INTRODUCTION
One hallmark of signal transduction events is the phosphorylation-induced transition of a protein from one activity state to another. Kinases, phosphatases, transcription factors, and enzymes all can be influenced by phosphorylation. However, it is becoming apparent that, in many cases, phosphorylation alone does not induce the change in activity state. Rather, it is the phosphorylation-induced association with 14-3-3 proteins that results in the transition to changes in activity.
The 14-3-3 proteins were characterized initially during a survey-and-catalog project of proteins that appeared to be specific to mammalian brain tissue (Moore and Perez, 1967). Proteins were given operational designations based on chromatography elution and starch–gel electrophoresis profiles. Several three-numbered proteins appeared to be brain specific, including 15-4-1, 4-4-2, and 14-3-3. For the 14-3-3 proteins, this three-numbered designation has become their de facto name, even though it conveys no functional information and causes continuing problems with nomenclature.
Because they were found at high concentrations in specific regions of the brain, it was concluded initially that 14-3-3s were involved in specific functions of the nervous system. A follow-up study found that 14-3-3s were transported along axons, supporting a relationship with neuronal activity that was supported further by the biochemical activity of a 14-3-3 protein as an activator protein of the brain enzymes Trp hydroxylase and Tyr hydroxylase (Yamauchi et al., 1981). Trp and Tyr hydroxylases are the rate-limiting enzymes in the biosynthesis of the catecholamine and indolamine neurotransmitters, and their activation clearly requires 14-3-3s, along with Ca2+, calmodulin, and calmodulin-dependent protein kinase (Ichimura et al., 1987, 1991, 1997). This initial identification of the kinase-related interaction of 14-3-3s and target molecules would emerge as a key aspect of 14-3-3 participation in cellular regulatory pathways and would transcend their expanded discovery in many tissues and organs other than the brain. In fact, as 14-3-3s were identified in plants, they also were being discovered in a large number of experimental contexts well beyond brains (Ferl, 1996).
Throughout this expansion of experimental contexts, which continues today, the main recurrent theme has been that 14-3-3 proteins interact physically with client proteins when the client is specifically phosphorylated and that the binding of 14-3-3s in this phosphorylation-dependent manner completes an important step in signal transduction. Indeed, there is increasing recognition that phosphorylation alone is not always sufficient to complete the transduction of the regulatory signal into a change in the activity state of the client. Physical association with additional regulatory proteins, particularly the 14-3-3s, can be absolutely required to complete the signal-induced transformation of the activity of the client. Thus, 14-3-3s now are viewed within a general context of signal transduction that is wide ranging and not at all limited to nervous tissue. Moreover, the discovery that kinases and phosphatases themselves can be bound and regulated by 14-3-3s (Aitken, 1995; Aitken et al., 1995b; Conklin et al., 1995; Camoni et al., 1998) firmly establishes 14-3-3s as integral components of signal transduction pathways. It should be noted, however, that 14-3-3s also can bind certain nonphosphorylated targets, indicating that they also have roles outside the context of phosphorylation-mediated signal transduction.
In this review, we examine several known clients of 14-3-3 phosphoregulation to place the basic models into operational contexts and consider the implications of the diversity of structure and function present within the 14-3-3 family. Finally, the possible implications of the full range of 14-3-3 influence are considered by examination of the potential number of 14-3-3 client proteins present in the Arabidopsis genome.
CELLULAR CONTEXTS OF 14-3-3 ACTIVITIES
A wide array of biological functions involving kinase-mediated signal transduction, growth and developmental regulation, and response to environmental stress have been attributed to members of the 14-3-3 family. The notable recurrent themes in these diverse systems are the involvement of protein–protein interactions, divalent cations, kinases, and phosphatases, and the role of 14-3-3s continues to center on direct participation in signal transduction events (Morrison, 1994, 1995). Although not exhaustively complete, this survey highlights these themes and roles.
Kinase-Mediated Signal Transduction
Signal transduction via protein phosphorylation is a common pathway for many organisms. Perhaps the best studied of these is the GTP-dependent Ras pathway. The stimulation of cell division by extracellular growth factors involves the receptor-based production of active Ras, which then turns on a series of protein kinases, including Raf-1, which activate enzymes in the nucleus that are critical for message transduction, including the transcription factors. In 1994, several investigators independently identified 14-3-3s within the Ras pathway as activators of Raf-1 (Fantl et al., 1994), and 14-3-3 proteins were found to associate with Raf-1 in vivo while completing its activation and recruitment to the membrane (Freed et al., 1994; Irie et al., 1994). Since these initial reports, other kinases have been shown to either bind to or be activated by 14-3-3s (Maru and Witte, 1991; Reuther et al., 1994; Liu et al., 1997), including the notable calcium-dependent protein kinase (CDPK) (Camoni et al., 1998).
Growth and Developmental Signaling
In several organisms, it has become apparent that 14-3-3s play a pivotal role in the growth and development of the cell. A clear example of this comes from the study of the yeast Saccharomyces cerevisiae 14-3-3s, known as BMH1 and BMH2 (van Heusden et al., 1995). Strains with disrupted BMH genes grow more slowly on minimal medium, and double mutants are lethal. Normal growth rate, however, can be rescued by heterologous expression of one of several 14-3-3s, including four different Arabidopsis isoforms (van Heusden et al., 1996). Examples of the involvement of 14-3-3s in higher order growth and development are available, especially for Drosophila melanogaster, in which the leonardo mutation demonstrates that 14-3-3s play a role in synaptic growth and learning (Skoulakis and Davis, 1996, 1998; Broadie et al., 1997; Li et al., 1997). The presence of an inherited variant of a human 14-3-3 has been associated with early-onset schizophrenia (Toyooka et al., 1999).
Response to Stress
The study of stress induction in different organisms has led to the identification of 14-3-3s as integral components of response pathways. Environmental conditions affect 14-3-3s directly, because external stimuli such as cold and increased salt or Suc have led to altered regulation of plant 14-3-3s (Kidou et al., 1993; Chen et al., 1994; Jarillo et al., 1994). Biological interactions between organisms also engage 14-3-3s, as in the Pseudomonas aeruginosa exotoxin S, which requires a cellular 14-3-3 for activity (Fu et al., 1993). A direct relationship exists between 14-3-3s and the receptor for the wilt-inducing phytotoxin fusicoccin (FC) (Oecking et al., 1994; Aducci et al., 1995, 1996; de Boer and Korthout, 1996). Interactions with 14-3-3s regulate the H+-ATPase under normal physiological conditions via blue light excitation in guard cells (Emi et al., 2001), and the complex between 14-3-3s and the H+-ATPase forms a binding site for FC.
Structure and Movement
There are examples of 14-3-3 proteins interacting with proteins that would not normally be considered enzymes or be subject to signal-induced transitions in activity. Keratin intermediate filaments are expressed in simple-type epithelia and are responsible for cell structural integrity, and 14-3-3s associate with keratins where they act as solubility factors (Liao and Omary, 1996). Other structural roles for 14-3-3s are indicated by the localization of 14-3-3s to the mitotic spindle apparatus, by associations with centrosomes (Pietromonaco et al., 1996), and by interactions with other cell scaffold–type proteins (Dellambra et al., 1995; Du et al., 1996). In certain contexts, 14-3-3s serve to alter the subcellular localization of their clients. The 14-3-3s contain a nuclear export signal, such that interaction between 14-3-3s and clients within the nucleus serves to assist in the nuclear export of the client (Rittinger et al., 1999; McKinsey et al., 2000).
14-3-3s RESOLVE SIGNAL TRANSDUCTION EVENTS
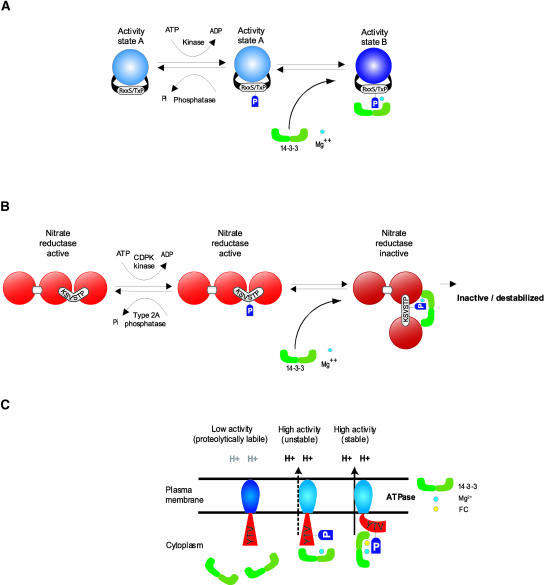
The underlying paradigm for 14-3-3 participation in signal transduction events is the phosphorylation-dependent association of 14-3-3s with their clients, and as a result of this binding, the client molecule undergoes a change in activity state (Figure 1). The key aspects of the model are as follows: (1) the change in activity state of the client molecule does not occur until the binding of the 14-3-3s, and (2) the binding of 14-3-3s is a requisite step in the process of signal-induced transition. This entire regulatory process would be contingent on the cellular levels of 14-3-3s, the kinase and phosphatase that act on the client enzyme, and divalent cations. This complex contingency, as well as the large number of possible 14-3-3 isoform combinations (see below), allows for multiple regulatory controls on the client activity.
Figure 1.
Consummated Signal Transduction.
(A) The basic paradigm for 14-3-3 regulation is a two-step process whereby the target protein is phosphorylated by a kinase but remains in its initial activity state until 14-3-3s bind in the presence of divalent cation to complete the transition to a new activity state.
(B) NR regulation by 14-3-3s is an extension of the basic paradigm. In its unphosphorylated state, NR is active. Phosphorylation by CDPK does not inactivate NR, but phosphorylation does tag the enzyme for 14-3-3 binding, which completes the signal-induced transition toward inactivation. NR that is phosphorylated and bound by 14-3-3s may be inactivated directly in a reversible fashion or may be destabilized and subjected to proteolysis.
(C) Aspects of the paradigm model apply to the apparently specialized case of 14-3-3 regulation of H+-ATPase, in which binding in the presence of FC locks the enzyme into a highly active state. In the absence of FC, 14-3-3s bind to increase the activity of H+-ATPase in a regulatable manner.
The most deeply explored biological function involving plant 14-3-3s is the regulation of metabolic enzymes, with nitrate reductase (NR) as the prime example (Figure 1B). NR is the rate-limiting enzyme in nitrogen fixation in plants (Campbell, 1996). NR regulation involves inactivation by phosphorylation, but phosphorylation itself does not inactivate NR. The complete inactivation process requires divalent cations and the binding of a 14-3-3 to the phosphorylated NR. The exact mechanism of the transition to inactivity of NR after binding of 14-3-3s is not understood entirely. In the basic paradigm model, NR, when phosphorylated and bound to divalent cations and 14-3-3s, is simply inactive, and that inactivity is reversible upon the removal of 14-3-3s and dephosphorylation (Huber et al., 1996). In a model extended from the paradigm, NR that is phosphorylated and bound by divalent cations and 14-3-3s is still active, but it is in an unstable state that is subject to proteolysis (Weiner and Kaiser, 1999, 2000; Kaiser and Huber, 2001). Regardless of the specific mode of NR inactivation, it is clear that the binding of 14-3-3s is a required part of the inactivation process. Although this conclusion is based largely on in vitro experiments, in extracts from spinach leaves, 14-3-3s are associated with NR only in the dark, in which the in vivo activity of NR is inhibited (Weiner and Kaiser, 1999), clearly supporting an in vivo role for the 14-3-3 regulation of NR.
One of the fundamental points that emerged from the primary work on NR and 14-3-3 interactions was the concept of binding sequence conservation. For example, a phosphopeptide based on the animal Raf-1 sequence can block the inactivation of NR by competition for 14-3-3 binding, and a consensus phosphoserine binding sequence can be derived from the observation of both NR and Raf-1 that extends to other 14-3-3–regulated proteins. This indicates that the core of the central paradigm model for 14-3-3 regulation (Figure 1) likely is consistent across a wide range of 14-3-3–regulated events (Huber et al., 1996; Sehnke and Ferl, 1996). This conclusion is strengthened by similar binding sites and regulation of other enzymes, perhaps most notably Suc phosphate synthase (Toroser et al., 1998). Yet, although the central themes of phosphorylation-dependent binding and regulation are largely consistent, the regulated outcome can be inactivation, activation, or some other alteration of the client molecule.
Such variation is necessary to explain 14-3-3 participation in the regulation of the plasma membrane H+-ATPase and the effects of FC (Jahn et al., 1997; Fullone et al., 1998; Olsson et al., 1998). In this case, 14-3-3s are positive regulators of H+-ATPase activity. When 14-3-3s and Mg2+ are bound to the C-terminal region of the H+-ATPase, pump activity is stimulated to a high-activity state that is unstable and revertable. In the presence of FC, however, the regulatory effect is a locked high-activity state (Figure 1C) that appears to account for the wilting syndrome induced by FC. The continuous H+-ATPase activity drives guard cell solute uptake, resulting in increased turgor pressure and leading to excessive transpiration. Proteolysis also may play a role in H+-ATPase regulation, in that 14-3-3s may maintain the activation of the H+-ATPase by protecting it directly from proteolytic removal of its regulatory C-terminal domain (Malerba and Bianchetti, 2001).
STRUCTURE OF 14-3-3s
The crystal structure has been solved for two mammalian 14-3-3s (Liu et al., 1995; Xiao et al., 1995). The protein sequences of 14-3-3s are highly conserved across evolutionary lineages, and the extreme conservation of the central core region of 14-3-3s makes the animal structure a very likely fit to the general features of plant 14-3-3s (Figure 2) (Ferl et al., 1994; Ferl, 1996). However, both of the known crystal structures fail to resolve the N and C termini, which are highly divergent among isoforms. Thus, it is possible to consider the paradigm regulatory features of 14-3-3s to be associated with the central conserved core while recognizing that the divergent termini might contribute to specific regulatory functions.
Figure 2.
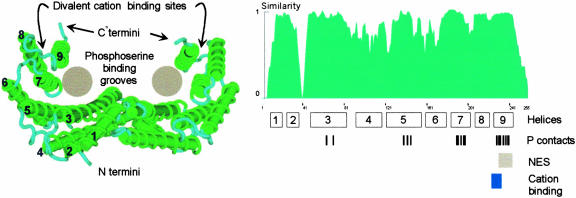
Model of the 14-3-3 Structure.
The structure of the main body of the protein is known to be a W-shaped clamp formed by two monomers, each of which is capable of binding phosphorylated peptides. The structure and positions of the extreme N and C termini are unresolved in the crystals, and the C termini likely form a movable flap that can seal the top of the clamp, perhaps depending on divalent cation binding. The conservation of the 14-3-3 protein sequences is evident when the similarity index among all Arabidopsis isoforms is plotted along the length of the molecule. Only three areas of the 14-3-3s show low degrees of similarity: the N terminus, the C terminus, and the region between helices 2 and 3. Shown below the similarity profile are the locations of the helices in the crystal structure as well as the locations of the residues responsible for contacting the phosphopeptide (P contacts) (Rittinger et al., 1999), the nuclear exclusion sequence (NES) (Rittinger et al., 1999), and the cation binding site (Lu et al., 1994). The crystal structure was taken from the protein database 1QJB and was modeled using Cn3D. The similarity profile among Arabidopsis 14-3-3 was calculated using the algorithms available in Vector NTI 7.0 (InforMax, Bethesda, MD).
The main feature of the 14-3-3 structure is a double-barreled, W-shaped clamp formed from the essentially antiparallel helices of the dimer pair. Each monomer produces a channel that is sufficient in size and shape to accommodate interaction with a phosphorylated peptide from a client protein (Yaffe et al., 1997; Petosa et al., 1998). Because 14-3-3s exist as dimers, it is possible that they could bring together two different proteins, or two different domains within one protein, allowing a direct interaction between the clients (Ferl, 1996; Yaffe et al., 1997). The divergent C terminus that lies over the channel could form a cap over the open end of the clamp, and there is a divalent metal binding site near the point at which the C terminus hinges to the main structure (Lu et al., 1994). The binding of divalent cations often is a requirement for 14-3-3 interaction with clients (Athwal et al., 1998a, 1998b), so the basic paradigm model in plants can be extended to include the C terminus and the metal binding hinge as a flexible structure that can regulate entry and exit from the phosphoserine binding grooves (Lu et al., 1994).
STRUCTURE OF THE 14-3-3–CLIENT COMPLEX
Subsequent to the solution of the 14-3-3 structures, two crystallographic studies were reported that characterized the specific binding of phosphoserine-containing target peptides to 14-3-3s (Yaffe et al., 1997; Rittinger et al., 1999). As predicted from previous studies, the central amphipathic channel created by the helices of the 14-3-3 is the site of phosphopeptide interaction, with the phosphoserine pointing toward the positively charged base of the channel. Specific ligand contacts also were clearly distinguishable for both types of peptides examined. However, because of the limited length of the target (15 amino acids), details regarding full-length client protein contacts and perturbations to the 14-3-3 structure were not identifiable.
The crystal structure of a 14-3-3 together with its client protein, serotinin N-acetyltransferase (AANAT), was reported recently, and the structure addresses some unresolved 14-3-3 features (Obsil et al., 2001). The structure of the complex confirms the essential aspects of the independently solved models of both AANAT and 14-3-3 ζ (Liu et al., 1995; Hickman et al., 1999) and essentially agrees with the previous 14-3-3/phosphoserine-containing peptide crystal models (Rittinger et al., 1999). The most significant difference between the models of the free and complexed 14-3-3s is the opening up of the central channel and the outward movement of helix 8 away from the channel. This movement involves repositioning of the loop between helices 8 and 9, the region in plants that is responsible for divalent cation binding (Lu et al., 1994).
MODE OF ACTION OF 14-3-3s
Although mechanistic information garnered from one 14-3-3– client structure has been established, it is not known if it is representative of all 14-3-3 interactions. The apparent presence of multiple binding sites within a client molecule further complicates this issue, because binding and regulation may be two separate events (Fantl et al., 1994; Jelinek et al., 1996; Clark et al., 1997). Furthermore, examples of 14-3-3 binding solely via nonphosphorylated sequences also exist, adding to the complexity of what would seem to be a uniform mechanistic model that would label 14-3-3s as simply phosphoserine/Thr binding proteins (Halbach et al., 2000; Zhai et al., 2001). However, structural and biochemical clues to the general 14-3-3 regulatory scheme and the consequences to the client proteins are mounting in the literature.
The structure of the AANAT–14-3-3 complex suggests that binding 14-3-3 involves conformational rearrangements brought about by substrate binding to AANAT before 14-3-3 association. The suggested outcome is that the AANAT–14-3-3 interaction may allow increased access of both substrates to the enzyme, thereby altering its dissociation constant and activity. Isothermal titration calorimetric measurements confirmed the enhanced substrate binding and decrease in the dissociation constant of AANAT when complexed with 14-3-3 (Obsil et al., 2001).
From biochemical studies of 14-3-3s and their clients, two major mechanisms have emerged that can account for the 14-3-3 regulation of clients in plants. The first is a shielding or stabilization effect. This effect appears to prevent accessibility to proteases and phosphatases. Examples of this effect in plants include the protection of glyceraldehyde-3-phosphate dehydrogenase, glutamyl-tRNA synthetase, Suc-phosphate synthase, CDPK 6, and NR from proteolytic degradation (Cotelle et al., 2000; MacKintosh and Meek, 2001), enhanced proteolysis of NR (Kaiser and Huber, 2001), protection from proteolysis in the regulation of the plasma membrane H+-ATPase (Malerba and Bianchetti, 2001), and prevention of the dephosphorylation of the 14-3-3 binding site in NR (Bachmann et al., 1996). The control of this protection is unclear, because it would seem that the 14-3-3s must first be dissociated before the client would be susceptible to phosphatases or proteolytic attack. However, the possibility of altered 14-3-3–client affinity in different physiological environments would provide additional 14-3-3 regulation based on dissociation constants. In addition, the protection can provide either activation or inhibition, as with H+-ATPase or NR, respectively. Usually, the proteolytic effects are not en mass degradation of the protein, such as that of the ubiquitin cascade, but rather simple removal of the portion of the peptide that contains the 14-3-3 binding sequence. This process of 14-3-3 regulation of cleavage also seems to be responsive to external stimuli such as sugar starving and hormonal release (Cotelle et al., 2000).
The second general mechanism for the modification of a client protein is the addition of a transient nuclear export signal, resulting in nuclear shuttling (Yang et al., 1999; Davezac et al., 2000; Igarashi et al., 2001). Again, the exact nature of this added shuttling function is not obvious. In the literature, there is support for two theories regarding how nuclear export occurs via 14-3-3 binding. Nuclear export signals have been postulated within the 14-3-3 that, when associated with the client protein, provide nuclear exclusion of the client protein (Dalal et al., 1999; Rittinger et al., 1999). The other belief is that the mere binding of 14-3-3s to client proteins that contain nuclear targeting signals masks this intrinsic signal and precludes nuclear import of the client protein via direct interference (Muslin and Xing, 2000).
It also appears likely that 14-3-3s themselves undergo modifications and structural changes that alter their functions and interactions with client proteins. In general, 14-3-3 proteins are known to be modified post-translationally. In animal systems, the N termini of many 14-3-3s are acetylated (Toker et al., 1992), and the animal α and δ isoforms are the phosphorylated versions of the β and ζ isoforms (Aitken et al., 1995a). In Arabidopsis, the ω isoform also can be phosphorylated and contains a demonstrable divalent cation binding site that binds calcium with an affinity higher than that of calpain II, the secondary calcium binding sites of calreticulins, and the regulatory sites of troponin C (Lu et al., 1994). The cation binding site is located in the C-terminal region, in the loop that connects helices 8 and 9 in the crystal structure (Figure 2) (Lu et al., 1994). In addition, the binding of cations changes the structure of 14-3-3s such that the C-terminal region becomes susceptible to proteolytic cleavage (Lu et al., 1994) and is altered in surface hydrophobicity (Athwal et al., 1998a). These data suggest that the modification and cation binding state of the 14-3-3s may play a role in the regulation of client binding, perhaps by gating access to the phosphoserine binding grooves of the central core (Lu et al., 1994; Sehnke and Ferl, 2000). It also has been reported that 14-3-3 dimerization is inhibited by calcium binding, thereby limiting any “clamping” capability of the 14-3-3 (Abarca et al., 1999).
ARABIDOPSIS 14-3-3s
The model for 14-3-3 function, together with the evolutionary conservation that is apparent through much of the 14-3-3 structure, leads to an initial presumption that all 14-3-3s have the same function. Support for this presumption comes from complementation studies in yeast, which indicate that several Arabidopsis 14-3-3s can substitute for the essential 14-3-3 functions in yeast (van Heusden et al., 1996), as well as studies that demonstrate that alien 14-3-3s can regulate NR (Moorhead et al., 1996). However, two lines of evidence support the hypothesis that there are important functional differences among the various 14-3-3s present in any multicellular organism. First, most higher organisms have fairly large and diverse 14-3-3 gene families. The central conserved areas of the sequences remain intact, but diversity does occur in several areas, especially at the termini (Figure 2). Second, biological phenotypes associated with altered 14-3-3 isoforms demonstrate properties that are difficult to explain in the absence of specificity, and various 14-3-3 isoforms have differential affinities for at least certain client proteins.
Diversity: The Arabidopsis Family of 14-3-3 Genes
Because of the complete nature of the Arabidopsis genome project, the Arabidopsis 14-3-3 family serves as an appropriate framework within which to examine 14-3-3 diversity in plants (Wu et al., 1997a; DeLille et al., 2001). Emerging data from other plant species indicate that there are no plant 14-3-3s that present obvious family members or branches of the 14-3-3 tree that are not represented in Arabidopsis (Ferl et al., 1994; Ferl, 1996; Wang and Shakes, 1996; Wu et al., 1997a; Rosenquist et al., 2000; DeLille et al., 2001).
The members of the Arabidopsis 14-3-3 family of proteins are named with Greek letter designations. This nomenclature in is keeping with the early 14-3-3 literature, which originally differentiated the 14-3-3 protein variants as isoforms that eluted differentially during column chromatography of brain extracts. It should be noted, however, that the current list of 14-3-3 isoforms from Arabidopsis is based on gene sequences rather than biochemical differentiation. In addition, the three-letter gene name for Arabidopsis 14-3-3s is GRF (general regulatory factor) (Rooney and Ferl, 1995), and Arabidopsis 14-3-3 gene and isoform designations often include the name GF14 (G-box factor 14-3-3 homolog) preceding the Greek letter (Wu et al., 1997a).
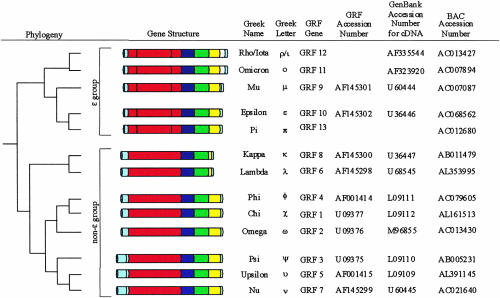
There are 15 members of the 14-3-3 gene family in Arabidopsis, bearing the gene names GRF1 through GRF15 (DeLille et al., 2001; Rosenquist et al., 2001). However, GRF14 and GRF15 appear to be remnant 14-3-3 genes in that they appear to be truncated, and the mRNA expression of GRF13 π has yet to be confirmed experimentally (Figure 3). Comparison of the Arabidopsis 14-3-3 family isoforms reveals that overall, ∼50% of the amino acids are identical in all isoforms, with large blocks of the central region conserved absolutely in all isoforms. However, the C and N termini are nearly unique for each isoform, supporting the potential for specific functions among isoforms (Figure 3) (Ferl et al., 1994; Ferl, 1996; Wu et al., 1997a). The correlation of deduced amino acid sequence and the intron placement indicate that different 14-3-3 domains may be encoded by separate exons, and what is essentially the highly divergent C terminus is encoded by its own exon (Daugherty et al., 1996).
Figure 3.
Arabidopsis 14-3-3 Gene Structure and Evolutionary Relationships.
The Arabidopsis 14-3-3 family is composed of two main evolutionary branches: the ɛ group and the non-ɛ group. The evolutionary relationships represented in the tree reflect the phylogenies derived from alignments of the central conserved regions of the Arabidopsis 14-3-3 derived amino acid sequences, and the main branches are supported by the genomic structure of the 14-3-3 genes. BAC, bacterial artificial chromosome. Individual exons are delineated by either different coloration or segmentation.
Phylogenetic analysis of Arabidopsis 14-3-3 isoforms based on sequence alignment of the central core region, with support from gene structure considerations, divides the Arabidopsis 14-3-3 family into two distinct fundamental groups, the ɛ group and the non-ɛ group. The ɛ and non-ɛ groups have been on distinct evolutionary tracks since before the separation of animals and plants (Ferl et al., 1994) (Figure 3). The ɛ group comprises the ɛ, π, ρ, ο, and μ isoforms, all of which demonstrate an exon structure distinct from that of the non-ɛ group isoforms. The non-ɛ group includes three organizational subgroups. The κ and λ isoforms form a group well isolated phylogenetically from the group containing ω, χ, and φ, but they all share a common genomic organization with conserved intron locations. The ν, υ, and ψ group contains an extra intron within the 5′ untranslated leader (Figure 3) (DeLille et al., 2001).
This well-characterized Arabidopsis 14-3-3 protein family provides a reference for understanding fundamental 14-3-3 functions that may be shared among isoforms, as well specialized functions that may be limited to specific isoforms or isoform subgroups. The fundamental principles derived from the 14-3-3 family likely will apply to the family trees of 14-3-3 isoforms in other plant species.
Localization: Hints of Specificity
The presence of a diverse gene family begs the question of specificity of gene expression. Expression patterns of the various 14-3-3 isoforms present in rat brain are regulated differentially and precisely according to the neuron type and the stage of cytodifferentiation (Watanabe et al., 1994). Analyses of mRNA distribution and 14-3-3 promoter:β-glucuronidase fusions demonstrate that Arabidopsis isoforms, like animal isoforms, can demonstrate a high degree of cell- and tissue-type specificity (Daugherty et al., 1996; Kuromori and Yamamoto, 2000; Rosenquist et al., 2001). Hence, at least some of the diversity in 14-3-3 genes is present to ensure their fundamental presence in certain cells, tissues, and organs.
The presence of a diverse gene family also creates the potential for biochemical specificity among the gene products and specificity of their subcellular organization. Subcellular localization could be directed by intrinsic 14-3-3 trafficking signals, or 14-3-3s might travel to subcellular locations based on specific physical interactions with client proteins that direct the localization of the 14-3-3–client complex. Specific 14-3-3s have been shown experimentally to occur within organelles, in addition to being found within the cytoplasm. Plant nuclei, plastids, and mitochondria all contain 14-3-3s (de Vetten and Ferl, 1994; Bihn et al., 1997; May and Soll, 2000; Sehnke et al., 2000, 2001; Bunney et al., 2001). The spectrum of 14-3-3 isoforms present within the organelle can be demonstrably different from the total spectrum present in the cell (Bihn et al., 1997; Sehnke et al., 2000). For example, υ and ν are the only two non-ɛ isoforms present in chloroplasts, indicating specificity among non-ɛ isoforms for their differential localization and suggesting that chloroplast-specific functions might be limited to υ and ν (Sehnke et al., 2000).
Diversity in subcellular localization among Arabidopsis 14-3-3s is demonstrated by 14-3-3–green fluorescent protein (GFP) fusions. GFP itself has a propensity for concentration in the nucleus, but it was found in other areas of the cells in Arabidopsis roots as well (Figure 4A). However, when GFP was tagged to the C-terminal end of 14-3-3 κ, the GFP signal was relocalized prominently to the plasma membrane region of the cell in addition to the nucleus (Figure 4B). If the GFP is tagged to 14-3-3 υ, the GFP is driven largely out of the nucleus and into the cytoplasmic parts of the cell (Figure 4C), whereas GFP tagged to 14-3-3 φ was present rather diffusely throughout the cell (Figure 4D). These data indicate that these different 14-3-3 isoforms occupy different cellular locations, either because the 14-3-3s are themselves directed to different locales or because the specific client proteins to which the 14-3-3s are bound are localized differentially.
Figure 4.
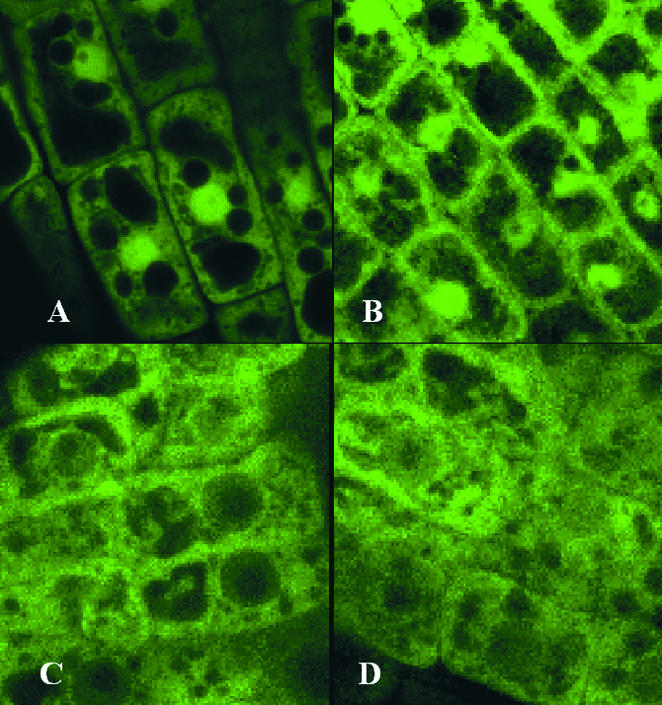
14-3-3–GFP Fusions Show Diverse Localization Patterns.
(A) GFP is localized to several areas of the Arabidopsis root cells and tends to concentrate to some extent within the nuclei.
(B) to (D) Fusion proteins in which GFP is located at the C terminus of diverse 14-3-3s tend to localize differentially within Arabidopsis root cells. The GFP κ signal is relocalized prominently to the plasma membrane region but remains concentrated in the nucleus (B). The GFP υ signal is largely driven out of the nucleus and into the cytoplasmic compartments (C), whereas the GFP φ signal is present rather diffusely throughout (D). Coding regions for the 14-3-3s were amplified by polymerase chain reaction, placed as translational fusions with GFP in pBI12135sGFP(S65T), and transformed into Arabidopsis Wassilewskija by vacuum infiltration. Five-day-old plants were visualized using an Olympus IX70 inverted microscope (Tokyo, Japan) mounted to a Bio-Rad MC 1024ES laser scanning system with 24-bit confocal imaging. Images were Kalman averaged four times.
Complicating the issues surrounding subcellular localization are the facts that 14-3-3 localization can be very dynamic and that 14-3-3s can be present as heterodimers or homodimers. In some systems, 14-3-3s function to enhance the nuclear export of their clients, whereas in other systems, the binding of 14-3-3s may interfere with the intrinsic signal of the client (Muslin and Xing, 2000). In either case, the 14-3-3 is localized dynamically along with the client to which it is bound. In fact, the random GFP tagging of 14-3-3 ω was recognized experimentally because the fusion altered the localization of GFP (Cutler et al., 2000). It is clear that different 14-3-3 isoforms can be present in different tissues and in different subcellular compartments. Given the size of the family, even in Arabidopsis, this sets the stage for a complex situation in which any given cell may have several 14-3-3 gene products present in some combination of subcellular compartments. Furthermore, as dimeric proteins, 14-3-3s can exist as homodimers or heterodimers. Heterodimerization occurs easily among Arabidopsis 14-3-3s in vitro (Wu et al., 1997b) and has been demonstrated in vivo (Hachiya et al., 1994), thus further complicating the already complex situation. The biological outcome of the entire 14-3-3 scenario, then, depends on there being specificity to the interactions and functions contributed by the individual 14-3-3 isoforms, so that their differential presence confers differential activity.
Specificity: Biological Phenotypes and Biochemical Functions
Given the size of 14-3-3 gene families and the wide range of pathways in which 14-3-3 regulation has been implicated, it would be difficult to predict the phenotypes of individual 14-3-3 mutations. In yeast, which have only two 14-3-3 genes, the gross phenotype of the 14-3-3 double disruptant is death. In D. melanogaster, which has at least two 14-3-3s that are modified by alternative splicing, 14-3-3 null mutations can result in defects that are embryo lethal (Li et al., 1997). These data suggest that the roles of 14-3-3s are so fundamental that complete or nearly complete loss of function is lethal. These data also seem to suggest that, at least in these rather simple gene family systems, the core activities of 14-3-3s are redundant. In yeast, for example, neither of the 14-3-3 single gene disruptants is lethal. In D. melanogaster, null mutations of the leonardo gene are embryo lethal, yet other mutations and alterations of expression can produce a series of rather severe, but somewhat variable, phenotypes largely connected to Ras/Raf signaling events (Li et al., 1997). Therefore, in complex higher organisms, alterations in phenotype caused by modulations of individual 14-3-3 genes are possible, even in the context of potential functional redundancy.
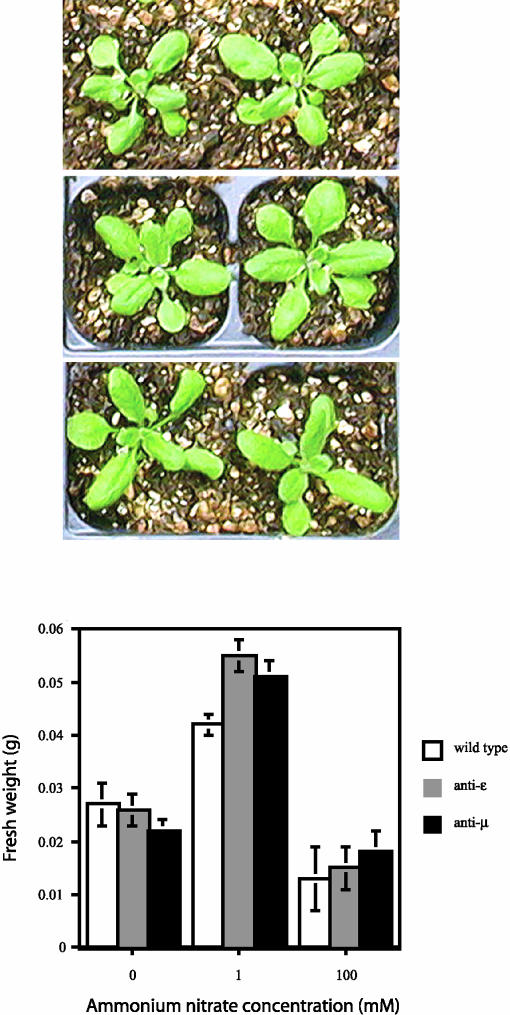
In plants, phenotypes associated with 14-3-3 genes have been examined solely by reverse genetic approaches. To date, T-DNA knockouts of Arabidopsis 14-3-3 genes have failed to produce any obvious phenotypes (Krysan et al., 1996), but overexpression and antisense constructions have produced phenotypic changes that allow insights into 14-3-3 function in plants. Work with overexpression and antisense constructions of 14-3-3s in potato suggested a role for 14-3-3s in plant development and senescence via an unknown mechanism involving an inverse correlation between 14-3-3 levels and senescence (Markiewicz et al., 1996; Wilczynski et al., 1998) that was attributed to nuclease activity reported previously as a function of 14-3-3s (Szopa, 1995). Arabidopsis plants expressing antisense ɛ or μ reduce or eliminate the expression of ɛ and μ in leaf starch grains (Sehnke et al., 2001). As a result, these antisense plants accumulate an unusual amount of starch but show no other obvious visible changes in the leaves.
This phenotype prompted the question of which granule-associated starch synthetic enzyme(s) might be regulated by 14-3-3s. A search of the NCBI nonredundant translated nucleotide database revealed that starch synthase IIIs contains a highly conserved 14-3-3 binding site, and experimental evidence suggests that 14-3-3s interact with the starch synthase III DulI, potentially to regulate starch accumulation (Sehnke et al., 2001). Because 14-3-3s are well-documented regulators of NR, these same antisense ɛ and μ plants with decreased 14-3-3 expression also were analyzed for their ability to assimilate nitrate. Different concentrations of ammonium nitrate were used to fertilize the antisense ɛ and μ plants along with wild-type controls under otherwise normal growing conditions (Figure 5). With no fertilizer treatment, wild-type and antisense plants grew to the same mass. However, with the addition of 1 mM ammonium nitrate, the antisense plants grew appreciably larger, as measured by both mass and canopy area. The increased efficiency for nitrate uptake was not observed when toxic levels of ammonium nitrate were provided, suggesting that antisense plants still are responsive to excess nitrate. Together, these data strongly suggest that the removal of specific NR inhibitors in vivo, namely ɛ and μ 14-3-3 isoforms, allows increased efficacy regarding nitrate assimilation, most likely via NR regulation.
Figure 5.
Enhanced Ammonium Nitrate Utilization of Antisense 14-3-3 Arabidopsis.
An ammonium nitrate response curve was generated for antisense 14-3-3 Arabidopsis versus the wild type. Transgenic Arabidopsis expressing antisense ɛ and μ 14-3-3 isoforms and wild-type plants were fed water and 1 and 100 mM ammonium nitrate. Plants were grown under identical environmental conditions. Images of wild-type (top), antisense ɛ (middle), and antisense μ (bottom) plants grown in 1 mM ammonium nitrate are shown at top, and the concentrations of at least six plants of each type were recorded for the graphs at bottom.
All of these data on the evolution, localization, and biological phenotypes indicate that the 14-3-3s contribute to fundamental biological processes and suggest that, in spite of the conservation of their core structure, isoforms have a degree of specificity with regard to the client proteins with which they interact. This potential for biological specificity is reflected most directly in the biochemical specificity and affinity of individual 14-3-3–client interactions. Widely differential binding of recombinant Arabidopsis 14-3-3 isoforms to substrate target peptides has been demonstrated in two different model 14-3-3–client systems, NR and H+-ATPase. In both systems, the ability of the recombinant proteins to bind the target peptide required phosphorylation of the target. A synthetic 18-mer phosphopeptide based on the regulatory sites of spinach NR was used in pulldown assays against several Arabidopsis 14-3-3 isoforms (Bachmann et al., 1996). Strong binding occurred between the NR phosphopeptide and ω, moderate binding occurred for both χ and ν, and little or no binding occurred for φ and ψ. Relative binding correlates directly with the ability to inactivate NR, with ω the strongest inactivating isoform and φ and ψ essentially incapable of inactivating NR.
A synthetic 16-mer phosphopeptide of the C-terminal amino acids of the AHA2 H+-ATPase was used for surface plasmon resonance spectroscopic analysis of the relative affinities of nine Arabidopsis 14-3-3 isoforms (Rosenquist et al., 2000). Binding to the AHA2 phosphopeptide ranged from very strong (for φ and χ) to extremely weak (for κ and λ), and the relative affinities among the nine isoforms largely aligned with the relative positions of the isoforms on the phylogenetic tree. In these different but well-characterized model systems, recombinant homodimeric 14-3-3 isoforms displayed specific and wide-ranging affinities for the target peptides, indicating that for the limited structural recognition peptide, differentially specific in vitro interactions clearly exist among isoforms.
14-3-3 CLIENTS: A PROTEOMICS PERSPECTIVE
The paradigm binding site sequence for 14-3-3 proteins is R/KxxpS/TxxP. This sequence is based on the consensus Raf binding site from animal systems, but observation and peptide competition studies show that the site is appropriate for plant 14-3-3s. It also is referred to as the mode-1 type site (Yaffe et al., 1997) and includes the largest number of characterized interactions. In many instances, it is a useful consensus for plant 14-3-3 interactions. However, a number of issues should be considered before adopting consensus sites in plants. First, the known binding sites within plants and especially Arabidopsis should be used to evaluate the current validity of the consensus. Then, the distribution of consensus targets sites within the genome should be evaluated.
An Initial Listing of Potential 14-3-3 Clients
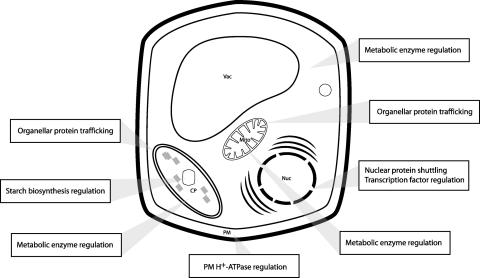
The 14-3-3 proteins bind to a wide array of client proteins. To date, the discovery of 14-3-3 involvement in many metabolic and signaling pathways has been coincidental, and most binding mechanisms are not characterized fully (Table 1). However, these interactions, together with less-well-understood interactions with enzymes, transcription factors, and chaperones that occur throughout the cell, serve to present an appropriate first impression of the tremendously broad range of client types under consideration (Figure 6). This impression is enhanced by biochemical studies designed to illuminate 14-3-3–dependent regulation of entire metabolic pathways, which bring additional and wide-ranging 14-3-3 clients under consideration (Moorhead et al., 1999).
Table 1.
Observed and Putative Interactions of 14-3-3 Client Proteins
| Protein | Binding Site(s) | Accession No. |
|---|---|---|
| R/KXXpSXP (mode-1) | ||
| Nitrate reductase | KKxSTP | AAA32830 |
| Vacuolar ATP synthetase catalytic subunit A | RKVSGP RGVSVP |
O23654 |
| ATP synthase β chain | RFLSQP | P19366 |
| Calcium-dependent protein kinase, AK1 | RTESKP | Q06850 |
| Starch synthase III | RYGSIP | CAA16796 |
| β-Amylasea | RYPSYP | P25853 |
| Protein kinase AFC2a | RNGSPP | P51567 |
| Protein kinase 54Ka | RQGSPP REQSIP |
S71169 |
| Magnesium-chelatase subunit CHLIa | RFDSNP | P16127 |
| COP1 regulatory proteina | RNISQP | P43254 |
| 2-cys peroxiredoxin BAS1a | RTLSSP | Q96291 |
| Homeobox-Leu zipper protein ATHB-5a | RSPSPP | P46667 |
| R/KXXXpSXP (mode-2) | ||
| 13-lipoxygenase Lox-2 | KPSDSKP | L37358 |
| Other | ||
| H+ ATPase | YpTV | P19456 |
Putative interaction.
Figure 6.
Summary of 14-3-3 Interactions and Locations.
Evidence of 14-3-3 participation in various plant physiological processes has arisen largely from the study of diverse client systems and most likely represents only a small proportion of their cellular involvement. Additionally, the localization of specific 14-3-3 isoforms is only now beginning to be elucidated with the production of isoform-specific antibodies. An up-to-date summary of 14-3-3–client localization is shown. CP, cytoplasm; Mito, mitochondrion; Nuc, nucleus; PM, plasma membrane; Vac, vacuole.
In the classic example of plant 14-3-3 enzymatic regulation, NR, the confirmed site of phosphorylation for Arabidopsis NR, is in the hinge region at Ser-534. The complete phosphorylation and 14-3-3 binding site is KSVpSTP and is essentially conserved among NR from other plants. This site conforms to the consensus mode-1 14-3-3 binding site, and studies with mutated spinach NR have illustrated the fidelity of the recognition system (Athwal et al., 1998b). The requirement for a basic residue preceding the pS also is required for phosphorylation by the kinase CDPK. To date, all of the clients characterized as interacting directly with 14-3-3s via phosphoserine/Thr fall into the same consensus, with the notable exceptions of the plasma membrane H+-ATPases and the lipoxygenases. The lipoxygenases have an additional residue between the pS and the K (Holtman et al., 2000a, 2000b), reflecting the mode-2 sites from the mammalian 14-3-3 binding sites (Yaffe et al., 1997). The H+-ATPases use a unique three-residue 14-3-3 binding site that is at the very C termini of the proteins. However, this is the only example of such binding; therefore, it may represent a specialized or smaller class of 14-3-3 clients.
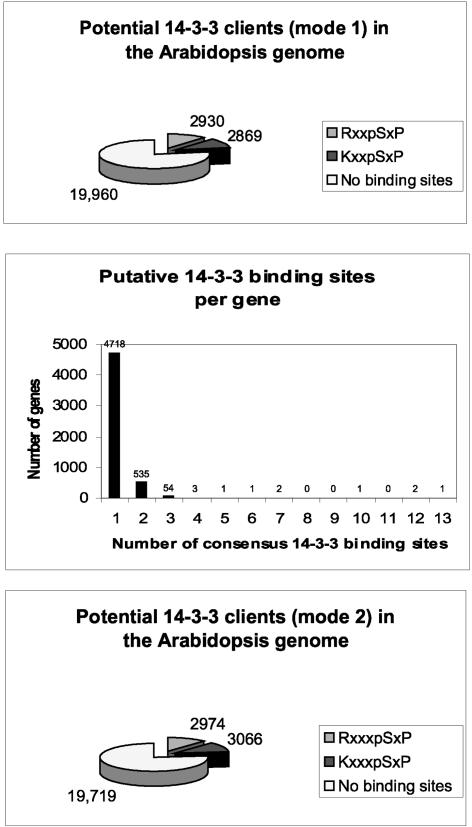
The mode-1 RxxpSxP and KxxpSxP consensus sequences implicate a large percentage of the potential proteins within the Arabidopsis genome as possible 14-3-3 clients (Figure 7). When these sites are presented in a search of the potential protein products deduced from the genome sequence, nearly 6000 predicted proteins are revealed as containing a 14-3-3 recognition sequence. (Pattern search analysis of the Nonredundant Arabidopsis Protein Database using PatMatch at The Arabidopsis Information Resource with the potential consensus 14-3-3 binding sites KxxSxP and RxxSxP identified 2869 and 2930 unique hits, respectively.) This means that approximately one in four proteins is potentially affected by 14-3-3 regulation.
Figure 7.
Potential 14-3-3 Clients within the Arabidopsis Genome.
More than 5799 gene products, or slightly >20% of the entire proteome, contain a mode-1 14-3-3 binding site. More than 500 gene products contain two or more mode-1 sites, and >6000 gene products contain mode-2 14-3-3 interaction sites. In total, nearly 40% of the proteins encoded by the Arabidopsis genome are potentially subject to regulation that involves 14-3-3 interaction.
The theoretical maximum number of targets for a hexameric binding site with residues fixed at three positions is 203, or 8000. This implies that there is some selection or limitation within the Arabidopsis genome of residues constituting the potential 14-3-3 binding sites, potentially constraining the actual consensus sequence. Closer analysis of representative potential targets demonstrates examples of this pattern and preference (Table 2). Clearly, the various potential sequences of the consensus are not represented equally in the Arabidopsis genome.
Table 2.
Number of Hits in Selected Sites
| Site | Hits |
|---|---|
| RKKSKP | 0 |
| RRRSSP | 7 |
| RSPSPP | 6 |
| REPSPP | 2 |
| RSVSPP | 6 |
| KRRSIP | 0 |
| KSASIP | 1 |
| KSPSPP | 3 |
| KSASTP | 2 |
| RSXSXP | 443 |
| RTXSXP | 182 |
To date, binding or regulation of plant proteins by 14-3-3s has required only a single 14-3-3 target sequence in the protein. However, in the mammalian 14-3-3 literature, there is growing interest in structural proteins that appear to bind 14-3-3s in multiple copies and perhaps sequester them. Interestingly, analysis of the potential Arabidopsis proteome 14-3-3 binding partners, with regard to the distribution of the number of putative mode-1 14-3-3 binding sites per protein, indicates that this system may exist in plants as well. Although the majority of the clients possess single 14-3-3 binding sites, >500 contain two sites. Several contain multiple sites, with 13 potential sites in one protein (Figure 7).
From these database analyses, it is apparent that the potential client list for mode-1 14-3-3 interaction is rather large. All of these potential clients contain at least one of the R/KxxSxP binding sites. But the mode-1 site is not the only site at which 14-3-3s interact with their clients. To gain a complete picture of the potential 14-3-3 interactions in Arabidopsis, mode-2 sites and nonphosphorylated interactions also must be considered.
Expanding the Potential Client List
The mode-2 consensus sequence is R/KxxpSxP. Pattern search analysis of the Arabidopsis database revealed that >6000 potential proteins contain at least one mode-2 binding site (Figure 7). Refined searches revealed that ∼900 mode-2 sites also are mode-1 sites, reducing the theoretical sum of potential 14-3-3–interacting proteins to ∼11,000. Some further reduction in this total is necessary because some proteins with multiple sites will have both mode-2 and mode-1 sites. A combined search of mode-1 and mode-2 sites (R/Kx2,3SxP) identified 10,521 targets. Therefore, the total number of potential 14-3-3 mode-1/mode-2 clients includes 40% of all potential Arabidopsis proteins. However, full consideration of the potential client list also must include interactions that are neither mode-1 nor mode-2. The C-terminal binding site of the H+-ATPases remains unique, but it opens the possibility for another family of clients. Nonphosphorylation-dependent interactions also have been reported (Petosa et al., 1998; Halbach et al., 2000; Zhai et al., 2001), creating yet another category of interactions outside of phosphorylated signal transduction. Thus, the potential client list could expand further.
Are we to conclude, then, that 14-3-3 proteins affect the regulated activity of half the proteins in Arabidopsis? Certainly, the potential exists. The mode-1 and mode-2 sites are present in the sequences of Arabidopsis proteins, and kinases that could phosphorylate them abound. There are several factors, however, that will reduce this client list. In terms of protein structure, sites that are buried within the structure of the protein are inaccessible to the 14-3-3s and to the kinases that would phosphorylate them. Biologically, the phosphorylated client would have to be present within the same cellular context and in the proper physiological environment (divalent cation levels, pH, etc.) for the interaction with 14-3-3s to occur. These restrictions undoubtedly will reduce the number of biologically relevant 14-3-3 clients and also will serve as biological checks for determining bona fide in vivo 14-3-3 clients.
PERSPECTIVES
The discovery of 14-3-3 proteins and the elucidation of their roles have prompted an evolution in the understanding of phosphorylation-induced signal transduction. What could be viewed at one time as a relatively simple phosphorylation event now must be viewed as the first step in a multistep process that is mediated by 14-3-3 proteins. Hence, the need to understand the role of 14-3-3s now necessarily accompanies the goals of understanding kinase-induced signal transduction. Historically, the discovery of roles for 14-3-3s in plant physiological regulation has been an ad-hoc, time-consuming, and rate-limiting step that arose from analyses of individual clients. This has led to some gaps in understanding that should be approached with interpretive care, such as the impression that 14-3-3s in animals are involved directly in kinase cascades, whereas 14-3-3s in plants are involved directly in metabolic enzyme regulation. However, the prospect of accelerating the identification process using proteomic analyses of defined groups of genes, coupled with comprehensive organ, tissue, and subcellular localization studies, should provide formidable tools for gaining an accurate understanding of global 14-3-3 interactions and regulation.
As the biological roles of 14-3-3 proteins continue to be elucidated, two fundamental questions should guide experiment design. First, is 14-3-3 diversity biologically relevant, or is diversity a tolerated result of the need to ensure 14-3-3 activity in all cells? Current data suggest that diversity is relevant, but they support no firm conclusion, in part because most client experiments have addressed diversity as a tangential aspect rather than a direct hypothesis. More experiments should assay not only the 14-3-3s that associate with a client but also those 14-3-3s that are present but not associated. Such experimental approaches are necessary to reduce anecdotal observation to tested fact. Second, are 14-3-3 proteins active regulatory molecules that impose another layer of regulation on top of the kinase signal, or are they passive cofactors that blindly attach to their clients and lead to inevitable and nonconditional conclusions? More information is needed on the in vivo state of 14-3-3 molecules with regard to their own modification in cellular contexts, their interaction with cations, and the structural and biochemical effects of these modifications and interactions. Only then can they be defined as relatively simple signaling cofactors or as complex focal points for signal integration and regulation.
Accession Number
The GenBank accession number for the predicted protein possessing 13 potential 14-3-3 target sites is AAF81360.
Acknowledgments
The authors thank the members of our laboratory, both past and present, who have worked directly or indirectly on 14-3-3 projects and who, together with the wider 14-3-3 community, have contributed to the discussion and development of our ideas on 14-3-3 regulation. We also thank Kathy Sehnke for her assistance with the production of the figures for this review. Our work on 14-3-3 proteins is supported by U.S. Department of Agriculture National Research Initiative Grant 00-35304-9601 and National Science Foundation Arabidopsis 2010 Grant MCB-0114501. This is article No. R-08659 of the Florida Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010430.
References
- Abarca, D., Madueno, F., Martinez-Zapater, J.M., and Salinas, J. (1999). Dimerization of Arabidopsis 14-3-3 proteins: Structural requirements within the N-terminal domain and effect of calcium. FEBS Lett. 462 377–382. [DOI] [PubMed] [Google Scholar]
- Aducci, P., Marra, M., Fogliana, V., and Fullone, M.R. (1995). Fusicoccin receptors: Perception and transduction of the fusicoccin signal. J. Exp. Bot. 46 1463–1478. [Google Scholar]
- Aducci, P., Ballio, A., Nasta, D., Fogliano, V., Fullone, M.R., and Marra, M. (1996). Fusicoccin and its receptors. Plant Growth Regul. 18 93–96. [Google Scholar]
- Aitken, A. (1995). 14-3-3 proteins on the MAP. Trends Biochem. Sci. 20 95–97. [DOI] [PubMed] [Google Scholar]
- Aitken, A., Howell, S., Jones, D., Madrazo, J., and Patel, Y. (1995. a). 14-3-3 alpha and delta are the phosphorylated forms of raf-activating 14-3-3 beta and zeta: In vivo stoichiometric phosphorylation in brain at a Ser-Pro-Glu-Lys motif. J. Biol. Chem. 270 5706–5709. [DOI] [PubMed] [Google Scholar]
- Aitken, A., Howell, S., Jones, D., Madrazo, J., Martin, H., Patel, Y., and Robinson, K. (1995. b). Post-translationally modified 14-3-3 isoforms and inhibition of protein kinase C. Mol. Cell. Biochem. 149–150 41–49. [DOI] [PubMed] [Google Scholar]
- Athwal, G.S., Huber, J.L., and Huber, S.C. (1998. a). Biological significance of divalent metal ion binding to 14-3-3 proteins in relationship to nitrate reductase inactivation. Plant Cell Physiol. 39 1065–1072. [DOI] [PubMed] [Google Scholar]
- Athwal, G.S., Huber, J.L., and Huber, S.C. (1998. b). Phosphorylated nitrate reductase and 14-3-3 proteins: Site of interaction, effects of ions, and evidence for an amp-binding site on 14-3-3 proteins. Plant Physiol. 118 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann, M., Huber, J.L., Athwal, G.S., Wu, K., Ferl, R.J., and Huber, S.C. (1996). 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 398 26–30. [DOI] [PubMed] [Google Scholar]
- Bihn, E.A., Paul, A.L., Wang, S.W., Erdos, G.W., and Ferl, R.J. (1997). Localization of 14-3-3 proteins in the nuclei of Arabidopsis and maize. Plant J. 12 1439–1445. [DOI] [PubMed] [Google Scholar]
- Broadie, K., Rushton, E., Skoulakis, E.M., and Davis, R.L. (1997). Leonardo, a Drosophila 14-3-3 protein involved in learning, regulates presynaptic function. Neuron 19 391–402. [DOI] [PubMed] [Google Scholar]
- Bunney, T.D., van Walraven, H.S., and de Boer, A.H. (2001). 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 98 4249–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni, L., Harper, J.F., and Palmgren, M.G. (1998). 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK). FEBS Lett. 430 381–384. [DOI] [PubMed] [Google Scholar]
- Campbell, W.H. (1996). Nitrate reductase biochemistry comes of age. Plant Physiol. 111 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Fu, H., Liu, D., Chang, P.F., Narasimhan, M., Ferl, R., Hasegawa, P.M., and Bressan, R.A. (1994). A NaCl-regulated plant gene encoding a brain protein homology that activates ADP ribosyltransferase and inhibits protein kinase C. Plant J. 6 729–740. [DOI] [PubMed] [Google Scholar]
- Clark, G.J., Drugan, J.K., Rossman, K.L., Carpenter, J.W., Rogers-Graham, K., Fu, H., Der, C.J., and Campbell, S.L. (1997). 14-3-3 zeta negatively regulates raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J. Biol. Chem. 272 20990–20993. [DOI] [PubMed] [Google Scholar]
- Conklin, D.S., Galaktionov, K., and Beach, D. (1995). 14-3-3 proteins associate with cdc25 phosphatases. Proc. Natl. Acad. Sci. USA 92 7892–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotelle, V., Meek, S.E., Provan, F., Milne, F.C., Morrice, N., and MacKintosh, C. (2000). 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 19 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal, S.N., Schweitzer, C.M., Gan, J., and DeCaprio, J.A. (1999). Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 19 4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty, C.J., Rooney, M.F., Miller, P.W., and Ferl, R.J. (1996). Molecular organization and tissue-specific expression of an Arabidopsis 14-3-3 gene. Plant Cell 8 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davezac, N., Baldin, V., Gabrielli, B., Forrest, A., Theis-Febvre, N., Yashida, M., and Ducommun, B. (2000). Regulation of CDC25B phosphatases subcellular localization. Oncogene 19 2179–2185. [DOI] [PubMed] [Google Scholar]
- de Boer, A.H., and Korthout, H.A. (1996). 14-3-3 protein homologues play a central role in the fusicoccin signal transduction pathway. Plant Growth Regul. 18 99–105. [Google Scholar]
- DeLille, J., Sehnke, P.C., and Ferl, R.J. (2001). The Arabidopsis thaliana 14-3-3 family of signaling regulators. Plant Physiol. 126 35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellambra, E., Patrone, M., Sparatore, B., Negri, A., Ceciliani, F., Bondanza, S., Molina, F., Cancedda, F.D., and De Luca, M. (1995). Stratifin, a keratinocyte specific 14-3-3 protein, harbors a pleckstrin homology (PH) domain and enhances protein kinase C activity. J. Cell Sci. 108 3569–3579. [DOI] [PubMed] [Google Scholar]
- de Vetten, N.C., and Ferl, R.J. (1994). Two genes encoding GF14 (14-3-3) proteins in Zea mays: Structure, expression, and potential regulation by the G-box binding complex. Plant Physiol. 106 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., Fox, J.E., and Pei, S. (1996). Identification of a binding sequence for the 14-3-3 protein within the cytoplasmic domain of the adhesion receptor, platelet glycoprotein Ib alpha. J. Biol. Chem. 271 7362–7367. [DOI] [PubMed] [Google Scholar]
- Emi, T., Kinoshita, T., and Shimazaki Ki, K. (2001). Specific binding of vf14-3-3a isoform to the plasma membrane H+-ATPase in response to blue light and fusicoccin in guard cells of broad bean. Plant Physiol. 125 1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl, W.J., Muslin, A.J., Kikuchi, A., Martin, J.A., MacNicol, A.M., Gross, R.W., and Williams, L.T. (1994). Activation of Raf-1 by 14-3-3 proteins. Nature 371 612–614. [DOI] [PubMed] [Google Scholar]
- Ferl, R.J. (1996). 14-3-3 proteins and signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 49–73. [DOI] [PubMed] [Google Scholar]
- Ferl, R.J., Lu, G., and Bowen, B.W. (1994). Evolutionary implications of the family of 14-3-3 brain protein homologs in Arabidopsis thaliana. Genetica 92 129–138. [DOI] [PubMed] [Google Scholar]
- Freed, E., Symons, M., Macdonald, S.G., McCormick, F., and Ruggieri, R. (1994). Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science 265 1713–1716. [DOI] [PubMed] [Google Scholar]
- Fu, H., Coburn, J., and Collier, R.J. (1993). The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 90 2320–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullone, M.R., Visconti, S., Marra, M., Fogliano, V., and Aducci, P. (1998). Fusicoccin effect on the in vitro interaction between plant 14-3-3 proteins and plasma membrane H+-ATPase. J. Biol. Chem. 273 7698–7702. [DOI] [PubMed] [Google Scholar]
- Hachiya, N., Komiya, T., Alam, R., Iwahashi, J., Sakaguchi, M., Omura, T., and Mihara, K. (1994). MSF, a novel cytoplasmic chaperone which functions in precursor targeting to mitochondria. EMBO J. 13 5146–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach, T., Scheer, N., and Werr, W. (2000). Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res. 28 3542–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, A.B., Namboodiri, M.A., Klein, D.C., and Dyda, F. (1999). The structural basis of ordered substrate binding by serotonin N-acetyltransferase: Enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell 97 361–369. [DOI] [PubMed] [Google Scholar]
- Holtman, W.L., Roberts, M.R., and Wang, M. (2000. a). 14-3-3 proteins and a 13-lipoxygenase form associations in a phosphorylation-dependent manner. Biochem. Soc. Trans. 28 834–836. [PubMed] [Google Scholar]
- Holtman, W.L., Roberts, M.R., Oppedijk, B.J., Testerink, C., van Zeijl, M.J., and Wang, M. (2000. b). 14-3-3 proteins interact with a 13-lipoxygenase, but not with a 9-lipoxygenase. FEBS Lett. 474 48–52. [DOI] [PubMed] [Google Scholar]
- Huber, S.C., Bachmann, M., and Huber, J.L. (1996). Post-translational regulation of nitrate reductase activity: A role for Ca2+ and 14-3-3 proteins. Trends Plant Sci. 1 432–438. [Google Scholar]
- Ichimura, T., Isobe, T., Okuyama, T., Yamauchi, T., and Fujisawa, H. (1987). Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett. 219 79–82. [DOI] [PubMed] [Google Scholar]
- Ichimura, T., Sugano, H., Kuwano, R., Sunaya, T., Okuyama, T., and Isobe, T. (1991). Widespread distribution of the 14-3-3 protein in vertebrate brains and bovine tissues: Correlation with the distributions of calcium-dependent protein kinases. J. Neurochem. 56 1449–1451. [DOI] [PubMed] [Google Scholar]
- Ichimura, T., Ito, M., Itagaki, C., Takahashi, M., Horigome, T., Omata, S., Ohno, S., and Isobe, T. (1997). The 14-3-3 protein binds its target proteins with a common site located towards the C-terminus. FEBS Lett. 413 273–276. [DOI] [PubMed] [Google Scholar]
- Igarashi, D., Ishida, S., Fukazawa, J., and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13 2483–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, K., Gotoh, Y., Yashar, B.M., Errede, B., Nishida, E., and Matsumoto, K. (1994). Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science 265 1716–1719. [DOI] [PubMed] [Google Scholar]
- Jahn, T., Fuglsang, A.T., Olsson, A., Bruntrup, I.M., Collinge, D.B., Volkmann, D., Sommarin, M., Palmgren, M.G., and Larsson, C. (1997). The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo, J.A., Capel, J., Leyva, A., Martinez-Zapater, J.M., and Salinas, J. (1994). Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol. Biol. 25 693–704. [DOI] [PubMed] [Google Scholar]
- Jelinek, T., Dent, P., Sturgill, T.W., and Weber, M.J. (1996). Ras-induced activation of Raf-1 is dependent on tyrosine phosphorylation. Mol. Cell. Biol. 16 1027–1034. Erratum Mol. Cell. Biol. 17, 2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, W.M., and Huber, S.C. (2001). Post-translational regulation of nitrate reductase: Mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 52 1981–1989. [DOI] [PubMed] [Google Scholar]
- Kidou, S., Umeda, M., Kato, A., and Uchimiya, H. (1993). Isolation and characterization of a rice cDNA similar to the bovine brain-specific 14-3-3 protein gene. Plant Mol. Biol. 21 191–194. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori, T., and Yamamoto, M. (2000). Members of the Arabidopsis 14-3-3 gene family trans-complement two types of defects in fission yeast. Plant Sci. 158 155–161. [DOI] [PubMed] [Google Scholar]
- Li, W., Skoulakis, E.M., Davis, R.L., and Perrimon, N. (1997). The Drosophila 14-3-3 protein Leonardo enhances Torso signaling through D-Raf in a Ras 1-dependent manner. Development 124 4163–4171. [DOI] [PubMed] [Google Scholar]
- Liao, J., and Omary, M.B. (1996). 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol. 133 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Bienkowska, J., Petosa, C., Collier, R.J., Fu, H., and Liddington, R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376 191–194. [DOI] [PubMed] [Google Scholar]
- Liu, Y.C., Liu, Y., Elly, C., Yoshida, H., Lipkowitz, S., and Altman, A. (1997). Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3-binding motif. J. Biol. Chem. 272 9979–9985. [DOI] [PubMed] [Google Scholar]
- Lu, G., Sehnke, P.C., and Ferl, R.J. (1994). Phosphorylation and calcium binding properties of an Arabidopsis GF14 brain protein homolog. Plant Cell 6 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh, C., and Meek, S.E. (2001). Regulation of plant NR activity by reversible phosphorylation, 14-3-3 proteins and proteolysis. Cell. Mol. Life Sci. 58 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba, M., and Bianchetti, R. (2001). 14-3-3 protein-activated and autoinhibited forms of plasma membrane H+-ATPase. Biochem. Biophys. Res. Commun. 286 984–990. [DOI] [PubMed] [Google Scholar]
- Markiewicz, E., Wilczynski, G., Rzepecki, R., Kulma, A., and Szopa, J. (1996). The 14-3-3 protein binds to the nuclear matrix endonuclease and has a possible function in the control of plant senescence. Cell. Mol. Biol. Lett. 1 391–415. [Google Scholar]
- Maru, Y., and Witte, O.N. (1991). The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell 67 459–468. [DOI] [PubMed] [Google Scholar]
- May, T., and Soll, J. (2000). 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T.A., Zhang, C.L., Lu, J., and Olson, E.N. (2000). Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, B.W., and Perez, V.J. (1967). Specific acidic proteins of the nervous system. In Physiological and Biochemical Aspects of Nervous Integration, F. Carlson, ed (Woods Hole, MA: Prentice Hall), pp. 343–359.
- Moorhead, G., Douglas, P., Morrice, N., Scarabel, M., Aitken, A., and MacKintosh, C. (1996). Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol. 6 1104–1113. [DOI] [PubMed] [Google Scholar]
- Moorhead, G., Douglas, P., Cotelle, V., Harthill, J., Morrice, N., Meek, S., Deiting, U., Stitt, M., Scarabel, M., Aitken, A., and MacKintosh, C. (1999). Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J. 18 1–12. [DOI] [PubMed] [Google Scholar]
- Morrison, D. (1994). 14-3-3: Modulators of signaling proteins. Science 266 56–57. [DOI] [PubMed] [Google Scholar]
- Morrison, D.K. (1995). Mechanisms regulating Raf-1 activity in signal transduction pathways. Mol. Reprod. Dev. 42 507–514. [DOI] [PubMed] [Google Scholar]
- Muslin, A.J., and Xing, H. (2000). 14-3-3 proteins: Regulation of subcellular localization by molecular interference. Cell. Signal. 12 703–709. [DOI] [PubMed] [Google Scholar]
- Obsil, T., Ghilando, R., Klein, D.C., Ganguly, S., and Dyda, F. (2001). Crystal structure of the 14-3-3ζ:serotinin N-acetyltransferase complex: A role for scaffolding in enzyme regulation. Cell 105 257–267. [DOI] [PubMed] [Google Scholar]
- Oecking, C., Eckerskorn, C., and Weiler, E.W. (1994). The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett. 352 163–166. [DOI] [PubMed] [Google Scholar]
- Olsson, A., Svennelid, F., Ek, B., Sommarin, M., and Larsson, C. (1998). A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 118 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petosa, C., Masters, S.C., Bankston, L.A., Pohl, J., Wang, B., Fu, H., and Liddington, R.C. (1998). 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 273 16305–16310. [DOI] [PubMed] [Google Scholar]
- Pietromonaco, S.F., Seluja, G.A., Aitken, A., and Elias, L. (1996). Association of 14-3-3 proteins with centrosomes. Blood Cells Mol. Dis. 22 225–237. [DOI] [PubMed] [Google Scholar]
- Reuther, G.W., Fu, H., Cripe, L.D., Collier, R.J., and Pendergast, A.M. (1994). Association of the protein kinases c-Bcr and Bcr-Abl with proteins of the 14-3-3 family. Science 266 129–133. [DOI] [PubMed] [Google Scholar]
- Rittinger, K., Budman, J., Xu, J., Volinia, S., Cantley, L.C., Smerdon, S.J., Gamblin, S.J., and Yaffe, M.B. (1999). Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4 153–166. [DOI] [PubMed] [Google Scholar]
- Rooney, M.F., and Ferl, R.J. (1995). Sequences of three Arabidopsis general regulatory factor genes encoding GF14 (14-3-3) proteins. Plant Physiol. 107 283–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist, M., Sehnke, P., Ferl, R.J., Sommarin, M., and Larsson, C. (2000). Evolution of the 14-3-3 protein family: Does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51 446–458. [DOI] [PubMed] [Google Scholar]
- Rosenquist, M., Alsterfjord, M., Larsson, C., and Sommarin, M. (2001). Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes: Expression is demonstrated for two out of five novel genes. Plant Physiol. 127 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke, P.C., and Ferl, R.J. (1996). Plant metabolism: Enzyme regulation by 14-3-3 proteins. Curr. Biol. 6 1403–1405. [DOI] [PubMed] [Google Scholar]
- Sehnke, P.C., and Ferl, R.J. (2000). Plant 14-3-3s: Omnipotent metabolic phosphopartners. Science's STKE. http://www.stke.org/cgi/content/full/OC_sigtrans;2000/56/pe1. [DOI] [PubMed]
- Sehnke, P.C., Henry, R., Cline, K., and Ferl, R.J. (2000). Interaction of a plant 14-3-3 protein with the signal peptide of a thylakoid-targeted chloroplast precursor protein and the presence of 14-3-3 isoforms in the chloroplast stroma. Plant Physiol. 122 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke, P.C., Chung, H.J., Wu, K., and Ferl, R.J. (2001). Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 98 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis, E.M., and Davis, R.L. (1996). Olfactory learning deficits in mutants for Leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron 17 931–944. [DOI] [PubMed] [Google Scholar]
- Skoulakis, E.M., and Davis, R.L. (1998). 14-3-3 proteins in neuronal development and function. Mol. Neurobiol. 16 269–284. [DOI] [PubMed] [Google Scholar]
- Szopa, J. (1995). Expression analysis of a Cucurbita cDNA encoding endonuclease. Acta Biochim. Pol. 42 183–189. [PubMed] [Google Scholar]
- Toker, A., Sellers, L.A., Amess, B., Patel, Y., Harris, A., and Aitken, A. (1992). Multiple isoforms of a protein kinase C inhibitor (KCIP-1/ 14-3-3) from sheep brain: Amino acid sequence of phosphorylated forms. Eur. J. Biochem. 206 453–461. [DOI] [PubMed] [Google Scholar]
- Toroser, D., Athwal, G.S., and Huber, S.C. (1998). Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 435 110–114. [DOI] [PubMed] [Google Scholar]
- Toyooka, K., Muratake, T., Tanaka, T., Igarashi, S., Watanabe, H., Takeuchi, H., Hayashi, S., Maeda, M., Takahashi, M., Tsuji, S., Kumanishi, T., and Takahashi, Y. (1999). 14-3-3 protein eta chain gene (YWHAH) polymorphism and its genetic association with schizophrenia. Am. J. Med. Genet. 88 164–167. [PubMed] [Google Scholar]
- van Heusden, G.P., Griffiths, D.J., Ford, J.C., Chin, A.W.T.F., Schrader, P.A., Carr, A.M., and Steensma, H.Y. (1995). The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229 45–53. [PubMed] [Google Scholar]
- van Heusden, G.P., van der Zanden, A.L., Ferl, R.J., and Steensma, H.Y. (1996). Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Lett. 391 252–256. [DOI] [PubMed] [Google Scholar]
- Wang, W., and Shakes, D.C. (1996). Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43 384–398. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., Isobe, T., Ichimura, T., Kuwano, R., Takahashi, Y., Kondo, H., and Inoue, Y. (1994). Molecular cloning of rat cDNAs for the zeta and theta subtypes of 14-3-3 protein and differential distributions of their mRNAs in the brain. Brain Res. Mol. Brain Res. 25 113–121. [DOI] [PubMed] [Google Scholar]
- Weiner, H., and Kaiser, W.M. (1999). 14-3-3 proteins control proteolysis of nitrate reductase in spinach leaves. FEBS Lett. 455 75–78. [DOI] [PubMed] [Google Scholar]
- Weiner, H., and Kaiser, W.M. (2000). Binding to 14-3-3 proteins is not sufficient to inhibit nitrate reductase in spinach leaves. FEBS Lett. 480 217–220. [DOI] [PubMed] [Google Scholar]
- Wilczynski, G., Kulma, A., and Szopa, J. (1998). The expression of 14-3-3 isoforms in potato is developmentally regulated. J. Plant Physiol. 153 118–126. [Google Scholar]
- Wu, K., Rooney, M.F., and Ferl, R.J. (1997. a). The Arabidopsis 14-3-3 multigene family. Plant Physiol. 114 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Lu, G., Sehnke, P., and Ferl, R.J. (1997. b). The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch. Biochem. Biophys. 339 2–8. [DOI] [PubMed] [Google Scholar]
- Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., and Gamblin, S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376 188–191. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.B., Rittinger, K., Volinia, S., Caron, P.R., Aitken, A., Leffers, H., Gamblin, S.J., Smerdon, S.J., and Cantley, L.C. (1997). The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91 961–971. [DOI] [PubMed] [Google Scholar]
- Yamauchi, T., Nakata, H., and Fujisawa, H. (1981). A new activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+-, calmodulin-dependent protein kinase: Purification and characterization. J. Biol. Chem. 256 5404–5409. [PubMed] [Google Scholar]
- Yang, J., Winkler, K., Yoshida, M., and Kornbluth, S. (1999). Maintenance of G2 arrest in the Xenopus oocyte: A role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 18 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J., Lin, H., Shamim, M., Schlaepfer, W.W., and Canete-Soler, R. (2001). Identification of a novel interaction of 14-3-3 with p190RhoGEF. J. Biol. Chem. 276 41318–41324. [DOI] [PubMed] [Google Scholar]