INTRODUCTION
Signaling pathways are rarely straightforward, and auxin signal transduction is no exception. The diverse events in which auxin is involved tell of the daunting complexity behind the plant's response to auxin. This one molecule can cause changes in rates of cell division and cell elongation, alter ion fluxes across membranes, cue changes in patterning and differentiation, and affect the expression of hundreds of genes (Davies, 1995; Berleth and Sachs, 2001). Does auxin act through multiple pathways to these diverse ends, or is there a single pathway that is dependent on the spatial, temporal, and environmental context in which auxin is received? Or is it both? Although the answers to these and other broad questions of auxin biology remain unclear, we are now in a position to connect at least some areas of research on auxin signal transduction into a more cohesive whole. Recent advances in our understanding of the apparent hub of auxin signaling, between perception and downstream auxin-induced gene expression, has developed from the mutational and molecular analysis of three main groups of proteins: the auxin/indoleacetic acids (Aux/IAAs), the auxin response factors, and components of the ubiquitin-mediated proteolytic pathway. It seems that regulated protein degradation is central to most aspects of the auxin response.
THE UBIQUITIN PATHWAY
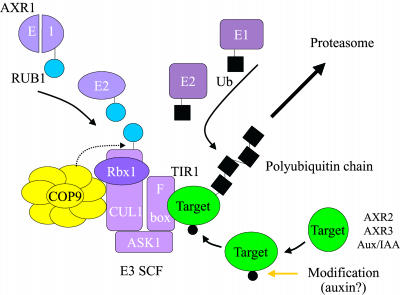
Ubiquitin-mediated proteolysis has emerged as being as fundamentally important as phosphorylation in terms of its involvement in diverse cellular events. Target proteins are condemned to degradation in the 26S proteasome by the addition of a polyubiquitin chain in what is essentially a three-step process (Figure 1) (Voges et al., 1999; Jackson et al., 2000; reviewed by Pickart, 2001). First, ubiquitin is activated by the ATP-dependent formation of a thiolester bond with a conserved Cys residue of a ubiquitin-activating enzyme, E1. Activated ubiquitin then is passed to one of a family of E2 ubiquitin-conjugating enzymes, from which it is conjugated to a Lys residue of the target protein, often with the assistance of an E3 enzyme, ubiquitin protein ligase. This process is reiterated such that one or two additional ubiquitins are polymerized upon the first ubiquitin molecule. Target proteins marked with less than four ubiquitins are poor substrates of the 26S proteasome and often escape degradation. In some cases, an additional step ensures efficient proteolysis by extending the ubiquitin chain, a process that requires a multiubiquitin chain assembly factor, or E4 (Koegl et al., 1999; Azevedo et al., 2001).
Figure 1.
Auxin Regulates the Ubiquitination of Target Proteins, Marking Them for Degradation by the 26S Proteasome.
Key components of this pathway are shown. Target proteins are recruited to the E3 SCF ligase by the F-box protein. Auxin-regulated modification of targets, which include the Aux/IAA proteins, is likely to be required before recognition by the F-box protein. Ubiquitin (Ub) is activated (via an E1 enzyme) and conjugated to the target (via E2–E3 interaction), a process that is reiterated to form a polyubiquitin chain. The ubiquitination activity of the SCF is enhanced by modification of the cullin subunit with the ubiquitin-like protein RUB1. RUB1 is activated by AXR1/ECR1, an E1-like enzyme. The COP9 signalosome regulates the deconjugation of RUB1 (dotted arrow) and possibly other processes relating to the efficient degradation of SCF targets.
Of course, ubiquitination must be directed against specific targets, and it is the E3 enzyme that mediates the critical step of substrate recognition, a fact reflected in the hundreds of E3s of various classes that are encoded by the eukaryotic genomes sequenced to date (Deshaies, 1999; Voges et al., 1999; Xiao and Jang, 2000; Pickart, 2001). Prominent among these, and certainly most relevant here, are the SCF ubiquitin protein ligases (for Skp1p, Cdc53p/cullin, and F-box protein in yeast and mammals) (reviewed by Deshaies, 1999; Xiao and Jang, 2000). SCF complexes are multimeric enzymes of at least four subunits built around a member of the cullin family of proteins (Mathias et al., 1996). In conjunction with a second subunit, RBX1 (also known as ROC1 or HRT1), the cullin catalyzes polyubiquitin chain formation by interacting with the E2 enzyme (Seol et al., 1999). The cullin also binds to a member of the SKP1 protein family, which, in turn, binds to an F-box protein, so called because it contains an N-terminal F-box motif required for its interaction with SKP1 (Skowyra et al., 1997; Kishi and Yamao, 1998; Patton et al., 1998). The C terminus of the F-box protein consists of any one of a number of protein–protein interaction domains that bind the relevant target protein, making it accessible to the E2-SCF complex (Kishi and Yamao, 1998; Patton et al., 1998).
Our knowledge of the SCF system is derived mainly from extensive work in yeast and mammals (Patton et al., 1998; Deshaies, 1999; Gray and Estelle, 2000; Jackson et al., 2000). There is an ever-growing number of examples of SCF activity in signal transduction, from the derepression of Glc transporter genes in response to Glc in Saccharomyces cerevisiae (Bernard and Andre, 2001) to the control of NF-κB activity in response to infection in mice (reviewed by Karin and Ben-Neriah, 2000). This work has established a paradigm for the involvement of the SCF in the regulation of gene expression that is as relevant to auxin signaling in plants as to Glc and amino acid signaling in budding yeast.
SCF-MEDIATED UBIQUITINATION MEDIATES THE AUXIN RESPONSE
The crucial evidence for the involvement of SCF-mediated protein degradation in auxin signaling came through the characterization of the tir1 mutants of Arabidopsis (Table 1) (Ruegger et al., 1998). Although the tir1 alleles were identified in a screen for resistance to inhibitors of auxin transport, the mutants were more markedly resistant to inhibitory concentrations of auxin, suggesting a defect in auxin response rather than transport (Ruegger et al., 1998). TIR1 was found to encode an F-box protein (Ruegger et al., 1998) and subsequently was shown to interact with either of the Arabidopsis Skp1-like proteins, ASK1 or ASK2, and the cullin AtCUL1 to form the functional SCFTIR1 (Gray et al., 1999, 2001). The idea that regulated protein degradation via SCFTIR1 is required for proper auxin signaling was strengthened by the analysis of mutation in ASK1 (Table 1). ask1-1 shows reduced auxin response, being slightly more auxin resistant than tir1-1 (Gray et al., 1999). This may reflect the fact that TIR1 has several close homologs in Arabidopsis, indicating the potential for redundancy in TIR1 function (Gray et al., 1999). The C-terminal domain of TIR1 and its homologs consists of Leu-rich repeats, which presumably are involved in target selection (Ruegger et al., 1998; Hsiung et al., 2001). What, then, are the targets for SCFTIR1? In the search for candidates, the Aux/IAA family of transcription factors are likely suspects.
Table 1.
Phenotypic Summary of Auxin Signaling Mutants
| Mutant/Gene | Protein | Mutant Phenotype | Reference |
|---|---|---|---|
| axr2-1/IAA7 | Aux/IAA | Reduced stature, short hypocotyl and expanded leaves in the dark, wavy leaves, agravitropic roots and shoots, reduced adventitious rooting, no root hairs |
Timpte et al., 1994; Wilson et al., 1990; Nagpal et al., 2000 |
| axr3-1/IAA17 axr3-3 | Aux/IAA | Reduced stature, increased shoot apical dominance, short hypocotyl and expanded leaves in the dark, curled leaves, agravitropic roots, increased adventitious rooting, no root hairs |
Leyser et al., 1996; Rouse et al., 1998 |
| bdl/IAA12 | Aux/IAA | Reduced stature, reduced shoot apical dominance, curled leaves, no embryonic root |
Hamann et al., 1999 |
| iaa28-1/IAA28 | Aux/IAA | Reduced shoot apical dominance, reduced lateral rooting | Rogg et al., 2001 |
| msg2-1/IAA19 msg2-2 msg2-3 msg2-4 | Aux/IAA | Agravitropic and aphototropic hypocotyl, reduced lateral rooting | Reed, 2001 |
| shy2-2/IAA3 | Aux/IAA | Reduced stature, short hypocotyl and expanded leaves in the dark, curled leaves, long root hairs |
Kim et al., 1996; Reed et al., 1998; Tian and Reed, 1999; Knox, K., Kepinski, S., Leyser, O., unpublished data |
| slr1-1/IAA14 | Aux/IAA | Agravitropic hypocotyl and root, no lateral roots, few root hairs | Reed, 2001 |
| mp/ARF5 | ARF | Defects in root meristem initiation in the early embryo, reduced auxin transport |
Berleth and Jürgens, 1993; Hardtke and Berleth, 1998; Mattsson et al., 1999 |
| nph4/ARF7 | ARF | Aphototropic hypocotyl with reduced gravitropic response, reduced auxin-inducible gene expression | Harper et al., 2000 |
| axr1-12/AXR1 axr1-3 | E1 RUB1- activating enzyme |
Auxin resistant, reduced stature, reduced shoot apical dominance, crinkled leaves, abnormal flowers, reduced lateral rooting, reduced auxin-inducible gene expression |
Estelle and Sommerville, 1987; Lincoln et al., 1990; Timpte et al., 1995 |
| tir1-1/TIR1 tir1-3 | F-box protein | Auxin resistant, reduced auxin-induced hypocotyl elongation, reduced lateral rooting |
Ruegger et al., 1998 |
| ask1-1/ASK1 | SKP protein | Auxin resistant, abnormal flowers, reduced lateral rooting |
Gray et al., 1999; Zhao et al., 2001 |
REGULATED Aux/IAA STABILITY IS CRITICAL FOR AUXIN SIGNALING
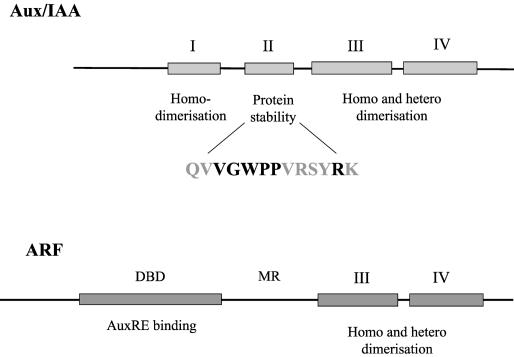
The Aux/IAAs are a family of extremely short-lived nuclear proteins (reviewed by Abel and Theologis, 1996). Aux/IAA genes are induced rapidly by auxin and are found throughout the higher plants. Their importance in auxin signal transduction is manifest in the severe auxin-related phenotypes arising from semidominant mutations in several of these genes, including AXR2/IAA7, AXR3/IAA17, SHY2/IAA3, BODENLOS/IAA12, SLR1/IAA14, MSG2/IAA19, and IAA28 (Table 1) (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001; reviewed by Reed, 2001). Aux/IAA proteins share a typical four-domain structure (Figure 2), and what is striking about these semidominant mutations is that they occur only within the highly conserved domain II, centered on a core GWPPV motif. The spectacular auxin-related phenotypic effects of these single–amino acid substitutions illustrate the critical importance of domain II to Aux/IAA function (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). Domain II is the site of the destabilization signal that confers instability on Aux/IAA members; measured half-lives range from as little as 6 min to >80 min (Abel et al., 1994; Ouellet et al., 2001).
Figure 2.
Protein Structures of Aux/IAAs and ARFs.
Aux/IAAs and ARFs share homology in their C-terminal domains III and IV, which mediate homodimerization and heterodimerization. Domain III contains a βαα motif that is important for dimerization. Domain II of Aux/IAAs is required for the auxin-regulated destabilization of the protein. The 13 amino acids that are sufficient to confer instability are shown, and those conserved across the Aux/IAA family are highlighted. ARFs have an N-terminal DNA binding domain (DBD) that binds to TGTCNC-type AuxREs of auxin-regulated genes. The amino acid composition of the middle region (MR) of ARFs affects their ability to activate transcription.
Translational fusions of Aux/IAA N-terminal regions containing domain II with reporter proteins such as β-glucuronidase (GUS) or luciferase have shown that this lability can be conferred on otherwise stable proteins (Worley et al., 2000; Gray et al., 2001). The transferable destabilization signal within Aux/IAAs has been finely mapped to a 13–amino acid region (or so-called degron) around the core GWPPV residues in domain II. Although this 13–amino acid degron fused to luciferase excludes an essential bipartite nuclear localization signal spanning domain II, the use of a heterologous nuclear localization signal has shown that in the correct subcellular context, these 13 amino acids are sufficient to confer instability (Ramos et al., 2001). Further direct and indirect measurements of Aux/IAA stability have shown that the phenotypic consequences of mutations in domain II may be the result of stabilization of the respective IAA proteins without affecting their nuclear localization (Worley et al., 2000; Gray et al., 2001; Ouellet et al., 2001). For example, the axr3-1 mutation, a Pro-to-Leu substitution in domain II, causes an almost sevenfold increase in protein half-life (Ouellet et al., 2001). Among many auxin-related phenotypes (Table 1), this mutation causes increased apical dominance, adventitious rooting, and complete agravitropism in roots (Leyser et al., 1996). Collectively, these data stress the importance of Aux/IAAs and their instability to auxin response. So, to reverse an earlier question, what are the regulators of Aux/IAA stability?
Aux/IAA PROTEINS ARE TARGETS OF SCFTIR1
Evidence of the involvement of SCFTIR1 in Aux/IAA degradation came from the analysis of AXR3/IAA17 reporter fusion proteins in the tir1 mutant background (Gray et al., 2001). This fusion protein consists of N-terminal domains I and II of AXR3, including the bipartite nuclear localization signal, fused translationally to GUS and expressed under the control of a heat-inducible promoter in transgenic Arabidopsis. After heat induction in a wild-type background, the fusion protein accumulates to relatively low levels and is depleted rapidly over time compared with controls. However, the stability of the same fusion protein was increased significantly when the construct was crossed into the tir1 background, indicating that TIR1 was required for the destabilization of AXR3 in planta (Gray et al., 2001). That TIR1 activity ultimately might result in AXR3 degradation in the 26S proteasome was supported by the finding that specific proteasome inhibitors stabilize AXR2-, AXR3-, and other Aux/IAA-reporter fusion proteins (Gray et al., 2001; Ramos et al., 2001).
The final step in defining a mechanistic link between Aux/IAA proteins and the SCFTIR1 ubiquitin ligase complex came with the direct demonstration of the interaction of AXR2 and AXR3 with SCFTIR1 in plant extracts. The use of glutathione S-transferase (GST)–tagged AXR2 or AXR3 in pulldown assays with extracts from plants expressing a c-myc epitope-tagged TIR1 yielded the entire SCFTIR1 complex, as identified by immunoblotting with c-myc, AtCUL1, and ASK2 antibodies (Gray et al., 2001). Further proof that this represents a biologically relevant interaction is indicated by the fact that when mutant GST–axr2-1 or GST–axr3-1 proteins were used in an identical pulldown assay, the interaction with SCFTIR1 was abolished or reduced severely (Gray et al., 2001). The latter data are in complete accord with the measured effect of the mutations on the stability of the axr2-1 and axr3-1 proteins in planta.
AUXIN FURTHER DESTABILIZES Aux/IAA PROTEINS
Genetic data have established the importance of regulating the stability of Aux/IAA proteins, suggesting that the plant might respond to auxin by regulating the turnover of Aux/IAAs. Although previous analysis had suggested that this was not the case (Abel et al., 1994), recent work examining the stability of Aux/IAA-reporter fusion proteins expressed from non-auxin-responsive promoters and using different time frames has reached different conclusions. These studies were able to uncouple any auxin-induced Aux/IAA protein destabilization from auxin-induced Aux/IAA gene expression to reveal a significant increase in the degradation of the Aux/IAA-reporter proteins in response to exogenously applied auxin (Gray et al., 2001; Zenser et al., 2001). This effect of auxin is dose dependent and extremely rapid, causing a considerable decrease in protein levels within 5 min (Gray et al., 2001; Zenser et al., 2001). In fact, it is possible that auxin might be absolutely required for Aux/IAA degradation, and if cells could be rid of basal levels of auxin, Aux/IAA proteins might be rendered extremely stable.
Perfectly consistent with the in planta measures of Aux/IAA stability, the in vitro pulldown assay with GST-AXR2 and GST-AXR3 showed an auxin-induced increase in the recovery of SCFTIR1, which also occurred in a dose-dependent manner (Gray et al., 2001). Indeed, the dose-response curves for the reduction in Aux/IAA stability and the increase in Aux/IAA–SCFTIR1 interaction are remarkably similar. Given the highly conserved nature of domain II across the Aux/IAAs, it is likely that many, if not all, members of the family will be susceptible to auxin-induced destabilization to some extent. Together, these observations convincingly support the idea that auxin stimulates the rapid degradation of Aux/IAAs by increasing their interaction with SCFTIR1 in some way.
REGULATION OF THE SCFTIR1–Aux/IAA INTERACTION BY AUXIN
The mechanism by which auxin influences SCFTIR1-mediated Aux/IAA turnover is not understood. However, clues to what might be happening may be gleaned from the many characterized examples of SCF-mediated signaling pathways in yeast and mammals. SCF-mediated ubiquitination usually is preceded by substrate modification. These changes turn stable proteins into susceptible targets of the SCF, and by far the most common modification is phosphorylation (Deshaies, 1999). A good example is that of NF-κB activation in mammals. NF-κB normally is sequestered in the cytoplasm, because its nuclear localization signal is masked by the IκB family of inhibitors. In response to inflammation and other stress stimuli, IκB is phosphorylated quickly, ubiquitinated by SCF E3RSIκB/β-TrCP, and ultimately degraded, freeing NF-κB to translocate to the nucleus to regulate gene expression (Karin and Ben-Neriah, 2000). Similarly, auxin might increase the affinity of Aux/IAAs for SCFTIR1 by stimulating their phosphorylation.
There is certainly an increasing body of biochemical and genetic evidence supporting a role for kinases in auxin signaling. A recent interesting example is the identification of an auxin-stimulated mitogen-activated protein kinase (MAPK) in Arabidopsis roots (Mockaitis and Howell, 2000). Because the auxin induction of the MAPK activity occurs over a time frame very similar to that of the auxin-induced destabilization of AXR3, these activities might be related. Furthermore, specific inhibitors of MAPKs abolish expression from the auxin-responsive BA3 promoter in the root elongation zone (Mockaitis and Howell, 2000). Although, in other systems, MAPKs tend not to be involved in phosphorylation before ubiquitination, these data indicate that MAPK activity can impinge on auxin signaling in some way. Another particularly relevant study has demonstrated that the N-terminal half of some Aux/IAA proteins is phosphorylated by phytochrome A in vitro (Colon-Carmona et al., 2000). Stabilizing gain-of-function mutations in several Aux/IAAs causes photomorphogenic development in the dark (Tian and Reed, 1999; Nagpal et al., 2000). The simple and perhaps naïve prediction would be that light-activated, phytochrome-mediated phosphorylation would contribute to a stabilizing rather than a destabilizing effect on Aux/IAA proteins. Although this is entirely possible, it is more likely that the phosphorylation(s) has other effects on Aux/IAA activity (see below). A third auxin-related kinase activity also has been described. Mutations in the PINOID Ser/Thr kinase cause a range of defects in auxin response (Christensen et al., 2000). Although the involvement of kinases in auxin signal transduction is not unexpected, their potential role in regulating the stability of Aux/IAAs is far from certain. This is because the 13–amino acid degron of domain II that is sufficient to confer auxin-inducible destabilization on a reporter protein does not contain any essential phosphorylation sites (Ramos et al., 2001). This means that the regulation of Aux/IAA stability is unorthodox, and if phosphorylation is a prerequisite of interaction with SCFTIR1, it must involve some kind of adaptor protein.
Examples of non-phosphorylation-dependent regulation of substrate–SCF interactions are beginning to emerge. One such case concerns the degradation of the human transcription factor, hypoxia-inducible factor (HIFα), by an SCF-related E3 in which substrate selection is mediated by the von Hippel–Lindau tumor-suppressor protein (VHL) (reviewed by Jackson et al., 2000). HIFα is required for the activation of a variety of responses to hypoxia that are repressed, when the oxygen supply is adequate, by the rapid destruction of HIFα via VHL-mediated ubiquitination. The interaction of HIFα with VHL was shown recently to be regulated through the hydroxylation of a HIFα Pro residue, a reaction that requires molecular oxygen (Ivan et al., 2001; Jaakkola et al., 2001). This modification of Pro may be relevant to the regulation of Aux/IAA–SCFTIR1 interaction, because the most striking feature of the consensus domain II degron is two highly conserved tandem Pro residues. These residues clearly are required for Aux/IAA instability, because their substitution is the most common cause of the dominant Aux/IAA-stabilizing mutations found in such alleles as axr3-1 and axr2-1 (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). The HIFα example of a divergence from the phosphorylation norm may tempt us to think of other novel modes of regulating the Aux/IAA–SCFTIR1 interaction. Perhaps the most exciting possibility is the simplest one, that auxin itself can be bound or conjugated within the degron, rendering the substrate susceptible to degradation by SCFTIR1. This may seem far-fetched, but it is a tantalizing prospect. Plant proteins can be modified in this way (Napier, 1995; Walz et al., 2002), and the example of HIFα regulation shows that the signal transduction chain can be very short, so that the environmental conditions limiting substrate modification and stability are the same conditions that demand a response.
Although without precedent elsewhere, a final possibility is that the interaction between Aux/IAAs and SCFTIR1 increases in response to auxin because of modification (perhaps phosphorylation) of the TIR1 protein.
REGULATION OF SCFTIR1 ACTIVITY
Another point at which the regulation of Aux/IAA ubiquitination could conceivably take place is at the level of SCF abundance, assembly, or activity. This suggestion comes from studies on SCFGrr1 of yeast, which is involved in cell cycle progression and Glc and amino acid signaling (reviewed by Johnston, 1999). The F-box protein Grr1 is structurally similar to TIR1, both containing C-terminal Leu-rich repeats. In yeast two-hybrid studies, it has been shown that the interaction between Grr1 and Skp1, part of the process of SCFGrr1 assembly, is enhanced significantly in response to Glc (Li and Johnston, 1997). Although this would seem to set a precedent for SCF assembly in response to environmental stimuli that might be extended to SCFTIR1, recent data have suggested other possible explanations. It has since been shown that F-box proteins are inherently unstable (Galan and Peter, 1999; Mathias et al., 1999). This instability has been shown to be the result of the autoubiquitination of the F-box protein while assembled in an SCF (Galan and Peter, 1999), and it seems that SCF substrate binding may confer protection on the F-box protein (Deshaies, 1999; Galan and Peter, 1999). Thus, the apparent increased interaction of Grr1 and Skp1 in response to Glc may reflect increased Grr1 stability as a result of a Glc-induced increase in the susceptibility of SCFGrr1 targets. Similarly, the stability and abundance of TIR1 might be important for normal auxin sensitivity, because overexpression of TIR1 in transgenic plants results in increased auxin response (Gray et al., 1999). These phenotypes are dependent on additional TIR1 participating in an SCF complex, because overexpression of a TIR1 variant in which the F-box had been mutated does not result in auxin response phenotypes (Gray et al., 1999). Furthermore, loss-of-function tir1 alleles appear to be semidominant, suggesting haploinsufficiency (Ruegger et al., 1998). Therefore, auxin responses could be brought about by changes in the abundance of TIR1. This explanation could account for some of the difference in auxin responsiveness observed between tissues, because TIR1 is not expressed uniformly throughout the plant (Gray et al., 1999).
Another important event affecting the activity, rather than the assembly, of SCFTIR1 has been shown to be crucial for normal auxin signaling. It involves the modification of the cullin subunit of the SCF by its conjugation with a ubiquitin-like protein called RUB1 (NEDD8 in humans and fission yeast) that is important for optimal SCF function (del Pozo et al., 1998). This modification with RUB1 (rubinylation) requires AXR1, one of the first mutationally defined auxin response genes. The axr1 mutants exhibit pleiotropic auxin-related phenotypes indicative of wide-ranging auxin resistance, which is more severe than that displayed by the tir1 alleles (Table 1) (Lincoln et al., 1990; Timpte et al., 1995; Gray et al., 1998; Ruegger et al., 1998; Stirnberg et al., 1999). Importantly, Aux/IAA-reporter protein fusions are stabilized in an axr1 background (Gray et al., 2001), and axr1 tir1 double mutants have a synergistic phenotype, indicating the AXR1 and TIR1 act in an overlapping pathway to destabilize Aux/IAA proteins (Ruegger et al., 1998).
AXR1 encodes an enzymatic subunit that dimerizes with a protein called ECR1 to form a RUB1-activating enzyme that is structurally and functionally homologous with the E1 ubiquitin-activating enzyme (Leyser et al., 1993; del Pozo and Estelle, 1999; del Pozo et al., 2002). Rubinylation involves the AXR1/ECR1 dimer and an E2-like protein, RCE1. Rubinylation proceeds in a manner analogous to the enzyme cascade that activates and conjugates ubiquitin to SCF targets, except that rubinylation does not seem to require an E3 ligase (del Pozo and Estelle, 1999). Thus, axr1 alleles show reduced conjugation of RUB1 to the AtCUL1 subunit of SCFTIR1, and this is the molecular basis of the stabilization of Aux/IAAs and auxin resistance in these mutants (del Pozo and Estelle, 1999; Gray et al., 2001). The small amount of RUB1 conjugation that does occur in axr1 mutants is likely to be through the activity of another Arabidopsis gene, AXL1, which is closely homologous with AXR1 (del Pozo et al., 2002). Most organisms that support the modification of cullins with ubiquitin-like proteins do so with only one or two RUB1/NEDD8-activating enzymes (Deshaies, 1999); therefore, it seems likely that in addition to being essential for normal auxin response, AXR1-mediated rubinylation will have a role in the degradation of diverse substrates in numerous other signaling pathways.
Recent results suggest that the removal of RUB1 from SCFTIR1 also is important for auxin signaling (Schwechheimer et al., 2001) and that this activity might be mediated by the COP9 signalosome (CSN) (Lyapina et al., 2001). The COP9 signalosome is a multiprotein complex that seems to be the enzymatic equivalent of a Swiss army knife, with attributed activities including phosphorylation, nucleocytoplasmic partitioning, interaction with the 26S proteasome, and now derubinylation/deneddylation (Kwok et al., 1999; Tomoda et al., 1999; Bech-Otschir et al., 2001; Schwechheimer et al., 2001; reviewed by Wei et al., 1998; Wei and Deng, 1999; Schwechheimer and Deng, 2001). Indeed, in the past 2 years of published research, the COP9 complex has gone from being only tangentially involved to being seemingly indispensable in the function of several E3-mediated degradation systems.
The COP9 signalosome was originally identified in screens for light signaling mutants in Arabidopsis (Chamovitz et al., 1996). Mutation in any one of its eight subunits leads to the loss of the entire complex and results in photomorphogenesis regardless of the presence or absence of light signals. In addition to this phenotype, null mutations in the COP9 subunits also result in seedling lethality. Using an antisense approach directed against the CSN5/JAB1 subunit, it has been possible to impose a less severe loss of COP9 function so that plants develop to adulthood despite reduced COP9 levels (Schwechheimer et al., 2001). Unexpectedly, the adult phenotype of these plants is similar to the axr1 mutant phenotype and includes auxin-resistant root elongation, reduced root branching, and increased shoot branching (Schwechheimer et al., 2001). When the antisense CSN5 construct was crossed into the axr1-3 background, a synergistic phenotype was observed, suggesting that the COP9 complex and AXR1 act in overlapping pathways (Schwechheimer et al., 2001).
Consistent with this idea, the stability of an Aux/IAA-luciferase fusion protein was increased in the CSN5 antisense background (Schwechheimer et al., 2001). Furthermore, the COP9 signalosome was found to coimmunoprecipitate with SCFTIR1, and AtCUL1 was shown to interact with the CSN2 subunit in a yeast two-hybrid assay (Schwechheimer et al., 2001). Despite the similar phenotypes resulting from AXR1 loss of function and COP9 partial loss of function, paradoxically, COP9 signalosome mutants accumulate AtCUL1-RUB1 conjugates, unlike axr1 mutants (Schwechheimer et al., 2001). This finding suggests that the removal of RUB1 from AtCUL1 and its addition is required for SCFTIR1-mediated ubiquitination and subsequent degradation by the 26S proteasome. Interestingly, each of the eight subunits of the COP9 signalosome is related to one of the eight subunits of the lid of the 26S proteasome, part of the 19S particle that feeds substrates to the proteolytic core of the complex. Perhaps, then, COP9 recognizes RUB1/NEDD8 in the same way that the lid of the 26S proteasome recognizes ubiquitin. RUB1/NEDD8 deconjugation by COP9 may be analogous to the deubiquitination activity of subunits of the 19S regulatory particle. Alternatively, COP9 may simply recognize the ubiquitinated target protein and possess an associated activity against RUB1/NEDD8.
So what is the function of RUB1/NEDD8 conjugation to cullin and the significance of its subsequent removal? It has been shown that NEDD8 modification of cullins promotes SCF ubiquitin ligase activity (Furukawa et al., 2000; Morimoto et al., 2000; Podust et al., 2000; Wu et al., 2000; Lyapina et al., 2001). Furthermore, it was demonstrated recently that NEDD8 is required for the efficient interaction of E2 ubiquitin with the SCF complex (Kawakami et al., 2001). As mentioned above, polyubiquitin chains of more than four ubiquitins are required for efficient proteolysis. Perhaps cycles of RUB1/NEDD8 conjugation and deconjugation are required to sequentially attract E2 ubiquitin, thereby promoting polyubiquitin chain formation. Alternatively, RUB1/NEDD8 deconjugation could be required to release oligoubiquitinated substrates (two to three ubiquitins) from the E2-SCF complex so that they can be polyubiquitinated efficiently by E4 multiubiquitin-promoting enzymes.
Although these are formal possibilities, the effects of CSN deficiency in fission yeast suggest that a reduction in the ubiquitin ligase activity of SCFTIR1 may not explain the increased stability of the Aux/IAA-reporter fusion protein and the auxin-related phenotypes in COP9-deficient backgrounds. Similar to the accumulation of RUB1-AtCUL1 conjugates in COP9-deficient Arabidopsis, CSN-deficient yeast accumulate NEDD8-modified cullin, but the effect is to increase rather than decrease SCF ubiquitin ligase activity (Lyapina et al., 2001). Because these data suggest that COP9-mediated RUB1/NEDD8 deconjugation affects a step between the initial ubiquitination of the target and its degradation, another possibility is that COP9 affects the delivery of ubiquitinated substrates to the 26S proteasome. Indeed, it has been shown that the export of the COP9 subunit CSN5 from the nucleus to the cytoplasm is associated with the destruction of p27kip1, a mammalian cyclin-dependent kinase inhibitor. p27kip1 is phosphorylated and ubiquitinated in the nucleus, but its degradation is dependent on it being escorted to the cytoplasm by CSN5/JAB1 (Tomoda et al., 1999; Podust et al., 2000), where it is likely surrendered to cytoplasmic proteasomes. Thus, the stabilization of Aux/IAA proteins in a CSN5-deficient background may be attributable to their poor delivery to the 26S proteasome rather than to their reduced polyubiquitination. Taking the proteasome delivery idea one step further, Schwechheimer and Deng (2001) recently proposed that COP9 could pass ubiquitinated substrates to the proteolytic core of the proteasome directly, supplanting its bona fide lid and thereby ensuring the efficient delivery of target proteins.
Studies in CSN5-deficient fission yeast have suggested that if COP9 does have a role in the nucleocytoplasmic partitioning of ubiquitinated substrates, this is not accompanied by any large change in the subcellular distribution of cullins (Lyapina et al., 2001). So, if ubiquitinated substrates are moved in this way but are not accompanied by an intact SCF, what would be the function of the RUB1/NEDD8 deconjugation of cullins? There are two possibilities. First, the RUB1/NEDD8 deconjugation may allow the release of the ubiquitinated substrate, possibly by prompting the disassembly of the SCF. Second, it is possible that COP9 mediates two functionally distinct events after ubiquitination: derubinylation/deneddylation and delivery of substrates to the proteasome. Recent data indicate that RUB1/NEDD8 modification is preceded by SCF assembly and possibly binding of the substrate by the F-box protein (Kawakami et al., 2001). Thus, the RUB1/NEDD8 deconjugation function of COP9 might be entirely unrelated to its substrate delivery function, acting to prevent the unnecessary interaction of ubiquitin-laden E2 with cullin, which was not part of a complete and ready SCF.
The activity of the RUB1 conjugation pathway in Arabidopsis apparently is unaffected by the addition of auxin, indicating that it is not a direct route for auxin signal transduction (del Pozo et al., 2002). Nevertheless, AXR1 clearly is crucial for normal auxin response, but is it important only in auxin response? Because it appears that all SCFs are subject to RUB1 modification, defects other than those relating to the auxin response might be expected, but these are not apparent in axr1 mutants. It is possible that axr1 mutants are so crippled by their defective auxin signaling that other phenotypes are masked. However, mutations in the F-box protein UFO, part of the putative SCFUFO, cause abnormal flower development that is not observed in an axr1 background (Samach et al., 1999; del Pozo et al., 2002). This reflects either a differential sensitivity for RUB1 modification in the UFO pathway or a greater dependence on rubinylation via the AXR1 homolog AXL1 (del Pozo et al., 2002). Although there is a differential requirement for RUB modification among SCFs in budding yeast (Lammer et al., 1998; Liakopoulos et al., 1998), it seems to be required more generally by SCFs in other eukaryotes (del Pozo and Estelle, 1999; Furukawa et al., 2000; Osaka et al., 2000; Gray et al., 2001). Analysis of the loss of AXL1 function should shed light on the extent to which RUB1 modification affects the responses mediated by diverse SCFs in Arabidopsis.
Aux/IAA PROTEIN FUNCTION
The defects in auxin signaling caused by mutations in Aux/IAAs and ubiquitin pathway components demonstrate the consequences of failing to regulate the stability of Aux/IAAs. But what is the function of the strict regulation of Aux/IAA abundance?
Aux/IAAs are nuclear proteins, and their activities as transcriptional regulators appear to be based on their ability to form a variety of dimers. First, individual Aux/IAAs are able to both homodimerize and heterodimerize with other members of the Aux/IAA family (Kim et al., 1997). These interactions require domains III and IV, and for some homodimerization events, domain I (Figure 2) (Kim et al., 1997; Ouellet et al., 2001). Although the function of Aux/IAA dimerization is not clear, evidence of its importance may be inferred from the fact that intragenic revertant mutations that suppress the axr3-1 phenotype (Table 1) compromise the ability of the protein to form dimers (Ouellet et al., 2001). Domain III contains a predicted βαα motif, which is characteristic of prokaryotic repressors that bind DNA as tetramers (Abel et al., 1994; Morgan et al., 1999). Indeed, in isolation, AXR3 domain III polypeptides were able to form dimers, trimers, and tetramers (Ouellet et al., 2001), indicating the possibility that full-length Aux/IAA proteins also may be able to form higher order multimers. Therefore, although the role of the domain III βαα motif may be only in the efficient formation of dimers/multimers, it is possible that Aux/IAAs can bind DNA directly and perhaps modulate transcription.
Aux/IAAs INTERACT WITH AUXIN RESPONSE FACTOR PROTEINS
Aux/IAAs can also heterodimerize with members of the auxin response factor (ARF) family of transcription factors (Kim et al., 1997; Ulmasov et al., 1997a, 1997b). ARFs share considerable homology in their C termini with the Aux/IAAs, possessing the conserved domains III and IV (Figure 2) (Ulmasov et al., 1997a). Interaction via these ARF domains allows dimerization with Aux/IAAs and also the formation of homodimers and heterodimers with other ARFs (Kim et al., 1997). Unlike Aux/IAAs, ARFs have been shown to bind DNA directly by virtue of a large B3-type DNA binding domain toward their N termini (Ulmasov et al., 1997a, 1999b). For the ARFs characterized to date, this binding is directed specifically to auxin response elements (AuxREs) containing TGTCNC motif(s) (typically TGTCTC) in various contexts that often involve coupling elements (Ulmasov et al., 1997a; Guilfoyle et al., 1998). Synthetic palindromic or direct repeats of these six nucleotides are sufficient to bind ARFs and confer auxin regulation on the transcription of a reporter gene (Ulmasov et al., 1997b, 1999a, 1999b). Hence, ARFs appear to mediate auxin-regulated gene expression through binding to AuxREs, and Aux/IAAs have the potential to alter the transcription of auxin-inducible genes by interacting with ARFs.
The number of interactions within and between Aux/IAAs and ARFs is potentially vast. Both the Aux/IAAs and ARFs constitute large families in Arabidopsis, with >25 Aux/IAA members and >20 ARFs (Reed, 2001). Tissue-specific and developmental variations in the expression of both families suggest that some of these proteins, although capable of dimerizing, will be separated spatially and temporally in the growing plant (Abel et al., 1995; Hardtke and Berleth, 1998). Nevertheless, the interactions are likely to be complex, reflecting the need for fine control of auxin induction of numerous genes.
Our understanding of the interactions of ARFs and Aux/IAAs that regulate transcription from auxin-inducible promoters is based largely on work with carrot cell suspension cultures, in which TGTCTC-containing AuxRE promoter–reporter fusions were used to monitor the effects of the coexpression of various ARF- and Aux/IAA-derived genes (Ulmasov et al., 1997b, 1999a). These experiments have shown that ARFs can act as both repressors and activators of transcription depending on qualitative differences in their middle regions between the N-terminal DNA binding domain and C-terminal domains III and IV (Figure 2) (Ulmasov et al., 1999a). ARF1, for example, which is P/S/T rich in its middle region, suppresses both basal and auxin-inducible expression from AuxRE-containing promoters. In contrast, expression of ARF5, ARF6, ARF7, or ARF8, which are Q rich in their middle regions, increases both basal and auxin-inducible expression from the AuxRE-containing promoters; on the other hand, expression of ARF2, ARF3, ARF4, or ARF9 has no effect. These regulatory characteristics were shown to be independent of AuxRE binding, because essentially identical results were obtained when the ARF DNA binding domain was replaced with the DNA binding domain from the budding yeast GAL4 gene and simultaneously the AuxRE–reporter gene fusion was replaced with a GAL4 binding element–reporter gene fusion (Ulmasov et al., 1999a). These data suggest that the auxin inducibility of AuxRE-containing promoters depends on the middle and C-terminal regions of the ARFs, because a Q-rich middle region with a C-terminal Aux/IAA-like dimerization domain is sufficient to confer auxin inducibility on a GAL4 DNA binding domain promoter element system.
When the DNA binding domain was removed from the various ARFs and these truncated versions were introduced into carrot protoplasts with an AuxRE-reporter construct, truncated ARFs with Q-rich middle regions still were able to activate transcription (Ulmasov et al., 1999a). These effects were abolished if the dimerization domains were removed as well. Because the localization of the truncated ARFs to the vicinity of the AuxRE likely is required, one explanation for these results is that ARFs can dimerize with endogenous ARFs that occupy the AuxREs through their DNA binding domains, and subsequently, their middle regions can regulate transcription (Ulmasov et al., 1999a). Because the overexpression of these truncated ARFs increases the basal level of reporter gene transcription, these data further indicate that the endogenous ARF with which they might interact is bound to the TGTCTC-AuxRE in the absence of additional auxin.
In contrast to the activating ARFs, ARF1 lost its ability to repress transcription when it was expressed without its DNA binding domain, suggesting that part of the repression is through the occupation of AuxREs (Ulmasov et al., 1999a). Domain-swapping experiments with truncated ARF1 and ARF7 provide further clues to the mechanism of ARF1 repression. The expression of the AFR1 middle region with ARF7 dimerization domains resulted in slight repression (Ulmasov et al., 1999a). This repression is likely the result of the chimeric protein titrating out endogenous activating components rather than possessing a repressing quality per se, reflecting a difference in the ability to interact with AuxRE-bound factors conferred by ARF1 and ARF7 dimerization domains.
Dimerization through their C-terminal domains clearly is important to the activities of ARFs on auxin-inducible promoters. Because Aux/IAAs are able to dimerize with ARFs, their potential to influence ARF-mediated transcription has also been examined in the carrot protoplast system (Ulmasov et al., 1997b). Four different Aux/IAAs were tested, and all were found to repress transcription from TGTCTC-containing AuxREs. This finding is consistent with the idea that Aux/IAA interaction with AuxRE-bound ARFs can reduce the ability of ARFs to activate transcription in response to auxin. An attractive hypothesis is that this occurs by Aux/IAAs competing with ARFs for dimerization through domains III and IV (Ulmasov et al., 1997b). However, the presence of endogenous ARFs on the AuxREs in this system has not been proven, and at present there is no direct evidence to show ARF-Aux/IAA association on a promoter.
A BASIC MODEL FOR AUXIN-INDUCED GENE EXPRESSION
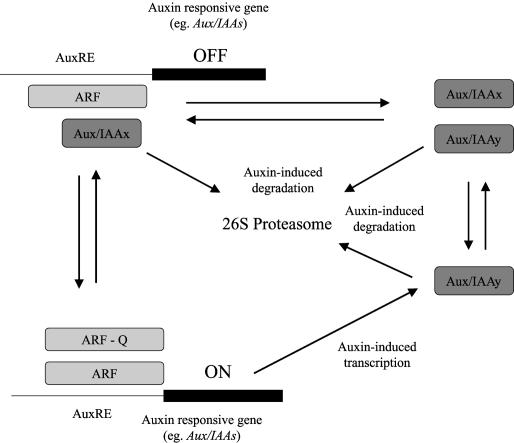
Combined with the data on the effect of auxin on Aux/IAA abundance, these observations have led to a model of auxin-regulated transcription from TGTCTC-containing AuxREs (Figure 3). At basal levels of auxin, Aux/IAAs are relatively stable and dimerize with ARF proteins on AuxREs, reducing transcription by preventing the recruitment of activating ARFs. Increasing auxin levels cause an increasing proportion of Aux/IAAs to be degraded, allowing the formation of a greater number of ARF-ARF dimers on AuxREs, and hence higher levels of transcription of auxin-responsive genes. Although this model lacks details, such as the level of multimerization of the interacting Aux/IAAs and ARFs, it seems a reasonable place to start: a point from which the unquestioned complexity can be built in.
Figure 3.
Model for Aux/IAA Action.
Aux/IAAs form a variety of dimers with both ARFs and other Aux/IAAs. The equilibrium of these dimerization events has the potential to alter gene expression from TGTCNC-containing AuxREs in auxin-responsive promoters. Auxin regulates both the degradation of Aux/IAA proteins and the transcription of Aux/IAA genes. Given that the diverse patterns of degradation and expression with the large Aux/IAA family would lead to considerable temporal variation in the relative abundance of different Aux/IAAs, the interactions are likely to be very complex.
How far does this model go toward explaining the existing genetic and molecular data on auxin-induced gene expression? Mutations in two of the Q-rich ARFs, MP/ARF5 and NPH4/ARF7 (Table 1), confer a range of auxin-related phenotypes, including reduced expression from TGTCTC-type AuxRE-containing auxin-inducible promoters (Hardtke and Berleth, 1998; Harper et al., 2000). This finding is consistent with the idea that Q-rich ARFs are required to activate the expression of auxin-inducible genes in vivo. There also is good evidence that transcription from TGTCTC AuxRE-regulated genes usually is kept inactive by very unstable auxin-inactivated repressor proteins, because transcription from AuxRE-containing promoters can be activated not only by auxin but also by inhibitors of protein synthesis such as cycloheximide (Koshiba et al., 1995). The possibility that these unstable transcriptional inhibitors are Aux/IAA proteins is supported by the observation that the dominant, stabilizing mutations in domain II of the Aux/IAA proteins often result in the constitutive repression of transcription from auxin-inducible AuxRE-containing promoters (Abel et al., 1995; Oono et al., 1998; Nagpal et al., 2000; Rogg et al., 2001).
Although these observations fit the general principle of the model, at first glance, other data do not. Aux/IAA genes were defined originally by their auxin inducibility, and most contain TGTCNC-type AuxREs in their promoters (reviewed by Abel and Theologis, 1996; Guilfoyle et al., 1998). Hence, at the same time that their protein levels are being depleted by auxin-induced SCFTIR1-mediated destabilization, they are being replenished by increased transcript accumulation. The auxin-induced increases in Aux/IAA transcript levels often persist for many hours and outlast more modest increases in Aux/IAA protein levels (Abel et al., 1995; Oeller and Theologis, 1995). This is not the pattern of message and protein accumulation that would be expected if Aux/IAA proteins repress their own transcription. On the contrary, if this were the case, then Aux/IAA mRNA levels should decrease as Aux/IAA protein levels increase.
One explanation might be that the model is based on transcriptional activation from TGTCTC-type AuxREs and does not include the contributions of other elements that affect the auxin induction of some Aux/IAAs (Ballas et al., 1995) and other auxin-responsive genes (Guilfoyle et al., 1998). These other elements may act as enhancers (Ballas et al., 1995) or may require other activating factors that have not been identified. If other factors are involved, the regulation of their activity would seem to require the involvement of the AXR1 pathway, because axr1 mutants are defective in virtually every measurable aspect of auxin signaling (Table 1). Therefore, although unproven, the possibility still exists that the βαα motifs of Aux/IAAs and ARFs might allow direct DNA binding. Because the important regulatory step of TGTCTC AuxRE induction seems to be the recruitment of activating ARFs with Q-rich middle regions (Ulmasov et al., 1999a), unorthodox ARF binding to other sites, possibly in cooperation with Aux/IAAs, also might bring about transcription. In any case, it is possible that other elements could contribute to longer term induction after the very rapid induction from TGTCTC-type AuxREs.
The Aux/IAAs are a large and diverse family; therefore, a second possibility is that in addition to the negative effect of some Aux/IAAs, others might indirectly have a positive effect on ARF-mediated gene expression. In the period after an auxin pulse, the balance of dimerization events could be biased toward allowing activating ARFs to interact on AuxREs. For example, newly synthesized individual Aux/IAAs that normally would block activating ARF dimers might themselves be sequestered and titrated out by other Aux/IAAs, which usually are at very low abundance when auxin levels are low. Similarly, nonactivating and repressing ARFs might be prevented from interfering with activating ARF dimerization at the AuxRE. This possibility would require significant temporal variation in the relative abundance of different Aux/IAAs.
Such variation is indeed reflected in the data. There are >25 different Aux/IAAs with varying temporal and dose responses (Abel et al., 1995). For example, some Aux/IAAs are induced very rapidly by auxin: SHY2/IAA3 message reaches almost maximal levels within 10 min, whereas the induction of AXR2/IAA7 is much slower, taking several hours to approach similar levels (Abel et al., 1995). Furthermore, the induction of IAA7 and IAA8 is insensitive to cycloheximide, suggesting that they form part of a secondary response to the auxin stimulus. Also, the limited data on Aux/IAA turnover show large differences in the stability of Aux/IAAs, the half-life of IAA7 being much shorter (∼11 min) than that of AXR3/IAA17 (∼80 min) (Gray et al., 2001; Ouellet et al., 2001). It also seems likely that there will be variation in the auxin-induced destabilization dynamics of Aux/IAAs (Gray et al., 2001).
Coupled with the possibility of different binding affinities among the interacting components, the functional significance of these differences may be to swing the balance of dimerization events between ARFs and Aux/IAAs initially toward, and then eventually away from, promoting the interaction of activating ARFs at the AuxRE. The idea that different Aux/IAAs do different things at different times in response to auxin certainly is supported by the genetic data. The similar stabilizing mutations of Aux/IAAs confer quite different and even opposite phenotypes. For example, the axr3-1 mutant has increased adventitious rooting, whereas the axr2-1 mutant has fewer adventitious roots than the wild type (Table 1) (Leyser et al., 1996; Nagpal et al., 2000).
It is possible that in the period after auxin stimulation, the activity of newly synthesized Aux/IAAs is affected by post-translational modifications that contribute to a prolonged response in the face of accumulating Aux/IAA protein. For example, phosphorylation of Aux/IAAs might alter their dimerization preferences to effect a bias toward the activation of AuxREs or even allow the direct stabilization and enhancement of activating ARF binding at the AuxRE. Indeed, if the phosphorylating activities of the auxin-induced MAPK, phytochrome A, or PINOID kinase act in any way through Aux/IAAs, it might be in modulating their activity in this manner. In these scenarios, there are parallels with the previously described activation and repression of the mammalian transcription factor NF-κB. Under normal stress conditions, the ubiquitin-mediated degradation of its inhibitors allows NF-κB to activate both the genes required to respond to stress and the expression of its own inhibitor, IκBα, ending the response (Karin and Ben-Neriah, 2000). However, in response to severe stress stimuli, such as Human immunodeficiency virus infection, for example, NF-κB activation is extended despite the accumulation of its inhibitors (DeLuca et al., 1999). This is because a hypophosphorylated version of another of its inhibitors, IκBβ, no longer inhibits but protects the binding of NF-κB to its DNA targets and prevents inhibition by IκBα (DeLuca et al., 1999).
AUXIN PERCEPTION
Clearly, auxin stimulation sets off a momentous chain of events in the nucleus, leading to the differential expression of hundreds of genes. However, despite valiant efforts, the mechanism by which the auxin signal is perceived is poorly understood. Several lines of evidence suggest the possibility of several modes of perception. Naturally, the search for auxin receptors has focused on proteins that are able to bind auxin (reviewed by Venis and Napier, 1995). In recent years, much attention has been directed toward the auxin binding protein ABP1 (reviewed by Napier, 1995). ABP1 binds auxin with high specificity and affinity, possessing a dissociation constant for the synthetic auxin naphthalene-1-acetic acid of 5 × 10−8 M (Napier, 1995). Although it shows no homology with any other known receptor family, ABP1 does seem to mediate several cellular responses to applied auxin, including tobacco mesophyll protoplast hyperpolarization (Leblanc et al., 1999a, 1999b), the expansion of tobacco and maize cells in culture (Jones et al., 1998; Chen et al., 2001), tobacco mesophyll protoplast division (Fellner et al., 1996), and stomatal closure (Gehring et al., 1998). It is clear that ABP1 acts at the cell surface to mediate these responses, because the exogenous addition of anti-ABP1 antibodies, which are unable to enter the cell, can interfere with the ability of auxin to bring about the responses. Paradoxically, the majority of ABP1 in the cell is retained in the endoplasmic reticulum, where the pH is too high for strong auxin binding (Tian et al., 1995; Henderson et al., 1997). Transgenic approaches to alter ABP1 levels have resulted in relatively modest phenotypes that in general seem to affect the balance between cell division and cell expansion (Jones et al., 1998; Bauly et al., 2000). Recently, however, an insertional mutant in Arabidopsis ABP1 conferring complete loss of ABP1 function resulting in embryo lethality has demonstrated an essential role for ABP1 in plant development (Chen et al., 2001).
Although ABP1 seems to act at the cell surface, evidence is accumulating for the intracellular perception of auxin. This is based largely on comparison of the effects of auxins that differ in their transport properties into and out of cells (Claussen et al., 1996). This idea has been strengthened by the characterization of Arabidopsis mutants that differ in their ability to transport auxins. For example, loss of function in the proposed auxin uptake carrier AUX1, a member of the amino acid permease family, results in a variety of phenotypes, including auxin-resistant root elongation and reduced root gravitropism (Pickett et al., 1990; Bennett et al., 1996; Marchant et al., 1999). The roots of aux1 are resistant to membrane-impermeable auxins such as 2,4-D but respond normally to the membrane-permeable auxin naphthylacetic acid, showing recovery of graviresponse (Yamamoto and Yamamoto, 1998; Marchant et al., 1999), suggesting that intracellular auxin is important for root growth inhibition. Intracellular auxin binding proteins that might mediate such responses have been identified in rice (Kim et al., 1998). One example is of a rice 57-kD soluble auxin binding protein that appears to interact directly with the plasma membrane proton-pumping ATPase, suggesting a very short signaling chain from auxin to increased proton pumping, cell wall acidification, and hence cell elongation (Kim et al., 2000, 2001).
In addition to the possible role of these auxin binding proteins, it has been proposed that proteins involved in auxin transport might be able to perceive auxin levels by monitoring the flux of auxin through transporters (Palme and Gälweiler, 1999). In Arabidopsis, there are four members in the auxin influx carrier family, typified by the amino acid permease-like AUX1 (Bennett et al., 1996), and eight members in the auxin efflux carrier family, including PIN1 and PIN2/EIR1/AGR1/WAV6 (reviewed by Palme and Gälweiler, 1999). It is possible that some of these auxin influx/efflux carriers might possess a specialized receptor function in addition to, or instead of, their transport activity. There are precedents here in the sensing of amino acids and Glc in the budding yeast S. cerevisiae. The perception of both amino acids and Glc has been shown to involve membrane proteins distinct from but highly homologous with amino acid or Glc transporters. For example, the amino acid permease-like sensor Ssy1 is structurally similar to amino acid transporters except that it contains a large N-terminal cytosolic tail that is essential for amino acid sensing (Didion et al., 1998; Iraqui et al., 1999). Similarly, the Glc transporter homologs Snf3 and Rgt2 are distinguished from normal transporters by their long C-terminal cytosolic tails, which are required for Glc perception (Özcan et al., 1998). Interestingly, the downstream signaling pathways of Ssy1 and Snf3/Rgt2 both require the involvement of SCFGrr1, the E3 ligase containing the F-box protein Grr1 that shows considerable homology with TIR1 (Li and Johnston, 1997; Ruegger et al., 1998; Bernard and Andre, 2001). Given the similarity of IAA to Trp, an attractive hypothesis is that auxin signaling may have evolved from an amino acid signaling pathway.
CONCLUSIONS
Undoubtedly, there is much left to learn about the plant's response to auxin. Although the model for auxin-induced gene expression provides a framework that allows the activities of SCFTIR1 and other SCFs, Aux/IAAs, and ARFs to be conceptualized, more details are required. Specifically, we need to understand the events that link auxin to changes in the stability of Aux/IAAs. We also need a better characterization of the consequences of the resulting fluctuations in Aux/IAA abundance for the balance of dimerization events between Aux/IAAs and ARFs. This will require an appreciation of the extent of functional diversity/redundancy among members of both families. It is relatively straightforward to compare spatial and temporal patterns of ARF and Aux/IAA induction and patterns of Aux/IAA instability. Addressing the extent, if any, of the differential binding preferences among the ARFs and Aux/IAAs in vivo is much more difficult. Large-scale ARF and Aux/IAA knockout programs and transcriptomic analyses are under way at present. These approaches should help us determine the activities of ARFs and Aux/IAAs that lead to the differential expression of hundreds of genes and coordinated developmental changes in response to auxin.
Acknowledgments
We thank Jerry Cohen and Jane Murfett for helpful discussions and Rebecca Garrod for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010447.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The Ps-IAA4/5-like family of early auxin-inducible messenger-RNAs in Arabidopsis thaliana. J. Mol. Biol. 251 533–549. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Santos-Rosa, M.J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6 354–358. [DOI] [PubMed] [Google Scholar]
- Ballas, N., Wong, L.-M., Ke, M., and Theologis, A. (1995). Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc. Natl. Acad. Sci. USA 92 3483–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauly, J.M., Sealy, I.M., Macdonald, H., Brearley, J., Droge, S., Hillmer, S., Robinson, D.G., Venis, M.A., Blatt, M.R., Lazarus, C.M., and Napier, R.M. (2000). Overexpression of auxin-binding protein enhances the sensitivity of guard cells to auxin. Plant Physiol. 124 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir, D., Kraft, R., Huang, X., Henklein, P., Kapelari, B., Pollmann, C., and Dubiel, W. (2001). COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 20 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P., Walker, A., Schulz, B., and Feldmann, K. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273 948–950. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Jürgens, G. (1993). The role of the MONOPTEROS gene in organizing the basal body region of the embryo. Development 118 575–587. [Google Scholar]
- Berleth, T., and Sachs, T. (2001). Plant morphogenesis: Long-distance coordination and local patterning. Curr. Opin. Plant Biol. 4 57–62. [DOI] [PubMed] [Google Scholar]
- Bernard, F., and Andre, B. (2001). Ubiquitin and the SCFGrr1 ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496 81–85. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.-W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121. [DOI] [PubMed] [Google Scholar]
- Chen, J.G., Ullah, H., Young, J.C., Sussman, M.R., and Jones, A.M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478. [DOI] [PubMed] [Google Scholar]
- Claussen, M., Luthen, H., and Bottger, M. (1996). Inside or outside? Localization of the receptor relevant to auxin-induced growth. Physiol. Plant. 98 861–867. [Google Scholar]
- Colon-Carmona, A., Chen, D.L., Yeh, K.C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.J., ed (1995). Plant Hormone Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Timpte, C., Tan, S., Callis, J., and Estelle, M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280 1760–1763. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1–ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCul1 is required for Auxin response. Plant Cell 14 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, C., Petropoulos, L., Zmeureanu, D., and Hiscott, J. (1999). Nuclear IκBβ maintains persistent NF-κB activation in HIV-1-infected myeloid cells. J. Biol. Chem. 274 13010–13016. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/RING-H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15 435–467. [DOI] [PubMed] [Google Scholar]
- Didion, T., Regenberg, B., Jorgensen, M.U., Kielland-Brandt, M.C., and Andersen, H.A. (1998). The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27 643–650. [DOI] [PubMed] [Google Scholar]
- Estelle, M., and Sommerville, C. (1987). Auxin resistant mutants of Arabidopsis thaliana with altered morphology. Mol. Gen. Genet. 206 200–206. [Google Scholar]
- Fellner, M., Ephritikhine, G., Barbier-Brygoo, H., and Guern, J. (1996). An antibody raised to a maize auxin-binding protein has inhibitory effects on cell division of tobacco mesophyll protoplasts. Plant Physiol. Biochem. 34 133–138. [Google Scholar]
- Furukawa, M., Zhang, Y., McCarville, J., Ohta, T., and Xiong, Y. (2000). The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20 8185–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J., and Peter, M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, C.A., McConchie, R.M., Venis, M.A., and Parish, R.W. (1998). Auxin-binding-protein antibodies and peptides influence stomatal opening and alter cytoplasmic pH. Planta 205 581–586. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jürgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, J., Bauly, J.M., Ashford, D.A., Oliver, S.C., Hawes, C.R., Lazarus, C.M., Venis, M.A., and Napier, R.M. (1997). Retention of maize auxin-binding protein in the endoplasmic reticulum: Quantifying escape and the role of auxin. Planta 202 313–323. [DOI] [PubMed] [Google Scholar]
- Hsiung, W.G., Chang, H.C., Pellequer, J.L., La Valle, R., Lanker, S., and Wittenberg, C. (2001). F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21 2506–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui, I., Vissers, S., Bernard, F., De Craene, J.O., and Boles, E. (1999). Amino acid signaling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan, M., Kondo, K., Yang, H.F., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J., Lane, W., and Kaelin, W., Jr. (2001). HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292 464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P., et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292 468–472. [DOI] [PubMed] [Google Scholar]
- Jackson, P.K., Eldridge, A.G., Freed, E., Furstenthal, L., Hsu, J.Y., Kaiser, B., and Reimann, J. (2000). The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10 429–439. [DOI] [PubMed] [Google Scholar]
- Johnston, M. (1999). Feasting, fasting and fermenting. Trends Genet. 15 29–33. [DOI] [PubMed] [Google Scholar]
- Jones, A.M., Im, K.H., Savka, M.A., Wu, M.J., DeWitt, N.G., Shillito, R., and Binns, A. (1998). Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282 1114–1117. [DOI] [PubMed] [Google Scholar]
- Karin, M., and Ben-Neriah, Y. (2000). Phosphorylation meets ubiquitination: The control of NF-kappa B activity. Annu. Rev. Immunol. 18 621–663. [DOI] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20 4003–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.C., Soh, M.S., Kang, B.J., Furuya, M., and Nam, H.G. (1996). Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 15 441–456. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.S., Kim, D.H., and Jung, J. (1998). Isolation of a novel auxin receptor from soluble fractions of rice (Oryza sativa L.) shoots. FEBS Lett. 438 241–244. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Kim, D., and Jung, J. (2000). Two isoforms of soluble auxin receptor in rice (Oryza sativa L.) plants: Binding property for auxin and interaction with plasma membrane H+-ATPase. Plant Growth Regul. 32 143–150. [Google Scholar]
- Kim, Y.S., Min, J.K., Kim, D., and Jung, J. (2001). A soluble auxin-binding protein, ABP(57): Purification with anti-bovine serum albumin antibody and characterization of its mechanistic role in the auxin effect on plant plasma membrane H+-ATPase. J. Biol. Chem. 276 10730–10736. [DOI] [PubMed] [Google Scholar]
- Kishi, T., and Yamao, F. (1998). An essential function of Grr1 for the degradation of Cln2 is to act as a binding core that links Cln2 to Skp1. J. Cell Sci. 111 3655–3661. [DOI] [PubMed] [Google Scholar]
- Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96 635–644. [DOI] [PubMed] [Google Scholar]
- Koshiba, T., Ballas, N., Wong, L.-M., and Theologis, A. (1995). Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J. Mol. Biol. 253 396–413. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9. J. Mol. Biol. 285 85–95. [DOI] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc, N., David, K., Grosclaude, J., Pradier, J.M., Barbier-Brygoo, H., Labiau, S., and Perrot-Rechenmann, C. (1999. a). A novel immunological approach establishes that the auxin-binding protein, Nt-abp1, is an element involved in auxin signaling at the plasma membrane. J. Biol. Chem. 274 28314–28320. [DOI] [PubMed] [Google Scholar]
- Leblanc, N., Perrot-Rechenmann, C., and Barbier-Brygoo, H. (1999. b). The auxin-binding protein Nt-ERabp1 alone activates an auxin-like transduction pathway. FEBS Lett. 449 57–60. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Lincoln, C.A., Timpte, C.S., Lammer, D., Turner, J.C., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 304 161–164. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 11 403–413. [DOI] [PubMed] [Google Scholar]
- Li, F., and Johnston, M. (1997). Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and cell cycle. EMBO J. 16 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos, D., Doenges, G., Matuschewski, K., and Jentsch, S. (1998). A novel protein modification pathway related to the ubiquitin system. EMBO J. 17 2208–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D., Wei, N., Shevchenko, A., and Deshaies, R. (2001). Promotion of NEDD8–CUL1 conjugate cleavage by COP9 signalosome. Science 292 1382–1385. [DOI] [PubMed] [Google Scholar]
- Marchant, A., Kargul, J., May, S.T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., and Bennett, M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias, N., Johnson, S., Byers, B., and Goebl, M. (1999). The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol. Cell. Biol. 19 1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias, N., Johnson, S.L., Winey, M., Adams, A.E.M., Goetsch, L., Pringle, J.R., Byers, B., and Goebl, M.G. (1996). Cdc53p acts in concert with cdc4p and cdc34p to control the G(1)-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16 6634–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991. [DOI] [PubMed] [Google Scholar]
- Mockaitis, K., and Howell, S.H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24 785–796. [DOI] [PubMed] [Google Scholar]
- Morgan, K.E., Zarembinski, T.I., Theologis, A., and Abel, S. (1999). Biochemical characterization of recombinant polypeptides corresponding to the predicted beta alpha alpha fold in Aux/IAA proteins. FEBS Lett. 454 283–287. [DOI] [PubMed] [Google Scholar]
- Morimoto, M., Nishida, T., Honda, R., and Yasuda, H. (2000). Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCFskp2 toward p27(kip1). Biochem. Biophys. Res. Commun. 270 1093–1096. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier, R.M. (1995). Towards an understanding of ABP1. J. Exp. Bot. 46 1787–1795. [Google Scholar]
- Oeller, P.W., and Theologis, A. (1995). Induction kinetics of the nuclear proteins encoded by the early indoleacetic acid-inducible genes Ps-IAA4/5 and Ps-IAA6 in pea (Pisum sativum L.). Plant J. 7 37–48. [DOI] [PubMed] [Google Scholar]
- Oono, Y., Chen, Q.G., Overvoorde, P.J., Kohler, C., and Theologis, A. (1998). age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka, F., Saeki, M., Katayama, S., Aida, N., Toh-e, A., Kominami, K., Toda, T., Suzuki, T., Chiba, T., Tanaka, K., and Kato, S. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17 2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palme, K., and Gälweiler, L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2 375–381. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14 236–243. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70 503–533. [DOI] [PubMed] [Google Scholar]
- Pickett, F.B., Wilson, A.K., and Estelle, M. (1990). The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94 1462–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust, V., Brownell, J., Gladysheva, T., Luo, R.-S., Wang, C., Coggins, M., and Pierce, J. (2000). A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. USA 97 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of Auxin/Indoleacetic Acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6 420–425. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Elumalai, R.P., and Chory, J. (1998). Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics 148 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Plant Cell 279 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Klenz, J., Kohalmi, S., Risseeuw, E., Haughn, G., and Crosby, W. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20 433–445. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2001). COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 11 420–426. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., and Deng, X.-W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292 1379–1382. [DOI] [PubMed] [Google Scholar]
- Seol, J.H., et al. (1999). Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91 209–219. [DOI] [PubMed] [Google Scholar]
- Stirnberg, P., Chatfield, S.P., and Leyser, H.M.O. (1999). AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H.C., Klambt, D., and Jones, A.M. (1995). Auxin-binding protein-1 does not bind auxin within the endoplasmic-reticulum despite this being the predominant subcellular location for this hormone-receptor. J. Biol. Chem. 270 26962–26969. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126 711–721. [DOI] [PubMed] [Google Scholar]
- Timpte, C., Wilson, A.K., and Estelle, M. (1994). The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte, C., Lincoln, C., Pickett, F.B., Turner, J., and Estelle, M. (1995). The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 8 561–569. [DOI] [PubMed] [Google Scholar]
- Tomoda, K., Kubota, Y., and Kato, J. (1999). Degradation of cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398 160–165. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Dimerization and DNA binding of auxin response factors. Plant J. 19 309–319. [DOI] [PubMed] [Google Scholar]
- Venis, M.A., and Napier, R.M. (1995). Auxin receptors and auxin binding proteins. Crit. Rev. Plant Sci. 14 27–47. [Google Scholar]
- Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68 1015–1068. [DOI] [PubMed] [Google Scholar]
- Walz, A., Park, S., Slovin, J.P., Ludwig-Muller, J., Momonoki, Y.S., and Cohen, J.D. (2002). A gene encoding a protein modified by the phytohormone indoleacetic acid. Proc. Natl. Acad. Sci. USA 99 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1999). Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15 98–103. [DOI] [PubMed] [Google Scholar]
- Wei, N., Tsuge, T., Serino, G., Dohmae, N., Takio, K., Matsui, M., and Deng, X.W. (1998). The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8 919–922. [DOI] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222 377–383. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21 553–562. [DOI] [PubMed] [Google Scholar]
- Wu, K., Chen, A., and Pan, Z.Q. (2000). Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1–CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275 32317–32324. [DOI] [PubMed] [Google Scholar]
- Xiao, W.Y., and Jang, J.C. (2000). F-box proteins in Arabidopsis. Trends Plant Sci. 5 454–457. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., and Yamamoto, K.T. (1998). Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol. 39 660–664. [DOI] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D., Yu, Q., Chen, M., and Ma, H. (2001). The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128 2735–2746. [DOI] [PubMed] [Google Scholar]