Abstract
Although it is well known that Tyr phosphatases play a critical role in signal transduction in animal cells, little is understood of the functional significance of Tyr phosphatases in higher plants. Here, we describe the functional analysis of an Arabidopsis gene (AtPTEN1) that encodes a Tyr phosphatase closely related to PTEN, a tumor suppressor in animals. The recombinant AtPTEN1 protein, like its homologs in animals, is an active phosphatase that dephosphorylates phosphotyrosine and phosphatidylinositol substrates. RNA gel blot analysis and examination of promoter-reporter constructs in transgenic Arabidopsis plants revealed that the AtPTEN1 gene is expressed exclusively in pollen grains during the late stage of development. Suppression of AtPTEN1 gene expression by RNA interference caused pollen cell death after mitosis. We conclude that AtPTEN1 is a pollen-specific phosphatase and is essential for pollen development.
INTRODUCTION
Reversible protein phosphorylation is the most common mechanism for signal transduction (Krebs, 1986; Hunter, 1995). Protein kinases and phosphatases that catalyze this process are categorized into two major classes based on their substrate specificity: the Ser/Thr kinases (phosphatases) and the Tyr kinases (phosphatases). Although both Ser/Thr and Tyr phosphorylation play a crucial role in cellular regulation in animal systems, only Ser/Thr phosphorylation has been established as an important regulatory mechanism in plant cells. Indeed, a large number of Ser/Thr kinases and phosphatases have been identified and shown to be important in various processes of plant growth and development (reviewed by Luan, 1998, 2000; Hardie, 1999). By contrast, a typical Tyr kinase has not been identified to date from a higher plant. It was thought that Tyr phosphatases might not exist either until recently, when several members of the protein Tyr phosphatase (PTP) family were characterized from Arabidopsis (Gupta et al., 1998; Xu et al., 1998; Fordham-Skelton et al., 1999; Ulm et al., 2001). These studies suggest that protein Tyr (de)phosphorylation performs critical functions in plants. In support of this hypothesis, a number of other genes that encode putative PTPs have been identified in the Arabidopsis genome.
All PTPs contain an active-site signature motif, (I/V)HCXAGXXR(S/T)G, that harbors the catalytic cysteinyl residue involved in the formation of a phosphoenzyme reaction intermediate (Guan and Dixon, 1990). The PTPs are categorized into three groups based on studies in animals: receptor-like PTPs, intracellular PTPs, and dual-specificity PTPs (DsPTPs) (Stone and Dixon, 1994). The receptor-like PTPs are found only in animals, not in fungi and plants. The other two categories exist in all eukaryotes, including plants (Gupta et al., 1998; Xu et al., 1998; Fordham-Skelton et al., 1999; Ulm et al., 2001). The DsPTPs constitute a special class of intracellular PTPs that are unique by hydrolyzing phosphoserine/phosphotyrosine in addition to phosphotyrosine. As a result, DsPTPs also are referred to as dual-specificity protein phosphatases. This subgroup of PTPs has been shown to regulate the mitogen-activated protein kinases (MAPKs) in a variety of signal transduction pathways in both animals and yeast (Keyse, 1998). Because a large number of MAPKs have been found in higher plants, PTP regulation of MAPKs may represent a mechanism common in all eukaryotic systems. Indeed, both AtPTP1 and DsPTP1 have been shown to dephosphorylate and deactivate a MAPK protein in vitro (Gupta et al., 1998; Huang et al., 2000). A putative DsPTP gene referred to as AtMKP1 is essential for UV resistance in Arabidopsis (Ulm et al., 2001). AtMKP1 mutants exhibit hyperactivation of plant protein kinase activities similar to animal ERK1/2, suggesting that AtMPK1 may function by regulating MAPK activity in Arabidopsis (Ulm et al., 2001).
PTEN (phosphatase and tensin homolog) is a unique DsPTP that contains the PTP catalytic core motif and shares significant similarity with the cytoskeleton-interacting protein tensin (Li et al., 1997; Liaw et al., 1997). PTEN not only functions as a DsPTP but also hydrolyzes phosphatidylinositol and inositol phosphates (Myers et al., 1997; Maehama and Dixon, 1998). In particular, phosphatidylinositol 3,4,5-triphosphate (PIP3) has been shown to be a physiological substrate of PTEN. PIP3 is the product of the phosphorylation of PIP2 by phosphatidylinositol 3-kinase (PI3K) and is essential for cell growth and cell cycle progression via the activation of protein kinase B (Maehama and Dixon, 1999). PTEN converts PIP3 to PIP2, thereby countering the action of PI3K and inhibiting cell growth (Stambolic et al., 1998; Wu et al., 1998; Maehama and Dixon, 1999; Sun et al., 1999; Maehama et al., 2001). Loss-of-function mutations in PTEN cause cancer in many cell types; therefore, PTEN is defined as a typical tumor suppressor (Maehama and Dixon, 1999). In addition to PIP3, studies have shown that PTEN dephosphorylates protein substrates, including focal adhesion kinase, and regulates MAPK pathways (Gu et al., 1998, 1999; Tamura et al., 1998). PTEN homologs are present and play critical roles in the development of other animals. For example, PTEN regulates cell number and cell size during eye development in Drosophila (Huang et al., 1999). In Caenorhabditis elegans, PTEN is involved in the insulin signaling pathway and regulates the life span of the animal (Ogg and Ruvkun, 1998). Most recently, studies have demonstrated a pivotal role for PTEN in chemotaxis in Dictyostelium (Funamoto et al., 2002). Here, we report that Arabidopsis, a flowering plant, also contain genes encoding PTEN-like phosphatases. One of them is expressed exclusively in pollen grains and is essential for pollen development.
RESULTS
Isolation and Sequence Analysis of a cDNA Encoding AtPTEN1
The identification of AtPTP1, AtDsPTP1, and AtMPK1 suggests that higher plants produce a number of PTPs that may function in a variety of cellular and developmental processes (Gupta et al., 1998; Luan, 1998, 2000; Xu et al., 1998; Fordham-Skelton et al., 1999; Luan et al., 2001; Ulm et al., 2001). We searched the Arabidopsis database for other PTPs and identified approximately a dozen DNA sequences that encode proteins with the catalytic core signature motif of PTPs, (I/V)HCXAGXXR(S/T)G (data not shown). Three such genes share highest homology with PTEN, a phosphatase with a tumor-suppressor function in animals (Maehama and Dixon, 1999). These include genes corresponding to GenBank accession numbers BAB11013, BAB02466, and T45864. We referred to these Arabidopsis PTEN-like genes as AtPTEN1 to AtPTEN3 (Figure 1). The present study focused on the functional analysis of AtPTEN1.
Figure 1.
Sequence Analysis of AtPTEN1.
(A) AtPTEN1 protein sequence deduced from the cDNA.
(B) Sequence alignments of Arabidopsis PTEN-like sequences and animal PTENs.
(C) Alignment of AtPTEN1 and a wild tomato pollen PTEN deduced from an EST clone.
In (B) and (C), the PTEN sequences shown are AtPTEN1, AtPTEN2, and AtPTEN3 from Arabidopsis, DmPTEN from Drosophila, HsPTEN from human, MmPTEN from rat, and LpPTEN from wild tomato. In (A) and (B), the numbers at right indicate the amino acid residue positions in the respective proteins, and the active site signature motif is underlined. In (B) and (C), the shaded boxes indicate identical amino acid residues. In (B), dashes represent gaps introduced to optimize the alignments.
Using genomic sequence information, the corresponding cDNA encoding AtPTEN1 was isolated and its sequence was analyzed (Figure 1). Compared with this cDNA sequence, the AtPTEN1 gene consists of seven exons and six introns, which span a 1.6-kb DNA region. The AtPTEN1 cDNA encodes a protein of 412 amino acid residues with a calculated molecular mass of 47.3 kD and a pI of 7.5. The N-terminal half of the deduced AtPTEN1 protein (Figure 1A) harbors the catalytic domain of the phosphatase and is highly conserved compared with animal PTEN proteins (Figure 1B). The C-terminal half of AtPTEN1 is very different from that of the animal PTENs and is not aligned here (Figure 1B). Recent structural analysis of PTEN indicates that this protein may be associated with membrane by the C-terminal half of the protein (Lee et al., 1999), consistent with its activity against a membrane phospholipid, PIP3. A unique C-terminal region in AtPTEN1 may suggest a distinct mode of action and/or a different subcellular localization. Searching the plant DNA databases using AtPTEN1 as a query identified several related genes in other plant species (data not shown). Among them, one EST from a wild tomato pollen cDNA library encodes a homolog of AtPTEN1. The EST-deduced partial protein sequence shares 75% identity (84% similarity) over a 100–amino acid region with AtPTEN1 (Figure 1C). This finding suggests that AtPTEN1 homologs are present in other plant species.
Recombinant AtPTEN1 Is an Active Phosphatase That Hydrolyzes Phosphotyrosine and PIP3
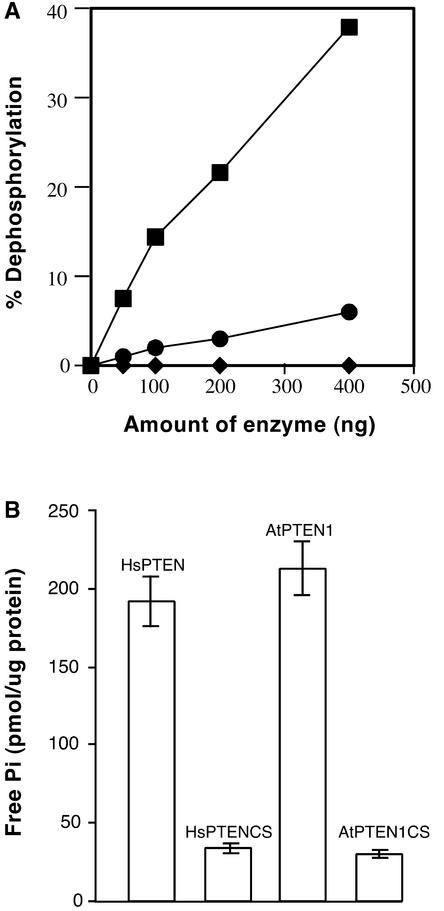
To address the function of AtPTEN1, we expressed the recombinant protein in Escherichia coli as a glutathione S-transferase–AtPTEN1 fusion protein and assayed its Tyr phosphatase activity using a 32P-labeled synthetic peptide (poly-Glu:Tyr, 4:1) as a substrate (Gupta et al., 1998). Like animal PTEN, AtPTEN1 effectively hydrolyzed the phosphotyrosine of this peptide substrate (Figure 2A). As shown in previous studies, the conserved Cys residue in the catalytic domain of all PTPs is essential for phosphatase activity. To determine whether the Cys residue in the putative catalytic core of AtPTEN1 is required for its activity, we produced a mutant protein (AtPTEN1C152S) by replacing the active-site Cys with Ser. This change completely abolished the Tyr phosphatase activity of AtPTEN1 (Figure 2A). Vanadate, a specific inhibitor for all PTPs, also inhibited AtPTEN1 activity against the peptide substrate. These results suggest that AtPTEN1 functions as a PTP in vitro.
Figure 2.
AtPTEN1 Is an Active Tyr and PIP3 Phosphatase.
(A) Tyr phosphatase activity of AtPTEN1 is proportional to enzyme concentration. The activity of AtPTEN1 (closed squares), AtPTEN1 in the presence of sodium vanadate (closed circles), and the AtPTEN1C152S mutant protein (closed diamonds) was measured as described in Methods. Relative activity is presented as a percentage of substrate dephosphorylation in 1 h.
(B) Dephosphorylation of PIP3 by AtPTEN1 and HsPTEN. The activity of wild-type or the Cys→Ser mutant of AtPTEN1 and HsPTEN against PIP3 was measured by the amount of free phosphate released from the substrate, as described in Methods.
Animal PTEN displays significant phosphatase activity against the phosphatidylinositol substrates, such as PIP3 (Maehama and Dixon, 1999). We conducted a similar in vitro phosphatase assay on AtPTEN1 and human PTEN (HsPTEN) using PIP3 as a substrate. As shown in Figure 2B, AtPTEN1 and HsPTEN displayed similar phosphatase activity on PIP3. As expected, the Cys-to-Ser mutation in the catalytic domain severely impaired the PIP3 phosphatase activity of both AtPTEN1 and HsPTEN (Figure 2B).
AtPTEN1 Is Expressed Specifically in Pollen Grains
To address the in vivo function of AtPTEN1, we examined its expression pattern in different organs of Arabidopsis by RNA gel blot analysis. The AtPTEN1 mRNA was highly abundant in flowers but was not detected in leaves, stems, or roots (Figure 3A). To examine AtPTEN1 expression during flower development, we collected flower buds and flowers at various developmental stages and analyzed the levels of AtPTEN1 transcript. As shown in Figure 3B, AtPTEN1 mRNA began to be detectable by RNA gel blot analysis after flower buds were separated from the cluster. It was most abundant in flower buds just before the opening of the flowers (Figure 3B). To determine which floral organ(s) expresses AtPTEN1, we dissected the flowers into various floral parts, and the total RNA from each of them was analyzed for AtPTEN1 expression by RNA gel blot analysis. Figure 3C shows that AtPTEN1 mRNA was detected only in stamens and pollen grains.
Figure 3.
AtPTEN1 Is an Anther- and Pollen-Specific Gene.
RNA gel blot analysis of AtPTEN1 mRNA levels in various plant organs (A), at different stages of flower development (B), and in separate floral parts (C) of Arabidopsis. In (A), the bottom gel shows ethidium bromide–stained rRNA as a loading control.
To further dissect the expression pattern of AtPTEN1, we generated transgenic Arabidopsis plants harboring the AtPTEN1 promoter–β-glucuronidase (GUS) reporter gene construct. We cloned the upstream promoter region of AtPTEN1 (−1082 to +1 in relation to ATG) and generated a translational fusion with the GUS coding sequence in the pBI101 vector (Jefferson et al., 1986). This construct was delivered into Arabidopsis by the floral dip method (Clough and Bent, 1998), and transformants were analyzed for GUS expression in the flowers of T1 plants. The pattern of GUS activity in the flower indicated that AtPTEN1 promoter was active in the anthers (Figure 4A). GUS activity often was detected on the stigma (Figure 4B). Further examination showed that the GUS staining on the stigma was attributable to the presence of pollen grains upon pollination (Figure 4B). When we examined the stained anthers more closely, GUS activity was associated only with pollen grains inside the anther (Figure 4C). Individual pollen grains were released from anthers and are shown in Figure 4D. To correlate AtPTEN1 promoter activity with the developmental stage of pollen grains, we stained anthers with both GUS substrate and 4′,6-diamindino-2-phenylindole (DAPI). In hundreds of pollen grains examined, GUS activity was observed only in trinuclear pollen grains (data not shown).
Figure 4.
AtPTEN1 Promoter Activity in Transgenic Arabidopsis Plants.
Transgenic plants harboring the AtPTEN1 promoter–GUS reporter construct were generated and analyzed histochemically for GUS activity in various floral organs. GUS activity (indicated by blue) was detected in the anthers of the whole flower (A), pollen grains on the stigma (B), pollen in the anther (C), and pollen grains released from the anther (D).
We also analyzed the young seedlings of T2 plants and detected no GUS activity in any of the vegetative tissues (data not shown). Based on the results from RNA gel blots and promoter-GUS activity assays in transgenic plants, we conclude that AtPTEN1 expression is associated exclusively with pollen grains at the trinuclear stage. This conclusion is consistent with the EST analysis discussed above. In all of the EST databases, we identified only one EST entry that represents a homolog of AtPTEN1, and this EST was isolated from a tomato pollen cDNA library (Figure 1C).
RNA Interference Silencing of AtPTEN1 Causes Pollen Cell Death during the Late Stage of Development
The pollen-specific expression of AtPTEN1 suggests that it may play a role in pollen development and/or function. To test this hypothesis, we disrupted the AtPTEN1 gene by reverse genetics procedures and examined changes in pollen development. Upon screening of ∼300,000 T-DNA insertional Arabidopsis lines, we were unable to identify a T-DNA insertion in AtPTEN1 (data not shown). This surprisingly low frequency of T-DNA insertion into AtPTEN1 appears to coincide with the results of a previous report (Bonhomme et al., 1998). That study indicated that the probability of finding a T-DNA insertion in a reproductive organ–specific gene is much lower than average, implying that many of these genes function in fertility (Bonhomme et al., 1998). In the case of AtPTEN1, it suggests that AtPTEN1 plays an essential role in fertility. We used an alternative approach to suppress the expression of AtPTEN1 by RNA interference (RNAi) that had been used successfully by others (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Gupta et al., 2002). To ensure a successful silencing, we constructed three different RNAi constructs that contained different promoters and/or different regions of the AtPTEN1 cDNA sequence. Because AtPTEN1 is a pollen-specific gene, we used pollen-specific promoters to drive the expression of the RNAi cassette. These included the native AtPTEN1 promoter (−1082 to +1 in relation to ATG) and the tomato Lat52 promoter (−666 to +1 in relation to ATG) (Twell et al., 1990; Muschietti et al., 1994). We also used different regions of the AtPTEN1 coding sequences in different RNAi constructs for the inverted repeat to express double-stranded RNA corresponding to AtPTEN1 (Figure 5A; see Methods for details). Transgenic Arabidopsis lines harboring these RNAi constructs and an empty vector control were generated by the floral dip method (Clough and Bent, 1998).
Figure 5.
Silencing of AtPTEN1 by the RNAi Procedure.
(A) Schemes of AtPTEN1 RNAi constructs. Transcription cassettes contain a partial AtPTEN1 cDNA (AtPTEN1), a partial GUS gene (GUS), the AtPTEN1 promoter or tomato Lat52 promoter (LeLat52 promoter), and the nopaline synthetase terminator (NOS). Shaded boxes depict the inverted repeat of AtPTEN1 cDNA. The hatched box in the bottom construct depicts the internal deletion in AtPTEN1 cDNA used to disrupt the open reading frame. Arrows point in the 5′-to-3′ direction.
(B) RNA gel blot analysis of AtPTEN1 transcripts from open flowers of control (lane 1) and AtPTEN1 RNAi lines containing different RNAi constructs (lanes 2 to 7). The top gel shows ethidium bromide–stained rRNA as a loading control. The bottom gel shows the RNA gel blot probed with AtPTEN1 cDNA.
(C) RT-PCR analysis of AtPTEN1 transcripts from open flowers in control and RNAi plants. Total RNAs isolated from open flowers of six independent AtPTEN1 RNAi lines from different constructs (lanes 2 to 7) and the empty vector line (lane 1) were used as templates in RT-PCR with either AtPTEN1-specific primers (bottom gel) or actin2-specific primers (top gel). M, 1-kb DNA ladder.
RNA gel blot analysis of AtPTEN1 mRNA detected a single mRNA species in the control plant (Figure 5B, bottom gel, lane 1) and a degrading pattern of multiple mRNA species in most of the RNAi lines from different constructs (Figure 5B, bottom gel, lanes 2 to 7). Multiple mRNA species in RNAi lines could represent the transcriptional products of the RNAi cassette and their degradation. Several lines harboring different RNAi constructs showed reduced levels of an mRNA species with a size comparable to that of the endogenous AtPTEN1 mRNA. However, it was difficult to conclude whether or not the mRNA species in the RNAi lines truly represent the level of endogenous AtPTEN1 mRNA, because both endogenous AtPTEN1 mRNA and RNAi products hybridized to the cDNA probe. Therefore, we examined the level of endogenous AtPTEN1 mRNA by reverse transcriptase–mediated (RT) PCR. We used a pair of oligonucleotides from the 5′ and 3′ untranslated regions that specifically recognize the endogenous mRNA but not the product of the RNAi cassette and/or its degraded products. All of the RNAi lines from different constructs (Figure 5C, bottom gel, lanes 2 to 7) showed a significant reduction in AtPTEN1 mRNA compared with the empty vector control line (Figure 5C, bottom gel, lane 1). As an internal control, the level of mRNA corresponding to the actin2 gene did not show a significant difference among different RNAi and control lines (Figure 5C, top gel).
We concluded that all three RNAi constructs worked as expected to effectively suppress AtPTEN1 expression. In most cases (Figure 5C), the mRNA level was reduced significantly but did not disappear completely, consistent with the pollen-specific expression pattern of the AtPTEN1 gene. Normally, an RNAi construct against a gene expressed in somatic cells causes suppression of the gene, even in heterozygous plants containing a single copy of the RNAi transgene (Chuang and Meyerowitz, 2000). However, the RNAi construct against a pollen-specific gene would not be expected to show 100% affected pollen in single-insertion events caused by the segregation of the RNAi construct during meiosis. Instead, a maximum of 50% reduction in mRNA levels would be observed. Most of the AtPTEN1 RNAi lines analyzed in this study contained multiple inserts (data not shown), which resulted in >50% reduction in AtPTEN1 mRNA (Figure 5C).
If AtPTEN1 is required for pollen development and/or function, its suppression in RNAi lines may show a phenotypic change in pollen grains. Indeed, microscopic analysis revealed that a large number of mature pollen grains in all RNAi plants were defective (Figure 6). Mature pollen grains from control plants were normal in shape (Figure 6A), in contrast to those of RNAi lines, which showed a mixture of both normal and aborted (collapsed) pollen grains (Figure 6B). The promoter-GUS experiments indicated that AtPTEN1 is expressed in the tricellular stage of pollen grains. To determine the stage of pollen grains affected in RNAi lines, flower buds were sorted according to various developmental stages (Piffanelli et al., 1998) and stained with DAPI. Consistent with the AtPTEN1 expression pattern, pollen cells at the tetrad, unicellular (data not shown), and bicellular (Figure 6C) stages were not affected, whereas tricellular pollen contained a mixture of normal and aborted/collapsed grains in RNAi plants (Figure 6B and data not shown).
Figure 6.
Phenotypic Analysis of Pollen from Control and AtPTEN1 RNAi Plants.
(A) Bright-field view of hydrated mature pollen from a control plant.
(B) Bright-field view of hydrated mature pollen from an AtPTEN1 RNAi plant.
(C) AtPTEN1 RNAi binuclear pollen.
(D) Propidium iodide–stained mature pollen from a control plant.
(E) to (G), (I), and (J) Rhodamine 123–stained mature pollen from control (E) and AtPTEN1 RNAi plants ([F], [G], [I], and [J]).
(H) Propidium iodide–stained mature pollen from an AtPTEN1 RNAi plant.
(I), (J), and (M) to (O) Enlarged views of pollen from AtPTEN1 RNAi plants show signs of cellular degeneration. Bright-field images in (M) and (N) are of the same pollen grain shown in (I) and (J), respectively. Arrows in (J) and (N) indicate the lesion in intine.
(K) and (O) Alexander vitality staining of pollen from a control plant (K) and an RNAi plant (O).
(L), (P), and (Q) Tinopal/DAPI double-stained mature pollen from a control plant (L) and RNAi plants ([P] and [Q]). Enlarged views of pollen from AtPTEN1 RNAi plants show signs of disintegration ([P] and [Q]).
To correlate the pollen defect with the silencing of AtPTEN1, we examined the percentage of defective pollen grains in several independent RNAi lines from different RNAi constructs using propidium iodide, a vitality stain that stains only the dead pollen grains (Huang et al., 1986). Control pollen grains generally were not stained with the dye (Figure 6D), whereas pollen from a RNAi line showed both viable pollen (unstained) and pollen stained brownish-purple, indicating aborted pollen even before collapsing (Figure 6H). The percentage of aborted pollen varied among the RNAi lines and was related closely to the reduction in AtPTEN1 mRNA (data not shown).
To further examine the cellular defects in RNAi pollen, mature tricellular pollen grains were stained with rhodamine 123, a stain used to measure metabolic activity in mitochondria. Mature pollen from control plants showed a uniform staining pattern (Figure 6E). By contrast, AtPTEN1 RNAi lines contained collapsed, aborted pollen grains lacking fluorescence (Figures 6F and 6G) or showing uneven fluorescence, which was a sign of cellular degeneration and loss of metabolic activity (Figure 6F). Closer examination of RNAi pollen detected some other cellular phenotypes. For instance, RNAi pollen grains have a receding plasma membrane (Figures 6I and 6M) or lesions in the intine (Figures 6J and 6N). Pollen viability and defects in intine structure in RNAi pollen were explored further using Alexander stain (a viability stain) or by double staining with tinopal (a cellulose stain) and DAPI (a nuclear stain). Viable mature pollen from wild-type plants was stained with Alexander stain, as shown in Figure 6K, whereas some pollen grains with a receding plasma membrane were observed in AtPTEN1 RNAi plants (Figure 6O). When mature pollen from AtPTEN1 RNAi plants were double stained with tinopal and DAPI, viable and collapsed pollen (data not shown) was observed as expected, along with aborted pollen grains clearly showing lesions (Figures 6P and 6Q). Control mature pollen double stained with DAPI and tinopal is shown as a reference (Figure 6L).
Using environmental scanning electron microscopy, pollen from control and AtPTEN1 RNAi plants was examined more closely. Pollen grains from a control plant appeared normal (Figures 7A and 7C), whereas a number of abnormal and collapsed pollen grains were detected in the AtPTEN1 RNAi anther (Figures 7B and 7D to 7F), consistent with the results observed using light microscopy (Figure 6). The collapsed pollen grains had shrunken, highly deformed exines and were reduced significantly in size (Figures 7D and 7E) compared with the normal pollen in the control plants. We frequently found invaginations (Figures 7D and 7F) on a number of AtPTEN1 RNAi pollen grains that were otherwise comparable to the control pollen in shape and size. More disruptions in the exine pattern were observed in pollen grains from RNAi plants. Such disruption of the exine wall led to larger gaps and lesions separating individual bacula (Figures 7D to 7F). Because such defects were not observed in the control pollen grains under the same conditions, we concluded that invagination formation and disruption of exine pattern in the mutant pollen were associated with the suppression of AtPTEN1 expression and may occur before pollen collapse.
Figure 7.
Analysis of Pollen from Control and AtPTEN1 RNAi Plants by Scanning Electron Microscopy.
Whole anthers from control (A) and RNAi (B) plants were examined by scanning electron microscopy. Pollen grains from a small area of anther surface are shown. Pollen grains from control (C) and AtPTEN1 RNAi (D) plants were photographed at a higher magnification to show invagination in RNAi pollen. The AtPTEN1 RNAi pollen also showed disruption in exine pattern ([E] and [F]). In (D) to (F), the arrows point to invaginations and bacula junctions. Bar in (A) = 10 μm for (A) and (B); bar in (C) = 5 μm for (C) to (E); bar in (F) = 2 μm.
In summary, pollen analysis in control and RNAi plants revealed an aborted pollen phenotype associated with AtPTEN1 suppression. The pollen cell death occurred at the beginning of the tricellular stage. We do not interpret other observations, such as the loss of metabolic activity and intine and exine wall defects, as unique symptoms attributable directly to the loss of AtPTEN1 function. Rather, these observations are consistent with an overall trend toward pollen cell death. We conclude from our phenotypic analysis that AtPTEN1 is essential for pollen maturation.
DISCUSSION
Our study characterized a PTEN homolog in a higher plant and identified its function in pollen development. The PTEN tumor suppressor is a phosphatase that can act on both phos-phorylated polypeptide and phosphoinositide substrates in vitro (Maehama and Dixon, 1999; Maehama et al., 2001). A number of studies using cell culture and knockout animal models have demonstrated that PIP3 is a physiological substrate for PTEN in the context of tumor suppression function in animals (Maehama and Dixon, 1999). PIP3 is a lipid second messenger produced by PI3K that is activated by growth factors (Wu et al., 1998; Sun et al., 1999). PIP3 binds to and activates the Akt/PKB protein kinase that in turn promotes cell cycle progression by activating cyclin-dependent protein kinase (Sun et al., 1999). PTEN suppresses cell division by dephosphorylating PIP3 and thereby inhibiting Akt/PKB activity. Although it is not known if AtPTEN1 uses PIP3 as a physiological substrate in pollen grains, in vitro activity assays indicate that AtPTEN1 enzyme activity is comparable to that of human PTEN in dephosphorylating PIP3. It is expected that PI-related molecules, including but not limited to PIP3, may serve as in vivo substrates for AtPTEN1 in pollen grains.
In contrast to our understanding of the PIP3 pathway in animal cells, little is known of PI function in cell cycle regulation in plants. None of the components in the animal PIP3 pathway has been identified in plant cells. For example, animal cells produce three types of PI3K, but only type-I enzymes participate in cell cycle regulation (Fruman et al., 1998). However, only type-III PI3K has been identified in plants (Hong and Verma, 1994; Welters et al., 1994). Although further analysis is required to determine whether PIP3 plays a similar role in plants as in animals, results from this study indicate that AtPTEN1 is involved in pollen development after the completion of meiosis and mitosis. The expression of AtPTEN1 became detectable only during late stages of pollen development, as demonstrated by both mRNA gel blot analysis (Figure 3B) and the promoter-GUS reporting system (Figure 4B). This expression pattern is consistent with the pollen defect detected in the transgenic plants containing the RNAi construct (Figure 6). In all of the plants examined, pollen cell death was observed after the formation of tricellular pollen grains. Therefore, a function for AtPTEN1 is likely to occur during the pollen maturation process after mitosis, suggesting that AtPTEN1 may not function in cell cycle regulation as PTEN does in animals.
Very little is known about the molecular mechanism underlying pollen maturation after cell division. Nevertheless, studies show that a large number of genes are transcribed after meiosis. The early phase of gene expression occurs after meiosis and decreases gradually during pollen development, whereas the late phase of gene expression starts at mitosis and increases as pollen matures (reviewed by Mascarenhas, 1989; McCormick, 1993). Many of the late gene transcripts may play a role during the germination of pollen, because the mRNAs are stored in the dry pollen grains and translated during germination (McCormick, 1993). It is clear that AtPTEN1 is a late phase gene and that its transcript is accumulated after mitosis. Therefore, AtPTEN1 might play a role in pollen germination and pollen tube growth in addition to its function in maturation. Although the function of PI-related compounds in pollen development has not been addressed, some studies have shown that PI metabolism is critical for pollen germination and tube growth (Franklin-Tong, 1999). PIP2 is a precursor of inositol 1,4,5-triphosphate (IP3), which has been shown to regulate calcium levels during pollen tube growth (Franklin-Tong et al., 1996). AtPTEN1 may dephosphorylate and convert PIP3 to PIP2 and potentially regulate calcium signaling in pollen germination.
Phosphoinositides play a critical role not only in cell cycle control and generating second messengers, such as IP3, for calcium signaling, but also in modulating a variety of cellular functions, including cytoskeletal organization and membrane trafficking (reviewed by Fruman et al., 1998; Takenawa and Itoh, 2001). Because mechanisms for cytoskeleton organization and cellular trafficking are highly conserved among eukaryotes, we speculate that PI metabolism may function in similar processes in plant cells, including pollen cells. During pollen maturation and germination, cellular trafficking is a vigorous process that is critical for cell wall deposition and cell shape remodeling (reviewed by Hepler et al., 2001). As one of the enzymes that are involved in the dephosphorylation of PIP3 and possibly other forms of PI metabolites, AtPTEN1 may regulate cellular trafficking and cell shape during pollen maturation by maintaining the concentration of certain PI metabolites, such as PIP2. AtPTEN1 function in PI metabolism also could be relevant to actin organization, which is critical for tip growth (Hepler et al., 2001).
Although PI-related molecules likely serve as physiological substrates for AtPTEN1 in pollen grains, we cannot exclude the possibility that AtPTEN1 may use protein substrates and regulate MAPK pathways, as indicated by studies in animal cells (Gu et al., 1998, 1999). Indeed, several MAPK genes are expressed in pollen grains (Wilson et al., 1995). At least one MAPK has been shown to activate during pollen rehydration and germination in tobacco (Wilson et al., 1997). Additional studies on the molecular mechanism of AtPTEN1 function will yield critical information on the regulation of pollen development.
METHODS
Cloning and Sequence Analyses of AtPTEN1 cDNA
A genome database search resulted in the identification of a genomic sequence encoding a protein that shares high homology with animal PTEN, especially in the N-terminal region. We named this gene AtPTEN1. This genomic sequence of AtPTEN1 was analyzed for the presence of introns and exons using the NetPlant gene program (Hebsgaard et al., 1996). Two primers, P68 (5′-CTTCCTCAACTT-CATGATCAATTC-3′) and P70 (5′-GTCTCAGCTGAGCTAGTCAGG-C-3′), were designed from the coding region of the putative AtPTEN1 gene. These primers were used to perform reverse transcriptase–mediated (RT) PCR on total RNA from flowers using the Calypso RT-PCR system (Tetra-Link International, Amherst, NY) according to the manufacturer's instructions. A 1371-bp AtPTEN1 cDNA was cloned and sequenced using fluorescent deoxynucleotide triphosphates, as described previously (Gupta et al., 1997).
Plant Materials and RNA Analyses
Arabidopsis thaliana plants (ecotype Columbia-gl) were grown in a greenhouse under long-day conditions (16-h/8-h light/dark cycle). Different organs were harvested, frozen in liquid nitrogen, and stored at −80°C if not used immediately. For RNA gel blot analysis, total RNA (10 μg) from different organs and pollen grains was isolated and resolved by electrophoresis on a 1.2% agarose-formaldehyde gel. The RNAs were transferred onto GeneScreen Plus nylon membranes (DuPont–New England Nuclear) and probed with 32P-labeled AtPTEN1 cDNA, as described previously (Gupta et al., 1997). To analyze the AtPTEN1 mRNA in RNA interference (RNAi) or control plants by RT-PCR, total RNA was extracted from open flowers from several independent lines at approximately the same developmental stage with Tripure reagent (Roche, Mannheim, Germany). Total RNA (1 μg) was heated to 65°C for 7 min and then subjected to reverse transcription reaction using Moloney murine leukemia virus reverse transcriptase (200 units per reaction; Life Technologies) and AmpliTaq (2 units per reaction; Perkin-Elmer) for 25 min at 48°C. PCR amplifications were performed with initial denaturation at 94°C for 2 min followed by 25 cycles of incubations at 94°C for 20 s, 55°C for 40 s, and 72°C for 1.5 min, and a final extension at 72°C for 10 min. Aliquots of individual PCR products were resolved by agarose gel electrophoresis and visualized with ethidium bromide under UV light.
Expression of the Recombinant AtPTEN1 Protein
The coding region of AtPTEN1 cDNA was amplified with Pfu polymerase (Stratagene) and cloned in frame with glutathione S-transferase (GST) in pGEX-KG vector (Amersham Pharmacia). The resulting clones were sequenced to ensure an in-frame fusion of AtPTEN1 with GST and to avoid clones that contain mutations introduced by PCR. The construct was transformed into Escherichia coli strain BL21 DE3 (Novagen, Madison, WI). Overexpression of the GST-AtPTEN1 fusion protein was performed as described previously (Gupta et al., 1998) with minor modifications. Briefly, bacterial culture was initiated and grown to mid-log phase. Expression of the GST fusion protein was induced by 0.1 mM isopropylthio-β-galactoside at 25°C for 2 h, and the bacterial cells were pelleted and resuspended in a buffer containing 100 mM NaCl, 50 mM Tris-HCl, pH 8.0, 2 μM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 2 mM EDTA. The GST fusion protein was purified by glutathione–Sepharose 4B according to the manufacturer's instructions (Amersham Pharmacia). All of the purification steps were performed at 4°C. Human PTEN (HsPTEN) and HsPTENCS mutant constructs in a GST vector were a kind gift from Hong Sun (Yale University, New Haven, CT) (Li et al., 1997). Expression and purification of HsPTEN was performed according to the procedure described above for AtPTEN1.
Site-Directed Mutagenesis
Two PCR primers, P81 (5′-CGCGGTTGTTCATTCCATGGCAGG-AAAAGG-3′) and P82 (5′-CCTTTTCCTGCCATGGAATGAACAACC-GCG-3′), were designed to mutagenize Cys-152 (TGC) to Ser-152 (TCC) in AtPTEN1. The site-directed mutagenesis was performed using the Quick-Change kit (Stratagene) according to the manufacturer's instructions. The mutation was confirmed by nucleotide sequencing before protein expression, and the mutant protein was purified as described for wild-type AtPTEN1.
Protein and Phosphatidylinositol 3,4,5-Triphosphate Phosphatase Assays
For the phosphatase assays using protein substrate, poly-Glu:Tyr (4:1) (Sigma) was labeled at Tyr residues. For Tyr labeling, 25 units of human c-Src Tyr kinase (Upstate Biotechnology, Lake Placid, NY) was incubated with 50 μg of protein substrate and 50 μCi of γ-32P-ATP (DuPont–New England Nuclear) in 100 μL of reaction buffer (25 mM Tris-HCl, pH 7.2, 5 mM MnCl2, 0.5 mM EGTA, 0.05 mM Na3VO4, and 25 mM Mg acetate) for 2 h at 30°C. The reaction mixture was loaded onto a Centri-Spin-10 column (Princeton Separations, Adelphia, NJ) to remove the free 32P-ATP and salts from the reaction. The purified labeled protein substrate was collected in the fluent after spinning at 750g for 3 min. Protein phosphatase activity was determined by measuring the release of free 32P from labeled substrates.
Phosphatidylinositol 3,4,5-triphosphate (PIP3) phosphatase activity was measured using a malachite green assay for colorimetric detection of inorganic phosphate release, as described previously (Maehama et al., 2000), with minor modifications. Phosphatase assays were performed with PIP3 (Biomol, Plymouth Meeting, PA) as a substrate for 12 min at 30°C (for AtPTEN1/AtPTEN1CS) or 37°C (for HsPTEN/HsPTENCS). The standard curve for estimating the released Pi was plotted as described previously (Maehama et al., 2000).
Generation of AtPTEN1 Promoter–β-Glucuronidase Reporter and AtPTEN1 RNAi Constructs and Arabidopsis Transformation
To study the expression pattern of AtPTEN1 in detail, the upstream promoter region (−1082 to +1 in relation to ATG) of AtPTEN1 was amplified by PCR and cloned as a translational fusion in frame with the β-glucuronidase (GUS) coding sequence in plant binary vector pBI101.1 (Jefferson et al., 1986). To silence AtPTEN1, we used the RNAi approach that has been shown to silence the endogenous genes successfully (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Gupta et al., 2002). We generated three different constructs using two different approaches. In one case, we used the partial GUS coding sequence between the inverted repeat of the partial AtPTEN1 cDNA sequence. To generate this construct, partial cDNA sequences encoding AtPTEN1 (nucleotides 1 to 500 of the cDNA) were amplified by PCR and cloned as inverted repeats with a partial GUS coding sequence (nucleotides 497 to 1530 in relation to the start codon) as a spacer. This RNAi cassette was flanked by either the AtPTEN1 promoter (−1082 to +1 in relation to ATG) or the tomato Lat52 promoter (−666 to +1 in relation to ATG) (Twell et al., 1990; Muschietti et al., 1994) and the nopaline synthetase terminator, respectively (Figure 5A). In another AtPTEN1 RNAi construct, the open reading frame of AtPTEN1 cDNA with an internal deletion (nucleotides 31 to 1265 of AtPTEN1 cDNA with an internal deletion of nucleotides 525 to 640) was cloned in the sense orientation followed by an inverted repeat of the 5′ end of the AtPTEN1 cDNA (nucleotides 31 to 510). The tomato Lat52 promoter (−666 to +1 in relation to ATG) (Twell et al., 1990) and the nopaline synthetase terminator (Figure 5A) flanked this RNAi cassette. Restriction digestion, PCR, and DNA sequencing confirmed the correct orientation of each fragment.
These AtPTEN1 RNAi constructs were transferred into Agrobacterium tumefaciens strain GV3101pMP9 to be used for Arabidopsis transformation. For transformation, Arabidopsis plants (ecotype Columbia) were grown in a greenhouse under long-day conditions (16-h/8-h light/dark cycle) for 4 weeks, and the bolts were cut. Approximately 1 week after clipping, the plants were transformed by the floral dip method (Clough and Bent, 1998). Briefly, Agrobacterium strains were grown in Luria broth for 24 h at 30°C. The cells were spun down and resuspended in infiltration medium (0.5 × Murashige and Skoog [1962] medium, 5% Suc, 1 × Gamborg's vitamins, 0.044 μM 6-benzylaminopurine, and 0.04% Silwet L77; Lehle Seeds, Round Rock, TX) to an OD600 of 1.5 to 2.0. Plants were dipped in this suspension for 15 s and grown in the greenhouse under long-day conditions. The seeds were harvested from these plants and screened on selection medium (0.5 × Murashige and Skoog [1962] medium, 1 × Gamborg's vitamins, and kanamycin [50 μg/mL]). The putative transformants were rescued from kanamycin-containing medium and grown in the greenhouse under long-day conditions.
Microscopy and Pollen Analysis
Samples for histochemical analysis of GUS activity were fixed under vacuum for 10 min in 50 mM sodium phosphate buffer, pH 7.0, 0.005% (v/v) Tween 80, and 0.3% (v/v) formaldehyde. Then, tissues were washed three times in 50 mM sodium phosphate and stained for GUS activity in 50 mM sodium phosphate, 10 mM EDTA, 0.01% (v/v) Tween 80, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Denville Scientific Inc., Metuchen, NJ). After staining overnight, the samples were washed three times with 50 mM sodium phosphate and fixed for 30 min at room temperature in 20% (v/v) ethanol, 5% (v/v) acetic acid, and 5% (v/v) formaldehyde. Chlorophyll-containing tissues were cleared in a series of ethanol:water mixtures up to 80% (v/v) ethanol, in which samples were stored.
For pollen staining, anthers were dissected to release pollen grains followed by incubation in different histochemical stains. To stain nuclei and cellulose, 4′,6-diamidino-2-phenylindole (Molecular Probes) and tinopal UNPA-GX (Sigma) were used, respectively. Mitochondria were stained to assay for metabolic activity with rhodamine 123 (Sigma). Solutions of 4′,6-diamidino-2-phenylindole, tinopal, and rhodamine 123 were prepared and used as described previously (Regan and Moffatt, 1990). Alexander stain and propidium iodide (Sigma) were prepared as described previously (Huang et al., 1986). Alexander stain stains viable pollen purple and aborted pollen blue, whereas propidium iodide penetrates only the dead cells. Microscopic analysis was performed using an Axiophot microscope (Zeiss, Jena, Germany) with either bright-field or epifluorescence optics. Images were photographed using a Spot digital camera (Diagnostic Instruments, Sterling Heights, MI) and processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
For environmental scanning electron microscopy, individual anthers and pollen grains from either control or AtPTEN1 RNAi plants were mounted on scanning electron microscopy stubs and coated with palladium-gold using standard techniques and vacuum desiccation, as described previously (Bozzola and Russell, 1999). Samples were examined at 4°C under 7 torr of pressure and 70% RH in the chamber of an E3 environmental scanning electron microscope (Electroscan) with an acceleration voltage of 10 kV. Digital images were taken with Image Analysis Acquisition Software (Sunnyvale, CA).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The AtPTEN1 cDNA sequence has been deposited in GenBank under accession number AJ490172. Accession numbers for the sequences shown in Figure 1 are as follows: AtPTEN1 (BAB11013), AtPTEN2 (BAB02466), AtPTEN3 (T45864), DmPTEN (AF144232), HsPTEN (U92435), MmPTEN (U92437), and LpPTEN (BG137001).
Acknowledgments
We thank Sheila McCormick for critical reading of the manuscript and Steve Ruzin, Denise Schichnes, and Gordon Vrdoljak for assistance in fluorescence and electron microscopy. We also thank Hong Sun for providing HsPTEN clones. This work was supported by U.S. Department of Agriculture Grant 98-35301 to S.L. and by the Torrey Mesa Research Institute, Syngenta Research and Technology, San Diego, CA.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005702.
References
- Bonhomme, S., Horlow, C., Vezon, D., De Laissardiere, S., Guyon, A., Ferault, M., Marchand, M., Bechtold, N., and Pelletier, G. (1998). T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol. Gen. Genet. 260, 444–452. [DOI] [PubMed] [Google Scholar]
- Bozzola, J.J., and Russell, L.D. (1999). Electron Microscopy: Principles and Techniques for Biologists. (Sudbury, MA: John and Bartlett), pp. 50–71.
- Chuang, C.-F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Fordham-Skelton, A.R., Skipsey, M., Eveans, I.M., Edwards, R., and Gatehouse, J.A. (1999). Higher plant tyrosine-specific protein phosphatases (PTPs) contain novel amino-terminal domains: Expression during embryogenesis. Plant Mol. Biol. 39, 593–605. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong, V.E. (1999). Signaling and the modulation of pollen tube growth. Plant Cell 11, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong, V.E., Drobak, B.K., Allan, A.C., Watkins, P.A.C., and Trewavas, A.J. (1996). Growth of pollen tubes of Papaver rhoeas is regulated by a slow-moving calcium wave propagated by inositol 1,4,5-trisphosphate. Plant Cell 8, 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman, D.A., Meyers, R.E., and Cantley, L.C. (1998). Phosphoinositide kinases. Annu. Rev. Biochem. 67, 481–507. [DOI] [PubMed] [Google Scholar]
- Funamoto, S., Meili, R., Lee, S., Parry, L., and Firtel, R.A. (2002). Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611–623. [DOI] [PubMed] [Google Scholar]
- Gu, J., Tamura, M., Pankov, R., Danen, E.H.J., Takino, T., Matsumoto, K., and Yamada, K.M. (1999). Shc and FAK differentially regulate cell motility and directionality modulated by PTEN. J. Cell Biol. 146, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J., Tamura, M., and Yamada, K.M. (1998). Tumor suppressor PTEN inhibits integrin- and growth factor-mediated mitogen-activated protein (MAP) kinase signaling pathways. J. Cell Biol. 143, 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K., and Dixon, J.E. (1990). Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science 249, 553–556. [DOI] [PubMed] [Google Scholar]
- Gupta, R., He, Z., and Luan, S. (2002). Functional relationship of cytochrome c(6) and plastocyanin in Arabidopsis. Nature 417, 567–571. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Huang, Y., Kieber, J., and Luan, S. (1998). Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 16, 581–589. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Webster, C.I., Walker, A.R., and Gray, J.C. (1997). Chromosomal location and expression of the single-copy gene encoding high-mobility-group protein HMG-I/Y in Arabidopsis thaliana. Plant Mol. Biol. 34, 529–536. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouze, P., and Brunak, S. (1996). Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24, 3430–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17, 159–187. [DOI] [PubMed] [Google Scholar]
- Hong, Z., and Verma, D.P.S. (1994). A phosphatidylinositol 3-kinase is induced during soybean nodule organogenesis and is associated with membrane proliferation. Proc. Natl. Acad. Sci. USA 91, 9617–9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.N., Cornejo, M.J., Bush, D.S., and Jones, R.L. (1986). Estimating viability of plant protoplasts using double and single staining. Protoplasma 135, 80–87. [Google Scholar]
- Huang, H., Potter, C.J., Tao, W., Li, D.-M., Brogiolo, W., Hafen, E., Sun, H., and Xu, T. (1999). PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development 126, 5365–5372. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Li, H., Gupta, R., Morris, P.C., Luan, S., and Kieber, J.J. (2000). ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 122, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, T. (1995). Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 80, 225–236. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Burgess, S.M., and Hirsh, D. (1986). β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83, 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse, S.M. (1998). Protein phosphatases and the regulation of MAP kinase activity. Semin. Cell Dev. Biol. 9, 143–152. [DOI] [PubMed] [Google Scholar]
- Krebs, E.G. (1986). The enzymology of control by phosphorylation. In The Enzymes, P.D. Boyer and E.G. Krebs, eds (New York: Academic Press), pp. 3–18.
- Lee, J.-O., Yang, H., Georgescu, M.-M., Di Cristofano, A., Maehama, T., Shi, Y., Dixon, J.E., Pandolfi, P., and Pavletich, N.P. (1999). Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323–334. [DOI] [PubMed] [Google Scholar]
- Li, J., et al. (1997). PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- Liaw, D., Marsh, D.J., Li, J., Dahia, P.L.M., Wang, S.I., Zheng, Z., Bose, S., Call, K.M., Tsou, H.C., Peacocke, M., Eng, C., and Parsons, R. (1997). Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16, 64–67. [DOI] [PubMed] [Google Scholar]
- Luan, S. (1998). Protein phosphatases and signaling cascades in higher plants. Acta Bot. Sin. 40, 883–889. [Google Scholar]
- Luan, S. (2000). Protein phosphatases: Structure, regulation and function. Adv. Bot. Res. 32, 67–107. [Google Scholar]
- Luan, S., Ting, J., and Gupta, R. (2001). Protein tyrosine phosphatases in higher plants. New Phytol. 151, 155–164. [DOI] [PubMed] [Google Scholar]
- Maehama, T., and Dixon, J.E. (1998). The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- Maehama, T., and Dixon, J.E. (1999). PTEN: A tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 9, 125–128. [DOI] [PubMed] [Google Scholar]
- Maehama, T., Taylor, G.S., and Dixon, J.E. (2001). PTEN and myotubularin: Novel phosphoinositide phosphatases. Annu. Rev. Biochem. 70, 247–279. [DOI] [PubMed] [Google Scholar]
- Maehama, T., Taylor, G.S., Slama, J.T., and Dixon, J.E. (2000). A sensitive assay for phosphoinositide phosphatases. Anal. Biochem. 279, 248–250. [DOI] [PubMed] [Google Scholar]
- Mascarenhas, J.P. (1989). The male gametophyte of flowering plants. Plant Cell 1, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. (1993). Male gametophyte development. Plant Cell 5, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Muschietti, J., Dircks, L., Vancanneyt, G., and McCormick, S. (1994). LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 6, 321–338. [DOI] [PubMed] [Google Scholar]
- Myers, M.P., Stolarov, J.P., Eng, C., Li, J., Wang, S.I., Wigler, M.H., Parsons, R., and Tonks, N.K. (1997). P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc. Natl. Acad. Sci. USA 94, 9052–9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S., and Ruvkun, G. (1998). The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2, 887–893. [DOI] [PubMed] [Google Scholar]
- Piffanelli, P., Ross, J.H.E., and Murphy, D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11, 65–80. [Google Scholar]
- Regan, S.M., and Moffatt, B.A. (1990). Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic, V., Suzuki, A., De La Pompa, J.L., Brothers, G.M., Mirtsos, C., Sasaki, T., Ruland, J., Rutland, J., Penninger, J.M., Siderovski, D.P., and Mak, T.W. (1998). Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39. [DOI] [PubMed] [Google Scholar]
- Stone, R.L., and Dixon, J.E. (1994). Protein-tyrosine phosphatases. J. Biol. Chem. 269, 31323–31326. [PubMed] [Google Scholar]
- Sun, H., Lesche, R., Li, D.-M., Liliental, J., Zhang, H., Gao, J., Gavrilova, N., Mueller, B., Liu, X., and Wu, H. (1999). PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA 96, 6199–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa, T., and Itoh, T. (2001). Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533, 190–206. [DOI] [PubMed] [Google Scholar]
- Tamura, M., Gu, J., and Yamada, K.M. (1998). Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: Involvement of focal adhesion kinase. Mol. Biol. Cell 9 (suppl.), 246A.. [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., and McCormick, S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109, 705–714. [DOI] [PubMed] [Google Scholar]
- Ulm, R., Revenkova, E., di Sansebastiano, G.-P., Bechtold, N., and Paszkowski, J. (2001). Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters, P., Takegawa, K., Emr, S.D., and Chrispeels, M.J. (1994). AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc. Natl. Acad. Sci. USA 91, 11398–11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C., Anglmayer, R., Vicente, O., and Heberle-Bors, E. (1995). Molecular cloning, functional expression in Escherichia coli, and characterization of multiple mitogen-activated-protein kinases from tobacco. Eur. J. Biochem. 233, 249–257. [DOI] [PubMed] [Google Scholar]
- Wilson, C., Voronin, V., Touraev, A., Vicente, O., and Heberle-Bors, E. (1997). A developmentally regulated MAP kinase activated by hydration in tobacco pollen. Plant Cell 9, 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Senechal, K., Neshat, M.S., Whang, Y.E., and Sawyers, C.L. (1998). The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA 95, 15587–15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Fu, H.-H., Gupta, R., and Luan, S. (1998). Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 10, 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]