INTRODUCTION
Biological nitrogen fixation is a process that can only be performed by certain prokaryotes. In some cases, such bacteria are able to fix nitrogen in a symbiotic relationship with plants. Bacteria of the genera Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium (collectively referred to as Rhizobium or rhizobia) are able to establish an endosymbiotic association with legumes. Under nitrogen-limiting conditions, the leguminous plants can form root nodules, in which the rhizobia are hosted intracellularly. There they find the proper conditions for reducing atmospheric nitrogen into ammonia. This review will focus on signal transduction events of Rhizobium-induced nodulation.
Nodule Formation
Basic research on the legume–Rhizobium interaction becomes more and more focused on two model legume species, Lotus japonicus and Medicago truncatula (Cook, 1999; Stougaard, 2001). Two model legumes have been selected because most legumes form nodules that belong either to the determinate or to the indeterminate nodule type and L. japonicus and M. truncatula, respectively, represent these two major legume groups (Pawlowski and Bisseling, 1996). Although the morphology and ontology of these two nodule types is different, the mechanisms underlying the formation of them are probably very similar. In this review, we will only give a description of nodule ontogeny of the indeterminate nodule type, as represented by M. truncatula.
The formation of a nodule requires the reprogramming of differentiated root cells to form a primordium, from which a nodule can develop. Furthermore, the bacteria must infect the root before the nitrogen-fixing root nodule can be formed. These steps in nodule formation involve changes in three root tissues, namely epidermis, cortex, and pericycle.
When rhizobia have colonized the root surface of their host, they induce morphological changes in the epidermis. These morphological changes are preceded by the induction of certain genes in a broad region of the epidermis. The best-studied examples are the early-nodulin genes ENOD12 and ENOD11, which are homologous and belong to a gene family encoding proline-rich proteins. ENOD12 and ENOD11 have a similar expression pattern and have been used as molecular markers to monitor signal transduction events in the epidermis (Scheres et al., 1990; Pichon et al., 1992; Journet et al., 2001).
Upon inoculation with rhizobia, root hairs will deform. This is caused by a reinitiating of tip growth in these cells, but with a changed growth direction (Heidstra et al., 1994; De Ruijter et al., 1998). These morphological changes are preceded by changes in the actin skeleton (Cardenas et al., 1998; De Ruijter et al., 1999). In some root hairs, the rhizobia induce deformations that resemble a so-called shepherd's crook, and such curled root hairs play an important role in the infection process. Root hair curling is probably caused by a gradual and constant reorientation of the growth direction of the hair by which a curl with a turn of ∼360° is formed (Van Batenburg et al., 1986; Ridge, 1993; Emons and Mulder, 2000). During the curling process, the bacteria become entrapped in the pocket of the curl. There the plant cell wall is modified in a very local manner, the plasma membrane invaginates, and new plant material is deposited. In this way a tube-like structure, the infection thread, is formed that contains the bacteria. The infection thread will grow toward the base of the root hair cell and subsequently to the nodule primordium (Figure 1A; Brewin, 1998).
Figure 1.
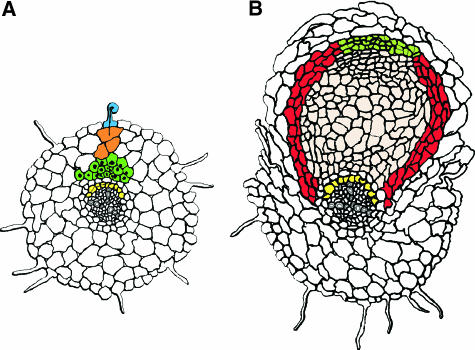
Schematic Drawing of Early Steps in Root Nodule Formation in Medicago truncatula Induced by Sinorhizobium meliloti.
(A) Nodule primoridum formation. The bacteria induce root hair curling and infection thread formation in epidermal cells (blue). Concomitantly, the inner cell layers are activated. Pericycle cells opposite a proto-xyleme pole express the ENOD40 gene and eventually will undergo a limited number of cell divisions (yellow). Cells in the cortex will loose their cell identity and enter the cell cycle. This results in the formation of a nodule primordium in the inner cortex (dark green). In these cells, ENOD12 and ENOD40 are expressed. Outer cortical cells also enter the cell cycle, but are arrested (orange) and will form preinfection threads.
(B) Root nodule differentiation. Cells of the nodule primordium obtain their final identity after rhizobial infection. At the base of the primordium, a radial pattern of a central tissue (pink) surrounded by peripheral tissues (red) is established. Concomitantly, cells at the apex of the primordium form a meristem (light green). These cells differ in their identity compared with primordium cells, in that they do not express ENOD12. The central tissue contains the cells that host the rhizobia.
Even before the infection thread has crossed the epidermis, cortical and pericycle cells respond in a local manner to the rhizobia. In pericycle cells, this is reflected by the rapid induction of ENOD40 in a zone opposite a protoxylem pole and by rearrangements of the microtubles (Yang et al., 1993; Timmers et al., 1999; Compaan et al., 2001). Eventually, these cells will undergo a limited number of cell divisions (Timmers et al., 1999). Following the activation of the pericycle, cells in the inner cortex dedifferentiate by entering the cell cycle. The group of dividing cortical cells is named the nodule primordium (Figure 1A).
Outer cortical cells, through which the infection threads will grow, form radially oriented, conical cytoplasmic columns. This organization of the cytoplasm takes place before the infection thread enters these cells; these therefore are named pre–infection threads (Van Brussel et al., 1992; Timmers et al., 1999; Van Spronsen et al., 2001). The pre–infection thread forming outer cortical cells also enter the cell cycle but are arrested before they can enter the M phase (Yang et al., 1994; Figure 1A). Therefore, it is assumed that pre–infection threads are in some way related to phargmoplasts (Van Brussel et al., 1992; Yang et al., 1994). The pre–infection threads will facilitate infection thread growth and direct it toward the nodule primordium. There the infection thread ramifies, followed by the release of bacteria into the primordial cells. Upon release, the bacteria remain surrounded by a membrane of plant origin and subsequently will differentiate into their symbiotic form and will start to fix nitrogen (for review, see Mylona et al., 1995).
In general, the transition from nodule primordium to young developing nodule occurs after infection of primordial cells. During this transition, cells at the base of the primordium establish a radial pattern consisting of a central tissue surrounded by peripheral tissues (Pawlowski and Bisseling, 1996). Concomitantly, cells at the apex of the primordium form a meristem that, by division, maintains itself and adds new cells to the different tissues according to the pattern established at the base of the primordium (Figure 1B). The identity of nodule primordium cells and nodule meristematic cell is different. For example, a meristematic cell is never infected by rhizobia, and genes that are activated in the primordium, e.g., ENOD12, are not transcribed in the nodule meristem (Scheres et al., 1990).
Specific signal molecules secreted by Rhizobium, named Nod factors, play a pivotal role in the induction of all early responses. For example, they are required for gene activation in the epidermis and pericycle cells, for the mitotic reactivation of the cortical cells, and for the formation of pre–infection threads.
Nod Factors
Proteins encoded by the rhizobial nodulation genes (nod, nol, and noe genes) are involved in the synthesis and secretion of Nod factors. The expression of these genes is activated when the bacteria perceive specific molecules, in general flavonoids that are secreted by the plant root (Zuanazzi et al., 1998). Flavonoids activate the bacterial transcriptional regulator NodD that in turn induces the transcription of the other bacterial nodulation genes involved in the synthesis of Nod factors.
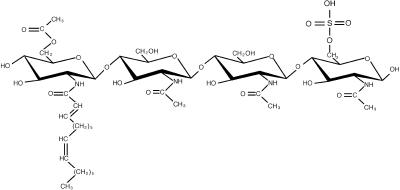
The basic structure of Nod factors produced by different rhizobial species is very similar. Generally, they consist of a β-1,4–linked N-acetyl-d-glucosamine backbone with 4 or 5 residues of which the non-reducing terminal residue is substituted at the C2 position with an acyl chain. Depending on the rhizobial species, the structure of the acyl chain can vary, and substitutions at the reducing and nonreducing terminal glucosamine residues can be present. The structure of Nod factors of different rhizobia and their function in nodulation has been reviewed recently (Mergaert et al., 1997; Cullimore et al., 2001). Here only the Nod factor structure of Sinorhizobium meliloti, the bacterium interacting with the Medicago species, is given (Figure 2).
Figure 2.
The Major Nod Factor Produced by Sinorhizobium meliloti.
The major Nod factor produced by Sinorhizobium meliloti contains four glucosamine units, an acyl chain of 16 C-atoms in length with two unsaturated bonds (determined by NodE and NodF), an acetyl group at the non-reducing terminal sugar residue (determined by NodL), and a sulfate group at the reducing terminal sugar residue (determined by NodH, NodP and NodQ) (Lerouge et al., 1990).
The vast majority of the Nod factors produced by S. meliloti are tetrameric and contain an acyl chain of 16 carbon atoms in length with two unsaturated bonds (C16:2). The terminal reducing glucosamine residue of this Nod factor is O-sulfated, whereas the other terminal glucosamine contains an O-acetyl group (Figure 2). In addition, low quantities of Nod factors containing C18-C26 (ω-1)-hydroxylated acyl chains and molecules that lack the O-acetyl group are formed (Lerouge et al., 1990; Roche et al., 1991; Schultze et al., 1992; Demont et al., 1993).
The differences in structure of Nod factors made by different rhizobial species are determined by the presence of species-specific nodulation genes or are due to allelic variation resulting in a different activity of the encoded enzymes. In general, the substitutions at the terminal residues and the structure of the acyl chain play a role in the ability of the bacterium to interact with its host plant. For example, the sulfate decoration of the S. meliloti Nod factor is a major determinant of host specificity because it is required for the induction of almost all symbiotic responses. In contrast, the O-acetate group as well as the structure of the acyl chain is especially required for efficient infection.
In the following section of this review, Nod factor perception and induced signal transduction events will be the central themes.
Nod Factor Binding
Nod factors induce responses in their hosts at picomolar concentrations. Therefore, it is often suggested that Nod factors are recognized by a high affinity receptor (e.g., Dénarié and Cullimore, 1993; Heidstra and Bisseling, 1996). Further, the amphiphylic nature of Nod factors, with their hydrophobic lipid tail and -hydrophilic sugar backbone, suggests that Nod factor receptors are located in the plasma membrane. The latter is supported by in vitro studies showing that Nod factors rapidly insert into membranes (Goedhart et al., 1999).
To identify putative Nod factor receptors, two approaches have been used: a direct approach to identify Nod factor binding sites in protein extracts, and a “candidate gene” approach to determine whether proteins encoded by known genes are able to bind Nod factors. Both approaches have led to the identification of Nod factor binding proteins.
Two Nod factor binding proteins, NFBS1 and NFBS2, have been identified using binding studies with protein extracts (Bono et al., 1995; Niebel et al., 1997). NFBS1 is found in a root extract of M. truncatula and has an affinity of 86 nM for Nod factors. However, it also binds with about the same affinity to the nonsulfated Nod factor, which is biologically inactive in Medicago species. Furthermore, a similar binding site occurs in tomato (Lycopersicon esculentum). Both observations make it unlikely that NFBS1 functions as a specific Nod factor receptor. NFSB2 has been identified in a M. varia cell suspension and is probably a plasma membrane–associated protein (Niebel et al., 1997). It has a higher affinity (Kd: 4 nM) for Nod factors than does NFBS1, but it also does not discriminate between sulfated and nonsulfated Nod factors. The lack of host-specific binding properties makes it unlikely that these Nod factor binding sites by themselves can function as a specific Nod factor receptor. Further, it remains to be demonstrated that NFBS2 occurs in roots. If it does, it might be part of a Nod factor binding complex, in combination with other components by which only Nod factors with a host-specific structure are recognized (Cullimore et al., 2001).
By using a “candidate gene” approach, it was shown that Nod factors bind to a lectin–nucleotide phosphohydrolase (LNP) named Db-LNP isolated from the roots of the legume Dolichos biflorus (Etzler et al., 1999). LNPs are members of the eukaryotic ATPase superfamily and catalyze the hydrolyses of the four nucleotide triphosphates as well as the nucleotide diphosphate ADP, which is a characteristic of Apyrases. It was shown that the ATPase activity of Db-LNP increases upon Nod factor binding to the lectin domain of the protein (Etzler et al., 1999).
Db-LNP, as well as its soybean (Glycin soja) ortholog GS52, is extracellular protein present at the surface of young root hairs. Upon inoculation of D. biflorus with Bradyrhizobium, the protein accumulates at the tip of the root hairs; this is not the case when non-host rhizobia are used. Furthermore, specific antibodies against the LNPs block root hair deformation as well as nodulation (Etzler et al., 1999; Day et al., 2000; Kalsi and Etzler, 2000). This strongly suggests that LNPs play a role in the early steps of nodulation. However, how it could function as a receptor (i.e., in amplifying the Nod factor signal) and why the ATPase activity is essential remain unclear.
Two other lectins have been shown to play a role in the early steps of nodulation. These are pea seed lectin PSL1 and Le1 of soybean. When these genes are expressed in heterologous plants, the host range of these plants is extended, but they also become more susceptible to their normal host rhizobial species. However, it is unlikely that the mode of action of these lectins depends on Nod factor binding (Diaz et al., 1995; Van Rhijn et al., 1998, 2001).
Behavior of Nod Factors and Implications for Nod Factor Perception
In vivo studies on the behavior of Nod factors in roots have provided additional insight concerning Nod factor perception. Nod factor behavior on the root surface was studied by using fluorescent, biologically active Nod factors, in combination with fluorescence correlation spectroscopy (FCS) (Goedhart et al., 2000). This is a very sensitive fluorescence microscopy technique allowing measurements at the single-molecule level that can be used to quantify molecular diffusion rates and to determine the concentration of fluorescent molecules. In FCS a focused laser beam illuminates a small volume element (∼1 μm3) and the number of fluorescent molecules that pass through this element as well as the average time required for this passage are determined. This allows a calculation of the concentration as well as the diffusion coefficient. Because the latter is related to the size of the molecule, it can be determined whether the fluorescently labeled molecule forms a complex with other components (for review, see Hink et al., 2002).
FCS studies showed that Nod factors, provided in an aqueous solution, accumulate in the cell wall of root hairs and reach concentrations of up to 50-fold higher than in the medium that was applied (Goedhart et al., 2000). Furthermore, Nod factors become highly immobilized, suggesting that they bind to (a) cell wall component(s). The accumulation of Nod factors in the cell wall as well as their immobilization is Nod factor structure independent and not host plant specific. Therefore, it is unlikely that this is caused by binding to a Nod factor receptor. However, rhizobia might use the immobilized accumulation of Nod factors in the cell wall to provide positional cues to the root hair.
So, Nod factors accumulate in cell walls at concentrations markedly higher than in the applied medium. This suggests that although Nod factors applied at picomolar concentrations induce responses, it is likely that a putative Nod factor receptor is exposed to markedly higher concentrations. Simple calculations suggest that in vivo, Nod factors are also present at a concentration markedly higher than picomolar levels. For example, within the pocket formed by a root hair curl, 20 to 50 bacteria are present and the total volume (including bacteria) is less than 100 μm3 (P. Smit and R. Geurts, unpublished data). If only a single Nod factor molecule were present in the pocket of the curl, the concentration would already be higher than 10−10 M. If the bacteria produce 100 Nod factor molecules each, the local concentration would be within the micromolar range. Therefore it seems unlikely that a putative Nod factor receptor would have such a high affinity for Nod factors as was originally proposed.
As demonstrated by studies involving FCS, Nod factors become immobilized in the plant cell wall. Therefore, it seems probable that secreted Nod factors will be present in the vicinity of the bacterial colony that secretes them. The resulting local occurrence of Nod factors could provide information to the host about the position of the bacteria that might be used to redirect root hair growth. This hypothesis is supported by the observation that local application of Nod factors at the root hair surface with a micropipette causes a redirection of growth toward the applied droplet (Esseling, et al., 2000). Thus, Nod factors may have a dual function, first as a ligand for a Nod factor receptor activating signal transduction cascades and second as a positional cue to redirect root hair growth.
The molecular basis of the redirection of root hair growth is not understood, but insight probably will be obtained via genetic studies. An example is the M. truncatula hair-curling (hcl) mutant that has lost the ability to form a tight curl, in which root hairs grow in a random direction although other responses are induced normally. Therefore, it has been proposed that the HCL protein is required to recognize/transfer the positional information provided by the rhizobia (Catoira et al., 2001).
Multiple Perception Mechanisms
The induction of early nodulin genes, the activation of the inner cell layers, and the establishment of root hair deformation and curling do not require a high degree of specificity to Nod factor structure. In Medicago species, all of these responses can be triggered by S. meliloti strains that have a mutation in nodFE and/or nodL, so neither the specific structure of the acyl chain nor the acetate substitution at the nonreducing terminal sugar residue are essential (Figure 2). In contrast, both structural characteristics are important for the formation of infection threads (Ardourel et al., 1994; Catoira et al., 2001).
Wild-type rhizobia will initiate tens or even hundreds of infection sites in the root, of which the majority will abort and only a limited number will infect a nodule primordium. S. meliloti nodL and nodFE mutants, respectively, induce markedly fewer infection events, although the number of nodules that is formed is about the same as in wild-type S. meliloti (Ardourel et al., 1994). Moreover, the S. meliloti nodFnodL double mutant has completely lost its ability to trigger infection thread formation, demonstrating that the non-acetylated C18:1 Nod factor is unable to trigger the formation of an infection thread. This has led to the hypothesis that two Nod factor perception mechanisms are operational in the epidermis: a mechanism involved in the bacterial entry that has a high stringency for the Nod factor structure; and a perception mechanism with a lower stringency involved in the induction of the other epidermal responses (Ardourel et al., 1994; Catoira et al., 2001).
A host gene that is specifically involved in controlling infection thread formation in relation to Nod factor structure is SYM2 (Geurts et al., 1997). SYM2 was identified in the pea (Pisum sativum) accession Afghanistan (SYM2A), where it inhibits infection by Rhizobium leguminosarum biovar viciae strains lacking nodX. NodX O-acetylates pentameric R. leguminosarum bv viciae Nod factors at the reducing terminal sugar residue (Firmin et al., 1993), which does not harbor a specific substitution in the absence of nodX (Spaink et al., 1991). Thus, the activity of SYM2 depends on the structure of the Nod factors secreted by the infecting rhizobia, and therefore, it may act in the mechanism that controls the entry of the bacteria.
The genomes of pea and M. truncatula are microsyntenic, and in M. truncatula, the SYM2 orthologous region has been identified (Gualtieri et al., 2002). However, whether a SYM2 ortholog occurs in M. truncatula is unknown.
Nod Factor–Activated Signal Transduction
Electrophysiological and cell biological approaches as well as pharmacological studies with specific blockers have provided insight into the signal transduction cascades that are activated in the root epidermis by Nod factors. One of the first responses is plasma membrane depolarization, which is observed ∼1 min after Nod factor addition. This depolarization is caused by a very rapid calcium influx, induced within seconds of Nod factor addition, followed by a chloride efflux. Upon depolarization of the membrane, an efflux of potassium ions is induced, by which the membrane can repolarize. The calcium influx is essential for Nod factor–induced membrane depolarization because the calcium channel antagonist nifedipine, as well as EGTA, inhibits this Nod factor–induced membrane depolymerization, whereas the calcium ionophore A23187 is able to mimic this response (Ehrhardt et al., 1992; Felle et al., 1995, 1996, 1998; Kurkdjian, 1995; Kurkdjian et al., 2000). Several minutes after Nod factor application, calcium oscillations occur in the perinuclear region of the root hairs and propagate radially through the cell (Ehrhardt et al., 1996).
Although the precise role of the induced changes in ion concentrations remains to be demonstrated, they have been useful as assays for the study of Nod factor signaling. In particular, calcium oscillation is a Nod factor response that occurs in several legume species, which adds to its usefulness as an assay for genetically dissecting Nod factor signaling (Ehrhardt et al., 1996; Wais et al., 2000; Walker et al., 2000).
Further insight into Nod factor signaling has been obtained by using specific drugs that mimic or interfere with certain Nod factor–induced responses. Within such pharmacological studies, a reporter gene driven by the promoter of the early-nodulin gene ENOD12 or ENOD11, has been a powerful tool. Upon inoculation with rhizobia or treatment with Nod factors, ENOD12 is induced in the root epidermis within 2 to 3 hr (Pichon et al., 1992; Pingret et al., 1998). To determine whether heterotrimeric GTP binding protein complexes (trimeric G-proteins) are part of a Nod factor–activated signaling pathway, the effect of mastoparan has been studied. Trimeric G-proteins have 3 subunits named Gα, Gβ, and Gγ, respectively, and are associated with G-protein–coupled receptors (GPCRs) that belong to the group of receptors containing 7 transmembrane helices. Mastoparan is a cationic peptide of 14 amino acids, and in animal systems, it has been shown to mimic the activated intracellular domain of GPCRs, leading to the activation of trimeric G-proteins (Law and Northrop, 1994; Ross and Higashijima, 1994). Mastoparan causes a spatial and temporal activation of MtENOD12::GUS in the root epidermis of M. truncatula that is comparable to the induction by Nod factors (Pingret et al., 1998). Furthermore, mastoparan induces root hair deformation in Vicia sativa (Den Hartog et al., 2001). Assuming that mastoparan activates plant trimeric G-proteins, it seems probable that a Nod factor–activated signaling pathway includes a trimeric G-protein (Pingret et al., 1998; Den Hartog et al., 2001). However, the degree to which plants and animals rely on trimeric G-proteins in their signal tranduction pathways is markedly different. In mammalian systems, hundreds of genes encoding GPCRs and tens of genes coding for the subunits of the trimeric G-proteins have been identified. In contrast, in plants, GPCRs and trimeric G-proteins are rare. For example, in the Arabidopsis genome, only one putative Gα- and Gγ-, and two putative Gβ-subunits, and very few putative GPCRs have been identified (Ellis and Miles, 2001). Likewise in M. truncatula, a similar low number of genes encoding putative GPCRs and trimeric G-protein subunits occur in over 125,000 expressed sequence tags (R. Geurts, unpublished data). This raises the question whether mastoparan-induced plant responses are mediated by the activation of a trimeric G-protein, and it remains unclear whether Nod factors activate such G-proteins.
Nod factor as well as mastoparan activates phospholipid signaling, causing an increase in the concentrations of phosphatidic acid (PA) and diacylglycerol pyrophosphate (DGPP). Changes in the concentrations of other phospholipids, such as phosphatidylinositol 4,5-bisphosphate, were not observed (Den Hartog et al., 2001). PA is probably a signal molecule in plants, and it is converted to DGPP by PA kinase. The Nod factor–induced increase of the PA concentration was caused by the activation of phospholipase C (PLC) as well as phospholipase D (PLD). The activation of both phospholipases was shown to be essential for the induction of root hair deformation. PLC activation was also shown to be essential for ENOD12 induction, but whether PLD is involved is not known (Pingret et al., 1998).
Genetic Dissection of Nod Factor Perception and Transduction
Legume crops have been used in genetic studies for decades and especially for pea a large collection of symbiotic mutants has been generated (Borisov et al., 2000). This collection has been screened for mutants that are blocked in the early steps of the symbiosis (for an overview of mutants mentioned in this review, see Table 1). In total, five complementation groups, named sym8, sym9, sym10, sym19, and sym30 have been identified (Duc and Messager, 1989; Kneen et al., 1994; Sagan et al., 1994). Recently, extensive screens were performed in L. japonicus and M. truncatula, the model legumes that are more amendable for positional cloning. In M. truncatula, this has resulted in the identification of three loci named dmi1 (doesn't make infections1), dmi2, and dmi3 (Catoira et al., 2000). The pea and M. truncatula mutants were analyzed in a similar way to genetically dissect the Nod factor signal transduction pathway and will be discussed here.
Table 1.
Overview of the Symbiotic Plant Mutants Described in this Reviewa
| Mutant | Phenotypic Characteristics |
|---|---|
| Lotus japonicus | |
| nin | Blocked in Nod factor–induced responses in the inner cell layers, but extensive curling of root hairs. |
| NIN encodes a putative transcription factor. | |
| Medicago sativa | |
| MNNN1008 (nn1) | Blocked in all Nod factor–induced responses. Myc−. |
| Putative ortholog of dmi2 and sym19. | |
| Medicago truncatula | |
| dmi1 | Blocked in all Nod factor–induced responses. Myc−. |
| dmi2 | Blocked in all Nod factor–induced responses. Myc−. Putative ortholog of nn1 and sym19. |
| dmi3 | Blocked in many Nod factor–induced responses, except Ca2+ oscillation for which it shows an |
| increased sensitivity. Myc−. | |
| hcl | Blocked in root hair curling. |
| nsp | Blocked in Nod factor–induced response in the inner cell layers. |
| skl | Ethylene insensitive. An increased number nodule primordia are formed opposite the proto phloem poles. Increased sensitivity to Nod factors in respect to Ca2+ oscillation. |
| Pisum sativum | |
| SYM2 | Controls infection thread formation in relation to Nod factor structure. |
| Occurs as natural variation. | |
| sym8 | Blocked in all Nod factor–induced responses. Myc−. |
| sym9 | Blocked in all Nod factor–induced responses, except Ca2+ oscillation. Myc−. |
| sym10 | Blocked in all Nod factor–induced responses. Myc+. |
| sym19 | Blocked in all Nod factor–induced responses. Myc−. Putative ortholog of nn1 and dmi2. |
| sym30 | Blocked in all Nod factor–induced responses, except Ca2+ oscillation. Myc−. |
(Myc+), able to establish a mycorrhizal interaction, and (Myc−), not able to establish a mycorrhizal interaction, for mutants blocked at an early stage of nodulation. For references, see text.
The above-mentioned pea and M. truncatula mutants are blocked in Nod factor–induced responses in the epidermis as well as in inner cell layers; the root hairs do not deform or curl, cortical cell divisions are not induced, and the early-nodulin genes ENOD12 and ENOD40 are not induced in the epidermis and pericycle, respectively (Markwei and LaRue, 1992; Albrecht et al., 1998; Schneider et al., 1999; Catoira et al., 2000; Walker et al., 2000). In fact, the only response observed upon application of Nod factors is the swelling of the root hair tips of the three M. truncatula dmi mutants (Catoira et al., 2000).
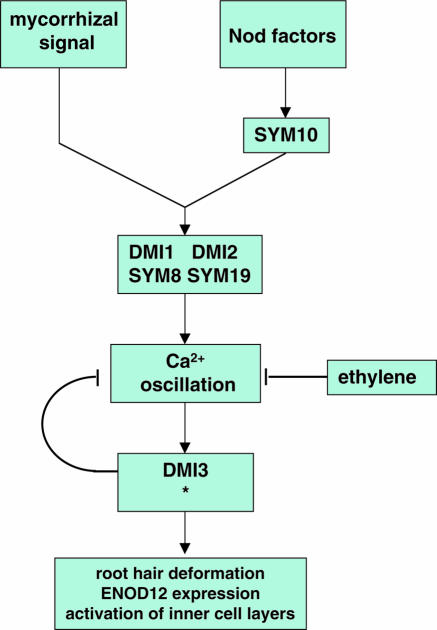
Nod factor–induced calcium oscillation turned out to be a useful response for classifying the mutants. This response could not be induced in the pea sym8, sym10, or sym19 mutants or in the M. truncatula dmi1 and dmi2 mutants. In contrast, calcium oscillation could be induced in sym9, sym30, and dmi3 mutants, respectively (Wais et al., 2000; Walker et al., 2000). This indicates that the first group of mutants is blocked at an earlier step of Nod factor signaling than the second group (Figure 3).
Figure 3.
Genetic Dissection of Nod Factor Signaling.
Mutations in sym8 and sym19 of pea and dmi1 and dmi2 of M. truncatula block Nod factor–induced calcium oscillation as well as mycorrhizal infection, whereas the pea sym10 mutation blocks calcium oscillation only. Therefore, it is likely that SYM10 acts upstream of SYM8 and SYM19 and probably is involved in a specific Nod factor perception mechanism (Wais et al., 2000; Walker et al., 2000). Calcium oscillation is negatively affected by ethylene as well as by DMI3 (Oldroyd et al., 2001a, 2001b).
(*), SYM9 and SYM30 of pea function downstream of calcium oscillation, but it was not investigated whether they have a control function like DMI3 (Walker et al., 2000).
It is known that some nodulation mutants are also blocked in their ability to interact with mycorrhizal fungi (Duc et al., 1989; Albrecht et al., 1999). All mutants described above, except the pea sym10 mutants, are blocked in mycorrhizal interaction (Duc et al., 1989; Catoira et al., 2000; Walker et al., 2000), so the group of mutants blocked in calcium oscillation can be further divided in two groups: sym10, which is myc+; and all others, which are myc−. Because the morphological responses induced by mycorrhizal fungi and rhizobia are different, it seems probable that the structure of the mycorrhizal signal molecules differs from that of rhizobial Nod factors. This is further supported by the fact that mycorrhizal symbiosis is very common and occurs in the vast majority of plant species, implying that non-legumes will also recognize a putative mycorrhizal signal, whereas they do not respond to rhizobial Nod factors. So it is probable that Nod factors and mycorrhizal signals are structurally different, and, therefore, different receptors are involved. For these reasons it seems probable that SYM10 is active upstream of the common steps of the Rhizobium- and mycorrhizal-activated signaling pathway, and that therefore it is a good candidate to encode a protein that is (directly) involved in Nod factor perception.
In M. truncatula as well as L. japonicus, positional cloning of symbiotic genes has been initiated. The genomes of pea, M. truncatula, and M. sativa are syntenic, and based on their map position and their comparable phenotypes, dmi2 and sym19 are probably orthologs of nn1 in M. sativa (Dudley and Long, 1989; Bradbury et al., 1991; Ehrhardt et al., 1996; Schneider et al., 1999; Walker et al., 2000; Endre et al., 2002). This locus was cloned and shown to encode a receptor-like kinase with leucine-rich repeats in a putative extracellular domain (Endre et al., 2002). The function of this receptor-like kinase in Nod factor signaling can now be addressed. If NN1 is involved in Nod factor perception, it probably forms part of a larger complex in which another component, e.g., SYM10, determines the specificity for Nod factors.
Nod Factor Signaling to Inner Cell Layers
Nod factor signaling is primarily studied using epidermal responses that are induced within minutes to hours after Nod factor application. In some cases, it is shown that these responses are induced in a cell-autonomous way. Also, the inner cell layers respond to Nod factors within hours. For example, ENOD40 gene expression is induced in the pericycle within 1 hr after application of Nod factors (Compaan et al., 2001). The question remains whether Nod factors are transported to these cells or whether plant signal molecules formed as a response to Nod factors induce the responses in the inner cell layers. Studies that addressed this question in an indirect way led to contradictory answers (Heidstra and Bisseling, 1996; Schlaman et al., 1997). Insight into the mechanisms leading to the activation of the inner cell layers may be obtained using genetic approaches. Many symbiotic mutants have been identified that are blocked in Nod factor–induced responses in the inner cell layers, whereas most responses in the epidermis are not affected. For example, the nsp (nodulation signalling pathway) mutant of M. truncatula and the nin (nodule inception) mutant in L. japonicus are both blocked in the mitotic activation of the cortex, whereas several epidermal responses are not affected (Schauser et al., 1999; Catoira et al., 2000). LjNIN has been cloned and encodes a putative transcription factor with a leucine-zipper domain in the carboxy-terminal half of the protein (Schauser et al., 1999). Further characterization of Ljnin and other mutants will most likely provide insight into the signaling pathways responsible for the activation of the cortical and pericycle cells of the plant root.
Feedback Regulation of Nod Factor Signaling
In general, signaling pathways are controlled by feedback regulatory mechanisms. Two mutants affected in feedback mechanisms regulating Nod factor signaling have been identified by using the calcium oscillation response. These are dmi3 and the ethylene-insensitive mutant Sickle (skl); both have an increased sensitivity to Nod factors, because calcium oscillation can be induced in these mutants by a ten-fold lower Nod factor concentration than needed to induce calcium oscillation in wild-type plants (Oldroyd et al., 2001a, 2001b). So, although DMI3 is essential for Nod factor signaling downstream of calcium oscillation, it is also involved in a negative regulation upstream of this response (Figure 3). Regarding ethylene, it has been known for a long time that it has a negative effect on nodule formation (e.g., Grobbelaar et al., 1971; Goodlass and Smith, 1979; Lee and LaRue, 1992), and the skl mutant suggests that ethylene is a negative regulator of Nod factor signaling. This is further supported by biochemical studies in which ethylene is applied to the plant root, resulting in cessation of the calcium oscillation within minutes (Oldroyd et al., 2001a). Strikingly, in the dmi3 mutant, ethylene can also block calcium oscillation, showing that ethylene and DMI3 could be part of two different regulatory mechanisms. (Figure 3; Oldroyd et al., 2001b).
Nod factor–induced responses in the inner cell layers are also controlled by ethylene. Ethylene is most likely produced in the pericycle cells opposite the phloem poles, because the gene encoding 1-aminocyclopropane-1-carboxylic acid oxidase, which catalyses the last step in ethylene biosynthesis, is expressed in these cells (Heidstra et al., 1997). Genetic studies show that the position as well as the number of nodule primordia is controlled by (the local production of) ethylene, because the skl mutant forms markedly more nodule primordia than do wild-type plants (Penmetsa and Cook, 1997). Furthermore, these nodule primordia are formed also opposite phloem poles (D.R. Cook, personal communication; R. Geurts, unpublished data). Similar results were obtained by treating wild-type plant roots with chemicals that interfere with either ethylene biosynthesis or the perception of ethylene (Heidstra et al., 1997). So, not only is ethylene a negative regulator of Nod factor signaling in the epidermis, but also the local production of this negative regulator controls where in the cortex nodule primordia are formed.
CONCLUDING REMARKS
In recent years, genetic analysis of signal transduction events in nodule formation has become a prominent strategy in nodulation research. These studies have been focused on M. truncatula and L. japonicus. Both legumes have now become mature model systems, as evidenced by the initiation of genome sequencing programs, the numerous expressed sequence tags, and the development of new tools like TILLING. These developments will make it possible to clone legume genes with great efficiency.
Recently, two genes involved in Nod factor signaling, NIN of L. japonicus and NN1 of M. sativa, were cloned. It is probable that in the coming few years many more genes will be cloned. It is to be expected that cloning of these genes will provide major new opportunities for cell biological, physiological, and biochemical approaches to finding answers to old but still intriguing questions concerning mechanisms that control a fascinating endosymbiosis.
Acknowledgments
We thank Carolien Franken for her contribution to Figure 1.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002451.
References
- Albrecht, C., Geurts, R., Lapeyrie, F., and Bisseling, T. (1998). Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J. 15 605–614. [DOI] [PubMed] [Google Scholar]
- Albrecht, C., Geurts, R., and Bisseling, T. (1999). Legume nodulation and mycorrhizae formation: Two extremes in host specificity meet. EMBO J. 18 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel, M., Demont, N., Debellé, F., Maillet, F., De Billy, F., Promé, J.C., Dénarié, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono, J.J., Riond, J., Nicolaou, K.C., Bockovich, N.J., Estevez, V.A., Cullimore, J.V., and Ranjeva, R. (1995). Characterization of a binding site for chemically synthesized lipo-oligosaccharidic NodRm factors in particulate fractions prepared from roots. Plant J. 7 253–260. [DOI] [PubMed] [Google Scholar]
- Borisov, A.Y., Barmicheva, E.M., Jacobi, L.M., Tsyganov, V.E., Voroshilova, V.A., and Tikhanovich, I.A. (2000). Pea (Pisum sativum L.) mendelian genes controlling development of nitrogen-fixing nodules and arbuscular mycorrhizae. Czech J. Genet. Plant Breeding 36 106–110. [Google Scholar]
- Bradbury, S., Peterson, R.L., and Bowley, S.R. (1991). Interactions between three alfalfa nodulation genotypes and two Glomus species. New Phytol. 119 115–120. [DOI] [PubMed] [Google Scholar]
- Brewin, N.J. (1998). Tissue and cell invasion by Rhizobium: The structure and development of infection threads and symbiosomes. In The Rhizobiaceae, H.P. Spaink, A. Kondorosi, and P.J.J. Hooykaas, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 417–429.
- Cardenas, L., Vidali, L., Dominguez, J., Perez, H., Sanchez, F., Hepler, P.K., and Quinto, C. (1998). Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol. 116 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira, R., Galera, C., De Billy, F., Penmetsa, R.V., Journet, E.P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Denarie, J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira, R., Timmers, A.C.J., Maillet, F., Galera, C., Penmetsa, R.V., Cook, D., Denarie, J., and Gough, C. (2001). The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128 1507–1518. [DOI] [PubMed] [Google Scholar]
- Compaan, B., Yang, W.C., Bisseling, T., and Franssen, H. (2001). ENOD40 expression in the pericycle precedes cortical cell division in Rhizobium-legume interaction and the highly conserved internal region of the gene does not encode a peptide. Plant Soil 230 1–8. [Google Scholar]
- Cook, D.R. (1999). Medicago truncatula—a model in the making! Curr. Opin. Plant Biol. 2 301–304. [DOI] [PubMed] [Google Scholar]
- Cullimore, J.V., Ranjeva, R., and Bono, J.J. (2001). Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 6 24–30. [DOI] [PubMed] [Google Scholar]
- Day, R.B., McAlvin, C.B., Loh, J.T., Denny, R.L., Wood, T.C., Young, N.D., and Stacey, G. (2000). Differential expression of two soybean apyrases, one of which is an early nodulin. Mol. Plant-Microbe-Interact. 13 1053–1070. [DOI] [PubMed] [Google Scholar]
- De Ruijter, N.C.A., Bisseling, T., and Emons, A.M.C. (1999). Rhizobium Nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol. Plant-Microbe Interact. 12 829–832. [Google Scholar]
- De Ruijter, N.C.A., Rook, M.B., Bisseling, T., and Emons, A.M.C. (1998). Lipochito-oligosaccharides re-initiate root hair tip growth in Vicia sativa with high calcium and spectrin-like antigen at the tip. Plant J. 13 341–350. [Google Scholar]
- Demont, N., Debellé, F., Aurelle, H., Dénarié, J., and Promé, J.C. (1993). Role of Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J. Biol. Chem. 268 20134–20142. [PubMed] [Google Scholar]
- Den Hartog, M., Musgrave, A., and Munnik, T. (2001). Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: A role for phospholipase C and D in root hair deformation. Plant J. 25 55–65. [DOI] [PubMed] [Google Scholar]
- Dénarié, J., and Cullimore, J. (1993). Lipo-oligosaccharide nodulation factors: A minireview new class of signaling molecules mediating recognition and morphogenesis. Cell 74 951–954. [DOI] [PubMed] [Google Scholar]
- Diaz, C.L., Logman, T.J.J., Stam, H.C., and Kijne, J.W. (1995). Sugar-binding activity of pea lectin expressed in white clover hairy roots. Plant Physiol. 109 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc, G., and Messager, A. (1989). Mutagenesis of pea (Pisum sativum L.) and the isolation of mutants for nodulation and nitrogen fixation. Plant Sci. 60 207–213. [Google Scholar]
- Duc, G., Trouvelot, A., Gianinazzi-Pearson, V., and Gianinazzi, S. (1989). First report of non-mycorrhizal plant mutants (Myc−) obtained in pea (Pisum sativum) and Fababean (Vicia Faba L.). Plant Sci. 60 215–222. [Google Scholar]
- Dudley, M.E., and Long, S.R. (1989). A nonnodulating alfalfa mutant displays neither root hair curling nor early cell division in response to Rhizobium meliloti. Plant Cell 1 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Atkinson, E.M., and Long, S.R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256 998–1000. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Ellis, B.E., and Miles, G.P. (2001). One for all? Science 292 2022–2023. [DOI] [PubMed] [Google Scholar]
- Emons, A.M.C., and Mulder, B. (2000). Nodulation factors trigger an increase of fine bundles of subapical actin filaments in Vicia root hairs: Implication for root hair curling around bacteria. In Biology of Plant-Microbe Interactions, P.J.G.M. De Wit, T. Bisseling, and J.W. Stiekema, eds (St. Paul, Minnesota: The International Society of Molecular Plant-Microbe Interaction), pp. 272–276.
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kaló, P., and Kiss, G.B. (2002). Cloning of a receptor kinase gene regulating symbiotic nodule development. Nature, in press. [DOI] [PubMed]
- Esseling, J.J., Lhuissier, F.G.P., Barker, D.G., and Emons, A.M.C. (2000). Local application of Nod factor on growing root hairs: Cellular and genetic responses. In The 4th Medicago Workshop, Abstract Book. (Madison, Wisconsin: www.medicago.org).
- Etzler, M.E., Kalsi, G., Ewing, N.N., Roberts, N.J., Day, R.B., and Murphy, J.B. (1999). A Nod factor binding lectin with apyrase activity from legume roots. Proc. Natl. Acad. Sci. USA 96 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1995). Nod signal-induced plasma membrane potential changes in alfalfa root hairs are differentially sensitive to structural modifications of the lipochitooligosaccharide. Plant J. 7 939–947. [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1996). Rapid alkalinization in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 10 295–301. [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1998). The role of ion fluxes in Nod factor signalling in Medicago sativa. Plant J. 13 455–464. [Google Scholar]
- Firmin, J.L., Wilson, K.E., Carlson, R.W., Davies, A.E., and Downie, J. (1993). Resistance to nodulation of c.v. Afghanistan peas is overcome by nodX, which mediates an O-acetylation of the Rhizobium leguminosarum lipo-oligosaccharide nodulation factor. Mol. Microbiol. 10 351–360. [DOI] [PubMed] [Google Scholar]
- Geurts, R., Heidstra, R., Hadri, A.E., Downie, A., Franssen, H., Van Kammen, A., and Bisseling, T. (1997). Sym2 of Pisum sativum is involved in a Nod factor perception mechanism that controls the infection process in the epidermis. Plant Physiol. 115 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart, J., Hink, M.A., Visser, A.J., Bisseling, T., and Gadella, T.W., Jr. (2000). In vivo fluorescence correlation microscopy (FCM) reveals accumulation and immobilization of Nod factors in root hair cell walls. Plant J. 21 109–119. [DOI] [PubMed] [Google Scholar]
- Goedhart, J., Rohrig, H., Hink, M.A., Van Hoek, A., Visser, A.J., Bisseling, T., and Gadella, T.W., Jr. (1999). Nod factors integrate spontaneously in biomembranes and transfer rapidly between membranes and to root hairs, but transbilayer flip-flop does not occur. Biochemistry 38 10898–10907. [DOI] [PubMed] [Google Scholar]
- Goodlass, G., and Smith, K.A. (1979). Effects of ethylene on root extension and nodulation of pea (Pisum sativum L.) and white clover (Trifolium repens L.). Plant Soil 51 387–395. [Google Scholar]
- Grobbelaar, N., Clarke, B., and Hough, M.C. (1971). The nodulation and nitrogen fixation of isolated roots of Phaseolus vulgaris L. Plant Soil (special vol.), 215–223. [Google Scholar]
- Gualtieri, G., Kulikova, O., Limpens, E., Kim, D.J., Cook, D.R., Bisseling, T. and Geurts, R. (2002). Microsynteny between pea and Medicago truncatula in the SYM2 region. Plant Mol. Biol., in press. [DOI] [PubMed]
- Heidstra, R., and Bisseling, T. (1996). Nod factor-induced host responses and mechanisms of Nod factor perception. New Phytologist 133 25–43. [Google Scholar]
- Heidstra, R., Geurts, R., Franssen, H., Spaink, H.P., Van Kammen, A., and Bisseling, T. (1994). Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 105 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra, R., Yang, W.C., Yalcin, Y., Peck, S., Emons, A.M., Van Kammen, A., and Bisseling, T. (1997). Ethylene provides positional information on cortical cell division but is not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development 124 1781–1787. [DOI] [PubMed] [Google Scholar]
- Hink, M.A., Bisseling, T., and Visser, A.J.W.G. (2002). Imaging protein-protein interactions in living cells. Plant Mol. Biol., in press. [DOI] [PubMed]
- Journet, E.P., El-Gachtouli, N., Vernoud, V., De Billy, F., Pichon, M., Dedieu, A., Arnould, C., Morandi, D., Barker, D.G., and Gianinazzi-Pearson, V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during my-corrhization in arbuscule-containing cells. Mol. Plant-Microbe Interact. 14 737–748. [DOI] [PubMed] [Google Scholar]
- Kalsi, G., and Etzler, M.E. (2000). Localization of a Nod factor-binding protein in legume roots and factors influencing its distribution and expression. Plant Physiol. 124 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen, B.E., Weeden, N.F., and LaRue, T.A. (1994). Non-nodulating mutants of Pisum sativum (L.) cv. Sparkle. J. Heredity 85 129–133. [Google Scholar]
- Kurkdjian, A.C. (1995). Role of the differentiation of root epidermal cells in Nod factor (from Rhizobium meliloti)-induced root-hair depolarization of Medicago sativa. Plant Physiol. 107 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkdjian, A., Bouteau, F., Pennarun, A.M., Convert, M., Cornel, D., Rona, J.P., and Bousquet, U. (2000). Ion currents involved in early Nod factor response in Medicago sativa root hairs: A discontinuous single-electrode voltage-clamp study. Plant J. 22 9–17. [DOI] [PubMed] [Google Scholar]
- Law, G.J., and Northrop, A.J. (1994). Synthetic peptides to mimic the role of GTP binding proteins in membrane traffic and fusion. Ann. N. Y. Acad. Sci. 710 196–208. [DOI] [PubMed] [Google Scholar]
- Lee, K.H., and LaRue, T.A. (1992). Exogenous ethylene inhibits nodulation of Pisum sativum L. cv. Sparkle. Plant Physiol. 105 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Promé, J.C., and Dénarié, J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344 781–784. [DOI] [PubMed] [Google Scholar]
- Markwei, C.M., and LaRue, T.A. (1992). Phenotypic characterization of sym8 and sym9, two genes conditioning non-nodulation in Pisum sativum ‘Sparkle’. Can. J. Microbiol. 38 548–554. [Google Scholar]
- Mergaert, P., Van Montagu, M., and Holsters, M. (1997). Molecular mechanisms of Nod factor diversity. Mol. Microbiol. 25 811–817. [DOI] [PubMed] [Google Scholar]
- Mylona, O., Pawlowski, P., and Bisseling, T. (1995). Symbiotic nitrogen fixation. Plant Cell 7 869–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel, A., Bono, J.J., Ranjeva, R., and Cullimore, J.V. (1997). Identification of a high affinity binding site for lipooligosaccharidic NodRm factors in microsomal fraction of Medicago cell suspension cultures. Mol. Plant-Microbe Interact. 10 132–134. [Google Scholar]
- Oldroyd, G.E., Engstrom, E.M., and Long, S.R. (2001. a). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E., Mitra, R.M., Wais, R.J., and Long, S.R. (2001. b). Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway. Plant J. 28 191–199. [DOI] [PubMed] [Google Scholar]
- Pawlowski, K., and Bisseling, T. (1996). Rhizobial and actinorhizal symbiosis: What are the shared features? Plant Cell 8 1899–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275 527–530. [DOI] [PubMed] [Google Scholar]
- Pichon, M., Journet, E.P., Dedieu, A., De Billy, F., Truchet, G., and Barker, D.G. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingret, J.L., Journet, E.P., and Barker, D.G. (1998). Rhizobium Nod factor signaling. Evidence for a G protein-mediated transduction mechanism. Plant Cell 10 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge, R.W. (1993). A model of legume root hair growth and Rhizobium infection. Symbiosis 14 359–373. [Google Scholar]
- Roche, P., Lerouge, P., Ponthus, C., and Promé, J.C. (1991). Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J. Biol. Chem. 266 10933–10940. [PubMed] [Google Scholar]
- Ross, E., and Higashijima, T. (1994). Regulation of G protein activation by mastoparans and other cationic peptides. Meth. Enzymol. 237 26–37. [DOI] [PubMed] [Google Scholar]
- Sagan, M., Huguet, T., and Duc, G. (1994). Phenotypic characterization and classification of nodulation mutants of pea (Pisum sativum L.). Plant Sci. 100 59–70. [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Scheres, B., Van de Wiel, C., Zalensky, A., Horvath, B., Spaink, H., Van Eck, H., Zwartkruis, F., Wolters, A.M., Gloudemans, T., Van Kammen, A., and Bisseling, T. (1990). The ENOD12 gene product is involved in the infection process during the pea-Rhizobium interaction. Cell 60 281–294. [DOI] [PubMed] [Google Scholar]
- Schlaman, W.R.M., Gisel, A.A., Quaedvlieg, N.E.M., Bloemberg, G.V., Lugtenberg, B.J.J., Kijne, J.W., Potrykus, I., Spaink, H.P., and Sautter, C. (1997). Chitin oligosaccharides can induce cortical cell division in roots of Vicia sativa when delivered by ballistic micro-targeting. Development 124 4887–4895. [DOI] [PubMed] [Google Scholar]
- Schneider, A., Walker, S.A., Poyser, S., Sagan, M., Ellis, T.H., and Downie, J.A. (1999). Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L.). Mol. Gen. Genet. 262 1–11. [DOI] [PubMed] [Google Scholar]
- Schultze, M., Quiclet-Sire, B., Kondorosi, E., Virelizier, H., Glushka, J.N., Endre, G., Géro, S.D., and Kondorosi, A. (1992). Rhizobium meliloti produces a family of sulfated lipo-oligosaccharides exhibiting different degrees of plant host specificity. Proc. Natl. Acad. Sci. USA 89 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink, H.P., Sheeley, D.M., Van Brussel, A.A.N., Glushka, J., York, W.S., Tak, T., Geiger, O., Kennedy, E.P., Reinhold, V.N., and Lugtenberg, B.J.J. (1991). A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 354 125–130. [DOI] [PubMed] [Google Scholar]
- Stougaard, J. (2001). Genetics and genomics of root symbiosis. Cur. Opin. Plant Biol. 4 328–335. [DOI] [PubMed] [Google Scholar]
- Timmers, A.C.J., Auriac, M.C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628. [DOI] [PubMed] [Google Scholar]
- Van Batenburg, F.H.D., Jonker, R., and Kijne, J.W. (1986). Rhizobium induces marked root hair curling by redirection of tip growth: A computer simulation. Physiol. Plant. 66 476–480. [Google Scholar]
- Van Brussel, A.A.N., Bakhuizen, R., Van Spronsen, P.C., Spaink, H.P., Tak, T., Lugtenberg, B.J.J., and Kijne, J.W. (1992). Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257 70–71. [DOI] [PubMed] [Google Scholar]
- Van Rhijn, P., Fujishige, N.A., Lim, P.O., and Hirsch, A.M. (2001). Sugar-binding activity of pea lectin enhances heterologous infection of transgenic alfalfa plants by Rhizobium leguminosarum biovar viciae. Plant Physiol. 126 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn, P., Goldberg, R.B., and Hirsch, A.M. (1998). Lotus corniculatus nodulation specificity is changed by the presence of a soybean lectin gene. Plant Cell 10 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Spronsen, P.C., Gronlund, M., Bras, C.P., Spaink, H.P., and Kijne, J.W. (2001). Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant Microbe Interact. 14 839–847. [DOI] [PubMed] [Google Scholar]
- Wais, R.J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R.V., Cook, D., Gough, C., Denarie, J., and Long, S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S.A., Viprey, V. and Downie, J.A. (2000). Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 97 13413–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., De Blank, C., Meskiene, I., Hirt, H., Bakker, J., Van Kammen, A., Franssen, H., and Bisseling, T. (1994). Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., Katinakis, P., Hendriks, P., Smolders, A., De Vries, F., Spee, J., Van Kammen, A., Bisseling, T., and Franssen, H. (1993). Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 3 573–585. [DOI] [PubMed] [Google Scholar]
- Zuanazzi, J.A.S., Clergeot, P.H., Quirion, J.-C., Husson, H.P., Kondorosi, A., and Ratet, P. (1998). Production of Sinorhizobium meliloti nod gene activator and repressor flavonoids from Medicago sativa roots. Mol. Plant-Microbe Interact. 11 784–794. [Google Scholar]