INTRODUCTION
Chloroplasts are essential for the unique photoautotrophic and sessile existence of higher plants. Chloroplasts account for >50% of the total soluble protein in leaves, and these proteins are encoded by both nuclear and chloroplast genomes. This separation of genetic information necessitates coordinated regulation in the expression of these genomes. The factors that influence the expression of photosynthetic genes include environmental (i.e., light quality and intensity) and developmental cues (McFadden, 1999). It is well known that the expression of chloroplast-encoded components of photosynthesis depends on nucleus-derived factors. A number of reports indicate that the expression of a set of nuclear genes that encode chloroplast-localized proteins is dependent on the functional state of the plastid via a process known as retrograde signaling. Blocking chloroplast development at an early stage by any of a variety of methods inhibits the transcription of numerous nuclear genes that encode chloroplast proteins, yet neither the signal nor its intracellular signaling pathway is known (Oelmüller, 1989). In this review, we discuss the recent genetic and physiological evidence for the existence of at least two independent retrograde signaling pathways, one mediated by tetrapyrroles, the other mediated by redox signaling, and suggest models for both pathways.
TETRAPYRROLES AND PHOTOSYNTHETIC GENE EXPRESSION
Tetrapyrroles Regulate Nuclear Transcription
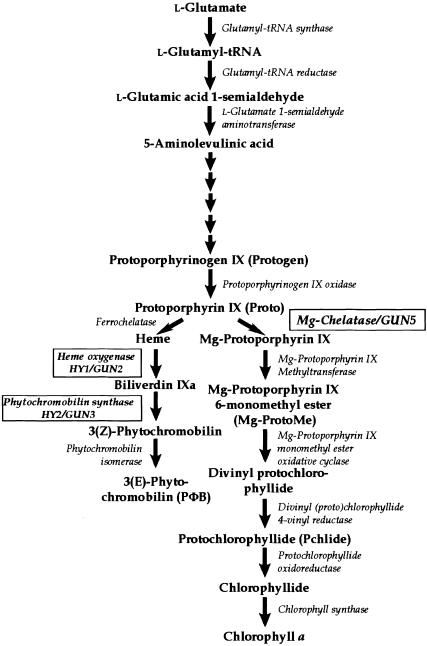
A number of candidates for chloroplast signal molecules have been proposed, such as plastid proteins and RNAs, plastid metabolic intermediates, and products of photosynthesis (Susek and Chory, 1992). An ever-growing body of evidence, both physiological and genetic, suggests that at least one of the plastid signals is a tetrapyrrole. Tetrapyrroles, which are synthesized within the plastid, are the intermediates and end products of heme, chlorophyll, and phytochromobilin (PΦB) biosynthetic pathways (Figure 1).
Figure 1.
The Plastid Tetrapyrrole Biosynthetic Pathway.
Five small arrows represent the five enzymatic steps that are required for the synthesis of protoporphyrinogen from 5-aminolevulinic acid. The reactions catalyzed by HY1/GUN2, HY2/GUN3, and GUN5 are boxed. Reduction of the 4-vinyl group can occur before or after the reaction catalyzed by protochlorophyllide oxidoreductase. The synthesis of common precursors and Mg-porphyrins is adapted from Beale (1999), and the synthesis of 3(E)-phytochromobilin from protoporphyrin IX is adapted from Terry et al. (1993).
Feeding studies with mutants and inhibitors of the chlorophyll biosynthetic pathway in Chlamydomonas reinhardtii suggest that chlorophyll biosynthetic pathway intermediates inhibit the expression of chloroplast protein genes that reside in the nucleus. In early experiments, conditions or compounds that are presumed to cause Mg-protoporphyrin IX monomethyl ester (Mg-ProtoMe) accumulation appear to be the most effective inhibitors of light-harvesting chlorophyll a/b binding protein of photosystem II (Lhcb) mRNA accumulation. Protoporphyrin IX (Proto) and protochlorophyllide (Pchlide) were found to be less effective than Mg-ProtoMe (Johanningmeier and Howell, 1984; Johanningmeier, 1988).
In Chlamydomonas, the intracellular pools of Mg-Proto and Mg-ProtoMe increase transiently during dark-to-light transitions, and this increase is correlated with increases in the expression of nuclear genes that encode the cytosolic and plastid heat shock proteins HSP70A and HSP70B, respectively (Kropat et al., 2000). Feeding Mg-Proto or Mg-ProtoMe to dark-grown mutants, cell types, or chemically treated cells that have reduced porphyrin levels induces HSP70A and HSP70B mRNA accumulation. Proto, Pchlide, and chlorophyllide did not affect the expression of HSP70A in similar experiments (Kropat et al., 1997, 2000). Mg-Proto and Mg-ProtoMe activate a heat- and light-responsive HSP70A promoter fragment, but they do not affect an HSP70A promoter fragment that is induced only by heat. This result suggests that Mg-Proto and Mg-ProtoMe can activate transcription through a light-responsive cis element (Kropat et al., 1997). Interestingly, Chlamydomonas cells take up exogenously supplied Proto and convert it to Mg-Proto, resulting in large intracellular Mg-Proto and Mg-ProtoMe pools, presumably in plastids. However, Proto feeding does not affect HSP70A or HSP70B expression. These data are consistent with a model in which Mg-Proto and/or Mg-ProtoMe are retained in the plastids of dark-grown Chlamydomonas cells. Mg-Proto and/or Mg-ProtoMe may be transported actively or passively out of the plastid in response to light, and these exported porphyrins activate HSP70 gene transcription (Kropat et al., 2000).
Chlorophyll biosynthetic intermediate levels correlate with the transcription of nuclear genes that encode chloroplast proteins in higher plants. For example, Mg-Proto and Mg-ProtoMe levels, as well as Mg-chelatase activity and Mg-Proto methyltransferase activity, all peak at dark-to-light transitions and decline throughout the subsequent 12-h-light period (Gibson et al., 1996; Jensen et al., 1996; Pöpperl et al., 1998; Papenbrock et al., 1999). The expression of Lhc genes is induced strongly by light, but Lhc mRNA levels peak after Mg-porphyrin levels peak (Gibson et al., 1996; Papenbrock et al., 1999). Procedures that presumably cause the accumulation of chlorophyll biosynthetic intermediates by increasing the level of 5-aminolevulinic acid (ALA) also inhibit gene expression in the nucleus. For example, the overproduction of ALA in transgenic tobacco increases chlorophyll content and reduces glutamate 1-semialdehyde aminotransferase mRNA levels, protein levels, and activity (Zavgorodnyaya et al., 1997), and ALA feeding represses Lhcb mRNA levels in Arabidopsis (Vinti et al., 2000) and cress seedlings (Kittsteiner et al., 1991). Moreover, compounds that affect chlorophyll biosynthesis also have been found to affect Lhcb gene expression. For example, thujaplicin, which abolishes Pchlide synthesis and causes the accumulation of porphyrins (particularly Mg-ProtoMe) upstream of Pchlide, specifically inhibits Lhcb transcription (Oster et al., 1996). The herbicide amitrole, which prevents normal prolamellar body development and causes Mg-Proto accumulation, inhibits the light induction of Lhcb and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (RbcS) mRNA levels and represses RbcS transcript levels in dark-grown seedlings (La Rocca et al., 2001).
Reduction in Mg-porphyrin pools also affects the expression of chloroplast protein genes that reside in the nucleus. Mg-chelatase inserts Mg2+ into the porphyrin ring of Proto IX and is composed of three subunits. These subunits are referred to commonly as ChlD, ChlH, and ChlI and have molecular masses of ∼70, 140, and 40 kD, respectively (Walker and Willows, 1997). Antisense tobacco plants with suppressed ChlH genes have reduced Mg-chelatase activity and chlorophyll content. ChlD and ChlI expression is not altered in ChlH antisense plants (Papenbrock et al., 2000). A lack of feedback regulation among Mg-chelatase subunit genes might be expected because these genes are expressed differently during the diurnal cycle (Papenbrock et al., 1999). However, compared with wild-type plants, the antisense ChlH plants showed reduced levels of ALA-synthesizing capacity, ALA dehydratase activity, Proto IX, and heme as well as a reduction in steady state mRNAs corresponding to glutamyl-tRNA-reductase, ALA dehydratase, and Lhcb genes (Papenbrock et al., 2000). Papenbrock et al. (2000) suggest that the Mg2+ branch of the tetrapyrrole biosynthetic pathway regulates the enzyme activities and nuclear genes for the enzymes in the early part of this biosynthetic pathway. If Mg-porphyrins in Chlamydomonas and higher plants affect the same plastid-to-nucleus signaling pathway as Mg-chelatase subunit underexpression, the signaling pathway might not be triggered by absolute levels of a chlorophyll biosynthetic intermediate(s). Instead, pathway flux, ratios of particular tetrapyrroles, bound or free levels of the porphyrin binding subunit of Mg-chelatase, and/or partitioning of intermediates into particular regions of the plastid might coordinate enzyme activity and nuclear gene expression.
Plastid-to-Nucleus Signaling Mutants Affect Tetrapyrrole Metabolism
Susek and Chory (1992) devised a genetic strategy for isolating mutants that do not repress Lhcb transcription completely in Arabidopsis seedlings in which chloroplast development is prevented by photooxidative damage. Using this approach, recessive mutations were isolated in five nuclear genes in which the normal coordinated expression of nuclear and chloroplast genomes is disrupted. This group of mutants is referred to as gun for genomes uncoupled (Susek et al., 1993; Mochizuki et al., 2001). The recessive nature of these mutations suggests that they disrupt a repressive signaling pathway. However, the possibility cannot be excluded that a positive signal accumulates in some of the gun mutants and that the positive signal does not reach an essential threshold in heterozygotes.
None of the gun mutations affects the tissue- and cell-specific, light-dependent, or circadian regulation of Lhcb genes. Therefore, these mutations appear to specifically impair the plastid regulation of nuclear transcription (Susek et al., 1993). Despite a clear interorganellar signaling defect, many of the gun mutants appear strikingly normal. However, a few of the gun mutants have defects in tetrapyrrole metabolism that cause paleness or light-signaling defects.
The gun1 mutant and the gun4 and gun5 mutants appear to define two partially redundant pathways, because gun1 gun4 and gun1 gun5 double mutants synergistically derepress Lhcb transcription in photobleached seedlings. In contrast, the gun phenotype is not significantly different among the gun4 gun5 double mutant and the corresponding single mutants (Mochizuki et al., 2001). Although gun1 mutants accumulate chlorophyll slowly after growing in the dark for several days and appear to be extremely light sensitive during deetiolation (Mochizuki et al., 1996), double mutant studies suggest that gun1 mutations do not affect tetrapyrrole metabolism (Vinti et al., 2000; Mochizuki et al., 2001).
In contrast, GUN2, GUN3, GUN4, and GUN5 are essential for proper tetrapyrrole metabolism (Vinti et al., 2000; Mochizuki et al., 2001). gun2 and gun3 are alleles of hy1 and hy2, respectively. HY1/GUN2 encodes heme oxygenase (Davis et al., 1999; Muramoto et al., 1999), and HY2/GUN3 encodes PΦB synthase (Kohchi et al., 2001). These enzymes are part of the “iron branch” of the tetrapyrrole biosynthetic pathway and are required for the synthesis of PΦB, the chromophore of phytochrome. The chlorophyll phenotypes among gun4, gun5, and hy1 alleles vary from slightly to extremely pale green, and the chlorophyll phenotypes of the hy1, gun4, and gun5 double mutants are severe (Vinti et al., 2000; Mochizuki et al., 2001). The chlorophyll phenotype of a double mutant prepared from gun5 and a phytochrome B (phyB) null mutant is indistinguishable from the phyB null parent, which indicates that the albino phenotype of the hy1 gun5 double mutant is not explained by a reduction in PΦB synthesis (Vinti et al., 2000). A better explanation probably comes from the observation that the chlorophyll biosynthetic pathway is perturbed in PΦB synthase and heme oxygenase mutants in Arabidopsis, tomato, and pea (Montgomery et al., 1999; Terry and Kendrick, 1999).
GUN5 encodes the ChlH subunit of Mg-chelatase (Mochizuki et al. 2001). Lhcb transcription also is derepressed in the photobleached maize Mg-chelatase mutant l-Blandy4 (Burgess and Taylor, 1988). Thus, gun2, gun3, and gun5 mutations affect tetrapyrrole biosynthesis directly. The pale phenotype of gun4 and double mutant studies suggest that gun4 also affects tetrapyrrole biosynthesis; once again, this implicates Mg-porphyrins in plastid-to-nucleus signaling (Vinti et al., 2000; Mochizuki et al., 2001). Together, these results imply that the albino phenotypes in the double mutants of hy1, gun4, and gun5 result from the misregulation of a photosensitizing tetrapyrrole(s) (Vinti et al., 2000).
The Arabidopsis long after far-red6 (laf6) mutant also appears to affect tetrapyrrole metabolism and nuclear transcription. laf6 was identified by its long hypocotyl in far-red light, which suggests a defect in phyA signal transduction. LAF6 encodes a soluble ATP binding cassette protein that localizes at the chloroplast periphery. These proteins typically are involved in the transport of various molecules across membranes. The long hypocotyl and attenuated gene expression phenotypes of laf6 are specific to far-red light. Proto levels are increased twofold in laf6. Although the laf6 mutant accumulates Proto, this mutant produces normal levels of PΦB (Møller et al., 2001). Protoporphyrinogen (Protogen) is synthesized from glutamate in the stroma. Protogen is transported to the envelope or thylakoid membrane, where it is oxidized to Proto by the membrane-bound Protogen oxidase. Protogen also is exported for mitochondrial heme synthesis (Beale, 1999; Che et al., 2000). LAF6 is proposed to participate in the ATP-dependent transport of Proto into the stroma, which presumably ensures that sufficient pools of Proto will be available for chloroplastic tetrapyrrole metabolism. In laf6, increased cytoplasmic levels of Proto are proposed to affect a phyA signaling pathway directly or indirectly (Møller et al., 2001). Lhcb expression is not derepressed in photobleached laf6 mutants (Å. Strand, unpublished results), which suggests that laf6 and the gun mutants affect different pathways. This result is surprising because laf6, gun2, gun3, gun4, and gun5 affect tetrapyrrole metabolism, but laf6 might have a different effect on pathway flux. It will be interesting to determine if double mutants prepared from laf6, gun2, gun3, gun4, and gun5 suffer from albinism like the double mutants prepared from hy1, gun4, and gun5.
The ChlH Subunit of Mg-Chelatase May Be a Tetrapyrrole Sensor
As discussed above, Mg chelatase contains three subunits: ChlH, ChlD, and ChlI. Two Arabidopsis ChlH mutants, gun5 and cch, have similar gun phenotypes. However, two different Arabidopsis ChlI mutants, cs and ch42, are not gun mutants. The effects of these four mutations on Mg-chelatase activity and chlorophyll synthesis vary dramatically. gun5 is slightly pale, and gun5 and the wild type have comparable amounts of Mg-chelatase activity. cs is more pale than gun5, but cs accumulates more chlorophyll and has higher levels of Mg-chelatase than cch (Mochizuki et al., 2001). ch42 does not accumulate chlorophyll and cannot grow without Suc (Koncz et al., 1990), even though there is a second copy of ChlI in the Arabidopsis genome (Asamizu et al., 1998). These results also suggest that, in addition to a role in the metallation of Proto, ChlH/GUN5 also serves as a porphyrin sensor for a plastid-to-nucleus signaling pathway (Mochizuki et al., 2001). This is not unreasonable, because ChlH/GUN5 binds porphyrins as a monomer in the absence of the other Mg-chelatase subunits (Karger et al., 2001).
Mg-chelatase appears to be localized at the chloroplast envelope (Beale, 1999), which is a reasonable location for a tetrapyrrole sensor for a plastid-to-nucleus signaling pathway. In fractionated chloroplasts, ChlH associates with envelope membranes in 5 mM Mg2+. However, in 1 mM Mg2+, ChlH is found in the stroma (Gibson et al., 1996; Nakayama et al., 1998). The association of ChlD with envelopes is similarly dependent on Mg2+ (Luo et al., 1999), but ChlI is found exclusively in the stroma when chloroplasts are fractionated with 4 mM MgCl2 (Nakayama et al., 1995). Because of the Mg2+-dependent association of ChlH with envelope membranes and the possible fluctuations in plastid Mg2+ concentrations (Leegood et al., 1985), immunocytochemical studies will be required to determine the location(s) of ChlH and other Mg-chelatase subunits within the chloroplast.
Summary and Model for Tetrapyrrole-Mediated Plastid-to-Nucleus Signaling
In summary, three observations are consistent with the idea that ChlH, the 140-kD Proto binding subunit of Mg-chelatase, is a tetrapyrrole sensor for an interorganellar signaling pathway and coordinates the expression of nuclear genes that encode chloroplast proteins with the functional state of the plastid, as depicted in Figure 2. First, nuclear transcription is uncoupled from the functional state of the plastid in two ChlH mutants, gun5 and cch, but not in two ChlI mutants, cs and ch42. Second, ChlH/GUN5 functions as a porphyrin binding protein in the absence of other Mg-chelatase subunits. Third, the modulation of Mg-porphyrin levels in Chlamydomonas and higher plants in dark-to-light transitions, feeding experiments, inhibitor treatments, and tetrapyrrole biosynthetic mutants correlates with changes in gene expression. We cannot exclude the possibility that intermediates that trigger a plastid-to-nucleus signaling pathway are simply perturbed in gun5 and that GUN5 does not participate directly in interorganellar communication.
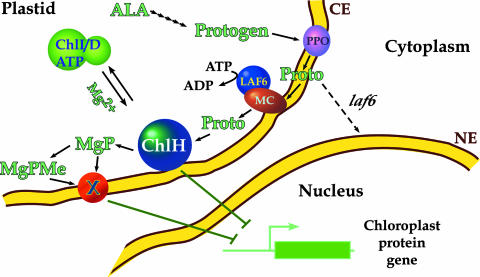
Figure 2.
Model for Plastid-to-Nucleus Signaling.
Protogen is synthesized from ALA in the stroma and transported to membranes where it is oxidized by PPO. The model of LAF6 and an integral membrane component (MC) participating in the ATP-dependent transport of Proto into the stroma is adapted from Møller et al. (2001). ChlH is required for Mg-chelation and may participate in plastid-to-nucleus signaling, as described in the text. The ChlI/ChlD/ATP complex may reside in the envelope, but it is shown in the stroma for clarity. Most nuclear genes that encode chloroplast proteins are repressed by Mg-porphyrins. However, Mg-porphyrins activate Chlamydomonas HSP70 genes. Factor X may transport Mg-Proto and Mg-ProtoME across the chloroplast envelope. CE, chloroplast envelope; MgP, Mg-Proto; MgPMe, Mg-ProtoME; NE, nuclear envelope.
If ChlH participates directly in interorganellar communication, a distinct ChlH complex may exist by itself or as a complex with other factors. A ChlH-tetrapyrrole complex might participate in signaling during diurnal bursts in Mg-porphyrin synthesis. A ChlH-tetrapyrrole complex might sense tetrapyrrole levels before essential ultrastructure is formed or as chloroplast ultrastructure is dismantled if, as suggested by La Rocca et al. (2001), photoreactive porphyrins accumulate when ultrastructure-dependent enzyme complexes dissociate. This tetrapyrrole signaling pathway may be important to prevent the accumulation of photosensitizing porphyrins and to coordinate the expression of nuclear genes that encode chloroplast proteins with the changes that occur in the photosynthetic apparatus during development and in response to the environment. Because heme, Mg-porphyrins, and other linear tetrapyrroles have been proposed to have regulatory functions in the plastid, it would be interesting to know the binding specificity of ChlH for other tetrapyrroles. Tetrapyrroles might interact with cytosolic signal transduction cascades, as suggested by Kropat et al. (1997)(2000) and Møller et al. (2001). ChlH could participate in the transport of tetrapyrroles across the plastid envelope, but it is possible as well that ChlH participates in plastid-to-nucleus signal transduction without affecting tetrapyrrole transport.
REDOX SIGNALING PATHWAYS AND PHOTOSYNTHETIC GENE EXPRESSION
Studies conducted in both Chlamydomonas and higher plants demonstrate that intermediary flux through the tetrapyrrole biosynthetic pathway in the chloroplast affects nuclear gene expression. However, numerous studies in green algae and higher plants have shown that the redox status of the plastoquinone (PQ) pool in the photosynthetic electron transport chain also exerts control over nuclear gene expression. This is a logical consequence of the fact that light induces changes in the stoichiometry of the two photosystems, thus changing the flow of electrons through the transport chain and modifying the redox status of the plant cell.
Redox Control in Photosynthetic Bacteria and Plastids
The abstraction of electrons from water and their eventual donation to NADP+ to generate NADPH require the participation of the two photosynthetic reaction centers, photosystems I and II (PSI and PSII). PSI and PSII are activated by different wavelengths of light, referred to as “light 1” and “light 2” (Duysens and Amesz, 1962; Myers, 1971). The activation of PSII by light 2 induces electron flow to PQ, reducing it to plastoquinol. Illumination with light 1 induces the oxidation of plastoquinol (through the cytochrome b6f complex) by PSI. Many of the studies described in this section were conducted using electron transport inhibitors. Electron flow from PSII to PQ is inhibited by DCMU. The inhibitor DBMIB (2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone) blocks electron flow from plastoquinol to PSI (Figure 3).
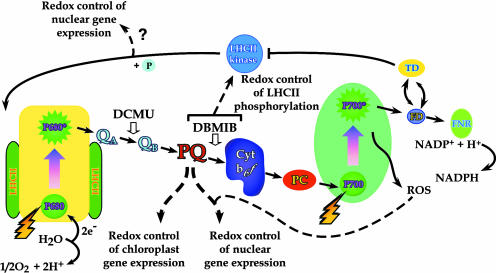
Figure 3.
Summary of Redox Activities That Control Gene Expression in Higher Plant Cells.
The PQ pool exerts control over both chloroplastic and nuclear transcription. ROS, in conjunction with the redox state of the PQ pool, influences the expression of antioxidant defense genes (e.g., APX genes). Cyt, cytochrome; FD, ferredoxin; FNR, ferredoxin-NADP+ reductase; PC, plastocyanin; QA and QB, primary and secondary electron-accepting plastoquinones of PSII; TD, thioredoxin.
Redox control of gene expression, of course, is bacterial in origin, and in bacteria it generally is mediated by two-component systems, such as the RegA-RegB system of Rhodobacter capsulatus (for review, see Bauer et al., 1999). Transcription, translation, and post-translational processes in the plastid itself are driven by redox chemistry.
At the post-translational level, the reduction of PQ to plastoquinol activates a protein kinase that phosphorylates PSII. Phosphorylated light-harvesting complex II (LHCII) migrates from the stacked to the unstacked regions of the thylakoid membrane, thus reducing PSII antenna size and favoring the absorption of light by PSI. As the PQ pool becomes oxidized, predominantly by the action of PSI, a phosphatase is activated. LHCII is dephosphorylated and migrates back to the appressed thylakoid membrane regions. This phosphorylation/dephosphorylation cycle permits the rapid distribution of light energy between the two photosystems (Allen, 1992).
Translation in chloroplasts is driven by the redox state of thioredoxin, which is controlled by the light-driven reduction of ferredoxin by PSI (Danon and Mayfield, 1994). Protein synthesis in chloroplasts increases ∼50- to 100-fold after exposure to light. However, there is no concomitant increase in chloroplast mRNA abundance, suggesting that light controls the rate of translation. In the chloroplast, nucleus-encoded activator proteins bind to the 5′ untranslated regions of messages and promote their translation. The binding of translational activating proteins to the 5′ untranslated region of psbA is controlled by the redox state of thioredoxin, which is reduced by PSI. The 5′ untranslated region of psbA mRNA forms a stem-loop structure that binds several of these proteins, forming a RNA-protein complex. The formation of the RNA-protein complex is favored by the addition of DTT, a dithiol reductant, but not by β-mercaptoethanol, a monothiol reductant, and is inhibited by the oxidizing agent dithiobenzoate. DTT reverses the action of oxidizing agents, and the addition of DTT and thioredoxin accelerates the rate of this reversal. Experiments with a Chlamydomonas mutant (cc703) that lacks the PSI reaction center showed that light could not promote wild-type levels of psbA translation. Together, these results suggest that the redox state of thioredoxin, and, by extension, PSI, controls translation in the chloroplast.
The control of the transcription of some chloroplast-encoded genes also is sensitive to redox signals generated by photosynthetic electron transport. Light conditions that predominantly excite one photosystem during the other cause an imbalance in excitation energy distribution between PSI and PSII. This situation decreases the efficiency of photosynthesis and may be counterbalanced in the short term by state transitions. During a longer time period, the plant cell adjusts the relative amounts of PSI and PSII by changes in gene expression. The rate of psaAB transcription is influenced by the quality of light. psaA and psaB are the central components of PSI, and the transcription of these genes decreases upon exposure to light 1 and increases upon exposure to light 2. Light 2 causes the reduction of the PQ pool. The addition of DCMU, an electron transport inhibitor that prevents the reduction of PQ, causes a decrease in the rate of psaAB transcription in isolated chloroplasts. Conversely, the addition of DBMIB, which prevents the oxidation of the PQ pool, increases the transcription of psaAB even in light 1 conditions. Therefore, the PQ pool regulates the transcription of a subset of chloroplast genes through a redox signal to adjust the relative stoichiometric amounts of the two reaction centers. The psbA gene, which encodes the D1 polypeptide of PSII, also is sensitive to regulation by redox poise (Pfannschmidt et al., 1999).
Redox Signals in Chloroplast-to-Nucleus Signal Transduction
All of the systems described above apply to redox control within the chloroplast. However, in eukaryotic photosynthetic cells, a number of genes that encode components of the photosynthetic apparatus are localized in the nucleus. This section describes the results of experiments that suggest that redox signals from the chloroplast influence the expression of a subset of nuclear genes involved in photosynthesis.
At different light intensities, the plant uses different redox signals to regulate photosynthetic gene expression. Under lower light levels, changes in light quality are relayed via signals that generally are believed to originate in or near the PQ pool. At higher irradiances capable of inducing photo-oxidative damage, redox signals are conveyed via the glutathione redox cycle and reactive oxygen species (ROS) (Pfannschmidt et al., 2001a). We will examine evidence for the involvement of both of these systems in plastid-nucleus signaling.
Redox control of nuclear gene expression is an adaptational, not a developmental, process. Light-regulated development is mediated via the phytochrome and cryptochrome photoreceptors. Redox signals, as generated by photosynthetic electron transport, are responsible for the photosynthetic adjustment to immediate environmental conditions, and their function is to allow the plant to adjust to changing light qualities and to maximize photosynthetic quantum yield.
Dark-light shift experiments with transgenic tobacco plants show that photosynthetic electron transport controls the transcription of the pea ferredoxin (FED1) gene as well as FED1 mRNA loading onto ribosomes (Petracek et al., 1998). The addition of DCMU destabilizes FED1 mRNA and leaves it susceptible to degradation. Changes in the chlorophyll a/b ratio of Dunaliella salina have been shown to correlate directly with changes in PSII excitation pressure regardless of whether the changes are attributable to varying irradiance or temperature (Maxwell et al., 1995). Throughout this series of experiments, cells were cultured under continuous irradiance, thus bypassing any phytochrome effect. The authors concluded that changes in the chlorophyll content of the cells, chlorophyll a/b ratios, and LHCII and Lhcb mRNA abundance are regulated by the redox poise of photosynthetic electron transport, as determined by changes in PSII excitation pressure. Escoubas et al. (1995) demonstrated that the redox status of the PQ pool influences nuclear photosynthetic gene expression and performed experiments that suggest the involvement of signaling components in the nucleus and/or cytoplasm that may participate in chloroplast-to-nucleus signal transduction. The green alga Dunaliella tertiolecta adjusts the abundance of LHCs to changing light intensities, a process known as photoacclimation. These alterations in the abundance of LHC are reversible and occur in fully differentiated cells, suggesting that this is not so much a chloroplast development pathway as an adjustment pathway.
Escoubas et al. (1995) also measured photoacclimation by measuring changes in cellular chlorophyll a content. A threefold increase in the amount of cellular chlorophyll occurred in Dunaliella tertiolecta cells within 24 h of transfer from high light (HL) to low light (LL). Changes in pigment content are a measure of LHC apoprotein abundance, and there was a threefold to fourfold increase in Lhcb mRNA within 9 h of a shift to LL.
The addition of DCMU to Dunaliella cultures mimics the effects of acclimation to LL. Cells grown in HL and DCMU induced a twofold increase in cellular chlorophyll a content within 12 h. Sublethal concentrations of DBMIB (PQ analog) had no effect on pigment levels. Conversely, when DBMIB was added to LL cells, the cellular levels of chlorophyll a decreased by 25%. To show that this effect is specific to the redox state of the PQ pool, uncouplers of photophosphorylation (carbonyl cyanide-m-chlorophyenylhydrazone, methylamine) and hydroxylamine, an uncoupler of water oxidation, were added to Dunaliella cultures. None of these inhibitors altered cellular chlorophyll levels, suggesting that neither ATP synthesis nor water splitting is involved directly in mediating Lhcb expression.
As discussed above, the redox state of the PQ pool controls a thylakoid protein complex that reversibly phosphorylates LHCII. It is of interest to know whether there are extraplastidic kinases that may be responsive to the redox state of the PQ pool. If the repression of Lhcb genes is mediated by a redox-controlled protein kinase, then inhibitors of protein phosphatases should affect the derepression (i.e., the enhancement of Lhcb gene expression at LL). The additions of okadaic acid, microcystin LR, and tautomycin to Dunaliella cultures inhibited the increase of chlorophyll a per cell by 33 to 56% within 24 h. Okadaic acid and microcystin LR specifically inhibit phosphatase activities found only in the cytoplasm and nucleus. Therefore, the inhibited phosphatases are signal transduction components outside of the chloroplast. Gel-shift experiments with protein extracts isolated from HL- and LL-treated cultures show that a G box–containing region from the Dunaliella Cab1 promoter binds a protein factor(s) present only in HL cells.
Illumination conditions that create predominantly excited PSI or PSII generate an imbalance in excitation energy distribution between PSI and PSII, which results in a decrease in photosynthetic efficiency. This is counterbalanced by an initial short-term response called “state transition” and then a long-term response that causes a readjustment of photosystem stoichiometry. As we have seen in green algae, this readjustment requires changes in photosynthetic gene expression. Photoacclimation is not the same in higher plants and green algae (Durnford and Falkowski, 1997). Whereas green algae face continuous drastic changes in light conditions, higher plants generally are adjusted to a narrower range of light conditions such as direct sun or primarily shady conditions, as would be found under a forest canopy. However, there are issues relating to the diurnal clock, and higher plants must be able to adjust photosynthetic capacity within these parameters.
Pfannschmidt et al. (2001b) demonstrated that the redox poise of the PQ pool also affects nuclear photosynthetic gene expression in higher plants. This study used nuclear PSI genes to measure photosystem stoichiometry adjustment and demonstrated that redox signals that control the transcription of genes localized to the chloroplast also affect the expression of photosynthetic genes in the nucleus. The promoters from four PSI genes, PETH, PSAF, PSAD, and PETE, were fused to the β-glucuronidase (GUS) reporter gene and used to construct transgenic tobacco lines. The lines were examined for their response to excitation pressures applied to the two photosystems. The PETH promoter had similar activities under all applied light regimens. The PSAF and PSAD promoters showed higher activity in PSII than PSI light, but there was no reduction after shifting from PSII to PSI. The PETE promoter, like the PSAF and PSAD promoters, had higher activity in PSII light compared with PSI light, but unlike the other two promoters, it was inactivated by a PSI-to-PSII shift.
The same tobacco lines also were treated with photosynthetic electron transport inhibitors. The PETH promoter did not respond to treatment with either DCMU or DBMIB. For the PSAD and PSAF promoters, DCMU inhibited activation by a PSI-to-PSII light shift and downregulated the expression of GUS in PSII-treated plants. DBMIB was effective only in conjunction with PSI light–driven oxidation, but not alone. This additive effect suggests that there may be a threshold requirement not fulfilled by the redox state of the PQ pool or PSI light alone. The PSAD and PSAF promoters showed a decrease in GUS activities, the same as after treatment with DCMU. Therefore, these two promoters seem to respond to redox signals that originate between the PQ pool and PSI, or to electron transport capacity in general. Because DBMIB prevents the PSI-induced repression of PETE, it appears that the redox status of the PQ pool regulates this promoter.
The majority of studies that implicate chloroplastic redox poise in the regulatory control of photosynthetic gene expression have involved inhibitor studies. As such, we should be cautious about possible side effects and artifacts induced by the altered metabolism of the treated cells. The Arabidopsis cue1 mutant offers evidence for the involvement of the redox state of the PQ pool in the regulation of nuclear photosynthetic genes (Streatfield et al., 1999). Arabidopsis cue1 mutants underexpress light-regulated nuclear genes that encode chloroplast-localized proteins. cue1 has mesophyll cell–specific chloroplast and cellular defects, which manifest in a reticulate phenotype. The severity of the cue1 phenotype is dependent on light intensity. CUE1 encodes the plastid inner envelope phosphoenolpyruvate/phosphate translocator. The biosynthesis of aromatic compounds is compromised in plants bearing the cue1 mutation. This results in reduced flux through the shikimate pathway, the source of phenolic UV light protectants such as flavonoids, hydroxycinnamic acids, and simple phenolics. PQ also is derived from the shikimate pathway, and cue1 plants show a decrease in the rapid induction kinetics of chlorophyll a fluorescence. The relative size of the PQ pool in the cue1-1 and cue1-6 mutant alleles is 10 to 50% lower than that estimated for wild-type plants. Furthermore, measurements of photosynthetic electron transport and photochemical and nonphotochemical quenching show that the primary electron-accepting PQ of PSII is more reduced transiently in the cue1 mutant and that altered redox poise may affect Lhcb expression. The reduced levels of phenolics in the mutant may make cue1 more susceptible to light intensity–reduced inductions in Lhcb transcription.
Most of the evidence that we have discussed suggests that the source of this signal is the PQ pool (Figure 3). However, Pursiheimo et al. (2001) provided evidence that LHCII kinase, and not the PQ pool, is the primary redox sensor that marks the beginning of a signaling cascade to the nucleus. The authors asked whether the chloroplast-to-nucleus signal transduction pathway shares components involved in reversible LHCII protein phosphorylation, which also is redox regulated. Pursiheimo et al. (2001) examined winter rye plants grown under a variety of light and temperature regimens. They increased PSII excitation pressure by shifting from a growth regimen (300 μmol·m−2·sec−1 at 20°C) to varying light and temperature combinations (e.g., 300 μmol·m−2·sec−1 at 5°C, 600 μmol·m−2·sec−1 at 20°C, 50 μmol·m−2·sec−1 at 20°C, and 50 μmol·m−2·sec−1 at 5°C). They monitored PSII excitation pressure, the phosphorylation level of LHCII, and the accumulation of Lhcb, rbcS, and psbA transcripts. LHCII protein phosphorylation and Lhcb transcript abundance showed a positive correlation. The authors argued that the inactivation of the LHCII kinase occurs via a “second loop” of redox regulation.
In pumpkin leaves, this cooperative regulatory loop involves the activation of LHCII kinase via reduction by the cytochrome b6f complex and its inactivation by the ferredoxin-thioredoxin system (Rintamäki et al., 2000). Thus, the regulation of Lhcb transcription, according to this model, is not under the direct control of the redox state of the PQ pool. Montané et al. (1998), using barley plants grown under a variety of light regimens and oxygen and carbon dioxide combinations, demonstrated that xanthophyll cycle pigments and early light-inducible proteins and mRNAs accumulate with increasing PSII excitation pressure. They found no correlation, however, between the reduction state of PSII and chlorophyll a/b ratios, or between LCHII protein or mRNA. They concluded that, in higher plants, the chlorophyll antenna size of PSII is not governed by the redox state of PSII. It is apparent that there are a number of different mechanisms that govern the expression of plastid components encoded in the nucleus, and redox control is but one of these mechanisms.
Oswald et al. (2001) investigated the relationship between photosynthetic electron transport and sugar signaling. Using Arabidopsis cell cultures and transgenic Arabidopsis lines carrying luciferase reporter genes driven by the cab2 promoter or the plastocyanin promoter, they found that the transcriptional activation of Lhcb and rbcS depends on photosynthetic electron transport, and not the sugar status of the cells. They concluded that a redox signal from the plastid overrides the sugar-related expression of nuclear photosynthetic genes.
Redox Signals and Antioxidant Defense Gene Expression
So far, this discussion has centered on the photoacclimation process, that is, adjusting light-harvesting capabilities and photosystem stoichiometries in response to changing light conditions. High levels of excess light energy, beyond what can be transduced by photosynthetic electron transport or dissipated as heat, can generate ROS, such as hydrogen peroxide, superoxide, singlet oxygen, and hydroxyl radicals. ROS activate antioxidant systems in both the plastid and the cytosol, which in turn can activate nuclear gene expression.
The Arabidopsis APX (ascorbate peroxidase) gene family is an important part of the antioxidant defense system. APX catalyzes the reduction of hydrogen peroxide to water using ascorbate as an electron donor. There are at least five described members of the Arabidopsis APX family, and a number of others have been annotated as such in the Arabidopsis Genome Project. Activities are localized to the chloroplast (APX4 and APX5), glyoxisomes, peroxisomes, mitochondria, and cytosol (APX1 and APX2). The expression of all of the APX genes is associated with signaling and stress responses (Mullineaux et al., 2000).
Increased light levels can lead to excess excitation energy (EEE). Protracted EEE results in the production of ROS. If the plant cannot neutralize the production of ROS via antioxidant systems, its cells will suffer irreversible photooxidative damage. Increases in foliar ROS are accompanied by an increase in the concentration of oxidized glutathione. Reduced glutathione is one of the major determinants of cellular redox state. In two significant articles, Karpinski et al. (1997)(1999) demonstrated that the expression of a number of nuclear genes involved in the antioxidant defense system of plant cells is controlled by the redox state of the chloroplast. An increase in oxidized glutathione induced the expression of APX1, and APX1 mRNA abundance increased ∼18-fold within the first hour of increased light treatment (Karpinski et al., 1997). Although levels declined in the post-stress period, they nonetheless remained higher than in control plants that had not been exposed to increased light. APX2 was not detectable before exposure to increased light but was induced very rapidly (∼7 min) after shifting to increased light and was downregulated rapidly during the first 2 h of the poststress period. If the conditions that produced EEE persisted, the plant cell underwent permanent photodamage and the expression of APX2 was inhibited.
DCMU has been shown to inhibit the induction of APX1 and APX2 by EEE. DBMIB, which inhibits the oxidation of PQ, induces the expression of APX1 and APX2 in LL. These data suggest that redox changes in PQ (or possibly in the second electron-accepting PQ of PSII) contribute to a regulatory system that controls APX gene expression. The treatment of Arabidopsis leaves with hydrogen peroxide can partially induce APX1 and APX2 expression and increase tolerance to increased light regimens. However, the addition of hydrogen peroxide to leaves exposed to increased light and DCMU does not override the inhibition of APX2 expression (Karpinski et al., 1999). This experiment suggests that there is a set sequence of events initiated by increased light treatment and culminating in nuclear gene expression, and it is not only an increase in ROS levels that control events in the nucleus. ROS may have a direct effect on the photosynthetic machinery, or they may work in tandem with a photosynthetic component.
How this signal is relayed to the nucleus is unknown. Mullineaux et al. (2000) have suggested three possibilities. First, hydrogen peroxide could diffuse down a concentration gradient and out of a light-stressed plastid and activate a cytosolic redox-sensitive regulator. Second, high-light conditions could increase photorespiratory rates, which would lead to increased hydrogen peroxide concentrations in the peroxisomes. A putative redox sensor in the peroxisome then could transmit a signal via increased flux through the glycolate pool. Third, Jäger-Vottero et al. (1997) have described an electron transport chain with cytosolic oxygen as the terminal electron acceptor. This redox chain could be a candidate for the transmission of a redox signal out of the chloroplast.
Plastid-Responsive Promoter Elements in Nuclear Genes
The cytoplasmic and nuclear proteins that participate in the plastid-to-nucleus signaling pathways described above are poorly understood. However, progress has been made on the identification of cis elements in nuclear genes that respond to positive signals from plastid tetrapyrroles in Chlamydomonas (Kropat et al., 1997) and repressive signals from photooxidized plastids in higher plants. As described above, the gun mutants suggest a role for tetrapyrroles in one of the repressive signaling pathways originating from photooxidized plastids.
Nuclear genes that encode chloroplast proteins are regulated by a diverse group of cis regulatory elements that act in a combinatorial fashion (Puente et al., 1996). Promoter:: reporter gene fusions have been used to map light and plastid response elements in promoters of several nuclear genes that encode chloroplast proteins. To date, it has not been possible to uncouple plastid- and light-responsive cis elements (Kusnetsov et al., 1996; McCormac et al., 2001). Mutation of five nucleotides from −59 to −55 in the spinach AtpC gene, which encodes the γ-subunit of the chloroplast ATP synthase, causes constitutive light- and plastid-independent expression, suggesting that these five nucleotides are part of a repressor binding site (Bolle et al., 1996). These five nucleotides are just upstream of a CAAT box. Currently, it is thought that repressors block AtpC transcription in etiolated and photobleached tissue by binding adjacent to the CAAT box and preventing the recruitment of a CAAT box binding factor. Also, activators of the AtpC promoter (e.g., light and cytokinin) are thought to inhibit repressor binding, which allows the CAAT box binding factor to assemble on the CAAT box (Bezhani et al., 2001).
Although light- and plastid-specific signals appear to act on the same cis elements, a number of different experiments suggest that these two signals are distinct. For example, etioplasts from dark-grown plants feature a prolamellar body and unstacked prothylakoid membranes, accumulate chlorophyll precursors, and are poised to become chloroplasts (Kirk and Tilney-Bassett, 1978). The herbicide amitrole prevents normal prolamellar body development and represses ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (RbcS) mRNA levels in dark-grown barley plants (La Rocca et al., 2001). A number of dark-grown photomorphogenic mutants in the det, cop, and fus class assemble prechloroplasts, although these prechloroplasts do not develop stacked thylakoids or synthesize chlorophyll because of a light-dependent step in the chlorophyll biosynthetic pathway. Nuclear genes that encode chloroplast proteins are overexpressed in the dark in many of these mutants. However, dark overexpression of these genes is prevented by plastid translation inhibitors, which also block prechloroplast development (Sullivan and Gray, 1999).
Two synthetic promoters, G-NOS101 and GATA-NOS101, respond to plastid signals; however, unlike bona fide light-inducible genes, they are not induced by light pulses that are too low in fluence to affect seedling morphology (Puente et al., 1996). Light also can enhance the translation efficiency of some mRNAs. In the case of glycolate oxidase, light induction of translational efficiency is independent of the plastid signal (Barak et al., 2001). To act on common promoter elements, light and plastid signaling pathways must converge at some point. Because a plastid signal is required for the overexpression of nuclear photosynthesis genes in the photomorphogenic mutants cop1 and lip1, the point of intersection appears to be downstream of COP1 (Sullivan and Gray, 1999, 2000). In the dark, COP1 is a nuclear protein that is required for proper photomorphogenesis, possibly by mediating targeted proteolysis through the 26S proteosome (Osterlund et al., 2000).
CONCLUSIONS
An emerging body of evidence shows that at least two types of chemical signals emanate from the chloroplast and contribute to control the expression of nuclear genes involved in photosynthesis. One of these signals originates in the tetrapyrrole biosynthetic pathway. The major issue in this field is this: exactly what intermediate leaves the chloroplast? Does any chemical entity actually exit the organelle? In Saccharomyces, mitochondrial heme regulates the expression of the nucleus-localized iso-1 cytochrome c (CYC1) gene. It has been shown that, under aerobic conditions that induce the biosynthesis of heme, the heme leaves the mitochondria and activates the HAP1 transcription factor, which in turn controls the transcriptional activation of the CYC1 gene (Guarente and Mason, 1983; Pfeiffer et al., 1989). The Arabidopsis genome has revealed no homologs of the yeast HAP1 protein, so it is possible that no cognate mechanism exists in higher plants. There is speculation (Kropat et al., 2000; Møller et al., 2001) that some porphyrins are exported from the chloroplast. What happens to them outside the confines of the organelle is unknown at present. It also is known that some of the enzymes of the magnesium branch of the tetrapyrrole biosynthetic pathway localize to the chloroplast envelope (Beale, 1999). It is possible that cytoplasmic signaling components recognize the composition of the plastid envelope and thus relay that information to the nucleus.
Much of chloroplast function is based on redox chemistry; thus, it stands to reason that there should be redox control of plastid genetic systems. We have discussed evidence that, in eukaryotic cells, the redox control of gene expression has been extended to the nuclear compartment. Again, at this time, we may only speculate about how redox signals are transmitted from the plastid, how they activate cytoplasmic sensors, and how, in turn, they activate or repress transcription factors in the nucleus. We look forward to further participation in this exciting field and to helping provide the experimental evidence that will further describe the components of retrograde signaling.
Acknowledgments
We thank Dr. Åsa Strand for helpful comments on the manuscript. Our work on intracellular signaling between the chloroplast and the nucleus is funded by a grant from the Department of Energy and The Howard Hughes Medical Institute. J.C. is an Associate Investigator of the Howard Hughes Medical Institute. M.S. received partial support from National Institutes of Health postdoctoral fellowship F32 GM18172, and R.M.L. was partially funded by a U.S. Department of Agriculture postdoctoral fellowship (97-35301-4656).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010446.
References
- Allen, J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098 275–335. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Sato, S., Kaneko, T., Nakamura, Y., Kotani, H., Miyajima, N., and Tabata, S. (1998). Structural analysis of Arabidopsis thaliana chromosome 5. VIII. Sequence features of the regions of 1,081,958 bp covered by seventeen physically assigned P1 and TAC clones. DNA Res. 5 379–391. [DOI] [PubMed] [Google Scholar]
- Barak, S., Nejidat, A., Heimer, Y., and Volokita, M. (2001). Transcriptional and post-transcriptional regulation of the glycolate oxidase gene in tobacco seedlings. Plant Mol. Biol. 45 399–407. [DOI] [PubMed] [Google Scholar]
- Bauer, C.E., Elsen, S., and Bird, T.H. (1999). Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53 495–523. [DOI] [PubMed] [Google Scholar]
- Beale, S.I. (1999). Enzymes of chlorophyll synthesis. Photosynth. Res. 60 43–73. [Google Scholar]
- Bezhani, S., Sherameti, I., Pfannschmidt, T., and Oelmüller, R. (2001). A repressor with similarities to prokaryotic and eukaryotic DNA helicases controls the assembly of the CAAT box binding complex at a photosynthesis gene promoter. J. Biol. Chem. 276 23785–23789. [DOI] [PubMed] [Google Scholar]
- Bolle, C., Kusnetsov, V.V., Herrmann, R.G., and Oelmüller, R. (1996). The spinach AtpC and AtpD genes contain elements for light-regulated, plastid-dependent and organ-specific expression in the vicinity of the transcription start sites. Plant J. 9 21–30. [DOI] [PubMed] [Google Scholar]
- Burgess, D.G., and Taylor, W.C. (1988). The chloroplast affects the transcription of a nuclear gene family. Mol. Gen. Genet. 214 89–96. [Google Scholar]
- Che, F.S., Watanabe, N., Iwano, M., Inokuchi, H., Takayama, S., Yoshida, S., and Isogai, A. (2000). Molecular characterization and subcellular localization of protoporphyrinogen oxidase in spinach chloroplasts. Plant Physiol. 124 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon, A., and Mayfield, S.P. (1994). Light-regulated translation of chloroplast messenger RNAs through redox potential. Science 266 1717–1719. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., Kurepa, J., and Vierstra, R.D. (1999). The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc. Natl. Acad. Sci. USA 96 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnford, D.G., and Falkowski, P.G. (1997). Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosynth. Res. 53 229–241. [Google Scholar]
- Duysens, L.N.M., and Amesz, J. (1962). Function and identification of two photochemical systems in photosynthesis. Biochim. Biophys. Acta 64 243–260. [Google Scholar]
- Escoubas, J.-M., Lomas, M., LaRoche, J., and Falkowski, P.G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92 10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, L.C., Marrison, J.L., Leech, R.M., Jensen, P.E., Bassham, D.C., Gibson, M., and Hunter, C.N. (1996). A putative Mg chelatase subunit from Arabidopsis thaliana cv C24: Sequence and transcript analysis of the gene, import of the protein into chloroplasts, and in situ localization of the transcript and protein. Plant Physiol. 111 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente, L., and Mason, T. (1983). Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32 1279–1286. [DOI] [PubMed] [Google Scholar]
- Jäger-Vottero, P., Dorne, A.-J., Jordanov, J., Douce, R., and Joyard, J. (1997). Redox chains in chloroplast envelope membranes: Spectroscopic evidence for the presence of electron carriers, including iron-sulfur centers. Proc. Natl. Acad. Sci. USA 94 1597–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, P.E., Willows, R.D., Petersen, B.L., Vothknecht, U.C., Stummann, B.M., Kannangara, C.G., von Wettstein, D., and Henningsen, K.W. (1996). Structural genes for Mg-chelatase subunits in barley: Xantha-f, -g and -h. Mol. Gen. Genet. 250 383–394. [DOI] [PubMed] [Google Scholar]
- Johanningmeier, U. (1988). Possible control of transcript levels by chlorophyll precursors in Chlamydomonas. Eur. J. Biochem. 177 417–424. [DOI] [PubMed] [Google Scholar]
- Johanningmeier, U., and Howell, S.H. (1984). Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardtii: Possible involvement of chlorophyll synthesis precursors. J. Biol. Chem. 259 13541–13549. [PubMed] [Google Scholar]
- Karger, G.A., Reid, J.D., and Hunter, C.N. (2001). Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry 40 9291–9299. [DOI] [PubMed] [Google Scholar]
- Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski, S., Reynolds, H., Karpinska, B., Creissen, G., and Mullineaux, P.M. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284 654–657. [DOI] [PubMed] [Google Scholar]
- Kirk, J.T.O., and Tilney-Bassett, J.A.E. (1978). The Plastids: Their Chemistry, Structure, Growth, and Inheritance. (Amsterdam, The Netherlands: Elsevier/North Holland).
- Kittsteiner, U., Brunner, H., and Rüdiger, W. (1991). The greening process in cress seedlings. II. Complexing agents and 5-aminolevulinate inhibit accumulation of cab messenger RNA coding for the light-harvesting chlorophyll a/b protein. Physiol. Plant. 81 190–196. [Google Scholar]
- Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A., and Lagarias, J.C. (2001). The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Reiss, B., Redei, G.P., and Schell, J. (1990). Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 9 1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat, J., Oster, U., Rudiger, W., and Beck, C.F. (1997). Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. USA 94 14168–14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat, J., Oster, U., Rudiger, W., and Beck, C.F. (2000). Chloroplast signaling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to the cytoplasm/nucleus. Plant J. 24 523–531. [DOI] [PubMed] [Google Scholar]
- Kusnetsov, V., Bolle, C., Lubberstedt, T., Sopory, S., Herrmann, R.G., and Oelmüller, R. (1996). Evidence that the plastid signal and light operate via the same cis-acting elements in the promoters of nuclear genes for plastid proteins. Mol. Gen. Genet. 252 631–639. [DOI] [PubMed] [Google Scholar]
- La Rocca, N., Rascio, N., Oster, U., and Rudiger, W. (2001). Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213 101–108. [DOI] [PubMed] [Google Scholar]
- Leegood, R.C., Walker, D.A., and Foyer, C.H. (1985). Regulation of the Benson-Calvin cycle. In Photosynthesis Mechanisms and the Environment, J. Barber and N.R. Baker, eds (Amsterdam, The Netherlands: Elsevier Science Publishers), pp. 190–258.
- Luo, M., Weinstein, J.D., and Walker, C.J. (1999). Magnesium chelatase subunit D from pea: Characterization of the cDNA, heterologous expression of an enzymatically active protein and immunoassay of the native protein. Plant Mol. Biol. 41 721–731. [DOI] [PubMed] [Google Scholar]
- Maxwell, D.P., Laudenbach, D.E., and Huner, N.P.A. (1995). Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol. 109 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac, A.C., Fischer, A., Kumar, A.M., Söll, D., and Terry, M.J. (2001). Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 25 549–561. [DOI] [PubMed] [Google Scholar]
- McFadden, G.I. (1999). Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 2 513–519. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., Susek, R., and Chory, J. (1996). An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 112 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N., Brusslan, J.A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, S.G., Kunkel, T., and Chua, N.H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané, M.-H., Tardy, F., Kloppstech, K., and Havaux, M. (1998). Differential control of xanthophylls and light-induced stress proteins, as opposed to light-harvesting chlorophyll a/b proteins, during photosynthetic acclimation of barley leaves to light irradiance. Plant Physiol. 118 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, B.L., Yeh, K.C., Crepeau, M.W., and Lagarias, J.C. (1999). Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiol. 121 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux, P., Ball, L., Escobar, C., Karpinska, B., Creissen, G., and Karpinski, S. (2000). Are diverse signaling pathways integrated in the regulation of Arabidopsis antioxidant defense gene expression in response to excess excitation energy? Philos. Trans. R. Soc. Lond. B 355 1531–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto, T., Kohchi, T., Yokota, A., Hwang, I., and Goodman, H.M. (1999). The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, J. (1971). Enhancement studies in photosynthesis. Annu. Rev. Plant Physiol. 22 289–312. [Google Scholar]
- Nakayama, M., Masuda, T., Sato, N., Yamagata, H., Bowler, C., Ohta, H., Shioi, Y., and Takamiya, K. (1995). Cloning, subcellular localization and expression of CHLI, a subunit of magnesium-chelatase in soybean. Biochem. Biophys. Res. Commun. 215 422–428. [DOI] [PubMed] [Google Scholar]
- Nakayama, M., Masuda, T., Bando, T., Yamagata, H., Ohta, H., and Takamiya, K. (1998). Cloning and expression of the soybean chlH gene encoding a subunit of Mg-chelatase and localization of the Mg2+ concentration-dependent ChlH protein within the chloroplast. Plant Cell Physiol. 39 275–284. [DOI] [PubMed] [Google Scholar]
- Oelmüller, R. (1989). Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochem. Photobiol. 49 229–239. [Google Scholar]
- Oster, U., Brunner, H., and Rudiger, W. (1996). The greening process in cress seedlings. V. Possible interference of chlorophyll precursors, accumulated after thujaplicin treatment, with light-regulated expression of Lhc genes. J. Photochem. Photobiol. 36 255–261. [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Oswald, O., Martin, T., Dominy, P.J., and Graham, I.A. (2001). Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 98 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock, J., Mock, H.-P., Kruse, E., and Grimm, B. (1999). Expression studies in tetrapyrrole biosynthesis: Inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208 264–273. [Google Scholar]
- Papenbrock, J., Mock, H.P., Tanaka, R., Kruse, E., and Grimm, B. (2000). Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek, M.E., Dickey, L.F., Nguyen, T.T., Gatz, C., Sowinski, D.A., Allen, G.C., and Thompsen, W.F. (1998). Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc. Natl. Acad. Sci. USA 95 9009–9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397 625–628. [Google Scholar]
- Pfannschmidt, T., Allen, J.F., and Oelmüller, R. (2001. a). Principles of redox control in photosynthesis gene expression. Physiol. Plant. 112 1–9. [Google Scholar]
- Pfannschmidt, T., Schütze, K., Brost, M., and Oelmüller, R. (2001. b). A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem. 276 36125–36130. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, K., Kim, K.-S., Kogan, S., and Guarente, L. (1989). Functional dissection and sequence of the yeast HAP1 activator. Cell 56 291–301. [DOI] [PubMed] [Google Scholar]
- Pöpperl, G., Oster, U., and Rudiger, W. (1998). Light-dependent increase in chlorophyll precursors during the day-night cycle in tobacco and barley seedlings. J. Plant Physiol. 153 40–45. [Google Scholar]
- Puente, P., Wei, N., and Deng, X.-W. (1996). Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Pursiheimo, S., Mulo, P., Rintamäki, E., and Aro, E.-M. (2001). Coregulation of light-harvesting complex II phosphorylation and Lhcb accumulation in winter rye. Plant J. 26 317–327. [DOI] [PubMed] [Google Scholar]
- Rintamäki, E., Martinsuo, P., Pursiheimo, S., and Aro, E.-M. (2000). Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 97 11644–11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streatfield, S.J., Weber, A., Kinsman, E.A., Husler, R.E., Li, J., Post-Beitenmiller, D., Kaiser, W.M., Pyke, K.A., Flügge, U.-I., and Chory, J. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (2000). The pea light-independent photomorphogenesis1 mutant results from partial duplication of COP1 generating an internal promoter and producing two distinct transcripts. Plant Cell 12 1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek, R.E., and Chory, J. (1992). A tale of two genomes: Role of a chloroplast signal in coordinating nuclear and plastid genome expression. Aust. J. Plant Physiol. 19 387–399. [Google Scholar]
- Susek, R.E., Ausubel, F.M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799. [DOI] [PubMed] [Google Scholar]
- Terry, M.J., and Kendrick, R.E. (1999). Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 119 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, M.J., Wahleithner, J.A., and Lagarias, J.C. (1993). Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 306 1–15. [DOI] [PubMed] [Google Scholar]
- Vinti, G., Hills, A., Campbell, S., Bowyer, J.R., Mochizuki, N., Chory, J., and Lopez-Juez, E. (2000). Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J. 24 883–894. [DOI] [PubMed] [Google Scholar]
- Walker, C.J., and Willows, R.D. (1997). Mechanism and regulation of Mg-chelatase. Biochem. J. 327 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavgorodnyaya, A., Papenbrock, J., and Grimm, B. (1997). Yeast 5-aminolevulinate synthase provides additional chlorophyll precursor in transgenic tobacco. Plant J. 12 169–178. [DOI] [PubMed] [Google Scholar]