop1-6 mutant.
(G) Expression profiles of eight representative genes from some of the cellular and metabolic pathways downregulated in the dark-grown cop1-6 mutant.
The expression ratios of genes between the 6-day-old dark-grown cop1-6 and wild-type seedlings shown in (B) to (G) were from seedlings grown at 30°C for 0, 0.5, 1, 1.5, 2, 3, 4, 5, and 6 days before being transferred to 22°C and/or analyzed. The color scale for (B) to (E) is shown at the bottom of (A).
We next examined the gene expression profiles from the cop1-6 and wild-type sample pairs grown for different time periods at 30°C before transfer to 22°C (Figures 3B to 3G). The number of differentially expressed genes decreased dramatically when seedlings were grown for >2 days at 30°C. After growth at 30°C for 4 days and then transfer to 22°C, only a small portion of genes exhibited differential expression (Figure 3B). To further demonstrate this drastic change in the number of differentially regulated genes, the number of genes showing twofold or greater change between cop1-6 and wild-type seedlings was recorded for each time point at which the seedlings were grown at 30°C (Figure 3B, bottom). The changes in the number of differentially expressed genes were most dramatic between days 2 and 3; these changes were essentially completed by day 4. This time scale is consistent with the phenotype changes (Figure 3A) (Hsieh et al., 2000). The induction and repression values for some representative genes are shown in Figures 3F and 3G.
Although the vast majority of genes reverted to wild-type expression after 4 days at 30°C, small subsets of genes were slower to revert. Figures 3C to 3E show data for three representative subclusters of genes whose expression patterns were significantly different. The expression of the genes in Figure 3C reverted to the wild-type level only after 5 days at 30°C before transfer to 22°C. Figures 3D and 3E show data for genes whose expression did not revert to the wild-type pattern even after being kept at 30°C for the entire 6-day period. However, only 28 genes exhibited similar differential expression (twofold or greater) between the cop1-6 and wild-type seedlings grown at 30°C in darkness for all 6 days (Figure 3B, lane 6). Those genes included 22 genes that were upregulated in cop1-6 mutant seedlings, such as HSP70 (At3g12580), DnaJ protein (At3g13310), metallothionein-like protein (At1g07610), and an abscisic acid–regulated protein, ATEM6 (At2g40170). Six genes were downregulated in the mutant seedlings, including endo-1,4-β-glucanase (At1g70710) and a myb-like protein (At3g48920). The reason why these genes remained differentially expressed is not clear.
COP1 Regulates the Expression of a Large Fraction of Arabidopsis Transcription Factors
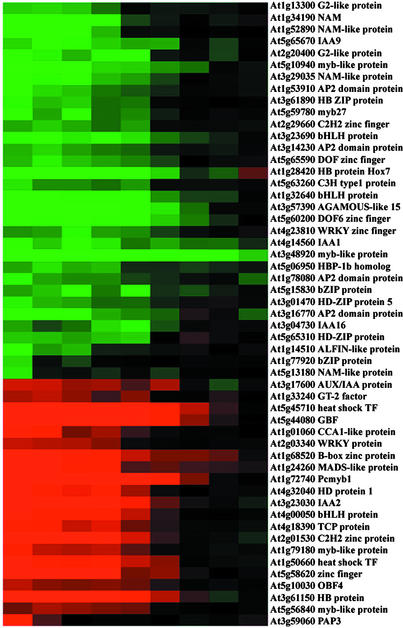
We examined the number of transcription factors whose expression was regulated by COP1. The microarray included 333 putative transcription factor genes, ∼20% of ∼1600 transcription factor genes in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000; Riechmann and Ratcliffe, 2000). A total of 53 transcription factor genes in our microarray exhibited consistent twofold or greater differential expression between dark-grown cop1-6 mutant and wild-type seedlings when grown at 22°C for 6 days. This percentage is similar to that of all genes showing differential expression between dark-grown cop1 mutant and wild-type seedlings in the microarray using the same twofold cutoff. These 53 transcription factor genes were further confirmed by sequencing: 21 of them were upregulated in the cop1-6 mutant seedlings, and 32 were downregulated.
The differential expression patterns of those 53 transcription factors shown in Figure 1B were extracted and are shown in Figure 4. The differentially expressed transcription factors exhibited distinct sensitivities and kinetics in response to the cop1-6 mutation. Some transcription factors, such as PAP3 (At3g59060) and a basic domain/Leu zipper factor (At1g77920), exhibited quick reversion to the wild-type state when the cop1-6 mutants were grown at 30°C for only 12 h, whereas the majority of transcription factors exhibited a similar expression pattern switch at day 2 to 3, like most of the genes regulated by COP1 (Figures 3B and 4). A few transcription factors, such as Pcmyb1 (At1g72740) and Hox7 (At1g28420), exhibited much slower kinetics in their expression pattern changes, and it took >4 or 5 days of growth of cop1-6 at 30°C to reach wild-type levels. Furthermore, one myb-like protein (At3g48920) still showed greater than twofold differential expression between cop1-6 mutant and wild-type seedlings even after growth at 30°C (the permissive temperature) for the full 6 days of COP1 activity recovery (Figure 4); at this time, the cop1-6 mutant and the wild type are phenotypically indistinguishable. This pattern of COP1 effect is consistent with a transcription cascade hypothesis (Tepperman et al., 2001), with early affected transcription factor genes affecting later transcription factors and then all other target genes.
Figure 4.
COP1 Regulates the Expression of a Large Group of Transcription Factors.
All of the transcription factors in our microarray that displayed twofold or greater differential expression in at least one of the nine sample pairs shown in Figure 3B were selected. The expression ratios of those 53 selected transcription factor genes between the 6-day-old dark-grown cop1-6 and wild-type seedlings were from seedlings grown at 30°C for 0, 0.5, 1, 1.5, 2, 3, 4, 5, and 6 days before being transferred to 22°C and/or analyzed. The color scale is shown at bottom.
HY5-Regulated Genes Are Largely Included in the Group Regulated by COP1
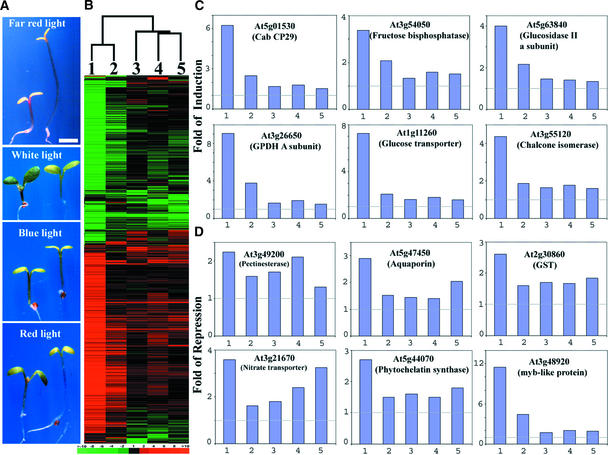
We have hypothesized that COP1 acts to degrade key transcription factors, such as HY5, in the dark, thus inhibiting light-dependent transcription in darkness (Osterlund et al., 2000). The failure of COP1 proteins to degrade their target transcription factors (in the cop1 mutants) would allow these transcription factors to drive photomorphogenic gene expression and development in the absence of light. This hypothesis predicts that each of those transcription regulators would be responsible for regulating a portion of the genes that are controlled by COP1. Thus, the expression profile of HY5, a key transcription factor that is a target of COP1 and a positive regulator for photomorphogenesis, should be largely included within the profile of COP1. Indeed, HY5-regulated genome expression in different light quality conditions (Figure 5) is largely included within that of COP1, although COP1 and HY5 act in an antagonistic manner. Among light qualities, the hy5 mutation affected the greatest number of genes expressed in far-red light, which is consistent with the most dramatic phenotypic difference observed in the same light condition (Figure 5A). However, the degrees of differential expression for most of the genes were much greater in the dark-grown cop1-6 mutant than those affected by the hy5 mutation in each light condition (Figure 5B). In fact, a large fraction of those COP1-regulated genes (70 to 90%) displayed differential expression less than the twofold cutoff in the hy5 mutant, and most of those genes were not affected significantly by the hy5 mutation. This is best illustrated in Figures 5C and 5D, in which the effects of cop1 and hy5 mutations on the expression of some representative cellular or metabolic pathway genes are shown. Among the pathways controlled by light, the expression of some of the genes involved in photosynthetic light and dark reactions, starch degradation, glycolysis, phenylpropanoid synthesis, cell wall degradation, water transport, and sulfate and nitrogen assimilation (Ma et al., 2001) also were affected by HY5, although to a reduced extent (see supplementary data at http://plantgenomics.biology.yale.edu/).
Figure 5.
Comparison of the Morphology and Genome Expression Profiles of hy5 Mutant and Wild-Type Seedlings Grown under Four Different Light Quality Conditions.
(A) Wild-type and hy5 mutant seedlings grown under different light quality conditions. In each panel, the seedling at left is the wild type and the seedling at right is the hy5 mutant. The seedlings were grown for 6 days in continuous far-red, white, blue, and red light. All seedlings were photographed at the same magnification. Bar = 1 mm.
(B) Hierarchical clustering display of expression ratios from wild-type versus hy5 mutant seedlings grown under different light quality conditions. The genome expression profile from dark-grown cop1-6 versus wild-type seedlings is included for comparison. Only those genes that exhibited twofold or greater differential expression in at least one sample pair among the five pairs examined here were included for comparison. A total of 1312 genes were included in the cluster analysis (see supplementary data at http://plantgenomics.biology.yale.edu/ for details). The color scale is shown at bottom. The dendrogram at top indicates the relationship among those data sets across all of the genes included in this clustering analysis.
(C) Expression profiles of six representative genes from some of the cellular and metabolic pathways upregulated in the dark-grown cop1-6 mutant.
(D) Expression profiles of six representative genes from some of the cellular and metabolic pathways downregulated in the dark-grown cop1-6 mutant.
The five bars in each graph of (C) and (D) correspond to the lanes in (B). Lane 1, expression ratios of dark-grown cop1-6 and wild-type seedlings; lane 2, expression ratios of far-red light–grown wild-type and hy5 seedlings; lane 3, expression ratios of white light–grown wild-type and hy5 seedlings; lane 4, expression ratios of blue light–grown wild-type and hy5 seedlings; lane 5, expression ratios of red light–grown wild-type and hy5 seedlings.
The overall genome expression profile attributable to HY5 was largely contained within the fraction of the genome controlled by COP1, whereas HY5 seemed to play the most prominent role in mediating the COP1 effect under far-red light (Figure 5B). However, there were a small number of exceptions in which gene expression was not regulated coordinately by COP1 and HY5. There were 17, 29, 37, and 26 genes that exhibited clearly distinct differential expression (twofold or greater) but that were not shared between the cop1 mutation and hy5 under far-red, white, blue, and red light, respectively. For example, some genes encoding ribosomal proteins were upregulated in the dark-grown cop1 mutants but either did not change or were downregulated in the hy5 mutant in different light conditions. A total of 12, 15, 11, and 19 genes exhibited clear expression changes in the hy5 mutant in far-red, white, blue, and red light conditions, respectively, but not in the cop1-6 mutant. This result suggests that HY5 also may play a role in processes not related to the COP1-mediated light control of gene expression.
Gene Expression Profile Controlled by COP1 in White Light
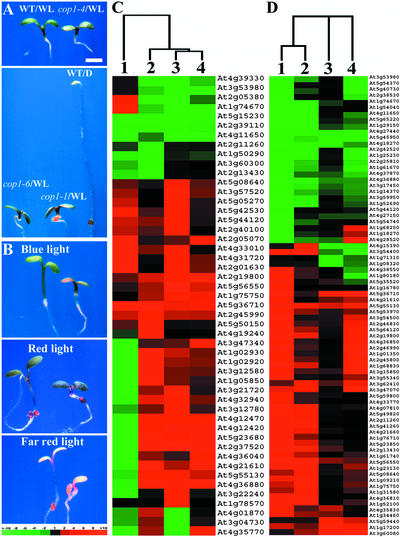
Because all cop1 mutants have exaggerated photomorphogenic development in the light (Deng and Quail, 1992) (Figures 6A and 6B), we wanted to define the basis of this phenotype on a genomic level with the same microarray system. We first grew different cop1 mutants and wild-type seedlings under the same white light conditions. Examination of the ratio of gene expression of each cop1 mutant versus wild-type seedlings revealed 275 genes that exhibited twofold or greater differential expression in at least one cop1 allele compared with the wild type under the same white light condition. Among these 275 genes, 49 genes exhibited threefold or greater differential expression in at least one cop1 mutant allele. To further characterize these 49 genes, we compared their expression patterns with the differential expression of light- and dark-grown seedlings in a cluster analysis (Figure 6C). Among these 49 genes, 39 (80%) exhibited similar expression patterns in all three cop1 mutant alleles. A small portion of genes (10) exhibited different, even opposite, regulation patterns among different cop1 alleles (Figure 6C). Most of these 49 genes, with the exception of 4 genes, are light-regulated genes. However, only approximately half of those differentially regulated genes (22) in the light-grown cop1 mutants were exaggerated in light regulation, whereas the other half (23) exhibited a differential expression pattern in light-grown cop1 mutants opposite that of light regulation in the wild type. For example, two CAB genes (At2g40100 and At2g05080) and two genes encoding enzymes for anthocyanin biosynthesis (At5g08640 for flavonol synthase and At5g05270 for chalcone isomerase) were upregulated by white light and upregulated even more dramatically in light-grown cop1 mutants (Figure 6C). In some other genes, such as the zinc finger protein Lsd1 (At4g21610) and a GAST1-like protein, cop1 mutations caused opposite regulation in white light compared with normal light regulation in the wild type.
Figure 6.
Comparison of the Morphology and Genome Expression Profiles of Three Representative cop1 Mutants and the Wild Type under Continuous White, Far-Red, Red, or Blue Light.
(A) Continuous white light–grown wild-type and three cop1 mutant Arabidopsis seedlings. D, darkness; WL, white light; WT, wild-type seedlings. Bar = 1 mm.
(B) Wild-type and cop1-6 mutant Arabidopsis seedlings grown under continuous blue, red, and far-red light. In each panel, the wild-type seedling is shown at left and the cop1-6 mutant seedling is shown at right.
(C) Hierarchical clustering display of expression profiles for the wild type and three representative cop1 mutants under white light. Differential expression ratios are shown for wild-type seedlings grown under white light and dark-grown seedlings (lane 1), white light–grown cop1-4 and wild-type seedlings (lane 2), white light–grown cop1-1 and wild-type seedlings (lane 3), and white light–grown cop1-6 and wild-type seedlings (lane 4). Only those genes that exhibited threefold or greater differential expression in at least one cop1 mutant were included for comparison. A total of 49 genes were included in the cluster.
(D) Hierarchical clustering display of expression ratios from light-grown cop1-6 mutant and wild-type seedlings. Differential expression ratios of selected genes are shown for blue light–grown cop1-6 mutant and wild-type seedlings (lane 1), red light–grown cop1-6 mutant and wild-type seedlings (lane 2), far-red light–grown cop1-6 mutant and wild-type seedlings (lane 3), and dark-grown cop1-6 mutant and wild-type seedlings (lane 4). Only those genes that exhibited threefold or greater differential expression in at least three light quality conditions were selected for comparison. A total of 77 genes were included in the cluster.
All seedlings shown in (A) and (B) were 6 days old and photographed at the same magnification. The color scale for (C) and (D) is shown at bottom of (B). The dendrograms at top in (C) and (D) indicate the relationship among those data sets across all of the genes included in the clustering analyses.
Interestingly, among 22 genes that exhibited upregulation in 6-day-old cop1-6 mutant seedlings grown at 30°C (Figure 3), 12 were included in the gene set that exhibited exaggerated upregulation in cop1 mutation alleles under white light. Those genes include the zinc finger protein Lsd1, a class I chitinase, two glutathione S-transferases, a DnaJ protein, HSP70, a Gln-dependent Asn synthetase, a seed imbibition–like protein, a pEARLI1-like protein, and three unknown proteins (At2g19800, At5g23680, and At1g75750). This result suggested that the expression of these genes was more sensitive to COP1 activity. Because we have shown that exaggerated light signaling by overexpressing photoreceptors resulted in both enhanced light regulation and contrasting regulation of light-regulated genes (Ma et al., 2001), it is reasonable to conclude that the light-grown cop1 mutant phenotype is caused by exaggerated light signaling and modulation of genome expression.
Gene Expression Profiles Controlled by COP1 in Distinct Light Qualities
We further examined the gene expression profiles of the cop1-6 mutants grown under different light qualities (blue, red, and far-red light; Figure 6B) compared with wild-type seedlings grown under the same conditions. To simplify the visualization of the result, only those genes (77) showing threefold or greater differential expression in at least one of three light qualities between cop1-6 mutant and wild-type seedlings sample pairs were selected. The expression patterns were subjected to cluster analysis with the corresponding gene expression profile of the dark-grown cop1-6 mutant versus dark-grown wild-type seedling pairs. As shown in Figure 6D, most of the genes (51 of 77) that had differential expression between cop1-6 mutant and wild-type seedlings grown in different light qualities also were regulated differentially by the cop1-6 mutation in darkness. For example, flavonol synthase, CAB genes, and the transcription factor CCA1 were included in those upregulated genes in cop1-6 mutant seedlings, whereas genes encoding the chlorophyll synthesis enzyme protochlorophyllide reductase and the tonoplast water channel protein aquaporin were among the downregulated genes in the cop1 mutant seedlings (Figure 6D). Under those three light quality conditions, it seems that the cop1 mutation simply exaggerated the light regulation for most of the genes represented in Figure 6D. In only 16 cases (of 77), the expression of the genes was not affected by cop1-6 in darkness but was regulated significantly under at least one light quality condition. However, there was great variation in the degree of the cop1-6 exaggerated effect on individual gene expression in different light quality conditions (Figure 6D). Again, the data are consistent with the conclusion that the hypersensitive photomorphogenic phenotype of the cop1-6 mutant under the three light quality conditions is the result of exaggerated light signaling and gene expression. However, the target genes exhibiting hypersensitivity to different light quality conditions in the cop1-6 mutant are not identical.
DISCUSSION
In this study, we systematically investigated COP1-controlled genome expression during Arabidopsis seedling development using a cDNA microarray. Because our study identified all previously known light- and COP1-regulated genes included in our microarray (Ma et al., 2001), it suggests that our microarray data are reliable. In addition, our results lend strong support to the working model built on molecular genetic studies of the past and provide several new insights regarding the COP1 control of gene expression and plant development pattern.
COP1-Regulated Genome Expression Overlaps Light-Regulated Expression
Light can activate a drastic and contrasting morphological change in seedling development, primarily by changes in genome expression (Terzaghi and Cashmore, 1995; Puente et al., 1996; Ma et al., 2001; Tepperman et al., 2001). Mutations in COP1 result in a dramatic phenotypic change in Arabidopsis seedlings. When grown in the dark, the cop1 mutant exhibits a photomorphogenic phenotype with a short hypocotyl, open cotyledons without an apical hook, and other aspects of light-grown phenotypes. It has been hypothesized that this dramatic change in developmental pattern is a consequence of an altered genome expression pattern, based on a limited test on ∼10 individual genes using traditional approaches (Deng et al., 1991, 1992; Ang et al., 1998; Chattopadhyay et al., 1998). In the present study, we examined the effect of COP1 on a set of >6126 genes during Arabidopsis photomorphogenesis. We used both strong and weak cop1 mutant alleles to profile genome expression controlled by COP1 in the dark. This work documented the vast extent of COP1 regulation of genome expression in higher plants. More than 1300 genes (>20%) showed differential expression between cop1 mutant and wild-type seedlings in the dark in our array (Figure 1) with a twofold cutoff. In addition, this set of COP1-controlled genes was verified further by controlling COP1 activity in planta through a temperature-shift assay using a temperature-sensitive cop1 allele (Figure 3). Furthermore, we showed that overexpression of a dominant-negative partial COP1 protein (N282) (McNellis et al., 1996; Torii et al., 1998) also led to a genome expression profile similar to that seen in the cop1 mutations (Figure 2). Therefore, it is reasonable to conclude that COP1 represses photomorphogenesis by regulating genome expression patterns.
It is important to note that the genome expression profile controlled by COP1 overlaps the profile controlled by light (Ma et al., 2001). Light appears to regulate a larger fraction (up to 30%) of the genome (Ma et al., 2001) compared with that regulated by COP1 (20%). However, this difference was attributable largely to the fact that a twofold cutoff was used in both cases, because the cop1 mutants have a quantitatively weaker effect on gene expression and the differential expression of many of the genes fell just below the twofold cutoff. Because none of the cop1 mutant alleles are null mutations, it is likely that their effect on gene expression was not complete. Thus, for a large fraction of light-regulated genes, their differential expression happens to fall below the twofold cutoff in dark-grown cop1 mutants. Taking this into account, 89, 90, and 85% of those light-regulated genes exhibited similar qualitative differential expression in the cop1-1, cop1-4, and cop1-6 mutants, respectively, in darkness (see supplementary data online).
In a previous study, we found that at least 26 fundamental cellular processes or metabolic pathways were regulated by light. Some of these were activated by light, whereas others were repressed by light (Ma et al., 2001). In the present work, we found that two additional metabolic pathways, the fatty acid biosynthesis and starch degradation pathways, also were activated by light and the cop1 mutation in darkness (Table 1). Light and COP1 antagonistically regulated expression patterns in all 28 cellular and metabolic pathways (Figure 1C). Therefore, it is evident that COP1 and light regulate the expression of the same set of genes in the genome, albeit in an antagonistic manner. It has been established that light can negatively regulate COP1 activity (von Arnim and Deng, 1994; Osterlund et al., 1999). Thus, it is plausible that light regulates genome expression largely by inhibiting COP1 activity. In our earlier study, we established that distinct photoreceptors act to perceive different light signals to regulate the expression of a largely common fraction of the genome (Ma et al., 2001). The findings presented here suggest that this common fraction of light-regulated genome control by different light signals could be achieved by modulating COP1 activity via distinct photoreceptor-initiated signaling events.
COP1 Controls Genome Expression and Plant Development by Targeted Degradation of Transcription Factors
How does COP1 achieve its control of expression over such a large portion of the genome? Recent molecular genetic analysis of the COP1–HY5 interaction suggested that COP1 may specifically target photomorphogenesis-promoting factors (such as HY5) for degradation via the proteasome (Ang and Deng, 1994; Hardtke et al., 2000; Osterlund et al., 2000; Holm et al., 2001). HY5 encodes a basic domain/Leu zipper transcription factor that binds to the G-box motif in the promoter of light-inducible genes such as RbcS1A and CHS1 and thereby plays a vital role in their activation during seedling photomorphogenesis (Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998; Hardtke et al., 2000). In the present study, we found that the COP1- and HY5-controlled gene expression profiles were largely overlapping. Essentially, HY5 seems to antagonistically regulate a portion of those genes controlled by COP1, and for most of those genes, HY5 contributes quantitatively toward only a fraction of the overall regulation exerted by COP1 (Figure 5). This result is consistent with the notion that COP1 controls gene expression and plant development through the targeted degradation of transcription factors such as HY5. Thus, COP1 negatively regulates those transcription factors, which normally bind directly to light-responsive promoters and regulate target gene expression.
Considering that many other transcription factors, in addition to HY5, are involved in plant photomorphogenesis (Quail, 2000; Holm et al., 2001; Tepperman et al., 2001) (Figure 4), we suggest that other transcription factors, which also are involved in promoting photomorphogenesis and parts of the genome expression, also might be targeted for degradation by COP1. It is through the combined regulation of the activities of all of these target transcription factors that COP1 achieves its control of light-regulated genome expression and developmental pattern. Our recent finding that the degradation of HYH, another basic domain/Leu zipper transcription factor, also was mediated by COP1 (Holm et al., 2002) supports this prediction. Further analysis of the COP1-controlled genes found in this study could help define the other COP1 target transcription factors that play a role in promoting photomorphogenesis.
METHODS
Experimental Materials
In all experiments, except as noted, the wild-type Arabidopsis thaliana ecotype was Columbia. All of the cop1 mutants (cop1-1, cop1-4, and cop1-6) and the hy5 mutant (hy5-221) used in this study were in the Columbia ecotype (Deng et al., 1991, 1992; Deng and Quail, 1992; McNellis et al., 1994a; Ang et al., 1998). The N282 overexpression line was in the Nossen ecotype (McNellis et al., 1996), and wild-type Nossen was used for its control. Surface sterilization and cold treatment of the seeds were performed as described previously (Ang and Deng, 1994). Arabidopsis seedlings were grown in growth medium on agar plates containing 1% Suc. The seedlings were grown in continuous white, red, far-red, and blue light or darkness for 6 days. The white light intensity was 152 μmol·m−2·s−1. The color light–emitting diode growth chambers (E-30LED2/3; Percival Scientific, Perry, IA) had intensities of 16.2 μmol·m−2·s−1 for blue light (470 nm), 108.5 μmol·m−2·s−1 for red light (670 nm), and 160.8 μmol·m−2·s−1 for far-red light (735 nm). For all temperature-shift experiments, seedlings were grown at 30°C in darkness for different periods before being transferred to a 22°C dark chamber for 6 days.
The microarray slide used in this study was described previously (Ma et al., 2001). For more information, see our World Wide Web sites (http:/plantgenomics.biology.yale.edu and http://info.med.yale.edu/wmkeck/dna_arrays.htm).
RNA Preparation and Fluorescence Labeling of Probe
Total RNA was extracted from whole seedlings using the Qiagen RNeasy Plant Mini prep kit (Valencia, CA). RNA preparations from at least two independent biological samples for each test were made and used for probe synthesis. At least two replicate microarray hybridizations were performed for each RNA sample; thus, each experiment produced at least four replicate data sets. Total RNA (50 μg) was first labeled with aminoallyl-dUTP (aa-dUTP; Sigma, St. Louis, MO) by direct incorporation of aa-dUTP (instead of Cy-3 or Cy-5 dUTP) during reverse transcription, as described previously (Ma et al., 2001). After 3 h of incubation at 42°C, the reaction was stopped by adding 5 μL of 0.5 M EDTA and incubating at 94°C for 3 min. The RNA then was hydrolyzed by the addition of 10 μL of 1 M NaOH followed by incubation at 65°C for 20 min. This reaction was neutralized by the addition of 6 μL of 1 M HCl and 2 μL of 1 M Tris-HCl, pH 7.5. The aa-dUTP–labeled cDNA was purified from the unincorporated aa-dUTP molecules by adding 400 μL of water and centrifugation through a Microcon YM-30 filter (Millipore, Bedford, MA) for 7 min at 11,000g. This washing step was repeated once. The purified, labeled probe was concentrated to a final volume of 7 μL and was labeled further with fluorescent dye by conjugating aa-dUTP and monofunctional Cy-3 or Cy-5 (Amersham Pharmacia Biotech, Piscataway, NJ) in a mixture containing 7 μL of cDNA solution, 0.7 μL of 1 M sodium bicarbonate (J.T. Baker, Philipsburg, NJ), and 1 μL of Cy-3 or Cy-5 dye (dissolved in DMSO). The mixture was mixed with a pipette tip and incubated at room temperature in the dark for 60 to 90 min. After incubation, the labeling reaction was stopped by adding 1 μL of 2 M ethanolamine (Sigma), followed by incubation at room temperature for 5 min. The dye-labeled probe was purified from the unincorporated dye molecules by washing three times through a Microcon YM-30 filter (Millipore) as described above. The purified labeled probes from sample pairs were combined and concentrated to a final volume of 7 μL for hybridization.
Hybridization, Washing, and Scanning
The protocols for hybridization to the Arabidopsis microarray, microarray slide washing, and scanning were as described previously (Ma et al., 2001). Separate TIFF images for Cy-3 and Cy-5 channels were obtained by scanning with an Axon GenePix 4000B scanner (Foster City, CA) at a resolution of 10 nm. Laser and photomultiplier tube voltages were adjusted manually to minimize background and the number of spots that had saturated signal values. The normalization of the two channels with respect to signal intensity was obtained by adjusting the photomultiplier tube and laser power settings. We chose the photomultiplier tube voltages to let the signal ratio of the majority of control genes be as close to 1.0 as possible.
Data Analysis
Most approaches were as described in our previous report (Ma et al., 2001) with minor modifications. Briefly, spot intensities were quantified using Axon GenePix image-analysis software. The channel ratio was measured using the GenePix median of ratio method and was normalized using the corresponding GenePix Pro 4.0 default normalization method. To merge the replicated GenePix output data files, a custom computer program, GPMERGE, was developed (http://bioinformatics.med.yale.edu/software.html). The source of annotation for converting EST accession number to gene locus identifier was a centralized database for Arabidopsis annotation, ATH1, at the TIGR Arabidopsis Genome Annotation Database (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/SEQUENCES/ATH1.seq). For those unique genes that have more than one EST clone, we developed a custom computer program to extract a ratio for the gene according to the following criteria. First, for those genes that have two EST clones and both show similar expression patterns, an average of the ratio value was used for the gene. If the ratio was in the opposite direction, implying an error in EST clone identity, we sequenced the EST clones to reveal the true identity for both clones. Second, for those genes that have three EST clones, if all of the EST clones show similar expression patterns, the median ratio was used for the gene; if two of them show similar expression patterns and the other one shows the opposite pattern, the average of the two similar ratios was used for the gene. Third, for those genes that have at least four EST clones, the median of the ratios was used.
Different kinds of expression pattern identification and pattern matching were conducted within or across these experimental groups. Within each group, hierarchical clustering analysis was performed, as described by Eisen et al. (1998). Only those genes that had more than twofold or threefold changes in expression (as specified above) in at least one of the experimental sets were used in the cluster analysis shown in the figures.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Magnus Holm, James Sullivan, and Haiyang Wang for reading and commenting on the manuscript. We are grateful to Kenneth Williams, Janet Hager, and the Yale DNA microarray laboratory of the Keck Biological Resource Center for the production of the microarray used in this study (http://info.med.yale.edu/wmkeck/dna_arrays.htm), which was supported in part by a National Institute of Diabetes and Digestive and Kidney Diseases Microarray Biotechnology Center grant (NIH 5 U24 DK58776; principal investigator, Kenneth Williams). Our research was supported by National Institutes of Health Grant GM-47850 to X.W.D., a grant from the National Program for Transgenic Plants of China (J99-A-001), and National Institutes of Health Grant GM59507 to H.Z. L.M. is a long-term postdoctoral fellow of the Human Frontier Science Program.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004416.
Footnotes
Online version contains Web-only data.
References
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., and Deng, X.W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Castle, L., and Meinke, D. (1994). A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6, 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.H., Puente, P., Deng, X.W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58, 991–999. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Caspar, T., and Quail, P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5, 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., et al. (2000). Unified nomenclature for the COP9 signalosome and its subunits: An essential regulator of development. Trends Genet. 16, 202–203. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2, 83–95. [Google Scholar]
- Deng, X.W., and Quail, P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19, 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., Hardtke, C.S., Gaudet, R., and Deng, X.W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., Ma, L., Qu, L., and Deng, X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev., 16, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, H.L., Okamoto, H., Wang, M., Ang, L.H., Matsui, M., Goodman, H., and Deng, X.W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Ma, L., Li, J., Qu, L., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., Torii, K.U., and Deng, X.W. (1996). Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8, 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. (1994. a). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., and Deng, X.W. (1994. b). Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misera, S., Mueller, A.J., Weiland-Heidecker, U., and Juergens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol. Gen. Genet. 244, 242–252. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Osterlund, M.T., Ang, L.H., and Deng, X.W. (1999). The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 9, 113–118. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., and Deng, X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16, 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente, P., Wei, N., and Deng, X.W. (1996). Combinatorial interplay of promoter elements constitutes minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15, 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2000). Phytochrome-interacting factors. Semin. Cell Dev. Biol. 11, 457–466. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Ratcliffe, O.J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2000). The COP/DET/FUS proteins: Regulators of eukaryotic growth and development. Semin. Cell Dev. Biol. 11, 495–503. [DOI] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop-Myer, C., Torii, K.U., McNellis, T.W., Coleman, J.E., and Deng, X.W. (1999). The N-terminal fragment of Arabidopsis photomorphogenic repressor COP1 maintains partial function and acts in a concentration-dependent manner. Plant J. 20, 713–717. [DOI] [PubMed] [Google Scholar]
- Su, Q., Frick, G., Armstrong, G., and Apel, K. (2001). POR C of Arabidopsis thaliana: A third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol. Biol. 47, 805–813. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi, W.B., and Cashmore, A.R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474. [Google Scholar]
- Torii, K.U., McNellis, T.W., and Deng, X.W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17, 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1992). COP9: A new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4, 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1999). Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15, 98–103. [DOI] [PubMed] [Google Scholar]
- Wei, N., Kwok, S.F., von Arnim, A.G., Lee, A., McNellis, T.W., Piekos, B., and Deng, X.W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in dark. Plant Cell 6, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.