Abstract
The androgen receptor, like other nuclear receptors, activates target genes by binding to hormone-responsive enhancers. Here we demonstrate that androgen induces robust recruitment of androgen receptor, members of the p160 coactivator family, and CREB-binding protein/p300 specifically at the distant enhancer of prostate-specific antigen (PSA) gene. Unexpectedly, we found that RNA polymerase II (Pol II) is directly recruited to the enhancer in a hormone-dependent manner, independent of the proximal promoter, and that the isolated PSA enhancer can mediate efficient androgen induction of transcription. Inhibition of the Pol II carboxyl-terminal domain kinase activity with low concentrations of flavopiridol blocks Pol II transfer from the enhancer to the promoter and selectively abolishes PSA induction by androgen. Moreover, elevated levels of the p160 coactivator ACTR/AIB1 increase both androgen-dependent and -independent PSA expression, by facilitating Pol II recruitment to the enhancer. These results support a model in which nuclear receptors and their coactivators mediate hormone induction by serving as a staging platform for Pol II recruitment.
Androgen is a key regulator of cell growth and differentiation in male sexual development and function as well as in the progression of prostate cancer. These hormonal effects are mediated by the androgen receptor (AR), a member of the nuclear receptor superfamily, which consists of ligand-dependent transcription factors (1). Recently, a growing number of nuclear proteins have been found to associate with AR and are postulated to mediate transcriptional control by the receptor (2, 3). Among them are members of the p160 coactivator family, including SRC-1, TIF2/GRIP1, and ACTR (AIB1/RAC3/TRAM1/pCIP). The p160 coactivators associate with nuclear receptors in a hormone-dependent fashion primarily through the central receptor–interaction domain that harbors several LXXLL motifs.
Although p160 coactivators possess intrinsic histone acetylase (HAT) activities, they are also capable of recruiting other HAT proteins such as CREB-binding protein (CBP), p300, and PCAF (4, 5), and the nuclear protein arginine methyltransferases CARM1 and PRMT (6, 7). Both the acetylases and methylases can regulate transcription by modifying nucleosomal and non-nucleosomal nuclear proteins (7, 8). However, how the p160 coactivators promote RNA polymerase II (Pol II) transcription is still not well understood. Domain mapping experiments suggest that transactivation by the p160 coactivators is largely attributable to their recruitment of CBP/p300 and the HAT activities (4, 9–12). Although earlier experiments indicate that CBP/p300 may interact directly with the Pol II complex (13, 14), this notion lost currency with the discovery of the importance of the HAT activity in transcriptional activation and the more recent demonstration that the simple recruitment and assembly of Pol II machinery in mammalian cells is not sufficient for productive transcription (15, 16).
Previously, we and others demonstrated that hormone induces histone hyperacetylation at the proximal promoters of nuclear receptor target genes presumably through the recruitment of p160 and p300/CBP coactivator complexes (8, 17). To further understand the molecular underpinnings of hormone induction of gene expression, we analyzed androgen-induced recruitment of AR and its associated coactivators to the entire 5′ regulatory region of the prostate-specific antigen (PSA) gene in hormone-responsive LNCaP cells. High-level androgen induction of PSA expression is conferred by an enhancer centered at ≈−4.2 kb on the PSA gene, which contains multiple tandem androgen response elements (18–20). Here, we show that on androgen stimulation, the p160 and CBP/p300 coactivators are preferentially recruited to the enhancer. This observation has led to our unexpected finding that Pol II is directly recruited to the enhancer in an androgen-dependent manner and that Pol II carboxyl-terminal domain (CTD) kinase activities are critical for the possible tracking of Pol II from enhancer to promoter.
Materials and Methods
Chromatin Protein Association Assays.
Chromatin immuno-precipitation (ChIP) assay was performed with LNCaP cells cultured in hormone-depleted medium for at least 3 days before treatment with ligands. In vitro protein association assay with immobilized reconstituted chromatin and details of the ChIP procedures and materials used are provided in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Transient Transfection.
LNCaP cells were transfected with luciferase reporter pGL3-basic, or its derivatives with PSA upstream sequences, pRL-SV40 Renilla luciferase, along with pCMX-ACTR or pSh-CMV-TIF2, by using the Lipofectin reagent. Cells were then treated with 10 nM 5α-dihydrotestosterone (DHT) for 24 h before harvest for luciferase assay. The pGL3-PSA reporters were constructed by inserting restriction fragments of the PSA upstream sequences with the indicated lengths as follows: 5.85-kb entire PSA upstream sequences, HindIII to HindIII; 630-bp PSA proximal promoter sequences, EcoRI to HindIII; 1.5-kb sequence containing the PSA enhancer, XbaI to BamHI; and 2.0-kb fragment containing the PSA enhancer and an immediate downstream sequence, XbaI to StuI.
Recombinant Adenovirus Vectors and Gene Expression Analysis.
ACTR cDNA was inserted into a modified pShuttle-CMV vector with 3× hemagglutinin tag sequence at the C terminus. The resulting constructs were used to generate recombinant adenovirus as described (8). Viruses were purified by centrifugation in CsCl step gradients. Viral titers were determined by end-point cytopathic effect assay with GFP adeno-vector as a reference. Gene expression was compared by using RT-PCR. Total RNA was prepared with the TRIzol reagent, and the cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase. Gene-specific primers for the PCR are provided in Supporting Text.
Results
AR and p160-CBP/p300 Coactivators Are Preferentially Recruited to the PSA Enhancer on Androgen Stimulation.
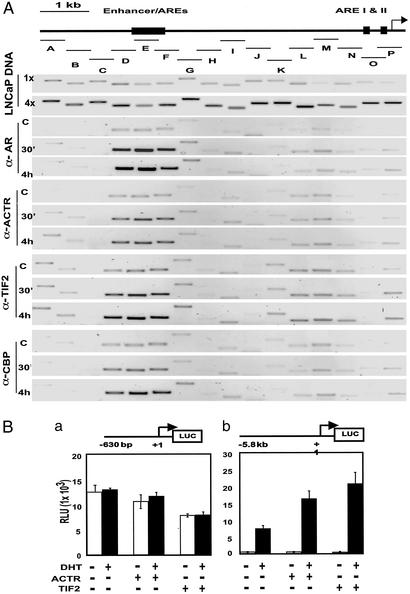
To investigate the role of AR-associated coactivators in PSA induction, we first examined androgen-induced, endogenous AR and coactivator occupancy over the entire 6-kb upstream regulatory region of PSA in LNCaP cells by ChIP assay. For semiquantitative measurement of the precipitated DNA, we used a panel of PCR primer pairs with approximately equal efficiency to amplify partially overlapping fragments (≈300–500 bp) encompassing the 6-kb region (Fig. 1A Top). The enhancer sequence was amplified as fragments D, E, and F, whereas the sequence of proximal AREs and initiation site was covered by fragments O and P. ChIP assay with anti-AR antibody demonstrated clearly that, on androgen induction, AR was strongly recruited to the enhancer region and this AR occupancy persisted hours after DHT addition (Fig. 1A, α-AR). Interestingly, the association of AR with the proximal promoter was only marginal after DHT stimulation (Fig. 1A and Fig. 6, which is published as supporting information on the PNAS web site). In the same experiments, we also measured the recruitment of p160 coactivators and coregulators CBP/p300. Indeed, robust recruitment of ACTR, TIF2, and CBP/p300 to the PSA 5′ regulatory region was observed when cells were treated with DHT. Importantly, as seen for AR, androgen-induced coactivator recruitment was highly restricted to the enhancer (Fig. 1A). Next, we transfected LNCaP cells with two reporters containing either the 6-kb upstream sequence or the 630-bp proximal sequence of PSA. Results in Fig. 1B indicate that the enhancer-containing 6-kb upstream sequence, not the proximal promoter, was capable of mediating androgen induction. In agreement with the ChIP results, cotransfection of coactivator ACTR and TIF2 constructs further potentiated androgen-stimulated transcription through the 6-kb sequence, but not the 630-bp promoter (Fig. 1B). Taken together, these results suggest that the androgen-responsive enhancer of the PSA gene mediates transcriptional activation through direct recruitment of AR and the p160-CBP/p300 coactivator complex.
Figure 1.
AR and p160-CBP/p300 coactivators are preferentially recruited to the PSA enhancer to mediate androgen activation of PSA promoter in LNCaP cells. (A) DHT induces the recruitment of AR and p160-CBP/p300 coactivators preferentially at the PSA enhancer. A diagram of the 6-kb upstream regulatory region of AR target gene PSA with androgen-responsive enhancer and elements indicated above and the various fragments amplified in the ChIP assay marked below as A to P. The amplification efficiency of the primer pairs for the different fragments was assessed by PCR under the same cycling condition by using two different volumes (1× and 4×) of diluted LNCaP cell genomic DNA prepared during the ChIP procedures. Indicated antibodies were used to perform ChIP assay with LNCaP cells treated with 10 nM DHT for 30 min or 4 h, or mock-treated (C) for 4 h, to measure chromatin occupancy of AR, ACTR, TIF2, and CBP along the 6-kb sequence of PSA. Input chromatin DNA taken before immunoprecipitation from different samples was analyzed by PCR and found to be identical (data not shown). (B) LNCaP cell transfection was performed to analyze the ability of PSA enhancer and/or promoter to mediate androgen transcriptional induction and coactivation function. LNCaP cells in hormone-depleted media were cotransfected with either the 630-bp PSA proximal promoter-Luc reporter (a) or the 5.85-kb entire PSA upstream regulatory sequence-Luc (b) and the ACTR or TIF2 expression plasmids. Transfected cells were then treated with 10 nM DHT (filled bars) or mock-treated (open bars) for 24 h before measuring luciferase activities.
Pol II Is Directly Recruited at PSA Enhancer, Independent of the Proximal Promoter, on Androgen Induction.
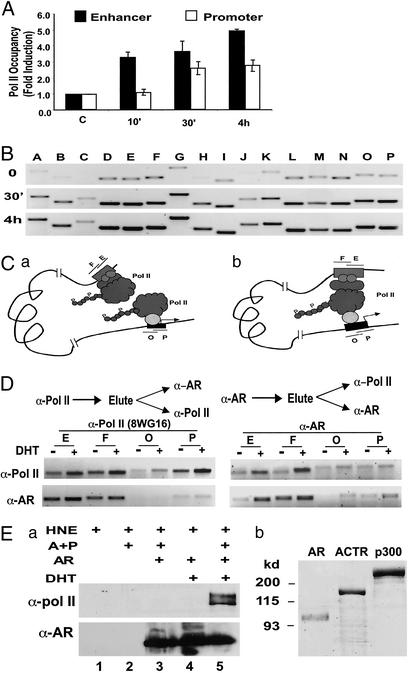
Transcription factors and their coregulators are believed to regulate transcription, in part, by controlling the recruitment and/or assembly of the general transcription machinery (16). We, therefore, examined how Pol II is recruited to the upstream sequence of the PSA gene in response to androgen stimulation. In agreement with our previous results that hormones induce Pol II recruitment to the promoters of estrogen receptor target genes (8), significant recruitment of Pol II to the PSA promoter region is observed on androgen induction (Fig. 2A, open bars). Intriguingly, strong androgen-dependent Pol II cross-linking to the enhancer was also detected. In fact, the DHT-induced Pol II cross-linking at the enhancer is more abundant than that at the promoter (Fig. 2A, filled bars). This unexpected result prompted us to extend the analysis to the other region of the PSA regulatory sequence. As shown in Fig. 2B, remarkably, a significant level of Pol II cross-linking was observed over the region between enhancer and promoter. However, when regions further upstream were analyzed, little Pol II occupancy was detected (Fig. 7, which is published as supporting information on the PNAS web site).
Figure 2.
Pol II is directly recruited to the PSA enhancer on androgen stimulation. (A) ChIP assay was performed with α-Pol II antibody (N-20) to analyze the Pol II occupancy at the enhancer (fragment E, filled bars) or the promoter (fragment P, open bars) of the PSA gene. The relative amount of Pol II occupancy in the presence versus absence of 10 nM DHT (fold induction) was determined by quantifying the PCR products obtained from three experiments. (B) Pol II occupancy over the entire PSA upstream regulatory sequence was analyzed by using ChIP assay as in A with the monoclonal α-Pol II antibody 8WG16. (C) Two possible modes of androgen-induced Pol II occupancy at the PSA 5′ regulatory region: Pol II is recruited to the enhancer and promoter independently (Left) or Pol II is recruited to the promoter and physically interacts with protein complex formed at the enhancer (Right). (D) Sequential ChIP assay was used to analyze the association of Pol II and AR at the PSA promoter (O and P) or the enhancer (E and F). LNCaP cells were treated with 10 nM DHT (+) or mock-treated (−) for 1 h before ChIP assay. Chromatin fragments were first immunoprecipitated with α-Pol II (Left) or α-AR antibody (Right). Immunocomplexes were eluted from the agarose beads and diluted for a second immunoprecipitation for AR or Pol II occupancy analysis. (Ea) Immobilized nucleosomal PSA enhancer fragment (−4500 to −3750) was incubated with HeLa nuclear extract (HNE), in the presence or absence of purified AR, ACTR and p300 (A+P) proteins shown in b, and DHT as indicated, followed by extensive washes and Western blotting with indicated antibodies. (b) Coomassie staining of purified AR (100 ng), ACTR (500 ng), and p300 (500 ng) proteins.
An attractive model of how an enhancer influences the function of the transcription machinery involves the physical interactions of protein complexes assembled at the enhancer with those at the promoter, leaving a DNA loop in between (i.e., the looping model). Formally, the cross-linking of Pol II to the enhancer can be attributed either by the contact of protein complexes formed at the enhancer with Pol II assembled at the promoter or by the occupancy of Pol II at the enhancer independent of promoter (Fig. 2C). The first scenario would predict an efficient cross-linking of AR and p160-CBP/p300 complex to the promoter sequence, which was not observed in our analysis (Fig. 1). To further discern the two possibilities, we took advantage of our observation that AR is recruited primarily to the enhancer. If Pol II is directly recruited to the enhancer, it would physically co-occupy the enhancer with AR. Therefore, fragmented chromatin–protein adducts precipitated by anti-Pol II antibody should contain the enhancer chromatin fragment that is also cross-linked with AR. As shown in Fig. 2D Left, when the eluate of anti-Pol II precipitates were reprecipitated with anti-AR antibody, indeed, the second precipitates contained the enhancer DNA. In contrast, no significant amount of promoter sequence can be detected, in accordance with the low occupancy of AR at promoter. When the same eluate of Pol II precipitates was reprecipitated with anti-Pol II antibody, strong hormone-dependent cross-linkings were seen at both enhancer and promoter, indicating that the failure to detect promoter sequence in the anti-AR reimmunoprecipitation is not caused by the preferential loss of promoter chromatin fragment during the elution. Again, consistent with the notion that AR and Pol II co-occupy enhancer but not promoter, a sequence from the enhancer but not promoter was detected after the anti-AR precipitates were subsequently immunoprecipitated with the Pol II antibody (Fig. 2D Right).
To determine whether the occupancy of Pol II at the enhancer is independent of the promoter sequence of PSA, we incubated the PSA enhancer chromatin assembled in vitro and immobilized on beads with HeLa nuclear extract in the presence or absence of purified AR, ACTR, and p300 proteins. After extensive washing, proteins associated with the chromatin were detected by Western blotting. Results in Fig. 2E show that both phosphorylated and unphosphorylated Pol II (top and bottom bands, respectively, in lane 5) was recruited to the enhancer only in the presence of DHT, and the AR, ACTR, and p300 proteins. Interestingly, AR is recruited to the enhancer independent of DHT. Taken together, these results indicate that, on androgen stimulation, Pol II is directly recruited to the enhancer, independent of the PSA promoter.
The Isolated Enhancer Mediates Strong Androgen Induction of Transcription Independent of PSA Proximal Promoter.
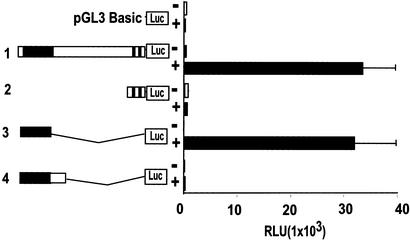
Our observation that Pol II is directly recruited to the PSA enhancer on androgen induction suggests that Pol II might function to initiate transcription through the enhancer. To test this idea, we performed transfection assay with promoterless reporter constructs containing different PSA 5′ upstream sequences. Remarkably, when transfected into LNCaP cells, the reporter linked to a PSA 5′ fragment containing essentially the core enhancer sequence (construct 3) mediated a robust DHT-induced reporter gene activity similar to the reporter that contains the entire 5′ PSA regulatory sequences (construct 1, Fig. 3). Surprisingly, however, a similar reporter containing 500-bp extra sequences downstream of the enhancer (construct 4) was completely inactive in response to DHT induction, suggesting that at least part of the enhancer function is controlled by an enhancer-context-dependent mechanism. By Northern analysis with a 1.5-kb fragment containing the enhancer sequence as a probe, we were unable to detect any mature transcripts initiated around the enhancer region in DHT-treated LNCaP cells (data not shown), indicating that the enhancer does not serve as a gene promoter in its natural chromatin context. Nevertheless, these results suggest that the androgen-responsive enhancer of the PSA gene not only can independently recruit Pol II, but also has the potential to mediate transcription.
Figure 3.
The PSA enhancer, in isolation, mediates androgen induction of transcription independent of PSA proximal promoter. Transient transfection was performed as in Fig. 1B with the promoterless pGL3 basic-Luciferase reporter, the 5.85-kb PSA upstream regulatory sequence linked to the Luc (lane 1), the 630-bp PSA proximal promoter-Luc (lane 2), the 1.5-kb sequence containing the PSA enhancer-Luc (lane 3), or the 2.0-kb fragment containing the PSA enhancer and immediate downstream sequence (lane 4). Transfected cells were treated with 10 nM DHT (filled bars) or mock-treated (open bars) for 24 h before measuring luciferase activities. RLU, relative luciferase unit.
Pol II CTD Phosphorylation Is Important in Mediating Pol II Transfer from the Enhancer to Promoter.
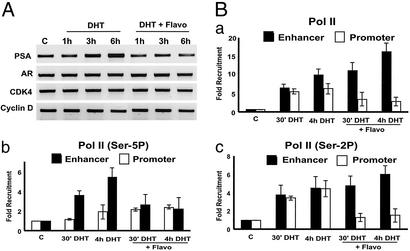
Phosphorylation of Pol II CTD at serines 5 and 2 of the heptad repeats is important in multiple stages of transcription, particularly the transition of Pol II function from promoter arrest to elongation (21). We thus examined whether Pol II–CTD phosphorylation could play a role in the conversion of Pol II recruited at the enhancer to the one functioning at the promoter. To this end, we resorted to the CTD kinases inhibitor flavopiridol (22, 23). We first analyzed the effect of flavopiridol on androgen-induced PSA expression by Pol II. Consistent with the previous reports that low concentration (<200 nM) of flavopiridol does not show inhibitory effect on most gene transcription (23), expression of AR itself and cell cycle genes such as Cdk 4 and cyclin D was not significantly affected by the treatment with 100 nM flavopiridol. In contrast, under the same condition, DHT-induced PSA transcription was completely blocked (Fig. 4A). We then examined the effect of flavopiridol treatment on androgen-induced Pol II recruitment/occupancy at PSA gene (Fig. 4B). Strikingly, treating cells simultaneously with DHT and flavopiridol strongly inhibited androgen-induced Pol II recruitment at the promoter while increasing the Pol II occupancy at the enhancer. Further analyses with antibodies recognizing Pol II phosphorylated at CTD Ser-2 and Ser-5 indicate that the decrease of recruitment in Ser-2 phosphorylated Pol II accounts largely for the overall decrease of Pol II recruitment at PSA promoter by flavopiridol treatment (Fig. 4B). This effect was not seen at the promoter of other genes whose expression was not affected by the low concentration of flavopiridol (data not shown). Because blocking transcriptional initiation or elongation would leave more Pol II assembled and stalled at the promoter, as demonstrated for the IL-8 gene (24), the decrease in Pol II occupancy of the PSA promoter by flavopiridol cannot be attributed to the blockade of transcription initiation or elongation at the promoter. Our finding that decreased Pol II occupancy at the promoter is accompanied by its simultaneously increased presence at the enhancer suggests that Pol II recruited at the enhancer of PSA transfers to the promoter and that flavopiridol perturbs this process by inhibiting CTD kinase activity.
Figure 4.
CTD kinases are involved in possible Pol II transfer from enhancer to promoter and androgen induction of PSA. (A) Flavopiridol selectively blocks DHT induction of PSA expression measured by RT-PCR. RNA was isolated from LNCaP cells treated with either 10 nM DHT or DHT plus 100 nM flavopiridol (DHT+Flavo) for the indicated times. (B) Flavopiridol blocks possible Pol II transfer from enhancer to promoter. LNCaP cells were treated as in A for the indicated times. ChIP assay using α-Pol II 8WG16 (recognizes nonphosphorylated and phosphorylated Pol II), H5, and H14 (recognize Pol II CTD phosphorylated at Ser-2 and Ser-5, respectively) was performed as in Fig. 2A to analyze the occupancy of the different phosphorylated forms of Pol II at the enhancer and the promoter.
ACTR Mediates Androgen Induction of PSA Through Facilitating Pol II Recruitment at Enhancer.
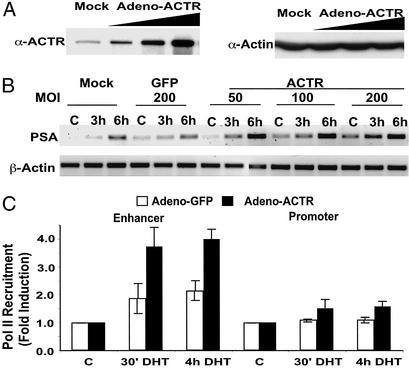
Because p160 coactivators ACTR and TIF2 are recruited primarily at the enhancer of PSA (Fig. 1), we decided to test the hypothesis that p160 coactivators might play a role in androgen-induced Pol II recruitment at the enhancer by ectopically expressing ACTR in LNCaP cells (Fig. 5A). When LNCaP cells were stimulated with a suboptimal concentration of DHT (1 nM), increasing the level of ACTR significantly enhanced both androgen-dependent and -independent PSA expression (Fig. 5B). Importantly, a marked increase in Pol II recruitment was detected by ChIP assay at the enhancer but not the promoter in cells with elevated level of ACTR (Fig. 5C). These results provide the evidence that p160 coactivators such as ACTR are involved in androgen induction of PSA expression by facilitating Pol II recruitment to the enhancer.
Figure 5.
ACTR mediates PSA transcription by increasing Pol II recruitment at the PSA enhancer. (A) LNCaP cells were infected with adenovirus vector expressing ACTR at different multiplicities of infection (mois) (10, 100, and 200). Whole-cell lysates were prepared 48 h after infection and analyzed by Western blot with antibodies against ACTR (BD Biosciences, San Diego) or β-actin. (B) RT-PCR was performed to measure PSA and β-actin expression in LNCaP cells infected with adeno-GFP or adeno-ACTR vectors at different mois and treated with 1 nM DHT for the indicated times before harvest. (C) Elevation of ACTR expression promotes Pol II recruitment to the PSA enhancer. Pol II occupancy at PSA enhancer and promoter was analyzed by ChIP assay with 8WG16 antibody. LNCaP cells were harvested 2 days after infection with adenovirus vectors at moi 100 for either GFP (open bars) or WT ACTR (filled bars) and treated with 1 nM DHT for the indicated times. C, without treatment.
Discussion
Enhancers are discrete DNA elements that mediate the transcriptional activation of DNA binding transcription factors at a distance from the promoter. It has long been speculated that enhancers may function as an entry site for RNA polymerase to relay the transcriptional activation signals impinged at the enhancer to the nearby promoter (25). By analyzing the occupancy of Pol II as well as AR and its coactivators over the entire PSA regulatory region, we found that androgen induces the recruitment of Pol II preferentially to the enhancer. We then demonstrated that AR and Pol II co-occupy strongly at the enhancer but not at the proximal promoter, thereby defying the explanation by the simple looping model in which the enhancer indirectly contacts Pol II complex assembled at the promoter. With highly focused analysis, Shang et al. (26) recently reported the detection of androgen-induced Pol II association with both PSA promoter and enhancer (26). Although their study did not address the molecular nature of such association, their result is consistent with our conclusion that Pol II is directly recruited at the enhancer. More strikingly, using in vitro reconstituted nucleosomal PSA enhancer DNA immobilized on the beads, we obtained results showing unequivocally that Pol II can be recruited to the enhancer independent of the promoter and that the Pol II recruitment at the enhancer entirely depends on the presence of DHT and the purified AR, ACTR, and p300 proteins. Further, the CTD kinase inhibitor flavopiridol promoted Pol II occupancy at the enhancer, with a concomitant block of Pol II occupancy at the promoter. Taken together, these results suggest that the recruitment of Pol II at the enhancer and the promoter may be controlled by distinct mechanisms. Recently, using ChIP assay, it was demonstrated that Pol II occupancy at the locus control region of β-globin genes is regulated by an unidentified mechanism that may differ from that at the β-globin promoter (27). Our finding that, on hormone induction, Pol II is recruited at a conventional enhancer suggests that direct Pol II recruitment may represent a general mechanism underlying enhancer function.
Conceptually, transcription factors and their coactivators or Pol II recruited at a distant enhancer could act on the promoter by either the looping mechanism or tracking along the chromatin (28). Enhancers in mammalian cells are often >5 or 10 kb away from their cognate promoters. Thus, it is less likely that protein–protein contact between enhancer and promoter is achieved simply by looping out the interval DNA. We tend to favor the “facilitated tracking” mechanism for the Pol II complex to interact with the promoter, based on the observations made in this study. Thus, we found that androgen-induced Pol II cross-linking appears to trail over the entire region of the 6-kb PSA upstream sequence. Intriguingly, the pattern of androgen-induced histone hyperacetylation resembles the pattern of Pol II cross-linking (data not shown). It is conceivable that the hyperacetylation of nucleosomal histones might help the transient association of Pol II with chromatin downstream of the enhancer. In this regard, it is tempting to postulate that one of the functions of Pol II CTD phosphorylation might be to facilitate the association of Pol II complex with chromatin remodeling factors.
In an attempt to assess directly the role of p160 coactivators in AR-mediated induction of gene expression, we observed that elevating ACTR level in prostate cancer cells could result in marked increase of both androgen-dependent and -independent PSA expression. Prostate cancer almost inevitably progresses from a hormone-dependent to a hormone-refractory state with elevated levels of PSA. Therefore, a thorough understanding of the mechanism that governs the expression of PSA will shed light on the mechanism underlying the androgen independence of prostate cancer. Recently, aberrant expression of p160 coactivators was identified in multiple human malignancies including prostate cancer (29). It would be of interest to study the involvement of p160 coactivators in the development of prostate cancer androgen independence.
Supplementary Material
Acknowledgments
We are grateful to Drs. Paul Gumerlock and Liang Xia for reagents. This work was supported in part by grants from the Department of Defense (DAMD17-01-1-0033) and the National Institutes of Health (RO1DK60019 and 9RO1DK57978-22). M.C.L. is supported by a National Institutes of Health training grant.
Abbreviations
- AR
androgen receptor
- PSA
prostate-specific antigen
- Pol II
RNA polymerase II
- CTD
carboxyl-terminal domain
- ChIP
chromatin immunoprecipitation
- DHT
5α-dihydrotestosterone
- CBP
CREB-binding protein
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 3.McKenna N J, O'Malley B W. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 5.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 6.Ma H, Baumann C T, Li H, Strahl B D, Rice R, Jelinek M A, Aswad D W, Allis C D, Hager G L, Stallcup M R. Curr Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 7.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson B M, Montminy M, Evans R M. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 9.Kraus W L, Manning E T, Kadonaga J T. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Wong J, Tsai S Y, Tsai M J, O'Malley B W. Proc Natl Acad Sci USA. 2001;98:12426–12431. doi: 10.1073/pnas.231474798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 12.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 14.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorris D R, Struhl K. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemon B, Tjian R. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 17.Shang Y, Hu X, DiRenzo J, Lazar M A, Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 18.Schuur E R, Henderson G A, Kmetec L A, Miller J D, Lamparski H G, Henderson D R. J Biol Chem. 1996;271:7043–7051. doi: 10.1074/jbc.271.12.7043. [DOI] [PubMed] [Google Scholar]
- 19.Cleutjens K B, van der Korput H A, Ehren-van Eekelen C C, Sikes R A, Fasciana C, Chung L W, Trapman J. Mol Endocrinol. 1997;11:1256–1265. doi: 10.1210/mend.11.9.9974. [DOI] [PubMed] [Google Scholar]
- 20.Huang W, Shostak Y, Tarr P, Sawyers C, Carey M. J Biol Chem. 1999;274:25756–25768. doi: 10.1074/jbc.274.36.25756. [DOI] [PubMed] [Google Scholar]
- 21.Orphanides G, Reinberg D. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 22.Rickert P, Corden J L, Lees E. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 23.Chao S H, Price D H. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 24.Barboric M, Nissen R M, Kanazawa S, Jabrane-Ferrat N, Peterlin B M. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 25.Muller H P, Schaffner W. Trends Genet. 1990;6:300–304. doi: 10.1016/0168-9525(90)90236-y. [DOI] [PubMed] [Google Scholar]
- 26.Shang Y, Myers M, Brown M. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 27.Johnson K D, Christensen H M, Zhao B, Bresnick E H. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 28.Blackwood E M, Kadonaga J T. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 29.Gregory C W, He B, Johnson R T, Ford O H, Mohler J L, French F S, Wilson E M. Cancer Res. 2001;61:4315–4319. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.