Abstract
The Mre11 protein is essential for the long-term genetic stability of the cell and acts to ensure the efficient repair of DNA damage. Vertebrate cells lacking Mre11 function are not viable. However, we report here that this is not the case in the model plant Arabidopsis. We have isolated two different Arabidopsis lines containing a T-DNA copy integrated at a different point in the MRE11 gene (AtMRE11). Both mutant plant lines were hypersensitive to DNA-damaging treatments but exhibited strikingly different developmental phenotypes. Furthermore, we also observed lengthened telomeres in these plant lines, showing that AtMre11 is involved in telomere maintenance. Thus, the lines we have isolated are unique tools with which to study in detail the role of AtMre11 in the mature plant.
INTRODUCTION
An organism's genetic material is damaged frequently by external agents, such as ionizing or solar radiation, aggressive metabolites, or during the process of DNA replication. DNA damage can be catastrophic to the cell, and its accurate repair is essential for the cell and its descendants to maintain genetic stability. Breaks in both strands of a duplex DNA molecule, a double-strand break (DSB), are among the most threatening for the cell. Two independent pathways have evolved for the repair of DSBs. The homologous recombination (HR) pathway for DSB repair is conservative and uses homologous sequences on the sister chromatid or elsewhere in the genome as a template to repair the DSB. The second pathway, nonhomologous end joining (NHEJ), rejoins the two ends of a DSB without using a homologous template. NHEJ can be mutagenic, because nucleotides can be lost or added at the repair site. The predominant DNA repair pathway (HR or NHEJ) varies in different cell types. Generally, HR is the major pathway in single-celled organisms such as bacteria and yeast, but NHEJ is the dominant DNA repair mechanism in most cell types of higher eukaryotes. However, there are exceptions, such as in the ES and DT40 cell lines of vertebrates and cells of the moss Physcomitrella patens, which use the HR pathway efficiently.
Cells have evolved an array of powerful enzymatic tools that are able to recognize DSBs, repair the DNA lesion, and prevent cell cycle progression until this is accomplished. In the yeast Saccharomyces cerevisiae, genes in the RAD52 epistasis group, which includes the MRE11 gene, are required. The Mre11 protein forms a complex with the Rad50 and Xrs2 proteins (the MRX complex), as shown in a yeast two-hybrid assay (Johzuka and Ogawa, 1995) and by protein coimmunoprecipitation (Usui et al., 1998). Orthologs of MRE11 and RAD50 have been identified in all organisms examined to date. Thus, this complex has been highly conserved during evolution. As in yeast, the Mre11 proteins in human and Arabidopsis cells also have been shown to interact with the respective Rad50 proteins (Dolganov et al., 1996; Daoudal-Cotterell et al., 2002). Although the NHEJ and HR pathways are essentially independent of one another, the MRX complex is involved in both pathways (Haber, 1998). However, the effect of null mutations on HR depends on the assay used, whereas such mutations always exhibit a strong effect on the efficiency of NHEJ. S. cerevisiae cells lacking Mre11 exhibit 50- to 100-fold reduced levels of NHEJ (Milne et al., 1996; Moore and Haber, 1996; Boulton and Jackson, 1998).
An insight into the possible biochemical role of Mre11 in the cell came from the observation that the Escherichia coli SbcC protein and the SbcD nuclease are homologous with the Rad50 and Mre11 proteins, respectively (Sharples and Leach, 1995). In vitro studies have reported that the Mre11 protein does have nuclease activity, including 3′-to-5′ single-stranded and double-stranded DNA exonuclease and single-stranded DNA endonuclease activity (Furuse et al., 1998; Paull and Gellert, 1998; Trujillo et al., 1998; Usui et al., 1998; Moreau et al., 1999). These activities are modified in efficiency and specificity by interactions with other members of the MRX complexes (Paull and Gellert, 1998, 1999; Trujillo et al., 1998). However, it is not clear whether the nuclease activity of Mre11 is necessary for NHEJ in vivo. Mutations in the phosphoesterase motifs of Mre11 have no effect on the levels of NHEJ or the sequence of the junctions obtained (Moreau et al., 1999; Wilson et al., 1999). One possibility may be that the MRX complex operates in NHEJ to recruit other factors to the broken DNA ends, such as the Ku heterodimer or another nuclease. Recent microscopy studies have suggested that the MRX complex also plays a structural role in NHEJ. The human Mre11/Rad50 complex was observed at the ends of linear DNA molecules and in some instances tethered one DNA molecule to another (de Jager et al., 2001). In another study, Chen et al. (2001b) also observed that the MRX complex was able to bridge DNA ends and visualized multiple linear DNA molecules tethered together.
The Mre11 protein is also essential during meiosis. During sexual reproduction, cells undergoing meiosis reduce their diploid chromosome content by half. To ensure the correct segregation of chromosomes to the respective gametes, chromosome sorting is followed by crossover recombination between homologous chromosomes. This, in collaboration with sister chromatid cohesion, allows the establishment of temporary connections between homologs, which allows them to orient toward the opposite poles of the meiosis I spindle (Moore and Orr-Weaver, 1998). Recombination between homologous chromosomes occurs at “hot spots,” which are sites at which DSBs are induced enzymatically. mre11 mutants are deficient in the formation of meiotic DSBs and the induction of meiotic recombination and thus do not form viable spores (Ajimura et al., 1993; Tavassoli et al., 1995). Mre11 may promote DSB formation at hot spots by altering the chromatin configuration (Ohta et al., 1998).
In yeast, Mre11, along with other NHEJ proteins such as the Ku heterodimer, also is necessary for the maintenance of telomere length. Mutations in any of these components result in shortened telomeres (Porter et al., 1996; Boulton and Jackson, 1998; Furuse et al., 1998; Nugent et al., 1998; Moreau et al., 1999; D'Adda di Fagagna, 2001). Mre11 may be required to process chromosome ends before the loading of telomeric proteins or may be necessary to maintain a stable telomere structure, such as the T-loop configuration of vertebrate telomeres. However, the effect of Mre11 on vertebrate telomeres is unclear, because no shortening of telomeres was observed in Mre11-deficient DT40 cells (Yamaguchi-Iwai et al., 1999).
The kinetics of MRX complex formation in response to DNA damage also has been studied. The MRX complex forms foci at DSBs induced by ionizing radiation or those associated with the process of V(D)J rearrangement during B and T cell development (Carney et al., 1998; Chen et al., 2000). The MRX complex, together with the ATM protein kinase, also has been implicated in the cell cycle response to DNA damage. A first indication for this came from the discovery that the human equivalent of the Xrs2 protein, Nbs1, was mutated in patients with the condition Nijmegen breakage syndrome (NBS) (Carney et al., 1998; Varon et al., 1998). Cells from patients with NBS fail to suppress DNA synthesis after exposure to ionizing radiation. Moreover, patients with an ataxia-telangiectasia–like disorder with cellular features similar to NBS were found to have mutations in the MRE11 gene (Stewart et al., 1999). In human cells, the MRX complex has been shown to suppress DNA replication through an interaction of Nbs1 with the E2F1 transcription factor (Maser et al., 2001).
Studying the role of MRE11 in multicellular organisms is difficult, given that it is essential in mammalian cells (Xiao and Weaver, 1997; Yamaguchi-Iwai et al., 1999). However, this is not the case in Caenorhabditis elegans, in which, as in yeast, mutations in CeMRE11 result in worms that are hypersensitive to ionizing radiation and that have reduced reproductive capacity (Chin and Villeneuve, 2001). Given the essential roles of Mre11 in many aspects of DNA metabolism and signaling in yeast and vertebrate cells, we were interested in studying its effects in Arabidopsis. We report here the isolation and characterization of two lines containing different mutations in the Arabidopsis MRE11 gene (AtMRE11). Plants homozygous for either mutation are viable. The T-DNA copy in the AtMRE11-1 line is inserted in the strongly conserved 5′ region of the gene. This mutation is semidominant, and plants homozygous for the T-DNA insertion were both dwarf and sterile and showed severe developmental defects. By contrast, the T-DNA copy in the AtMRE11-2 mutant line is inserted in the 3′ end of the gene, which is conserved less strongly between different Mre11 orthologs. This mutation is recessive and gives rise to phenotypically normal plants that are fertile. Both plant lines are hypersensitive to genotoxic agents. Furthermore, we found that both mutations affected the length of plant telomeres. In contrast to the findings in yeast, the telomeres in these AtMre11-deficient plants were longer than those in wild-type plants. This result suggests that plants possess an alternative mechanism for telomere length maintenance.
RESULTS
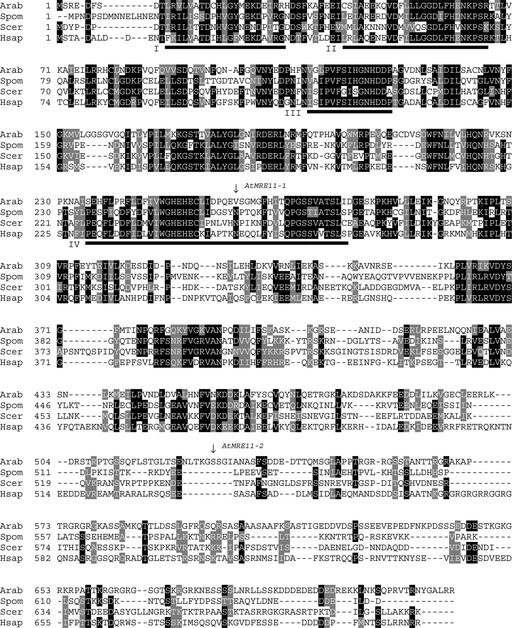
The MRE11 gene has been identified in the genomes of all of the eukaryotes sequenced to date. Recently, the sequencing of the genome of Arabidopsis allowed the identification of a plant MRE11 ortholog (Hartung and Puchta, 1999). A comparison of the Arabidopsis Mre11 protein (AtMre11) with Mre11 proteins from several other organisms is shown in Figure 1. The homology between the different Mre11 orthologs is strongest in the four conserved phosphoesterase domains but is less pronounced in the C terminus of the proteins. We screened the Arabidopsis Knockout Facility (Krysan et al., 1999) to identify Arabidopsis lines containing T-DNA insertions in AtMRE11. After several rounds of screening, we were able to isolate two Arabidopsis lines that contain such insertions and named them AtMRE11-1 and AtMRE11-2.
Figure 1.
Alignment of Mre11 Proteins from Arabidopsis (Arab), Schizosaccharomyces pombe (Spom), S. cerevisiae (Scer), and Humans (Hsap).
The phosphoesterase motifs (I to IV) conserved between the Mre11 proteins and the E. coli SbcC nuclease are indicated with black boxes (Sharples and Leach, 1995). The positions at which the Arabidopsis Mre11 protein is truncated by the T-DNA insertions in the AtMRE11-1 and AtMRE11-2 lines are indicated with arrows.
Isolation and Characterization of the AtMRE11-1 Mutant
A reverse genetics approach was taken to identify Arabidopsis lines containing mutations in AtMRE11. The gene-specific primer TS1 in combination with a T-DNA–specific primer (JL-202) were used to screen DNA pools from a collection of T-DNA–transformed Arabidopsis plant lines. A 1.7-kb PCR product was amplified using these primers. Sequencing of this product revealed that a plant line was present in the collection containing a T-DNA copy inserted into exon 9 of the AtMRE11 gene. After several further PCR rounds on smaller DNA pools, we eventually isolated a single plant containing this T-DNA insertion and named the line AtMRE11-1.
The single original plant of the AtMRE11-1 line we isolated appeared normal and produced seeds. On a DNA gel blot, this plant was heterozygous for the T-DNA insertion in AtMRE11 (Figure 2C, lane 3). Seeds from this original plant were germinated on medium containing kanamycin. The seedlings segregated 3:1 for kanamycin resistance:sensitivity (203 resistant:69 sensitive), indicating that this line contained a single T-DNA locus. This finding was confirmed by DNA gel blot analysis using a NPTII probe to detect the T-DNA (data not shown). Six days after germination, approximately one-third (64) of the kanamycin-resistant seedlings started to show a dwarf phenotype (Figure 3A). DNA from a pool of these small seedlings (60 in total) was isolated and analyzed on a DNA gel blot (Figure 2C, lane 2). All of these dwarf plants were homozygous for the T-DNA insertion, as demonstrated by the absence of a characteristic 5.6-kb EcoRI fragment. Furthermore, this result shows that the dwarf phenotype was linked to the T-DNA insertion.
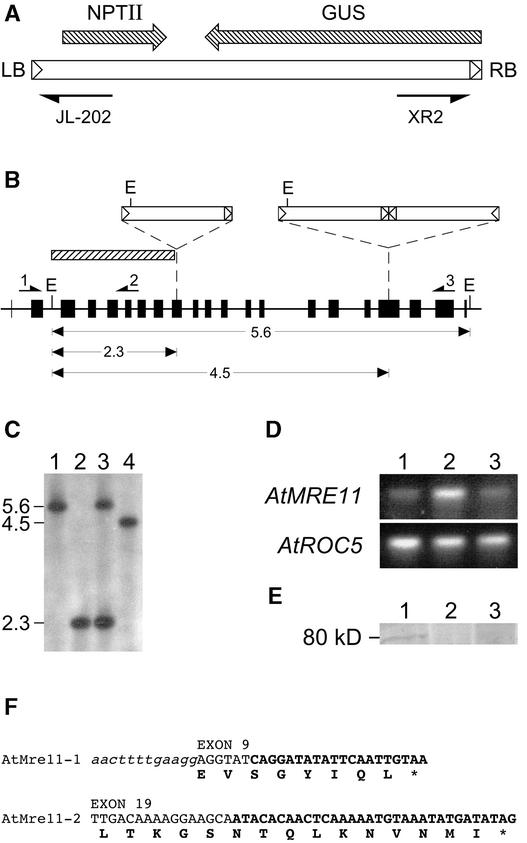
Figure 2.
Molecular Analysis of the T-DNA Insertions.
(A) Scheme of the T-DNA used for mutagenesis. The orientations of the NPTII and β-glucuronidase (GUS) transcription cassettes are shown. The primers JL-202 and XR2 also are shown. LB, left border; RB, right border.
(B) The genomic organization of AtMRE11 is shown (22 exons) with the positions of the integrated T-DNAs. The shaded rectangle represents the probe used for the DNA gel blot. The primers used for reverse transcriptase–mediated (RT) PCR are also shown (1, RT-MRE1; 2, RT-MRE2; 3, PRE3). In the AtMRE11-1 line, the left border of the T-DNA was linked to the AtMRE11 sequence at position 953. The right border end of the T-DNA was inserted at position 979. This results in a 26-bp deletion of genomic DNA in exon 9 of the gene. Two base pairs were lost from the left border T-DNA, whereas the right border end remained intact. In the AtMRE11-2 line, filler DNA was found on both sides of the inverted repeat. This filler DNA (65 and 59 bp) was linked to positions 1757 and 1759, respectively, in the AtMRE11 sequence. Therefore, in the AtMRE11-2 line, the integration of the T-DNA inverted repeat resulted in the deletion of a single nucleotide from the AtMRE11 locus (G at position 1758). E, EcoRI restriction site.
(C) DNA gel blot of the AtMRE11-1 and AtMRE11-2 lines. Lane 1, wild- type seedlings (Wassilewskija); lane 2, dwarf kanamycin-resistant AtMRE11-1 seedlings; lane 3, normal kanamycin-resistant AtMRE11-1 seedlings; lane 4, normal kanamycin-resistant AtMRE11-2 seedlings. All DNA samples were digested with EcoRI.
(D) RT-PCR performed on total RNA isolated from the two mutant lines. RT-PCR was performed using the primers RT-MRE1 and RT-MRE2 (top) to amplify the AtMRE11 cDNA. Amplification of the cyclophilin AtROC5 cDNA (bottom) was performed using primers RT-ROC3.1 and RT-ROC5.1. Lane 1, wild type (Wassilewskija); lane 2, AtMRE11-1−/−; lane 3, AtMRE11-2−/−.
(E) Protein gel blot analysis using a polyclonal antibody raised against the N terminus of the AtMre11 protein. Lane 1, wild-type seedlings (Wassilewskija); lane 2, AtMRE11-1−/− plants; lane 3, AtMRE11-2−/− plants.
(F) Sequence analysis of the T-DNA insertions in the AtMRE11-1 and AtMRE11-2 lines. The top lines show the genomic sequence of the AtMRE11 locus. Intron sequences are shown in lowercase italic letters. Exon sequences are shown in uppercase letters. The nucleotides derived from the T-DNA insertion are shown in uppercase boldface letters. The bottom lines show the predicted amino acid sequence as a result of the T-DNA insertions. Ten amino acids (NTQLKNVNMI) are derived from the filler DNA and form the C terminus of the predicted protein in the AtMRE11-2 line.
Figure 3.
Phenotypes of the AtMRE11-1 and AtMRE11-2 Lines.
(A) At left, AtMRE11-1−/− seedlings; at right, AtMRE11-1+/− seedlings.
(B) Mature plants. From left to right, AtMRE11-1−/−, AtMRE11-1+/−, AtMRE11-2−/−, and the wild type.
(C) Fasciation in mature AtMRE11-1−/− plants.
(D) to (G) AtMRE11-1−/− seedlings.
(H) Main root of an AtMRE11-1+/− seedling.
(I) Main root of an AtMRE11-1−/− seedling.
(J) Lateral root of an AtMRE11-1−/− seedling.
(K) Cotyledon epidermis of an AtMRE11-1−/− seedling.
(L) Dark-field view of a leaf from an AtMRE11-1−/− seedling.
To establish the effect of this T-DNA insertion on plant development, a detailed phenotypic analysis of the AtMRE11-1 line was performed. Because the dwarf seedlings homozygous for the AtMRE11-1 mutation (AtMRE11-1−/−) grew so slowly, we left them to grow on half-strength Murashige and Skoog (1962) (MS) medium for up to 6 weeks. After this period, it was apparent that these plants had severe developmental abnormalities, but the severity of the phenotypes varied between different seedlings (Figures 3D to 3G). In all of these seedlings, the hypocotyls were thickened, the roots became agravitropic, and the plants had an altered leaf morphology and an obvious defect in radial symmetry. The cotyledons also showed early necrosis. This often began as a brown spot (Figure 3E), which then spread to the entire cotyledon. This process often was very fast, with the first signs of necrosis to complete cotyledon necrosis occurring within 24 to 36 h. The necrosis occurred earlier in smaller seedlings with the more severe developmental phenotypes. Cotyledons and leaves from plants showing the mild phenotypes were cleared and studied using dark-field microscopy. Every leaf studied had an altered morphology and defects in the vasculature (Figure 3L). In a detailed analysis of the leaf epidermis, we also often observed clustering of stomata (Figure 3K). Examination of the root meristems also revealed some severe defects. As a control, root meristems from AtMRE11-1+/− plants were examined (Figure 3H). These showed the distinctive precise organization of cell files characteristic of the Arabidopsis root meristem. By contrast, the root meristem of 6-week-old AtMRE11-1−/− seedlings had lost all organization and were composed of large cells (Figure 3I). Development of lateral roots seemed to begin normally, but this broke down completely as the lateral roots matured (Figure 3J).
Only the fittest AtMRE11-1−/− seedlings (Figure 3D) survived in soil to form mature plants (Figure 3B). In fact, the most severely affected seedlings eventually died on the half-strength MS medium after approximately 8 weeks. Plants with the milder phenotype that grew into mature plants in soil often showed fasciation in the stems or in the floral organs and had very few small, thin leaves (Figure 3C). The components of the MRX complex are essential for gametogenesis (Ajimura et al., 1993; Chin and Villeneuve, 2001; Gallego et al., 2001). In line with this fact, we found that AtMRE11-1−/− plants did produce flowers, but these were sterile. On closer inspection, it was found that the plants did not produce any pollen. In C. elegans, the MRE11 gene also is not essential. However, worms homozygous for the ok179 mutation in CeMRE11 show a progressive loss of viability over several generations, indicating that CeMre11 is essential for maintaining reproductive capacity in this species (Chin and Villeneuve, 2001). Like the AtMRE11-1 mutation, the ok179 mutation in the CeMRE11 ortholog also deletes the fourth conserved phosphoesterase domain.
When we germinated seed from AtMRE11-1+/− plants on medium, we observed a 3:1 segregation of the dwarf phenotype, suggesting that the AtMRE11-1 mutation was recessive. When AtMRE11-1+/− seedlings were germinated on half-strength MS medium and then transferred to soil, they appeared normal. However, when seed from AtMRE11-1+/− plants was sown directly in soil, the plants that were heterozygous for the AtMRE11-1 mutation presented several phenotypes, such as fasciation and altered leaf morphology characteristic of the mature AtMRE11-1−/− plants (data not shown). This finding suggested that the AtMRE11-1 mutation can, under certain stress conditions, act in a semidominant manner. In these experiments, we never observed mature AtMRE11-1−/− plants, suggesting that they are unable to develop directly in soil.
We attempted to complement the developmental phenotypes of the AtMRE11-1 line by expressing the AtMRE11 cDNA under the control of the constitutive 35S promoter. T2 AtMRE11-1−/− and AtMRE11-1+/− seedlings from several independent lines expressing the AtMRE11 cDNA were checked for developmental phenotypes. The complemented AtMRE11-1−/− seedlings were intermediate in size and developed normally (data not shown). However, the fertility of such plants was reduced compared with that of complemented AtMRE11-1+/− plants, which produced wild-type amounts of seed. The siliques were small and contained only a few viable seeds. Therefore, in the AtMRE11-1 line, we were able to complement the fasciation phenotypes, but the sterility and growth defects of AtMRE11-1−/− were complemented only partially. This finding is not surprising, given the semidominant nature of the AtMRE11-1 mutation.
Isolation and Characterization of the AtMRE11-2 Mutant
Screening of the plant population detected a second plant that contained a T-DNA insertion in AtMRE11. In a PCR screen of pooled DNA, the primers TS1 and JL-202 amplified a 4-kb fragment. Sequencing of this fragment showed that a plant line was present in the collection that contained a T-DNA insertion in exon 19. A single plant with this T-DNA insertion was isolated, and this line was named AtMRE11-2. On a DNA gel blot, we showed that it was homozygous for the T-DNA insertion (Figure 2C, lane 4). This line showed two bands on a DNA gel blot when a NPTII probe was used to detect the T-DNA copy number (data not shown), suggesting that the AtMRE11-2 line contained two T-DNA copies. The T-DNA copies were linked, because seedlings resulting from a backcross of the AtMRE11-2 line with the wild-type line (Wassilewskija) segregated 3:1 for kanamycin resistance:sensitivity. We mapped both ends of the T-DNA insertion and found that the two T-DNA copies were present in the AtMRE11-2 line as an inverted repeat (Figure 2B).
The AtMRE11-2−/− plants developed normally and showed none of the developmental defects observed in AtMRE11-1−/− plants (Figure 3B). Unexpectedly, AtMRE11-2−/− plants also were fertile. AtMRE11-2−/− plants were grown for six generations to test any possible effect of the mutation on reproductive capacity. No difference in the phenotype of plants or the number of seeds they produced was observed between plants from the first or later generations. Therefore, we propose that the missing 191 amino acids from the C terminus of the AtMre11 protein are not necessary for gametogenesis in Arabidopsis.
Expression Analysis
The nature of the AtMRE11-1 and AtMRE11-2 mutations was studied further using reverse transcriptase–mediated (RT) PCR. As shown in Figure 2D, using this semiquantitative RT-PCR method, a 429-bp fragment from the 5′ region of the AtMRE11 cDNA was detected in cDNA pools synthesized from total RNA isolated from both wild-type and mutant seedlings. In several independent experiments, we always observed more product from AtMRE11-1−/− seedlings. Consequently, we conclude that the T-DNA insertions do not lead to instability of the AtMRE11 mRNA. We investigated this further and performed RT-PCR using primers annealing to the T-DNA and a primer located in the 3′ region of the AtMRE11 cDNA. These primers also amplified a product of the expected size (data not shown), suggesting that the mutant lines produced a AtMRE11 mRNA that included the T-DNA sequences. As a result of the orientation of the transcription cassettes on the T-DNA (Figure 2A), it is unlikely that transcription begins at expression signals in the T-DNA and proceeds toward the 3′ region of AtMRE11. Control experiments amplified a 400-bp fragment of the cyclophilin AtROC5 cDNA that was present in equal amounts in both the wild-type and mutant cDNA populations. Protein gel blot analysis was performed on wild-type, AtMRE11-1−/−, and AtMRE11-2−/− plants (Figure 2E) using a polyclonal antibody raised against the N terminus of AtMre11. An 80-kD band, the predicted size of the AtMre11 protein, was present only in wild-type plants.
AtMRE11-2−/− Plants Are Hypersensitive to DNA-Damaging Treatments
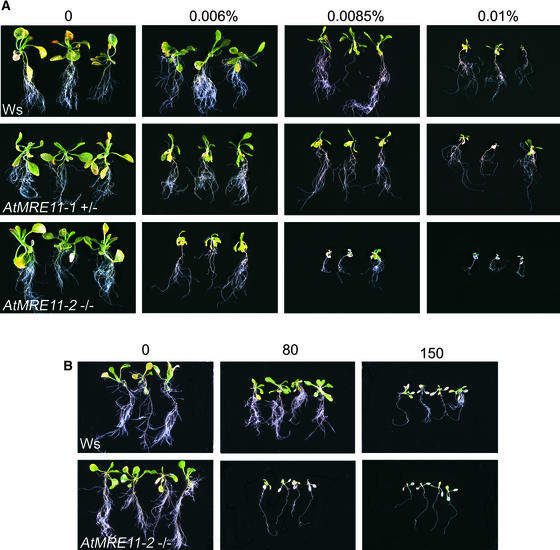
We tested whether plants with the AtMRE11-1 and AtMRE11-2 mutations were hypersensitive to DNA-damaging treatments. The AtMRE11-2−/− plants were hypersensitive to both methyl methane sulfonate and x-ray treatments (Figure 4). This phenotype was shown to be recessive, because we observed a 3:1 segregation of MMS resistance:sensitivity in F2 plants resulting from a backcross of AtMRE11-2−/− plants with the wild type. From studying the developmental phenotypes caused by the AtMRE11-1 mutation, we suspected that this mutation was semidominant. To test this idea, we checked the MMS sensitivity of AtMRE11-1+/− plants. These plants also were hypersensitive to MMS (Figure 4), although not to the same extent as AtMRE11-2−/− plants. Finally, the MMS hypersensitivity phenotype was complemented in AtMRE11-2−/− plants when the AtMRE11 cDNA was expressed by the constitutive 35S promoter (data not shown).
Figure 4.
Response of the AtMRE11-1 and AtMRE11-2 Lines to DNA-Damaging Treatments.
(A) Hypersensitivity of the AtMRE11-1 and AtMRE11-2 lines to MMS. The MMS concentrations tested are given above the top row. Ws, Wassilewskija.
(B) Hypersensitivity of AtMRE11-2−/− plants to x-rays. The x-ray dosage (Gray) is shown above the top row.
Lengthened Telomeres in the AtMRE11-1 and AtMRE11-2 Mutants
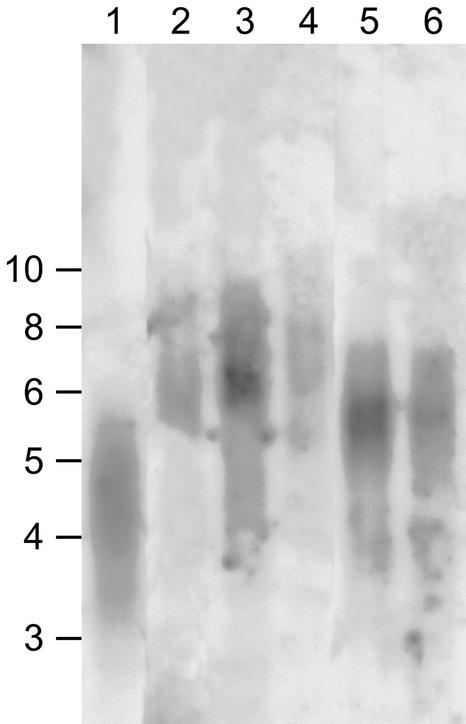
In other organisms, mutations in components of the NHEJ pathway shorten the telomeres. To investigate this possibility in plants, and in view of the observed developmental abnormalities in the AtMRE11-1 line, we determined the length of the telomeres in AtMRE11-1−/−, AtMRE11-1+/−, and AtMRE11-2−/− plants. Because the AtMRE11-2−/− plants are fertile, we also were able to determine whether the telomere length changed during six successive generations. We isolated DNA from seedlings from each generation and performed DNA gel blot analysis using a telomere-repeat probe to detect the characteristic telomere smears. To our surprise, in all of the mutant plants analyzed, the telomeres were longer than those in the wild-type plant (Figure 5). In the AtMRE11-2−/− plants, the telomeres also were maintained at this longer size during all six generations tested (data not shown). In addition, we found that the AtMRE11-1 mutation is dominant for increased telomere length, because the AtMRE11-1+/− plants also showed lengthened telomeres, although in this case the telomeres were not lengthened to the extent seen in AtMRE11-2−/− plants. These results show that although the MRE11 genes are conserved in many organisms, in plants it functions to maintain shortened telomeres, and the C terminus of the protein is essential for this process. Recently, we and others have isolated a plant line containing a mutation in the Arabidopsis KU70 (AtKU70) gene. In line with the data presented here, plants homozygous for this mutation also show lengthened telomeres, but in this case, their length is increased to greater than ∼30 kb (Bundock et al., 2002; Riha et al., 2002). Thus, although plants contain orthologs of the MRE11 and KU70 genes, these proteins have a different effect on telomere length than that observed in yeast and animals.
Figure 5.
Telomere Length in the AtMRE11-1 and AtMRE11-2 Lines.
Lane 1, wild type (Wassilewskija); lanes 2 to 4, three successive generations (T2 to T4) of AtMRE11-2−/− plants; lane 5, AtMRE11-1+/− plants (T3 generation); lane 6, AtMRE11-1−/− plants.
DISCUSSION
Previous studies have revealed that Mre11, as part of a complex with the Rad50 and Xrs2/Nbs1 proteins, functions in many diverse mechanisms of DNA repair and metabolism. Here, we report the isolation and characterization of two Arabidopsis lines containing mutations in the AtMRE11 gene. The differences between the two lines are striking. Although AtMRE11-1−/− plants are dwarf, sterile, and show numerous developmental defects, AtMRE11-2−/− plants are normal and fertile. The developmental, sensitivity to DNA-damaging agents, and telomere length phenotypes of the AtMRE11-1 mutation demonstrate that it is semidominant. Expression analysis showed that the mutant plant lines express AtMRE11 mRNA to wild-type levels; however, because of the T-DNA insertions, we were unable to detect the full-length AtMre11 proteins. The antibody also recognized other proteins, present in all samples analyzed, that were identical in size to the predicted truncated proteins in the AtMRE11-1−/− and AtMRE11-2−/− plants. Therefore, we were unable to detect such truncated proteins, although given the semidominant nature of the AtMRE11-1 mutation, we would expect a truncated protein to be produced in this line. A truncated AtMre11 protein in AtMRE11−/− plants may function in a dominant manner by titrating out cellular components involved in development, MMS resistance, and telomere length maintenance. This truncated protein may prevent the formation of AtMre11 homodimers or the interaction with AtRad50 or with other unidentified proteins. Similarly, the phenotypes of AtMRE11-2−/− plants could be attributable to altered protein–protein interactions. It will be interesting to perform coimmunoprecipitation experiments with extracts from the mutant plants to identify proteins that copurify with any truncated forms of AtMre11 produced in the mutant lines. Such experiments also would allow us to map the interaction domains of AtMre11 with itself and with AtRad50. Alternatively, the developmental phenotypes may be characteristic of an accumulation of inaccurately repaired DNA breaks during cell growth. However, if this were the case, such phenotypes would be apparent in AtMRE11-2−/− plants, which also are affected in DNA repair, as shown by the hypersensitivity to MMS.
The data presented here indicate that AtMre11 influences the length of plant telomeres. Telomeres are specialized structures at the ends of chromosomes that are essential for genomic stability. Telomeres are made up of short G-rich repeats that conform to the consensus sequence Tx(A)Gy. The length of the telomeres varies by organism and cell type. In Arabidopsis, telomeres are 2 to 4 kb in length and consist of repeats of the sequence TTTAGGGn. During mitosis, the DNA replication machinery is unable to replicate the ends of chromosomes, leading to a progressive shortening of chromosomes during successive cell divisions. This had led to speculation that telomere shortening may be linked to cellular senescence. This end replication problem is overcome by the activity of the reverse transcriptase telomerase, which is able to add telomere sequences to the ends of chromosomes. In mammals, telomerase is regulated developmentally in different cell types and shows high expression in reproductive tissues but is inactivated in somatic tissues. The telomerase activity in different plant tissues also has been measured. The highest activity is found in proliferating tissues, such as meristems, but telomerase activity is low or undetectable in nonmeristematic tissues, such as leaves (Rhia et al., 1998; Fitzgerald et al., 1999).
Recently published reports (Riha et al., 2001) have described the characterization of an Arabidopsis telomerase mutant (tert). Such plants have a mutation in the reverse transcriptase subunit of telomerase, resulting in shortening of the telomeres (∼500 bp per generation). The plants grow normally for five generations, at which point subsequent generations show increasingly severe developmental defects, presumably as a result of the loss of telomeres that is associated with genomic instability. Telomere shortening also has been reported in cells lacking components of the MRX complex or the Ku heterodimer (Porter et al., 1996; Boulton et al., 1998; D'Adda di Fagagna, 2001). Because telomeres are a source of free DNA ends in the cell, it is not surprising that proteins involved in DSB repair are involved in their maintenance. Given that many studies have described telomere shortening in NHEJ mutants, it is especially surprising that we observed lengthening of telomeres in the AtMRE11 mutants.
Several hypotheses may explain this result. In the cell, telomere length is thought to be governed by a homeostasis mechanism that operates via the influence of positive and negative factors on the enzyme telomerase. One possible role of AtMre11 at Arabidopsis telomeres may be its action on proteins that inhibit telomere elongation. In mammalian cells, the telomere binding proteins TRF1 and TRF2 have been identified. Cells expressing truncated dominant-negative forms of these proteins have lengthened telomeres (van Steensel and de Lange, 1997; Smogorzewska et al., 2000). In human cells, the TRF2 protein copurifies with components of the MRX complex (Zhu et al., 2000). The TRF1 and TRF2 proteins are thought to promote the formation of a large duplex loop (T loop) at mammalian telomeres, whereby the telomere is looped backward so that the single-stranded tail invades duplex telomeric sequences, making it inaccessible to telomerase. Zhu and colleagues propose that TRF2 recruits the MRX complex to the telomeres to facilitate T-loop formation by stabilizing the interaction of the invading strand with the telomeric sequences, analogous to its proposed role in recombinational repair. In Arabidopsis, a telomere binding protein (AtTRP1) homologous with TRF1 has been identified (Chen et al., 2001a). As we have proposed for the action of AtKu70 on telomere length in Arabidopsis, AtMre11 may bind normally to AtTRP1 and regulate its proposed negative effect on telomere length. As we have observed previously in the AtKU70-1 mutant plant line, all of the telomeres in the cell appear to be lengthened in the AtMRE11 mutant plants. This observation suggests a model in which telomere homeostasis has been affected.
Our data are in contrast to recent data reporting that no change in telomere length was seen in plants lacking AtRad50. In addition, cultured cell lines lacking AtRad50 showed a progressive shortening of telomeres (Gallego and White, 2001). In human cells, MRE11, RAD50, and NBS1 all coimmunoprecipitate with the telomeric protein TRF2 and colocalize at the telomeres (Zhu et al., 2000). Assuming that such a colocalization also occurs in plants, it is possible that it is mediated by a direct interaction between AtMre11 and a TRF2 ortholog and that only AtMre11 is required for the maintenance of short telomeres. The different extents to which the telomeres lengthened in AtMRE11-1−/− and AtMRE11-2−/− plants may be explained by the dominant nature of the AtMRE11-1 mutation. Like the developmental defects, any truncated AtMre11 produced may only partially titrate out other telomere length-controlling components, resulting in the intermediate effect we observed. In S. cerevisiae, a mutation in the first phosphoesterase domain (Mre11D16A) leads to intermediate telomere shortening, whereas the C terminus of Mre11 (amino acids 643 to 692) is not necessary for telomere maintenance (Furuse et al., 1998).
It is important to stress the differences found in this study between the proposed functional domains of the Mre11 proteins of S. cerevisiae and Arabidopsis. Many studies have attempted to map the regions of the S. cerevisiae Mre11 protein that are required for meiosis. Many mutations in the conserved phosphoesterase motifs (e.g., P84S, T188I, H213Y, and H125N), in addition to deletion of the C terminus of Mre11, cause defects in meiosis (Nairz and Klein, 1997; Bressan et al., 1998; Furuse et al., 1998; Tsubouchi and Ogawa, 1998; Usui et al., 1998; Moreau et al., 1999). The C terminus of AtMre11 is dispensable for gametogenesis in Arabidopsis. Therefore, the regions of AtMre11 required for meiosis appear to be different from those mapped in the yeast Mre11 protein. The S. cerevisiae mre11-5 mutant (Usui et al., 1998) also shows that functional comparisons between yeast and Arabidopsis Mre11 proteins are not always possible. The mre11-5 mutant is missing 136 amino acids of the C terminus of Mre11, producing a truncated protein similar to that in the AtMRE11-2 mutant; however, in contrast to the AtMRE11-2−/− plants, these yeast mutants do not sporulate but are resistant to genotoxic agents.
In conclusion, we have demonstrated that AtMRE11 plays a role in DNA repair, plant development, and telomere maintenance in Arabidopsis. Plants seem to be able to tolerate mutations in nonhomologous end joining (NHEJ) genes much more readily than mammalian cells. This makes plants an ideal system in which to investigate the roles of NHR genes in multicellular organisms by studying lines that have multiple mutations in NHR genes, as has been done previously in yeast.
METHODS
Constructs
The Arabidopsis thaliana MRE11 cDNA was amplified from a pool of seedling cDNA and inserted into pGEM-T Easy (Promega) to generate pSDM2011. A NcoI-SpeI fragment then was cloned into pCAMBIA3301 cut with NcoI-NheI to form pSDM2012, which was electroporated to Agrobacterium tumefaciens strain LBA1115. Plants were transformed using the floral dip method (Clough and Bent, 1998). Seeds from these plants were collected and surface-sterilized. Transformants were selected on half-strength Murashige and Skoog (1962) (MS) medium containing glufosinate-ammonium (20 mg/L) and timentin (100 mg/L).
Isolation and Characterization of the Plant Lines
The primer TS1 (5′-TGATACACTTCGAGTACTTGTTGCAACTG-3′) was used in combination with a T-DNA left border primer (JL-202, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) in a PCR screen on pooled plant DNA to identify plant lines containing a T-DNA copy inserted into the AtMRE11 gene. Using primers TS1 and JL-202, specific 1775-bp (AtMRE11-1) and 4009-bp (AtMRE11-2) PCR fragments were amplified from the largest DNA pools and detected in consecutive smaller DNA pools until single positive plants were identified. PCR was performed on DNA from the AtMRE11-1 line using primers TS2 (5′-ATCCTACCCTAAACTGGTTAAATGTGCTG-3′) and XR2 (5′-TGGGAAAACCTGGCGTTACCCAACTTAAT-3′) to map the insertion point of the T-DNA right border. In the AtMRE11-2 line, the primers JL-202 and MRET1 (5′-AGCCAATCAGGAAACAGCTTTGGA-3′) were used to map the other end of the T-DNA inverted repeat. DNA for blotting was isolated using the DNeasy Mini Kit (Qiagen, Valencia, CA) and digested with EcoRI. The probe used on the DNA gel blot was a 1-kb genomic fragment amplified using the primers PRE1 (5′-ACTACGTTAGTTAAAGCC-3′) and MR3 (5′-GGCAAGTAACATTGTGGCTCACTC-3′). DNA gel blot analysis to detect telomere length was performed as described previously (Gallego and White, 2001). Briefly, DNA was isolated from 12-day-old seedlings using the DNeasy Plant Mini Kit (Qiagen), digested with MboI, separated on an agarose gel, and blotted. An end-labeled telomere repeat primer [5′-(TTTAGGG)6-3′] was used as a probe. Protein gel blot analysis was performed as described previously (Goedecke et al., 1999).
Hypersensitivity to Methyl Methane Sulfonate and X-Rays
Seeds from AtMRE11-2−/− or AtMRE11-1+/− plants were surface-sterilized as described previously (Weijers et al., 2001) and germinated on solid half-strength MS medium containing kanamycin (100 mg/L). Four days after germination, the kanamycin-resistant seedlings were transferred to liquid half-strength MS medium. Methyl methane sulfonate (Sigma) then was added to this medium to the stated concentrations (Figure 4). Plants were scored after 3 weeks. For x-ray sensitivity, 4-day-old seedlings in liquid half-strength MS medium were irradiated with the stated dosage of x-rays at 6 Gray/min using a 225 SMART x-ray machine as a source (Andrex SA, Copenhagen, Denmark) operated at 200 kV and 4 mA with a 1-mm aluminum filter. Plants were scored after 3 weeks of additional growth.
Reverse Transcriptase–Mediated PCR
RNA was isolated from 12-day-old seedlings using the RNeasy Mini Kit (Qiagen). Total RNA (1 μg) isolated from wild-type seedlings or homozygous mutant plants was used for the first-strand cDNA synthesis reaction, which was performed as described previously (Weijers et al., 2001). Fragments were amplified from these cDNA pools using Taq polymerase (Roche Diagnostics, Almere, The Netherlands). The primers RT-MRE1 (5′-TGCCACTTGGGCTACATGGAGA-3′) and RT-MRE2 (5′-GCCAACACCAGAACCACCAAGA-3′) were used to amplify a 429-bp fragment of the AtMRE11 cDNA. The T-DNA primers XR2 and JL-202 were used in combination with PRE3 (5′-CTTTAGTGCTCTCATCG-3′) to amplify 3′ regions of the AtMRE11 cDNA. The primers RT-ROC5.1 (5′-CGGGAAGGATCGTGA-TGGA-3′) and RT-ROC3.1 (5′-CCAACCTTCTCGATGGCCT-3′) were used to amplify a 400-bp fragment of the AtROC5 cDNA. PCR conditions were as follows: 2 min at 94°C and 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Number
The accession number for the AtMRE11 sequence is AJ243822.
Acknowledgments
We thank Hildo Offenberg for the kind gift of the anti-AtMre11 antibody, Haico van Attikum for our many useful discussions, and Peter Hock for his work on the figures. This work was supported by the Stichting Binair Vector Systeem and Framework V European Union project Plantrec (QLRT-2000-01397).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005959.
References
- Ajimura, M., Leem, S.H., and Ogawa, H. (1993). Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S.J., and Jackson, S.P. (1998). Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, D.A., Olivares, H.A., Nelms, B.E., and Petrini, J.H.J. (1998). Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., van Attikum, H., and Hooykaas, P. (2002). Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney, J.P., Maser, R.S., Olivares, H., Davies, E.M., Le Beau, M., Yates, J.R., Hays, L., Morgan, J.F., and Petrini, J.H. (1998). The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477–486. [DOI] [PubMed] [Google Scholar]
- Chen, C.M., Wang, C.T., and Ho, C.H. (2001. a). A plant gene encoding a Myb-like protein that binds telomeric GGTTTAG repeats in vitro. J. Biol. Chem. 276, 16511–16519. [DOI] [PubMed] [Google Scholar]
- Chen, H.T., et al. (2000). Response to RAG-mediated V(D)J cleavage by NBS1 and γ-H2AX. Science 290, 1962–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Trujillo, K., Ramos, W., Sung, P., and Tomkinson, A.E. (2001. b). Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/ Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Chin, G.M., and Villeneuve, A.M. (2001). C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G2 DNA damage checkpoint. Genes Dev. 15, 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- D'Adda di Fagagna, F., Hande, M.P., Tong, W.-M., Roth, D., Lansdorp, P.M., Wang, Z.-Q., and Jackson, S.P. (2001). Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11, 1192–1196. [DOI] [PubMed] [Google Scholar]
- Daoudal-Cotterell, S., Gallego, M.E., and White, C.I. (2002). The plant Rad50-Mre11 protein complex. FEMS Lett. 516, 164–166. [DOI] [PubMed] [Google Scholar]
- de Jager, M., van Noort, J., van Gent, D.C., Dekker, C., Kanaar, R., and Wyman, C. (2001). Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Dolganov, G.M., Maser, R.S., Novikov, A., Tosto, L., Chong, S., Bressan, D.A., and Petrini, J.H.J. (1996). Human Rad50 is physically associated with human Mre11: Identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol. Cell. Biol. 16, 4832–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, M.S., Riha, K., Gao, F., Ren, S., McKnight, T.D., and Shippen, D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., Nagase, Y., Tsubouchi, H., Murakami-Murofushi, K., Shibata, T., and Ohta, K. (1998). Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17, 6412–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., Jeanneau, M., Granier, F., Bouchez, D., Bechtold, N., and White, C.I. (2001). Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 25, 31–41. [DOI] [PubMed] [Google Scholar]
- Gallego, M.E., and White, C.I. (2001). RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedecke, W., Eijpe, M., Offenberg, H.H., van Aalderen, M., and Heyting, C. (1999). Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat. Genet. 23, 194–198. [DOI] [PubMed] [Google Scholar]
- Haber, J.E. (1998). The many interfaces of Mre11. Cell 95, 583–586. [DOI] [PubMed] [Google Scholar]
- Hartung, F., and Puchta, H. (1999). Isolation of the complete cDNA of the Mre11 homologue of Arabidopsis (accession No. AJ243822) indicates conservation of DNA recombination mechanisms between plants and other eukaryotes. Plant Physiol. 121, 312. [Google Scholar]
- Johzuka, K., and Ogawa, H. (1995). Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139, 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R.S., Mirzoeva, O.K., Wells, J., Olivares, H., Williams, B., Zinkel, R.A., Farnham, P.J., and Petrini, J.H. (2001). Mre11 complex and DNA replication: Linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21, 6006–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, G.T., Jin, S., Shannon, K.B., and Weaver, D.T. (1996). Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, D.P., and Orr-Weaver, T.L. (1998). Chromosome segregation during meiosis: Building an unambivalent bivalent. Curr. Top. Dev. Biol. 37, 263–299. [DOI] [PubMed] [Google Scholar]
- Moore, J.K., and Haber, J.E. (1996). Cell cycle and genetic requirements for two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, S., Ferguson, J.R., and Symington, L.S. (1999). The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nairz, K., and Klein, F. (1997). mre11S: A yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis. Genes Dev. 11, 2272–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, C.I., Bosco, G., Ross, L.O., Evans, S.K., Salinger, A.P., Moore, J.K., Haber, J.E., and Lundblad, V. (1998). Telomere maintenance is dependent on activities required for and repair of double-strand breaks. Curr. Biol. 8, 657–660. [DOI] [PubMed] [Google Scholar]
- Ohta, K., Nicolas, A., Furuse, M., Nabetani, A., Ogawa, H., and Shibata, T. (1998). Mutations in the MRE11, RAD50, XRS2 and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc. Natl. Acad. Sci. USA 95, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T.T., and Gellert, M. (1998). The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double strand breaks. Mol. Cell 1, 969–980. [DOI] [PubMed] [Google Scholar]
- Paull, T.T., and Gellert, M. (1999). Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, S.E., Greenwell, P.W., Ritchie, K.B., and Petes, T.D. (1996). The DNA-binding protein Hdfp1 (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhia, K., Fajkus, J., Siroky, J., and Vyskot, B. (1998). Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 10, 1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha, K., Watson, J.M., Parkey, J., and Shippen, D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples, G.J., and Leach, D.R. (1995). Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17, 1215–1217. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, G.S., Maser, R.S., Stankovic, T., Bressan, D.A., Kaplan, M.I., Jaspers, N.G., Raams, A., Byrd, P.J., Petrini, J.H., and Taylor, A.M. (1999). The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577–587. [DOI] [PubMed] [Google Scholar]
- Tavassoli, M., Shayeghi, M., Nasim, A., and Watts, F.Z. (1995). Cloning and characterization of the Schizosaccharomyces pombe rad32 gene: A gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo, K.M., Yuan, S.S., Lee, E.Y., and Sung, P. (1998). Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11 and p95. J. Biol. Chem. 273, 21447–21450. [DOI] [PubMed] [Google Scholar]
- Tsubouchi, H., and Ogawa, H. (1998). A novel mre11 mutation impairs processing of double strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 18, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., Ohta, T., Oshiumi, H., Tomizawa, J., Ogawa, H., and Ogawa, T. (1998). Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95, 705–716. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., and de Lange, T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–742. [DOI] [PubMed] [Google Scholar]
- Varon, R., et al. (1998). Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93, 467–476. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Franke-van Dijk, M., Vencken, R.-J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299. [DOI] [PubMed] [Google Scholar]
- Wilson, S., Warr, N., Taylor, D.L., and Watts, F.Z. (1999). The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 27, 2655–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y., and Weaver, D.T. (1997). Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25, 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., Sonoda, E., Sasaki, M.S., Morrison, C., Haraguchi, T., Hiraoka, Y., Yamashita, Y.M., Yagi, T., Takata, M., Price, C., Kakazu, N., and Takeda, S. (1999). Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.-D., Kuster, B., Mann, M., Petrini, J.H.J., and de Lange, T. (2000). Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25, 347–352. [DOI] [PubMed] [Google Scholar]