Abstract
The RPS2 gene in Arabidopsis thaliana governs resistance to strains of the bacterial pathogen, Pseudomonas syringae pv. tomato, that express the avrRpt2 gene. The two loci are involved in a gene-for-gene interaction. Seventeen accessions of A. thaliana were sequenced to explore the diversity present in the coding region of the RPS2 locus. An unusually high level of nucleotide polymorphisms was found (1.26%), with nearly half of the observed polymorphisms resulting in amino acid changes in the RPS2 protein. Seven haplotypes (alleles) were identified and their evolutionary relationships deduced. Several of the alleles conferring resistance were found to be closely related, whereas susceptibility to disease was conferred by widely divergent alleles. The possibility of selection at the RPS2 locus is discussed.
The resistance of plants to pathogen attack is often triggered by the ability of a plant to recognize invading pathogenic organisms. Pathogen recognition is controlled at the genetic level by resistance genes in the plant as well as by avirulence (avr) genes in the pathogen. Single-locus plant resistance genes govern the recognition of pathogens expressing specific recognition factors, whose production is in turn controlled by single avr genes in the pathogen (1, 2). The prevalence of avr genes suggests that these might be of some benefit to the pathogen (refs. 3 and 4; A. Kloek and B.N.K., unpublished data). Plant disease resistance often involves the interaction of these single, dominant, or semidominant genes in a specific “gene-for-gene” relationship; if either member of such a gene pair is not functional or is absent, the interaction may result in disease (1, 2). Although the genetic basis of disease resistance is well established for a large number of plant–pathogen interactions, the fundamental questions of how plant disease resistance genes originate and evolve are the subject of much speculation (2, 5–7).
Several disease resistance loci have been isolated from a variety of plant species (2, 5). Among these is the RPS2 gene of Arabidopsis thaliana, which mediates resistance to strains of the bacterial pathogen Pseudomonas syringae pv. tomato expressing the avr gene avrRpt2 (8, 9). This interaction follows a gene-for-gene relationship in which A. thaliana plants possessing a functional allele at the RPS2 locus are resistant to strains of P. syringae carrying avrRpt2, whereas plants lacking a functional copy of RPS2 are susceptible to P. syringae, regardless of whether the strain carries the avr gene.
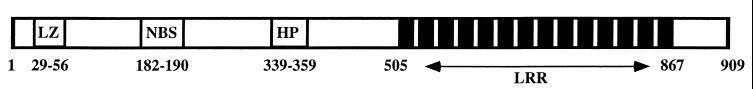
The predicted RPS2 gene product is a protein of 909 amino acids that contains several interesting protein motifs, including a Leu zipper region, a nucleotide-binding site, a small internal hydrophobic domain, and a series of 14 imperfect Leu-rich repeats (Fig. 1; refs. 10 and 11). The presence of these motifs in RPS2 suggests that this protein may be a component of a signaling pathway coupling pathogen recognition to expression of defense responses. RPS2 is a member of a growing class of resistance genes that share a striking amount of structural and organizational similarity (2, 5). The fact that this Leu-rich repeat class of resistance genes includes members from several different plant species that govern resistance to diverse pathogens suggests that disease resistance in a wide variety of plants might be mediated through a common mechanism and that the resistance genes may have a common origin.
Figure 1.
Putative functional motifs found in the predicted RPS2 protein product. The protein is 909 aa long and contains the following motifs: Leu zipper (LZ), nucleotide-binding site (NBS), hydrophobic region (HP), and a series of 14 Leu-rich repeats (LRR). The Leu zipper and Leu-rich repeats are proposed to mediate protein–protein interactions. The position of each motif (amino acid number) is indicated.
Alleles that result in disease-susceptible phenotypes are often thought to be single mutational variants of a wild-type resistance allele. However, there is no direct evidence that susceptible alleles are, in fact, derived from resistant alleles. Little is known about the evolutionary dynamics of these plant-pathogen genetic systems in A. thaliana (or in any plant, for that matter), and the molecular evolution of resistance genes remains largely unexplored. There is scant information about the level of polymorphism and about the evolutionary relationships among the alleles at a given resistance locus. Furthermore, in cases in which multiple alleles for resistance have been found, there is little knowledge about the underlying DNA sequences that yield this variation. Recent studies in the area, however, are increasingly focusing on questions of allelic diversity (12).†
In the current study we examined the molecular evolution of the RPS2 locus in A. thaliana. The study had four objectives: (i) to evaluate the level of polymorphism of the RPS2 gene at the DNA sequence level, (ii) to determine the evolutionary relationships among alleles at this locus by the construction of a gene tree, (iii) to determine the correspondence between resistance/susceptibility phenotypes of plants and the underlying allele sequence, and (iv) to address the possibility of selection at the RPS2 locus.
MATERIALS AND METHODS
Plant Material.
Sixteen accessions (ecotypes) of A. thaliana were analyzed in this study. Six were obtained from the Arabidopsis Biological Research Center, located at Ohio State University. These six were Columbia (Col-0, accession no. CS1092), from the United States; Landsberg erecta (Ler-0, CS20), from Germany; Tsu (Tsu-0, CS1564), from Japan; Wassillewskija (Ws-0 and CS1602), from Russia; Würzburg (Wü-0, CS1614) from Germany; and Zu-0 (CS1626), from an unspecified location. A seventh, unidentified accession, designated UIE132, was isolated as a contaminant in a mutant screen (B.N.K., unpublished data). Preliminary sequence analysis revealed more than one RPS2 allele in the Zu-0 accession sampled. Thus, nine inbred lines (Zu-0-1 to Zu-0-9) from this accession were generated for further analysis by planting selfed seeds from each of nine individual plants derived from the original Zu-0 analyzed. Seeds from the original Zu-0 stock were also regrown to check for the presence of multiple RPS2 alleles. In addition, the disease-susceptible mutant, rps2-201C (derived from Col-0; ref. 8), was included in this analysis. A minimum of nine seeds from each A. thaliana accession was planted in 3-inch pots in soil consisting of 60% Redi-earth (Scotts) and 40% vermiculite and subjected to a 48-h cold treatment. Plants were transferred to a 24°C growth chamber and grown for 4 to 5 weeks under an 8-h photoperiod before analysis.
DNA Sequencing.
Leaves from 4- to 5-week-old plants were harvested and either frozen in liquid nitrogen or dried in silica gel. DNA extraction was carried out with a modified cetyltrimethylammonium bromide mini-prep procedure (14). The RPS2 gene was amplified in two overlapping segments. Two primers external to the ORF and two primers internal to the ORF were designed for this purpose. The two sets of primers used were flank1 (5′-CCTTTAATCTTTATGAGTCAACACCTC-3′) and intern2 (5′-GCTGTTCTGTTGGAGCATCAGTG-3′) to amplify the 5′ end of the gene and intern1 (5′-ATGGCATCTGAACAGGGGAC-3′) and flank2 (5′-TCTCTAGTTTTGTGGCTATGTGGAA-3′) to amplify the 3′ end of the gene. The PCR products were purified by a variation of the method described by Vogelstein and Gillespie (15).
Sequencing was carried out with the cycle-sequencing fmol DNA Sequencing System (Promega). DNA fragments were labeled with 35S-dATP. The fragments were initially denatured for 1 min at 95°C and then subjected to 30 cycles of the following PCR conditions: 30 s at 95°C, 30 s at 55°C, and 1 min at 70°C. A final cycle of 10 min at 72°C was included. A suite of sequencing primers derived from the known Col-0 sequence (GenBank accession no. U14158) and the four amplification primers described above were used to prime DNA synthesis in the sequencing reactions.
Data Analysis.
Alignment of the DNA sequences was done by eye. The relationship between each of the haplotypes (alleles) was used to generate a gene, or haplotype, tree, which graphically illustrates the number of mutational differences separating each haplotype. Each “step” in the tree represents a change of a single nucleotide. Haplotypes are related to each other by parsimony. Lack of homoplasies (character states that have evolved more than once) facilitated the inference of the haplotype tree. Haplotype trees are unrooted, i.e., they have no evolutionary direction, because the events leading to the origin of each haplotype cannot be inferred solely from the available sequence data.
Pathogen Inoculation.
To determine the resistance phenotype conferred by each RPS2 allele, plants from each accession were inoculated with two different strains of P. syringae pv. tomato (Pst): strain PstDC3000, which does not carry avrRpt2, and strain PstDC3000 carrying avrRpt2 on plasmid pV288 [PstDC3000(avrRpt2)] (16). The PstDC3000 strain without the avr gene contained the control vector pVSP61 (16). Plants were inoculated by dipping entire leaf rosettes in a bacterial suspension containing 2–4 × 108 cells per ml (OD600 = 0.2–0.4) and 0.02% Silwet L-77 (OSi Specialties, Danbury, CT) as previously described (8).
RESULTS AND DISCUSSION
Polymorphism at RPS2.
A total of 2,857 nucleotides of the RPS2 gene were sequenced for each of the 17 accessions of A. thaliana. This encompassed the 2,727 nucleotides containing the RPS2 protein-coding region, which contains no introns (10, 11), and a portion of the flanking DNA, including 18 nucleotides at the 5′ end and 107 nucleotides at the 3′ end of the ORF. For two accessions, even though the entire RPS2 gene was present, the complete sequence was not obtained: the Wü-0 sequence is missing 17 nucleotides (2,128–2,144), and the UIE132 sequence is missing 4 nucleotides (2,419–2,422).
Within the entire sequenced region 36 polymorphic sites were observed (Table 1); of these, 19 result in silent substitutions, 5 involve conservative amino acids changes, 11 involve nonconservative amino acid changes, and 1 (Zu-0-7, Zu-0-8) results in a change from tryptophan to a stop codon. Mutations occur throughout RPS2 but are more frequent in the second half of the gene, in the region encoding the Leu-rich repeats (Fig. 1). One of the observed mutations falls outside of the RPS2-coding region (nucleotide 2745; Ws-0). The positions of all nucleotide changes found among the alleles are listed in Table 1. Fourteen of the accessions yielded seven alleles. Three accessions (Zu-0-2, Zu-0-5, and Zu-0-9) contain two different RPS2 alleles, indicating that some original Zu-0 accessions are heterozygous at RPS2. These lines were not used in subsequent analyses.
Table 1.
Nucleotide polymorphisms at RPS2†
| Ecotype‡ | 311*§ | 426 | 461* | 704* | 1092 | 1233 | 1245 | 1255* | 1311 | 1315* | 1326 | 1359 | 1374 | 1438 | 1440 | 1458 | 1543* | 1548 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Col-0¶ (R) | A | A | C | G | A | C | A | T | C | C | C | T | T | T | G | G | G | C |
| Ler-0 (R) | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Tsu-0 (R) | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| Ws-0 (P) | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . |
| Wü-0 (S) | G | C | . | . | T | . | . | C | T | A | . | C | C | C | A | A | . | T |
| Zu-0 (S) | G | C | . | . | T | T | T | C | T | A | . | C | C | C | A | A | . | T |
| Zu-0-1 (S) | G | C | . | . | T | T | T | C | T | A | . | C | C | C | A | A | . | T |
| Zu-0-3 (S) | G | C | . | . | T | T | T | C | T | A | . | C | C | C | A | A | . | T |
| Zu-0-4 (S) | G | C | . | . | T | T | T | C | T | A | . | C | C | C | A | A | . | T |
| Zu-0-6 (S) | G | C | . | . | T | T | T | C | T | A | . | C | C | C | A | A | . | T |
| Zu-0-7 (S) | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Zu-0-8 (S) | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| UIE132 (P) | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| rps2-210C(S) | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 1569* | 1632* | 1634* | 1698 | 1815 | 1911 | 1923 | 1926 | 1931* | 1946* | 1958* | 2002* | 2109* | 2250 | 2334* | 2353* | 2498* | 2745* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | C | C | G | A | C | C | T | A | G | C | A | A | G | C | C | G | G |
| . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . |
| . | . | . | . | . | . | . | . | . | . | . | . | T | . | G | . | C | . |
| . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | C |
| T | A | A | T | G | T | G | A | G | A | T | . | T | T | . | G | C | . |
| T | A | A | T | G | T | G | A | G | A | T | . | T | T | . | G | C | . |
| T | A | A | T | G | T | G | A | G | A | T | . | T | T | . | G | C | . |
| T | A | A | T | G | T | G | A | G | A | T | . | T | T | . | G | C | . |
| T | A | A | T | G | T | G | A | G | A | T | . | T | T | . | G | C | . |
| T | A | A | T | G | T | G | A | G | A | T | . | T | T | . | G | C | . |
| . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| . | . | . | . | . | . | . | . | . | . | . | . | T | . | G | . | C | . |
| . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . |
Nucleotides are numbered beginning at the start of the ORF. Accessions possessing the same nucleotide as Col-0 are marked by dots at that site. Nucleotide changes are indicated by the appropriate letter.
Phenotypes corresponding to each ecotype are indicated in parentheses; R, resistant; S, susceptible; P, partially resistant.
Nucleotides marked by an asterisk indicate sites at which amino acid changes occur.
The Col-0 sequence is used as reference for convenience only.
The RPS2 gene exhibits one of the highest levels of intraspecific sequence polymorphism found in a plant gene to date; 1.26% of nucleotides are polymorphic within RPS2. Moreover, nearly half of the observed polymorphisms result in a change in amino acid composition and, of these, 70% (12 of 17) are nonconservative changes. Comparable levels of sequence polymorphism are rarely found in plants and are most often confined to introns of genes (e.g., ref. 17) or to the ancient, selectively maintained polymorphisms of self-incompatibility alleles (18). Bergelson et al. (19) recently examined levels of polymorphism for a mitochondrial locus and three nuclear genes within a large sample of 115 field collected lines and seven ecotypes of A. thaliana. No variation was found at the mitochondrial locus and a total of 14 polymorphic sites were detected in the 5,346 nucleotides of the three nuclear loci examined in the study. For these combined nuclear genes, the percentage of polymorphic loci within A. thaliana is 0.26% (19), 1 order of magnitude lower than for the RPS2 locus. Thus, the RPS2 sequence appears to be evolving rapidly, both in relation to other plant genes and to other genes in the A. thaliana genome.
RPS2 Gene Tree and Molecular Evolution.
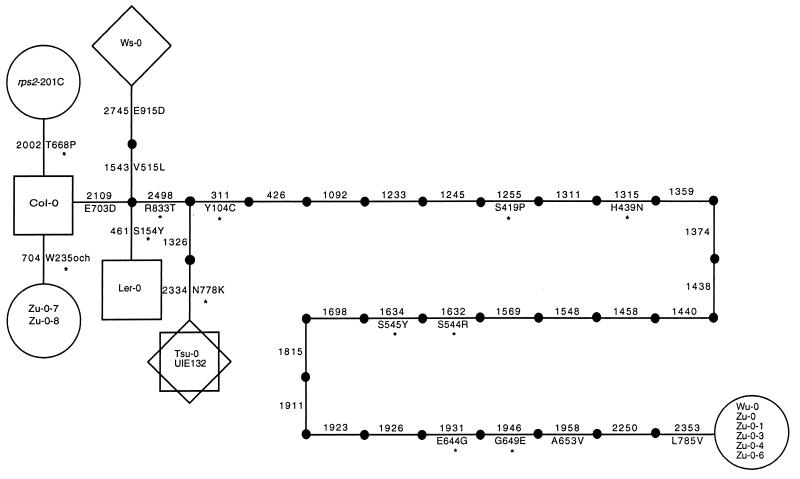
A very straightforward relationship, with no observed homoplasies (convergent or parallel mutations that confound true relationships between genes), is observed among the A. thaliana RPS2 alleles. Fig. 2 depicts the most parsimonious gene tree, which shows point mutational steps between the haplotypes. The tree is unrooted, meaning that no specific allele is identified as the ancestral allele from which other mutational variants are derived. Nearly all accessions yielded a single RPS2 haplotype, suggesting that most accessions are homozygous for a given RPS2 allele. The exception is the original Zu-0 analyzed, which segregates two distinct haplotypes (represented by Zu-0-1 and by Zu-0-7). In two cases, different accessions have the same sequence and, thus, appear to have the same alleles; the Zu-0-1 and Wü-0 sequences are identical, and the Tsu-0 and UIE132 sequences are identical. However, because only the sequence for the coding region was determined, the possibility for differences in the regulatory region of these alleles remains open. It is clear from differences in morphology and phenotypic responses to pathogen infection (see below) that Tsu-0, UIE132, Zu-0-1, and Wü-0 are distinct accessions.
Figure 2.
Gene tree depicting the nucleotide differences between all haplotypes. Each step represents a point mutation leading to a nucleotide change. Numbers refer to the nucleotide where a change occurred (where number 1 defines the first nucleotide in the coding sequence). Amino acid changes are indicated along with the amino acid number. Nonconservative changes are marked by an asterisk. For convenience, the Col-0 sequence is used as a reference for amino acid changes. The tree is unrooted and no evolutionary direction is implied. Phenotypes corresponding to each haploytpe are classified as follows: ○, susceptible; □, resistant; ⋄, partially resistant.
The allele tree is nonsymmetrical. It consists of a cluster of six alleles separated by several short branches of one to four mutational steps, linked to another single allele by a very long branch of 27 mutational steps. This distant allele is found in two distinct accessions, Wü-0 and those represented by Zu-0-1. Several aspects of RPS2 evolution are apparent from the gene tree. First, alternative allelic forms of RPS2 are not necessarily separated by single mutational steps. In this study Col-0 and the allele represented by Zu-0-7 are the only two naturally occurring alleles separated by a single mutation (the rps2-201 allele is derived by mutagenesis from Col-0). The allele present in Wü-0 and Zu-0-1 is highly divergent from other alleles; the mutations that separate this allele from other alleles result in silent, conservative, and nonconservative amino acid changes. Moreover, these data indicate that single accessions of the primarily self-fertilizing A. thaliana, which are presumed to be homozygous, might in fact segregate for evolutionarily distant RPS2 alleles.
To determine whether the types of mutations (synonymous, conservative, nonconservative) are distributed at random along short and long branches of the tree, a Fisher’s exact test was carried out to determine whether branch length is independent of the type of mutation. The type of mutation is not independent of branch length (P = 0.0066 < 0.05). The ratio of nucleotide changes that lead to amino acid substitutions versus silent mutations is higher in short branches than in the long branch (8:1 vs. 9:18). The tree structure indicates that a significantly high level of amino acid replacements cluster together and generate closely related alleles.
Relationship Between Resistance Phenotypes and RPS2 Genotypes.
To determine the resistance phenotype conferred by each RPS2 allele, plants from each accession were inoculated with PstDC3000 and PstDC3000(avrRpt2). All 17 accessions are fully susceptible to PstDC3000 and exhibit typical disease symptoms consisting of small, individual gray lesions surrounded by an area of chlorosis (data not shown). In contrast, the accessions show a variety of different responses when inoculated with PstDC3000(avrRpt2). Three accessions, Col-0, Ler-0, and Tsu-0, are fully resistant to PstDC3000(avrRpt2) and exhibit no disease symptoms. Twelve accessions, rps2-201C, Wü-0, Zu-0, and all nine lines derived from Zu-0, are fully susceptible and exhibit typical disease symptoms. Plants from two accessions, Ws-0 and UIE132, exhibit unusual symptoms when inoculated with PstDC3000(avrRpt2). These consist of large necrotic patches of tissue lacking the characteristic small lesions typical of susceptible plants. These symptoms appear to be indicative of partial resistance in these accessions, because inoculation with high doses of P. syringae expressing avrRpt2 results in delayed activation of defense responses in these plants (data not shown). Additionally, the level of bacterial growth of PstDC3000(avrRpt2) in these plants is intermediate between the high levels of growth observed in fully susceptible tissue and the low levels of growth observed in fully resistant plants (data not shown).
The disease-resistance phenotype of each accession was mapped onto the haplotype tree (Fig. 2). The distribution of phenotypes on the tree is nonrandom. All of the haplotypes conferring resistance belong to a cluster of closely related alleles, which also includes susceptible types. The resistant alleles are separated by only two to four nucleotide changes, and most of these changes result in amino acid substitutions (Table 1 and Fig. 2). Of these, three cases of substitution are conservative and three are nonconservative. Two alleles that confer susceptible phenotypes also fall within this cluster. The first of these, rps2-201, a mutant derived from Col-0, was isolated in a screen designed specifically to identify genes involved in disease resistance (8). The other susceptible haplotype in the cluster is present in five of the accessions derived from Zu-0 (including the three heterozygous Zu accessions) and contains a stop codon at amino acid 235, creating a truncation that disrupts gene function. A third allele that confers a susceptible phenotype is present in Wü-0 and Zu-0-1, -3, -4, and -6. As with the other susceptible phenotypes, there is strong evidence that susceptibility in accessions Wü-0 and Zu-0-1 is specifically caused by a defective RPS2 allele (8). Thus, lack of resistance in all three susceptible classes of accessions can be explained by a defect at RPS2.
Several alternative alleles confer the same phenotypic response, either resistance or susceptibility, on infection with P. syringae. Whereas resistant haplotypes are closely related, susceptible haplotypes can be widely divergent. These results are consistent with the intuitive notion that resistant alleles can tolerate little mutation before compromising their functionality, whereas there are many ways of rendering an allele nonfunctional. However, it is clear that the RPS2 protein can tolerate a significant amount of substitution while retaining function. In the future, as additional A. thaliana RPS2 alleles are analyzed, the distribution of observed resistant and susceptible phenotypes could change. Most interesting would be identification of alleles that can be placed along the long branch leading to susceptibility and determination of whether they are functional or not. One final aspect of phenotypic variation is worth noting. In the case of Tsu-0 and UIE132, the accessions encode the same functional RPS2 protein. However, the response of the plants to infection by PstDC3000(avrRpt2) is substantially different. Tsu-0 is fully resistant, whereas UIE132 is only partially resistant. The presence of such phenotypic variation between these two accessions suggests either that the two RPS2 alleles are regulated differently or that the resistance response in UIE132 is modified by other genetic loci. In support of the latter hypothesis, preliminary genetic analysis of resistance to PstDC3000(avrRpt2) in UIE132 indicates that this accession contains a fully functional allele at the RPS2 locus and that partial resistance is caused by one or more loci that modify RPS2 function (R. Zentella, D. Brooks, and B.N.K., unpublished data).
Selection at the RPS2 Locus.
The presence of a single long branch in a gene tree is not predicted under a model of neutral mutation, where all mutations are selectively equivalent with no differences in fitness. It is possible that the structure of the tree is attributable to sampling artifact. However, the presence of the extremely long branch of the RPS2 gene tree also raises the issue of selection on mutations. Selection is usually difficult to detect, but an attempt was made to use Tajima’s D statistic (20) to test polymorphic sites at the RPS2 locus for neutral mutations. The D statistic compares the average number of pairwise nucleotide differences between DNA sequences (π) with the average number of segregating sites (θ). Both of these measures estimate the amount of genetic variation at a locus, but π takes into account the frequency of each allele in the sample, because pairwise comparisons are done between all sequences. On the other hand, θ is independent of frequency. The expectation is that DNA sequences under selection should show a decline in frequency of deleterious mutants, thus affecting the measure of π. The two measures of genetic variation are expected to be similar only in the absence of selection and other factors that may affect allele diversity (20, 21).
Tajima’s D was calculated with the entire suite of detected mutations. The difference between the estimates of π and θ were nonsignificant (π = 13.444, θ = 13.246, D = 0.076), indicating that the overall level of polymorphism found at the RPS2 locus is consistent with the neutral mutation hypothesis. However, the possibility of selection occurring at this locus cannot be dismissed. This sample is not made up of individuals from a population but from a random sample of accessions from a species. Thus, the accessions do not represent an interbreeding coherent group in which selection processes affect polymorphisms. The accurate measurement of selection awaits a population level study of resistance genes.
There is another reason not to discard the possibility of natural selection occurring at the RPS2 locus. High nucleotide diversity and intermediate frequencies of alternative alleles, as found here, are consistent not only with neutral mutations. Such patterns can also be generated in loci where there is selection for diversity, i.e., where common haplotypes are selected against. In RPS2, the occurrence of selection at the nucleotide level would not be unexpected, because the locus is directly involved with disease resistance, a trait that must have some fitness consequences for the plant. Although the possible nature of selection at disease resistance loci is a matter of speculation, plants and their pathogens form a coevolutionary system, where evolutionary changes in one species leads to evolutionary changes in another. Thus, possible scenarios include frequency-dependent selection or an “evolutionary arms race,” for which selection might favor increased phenotypic and allelic diversity at disease resistance loci to deter or slow selection for loss of avr genes or gain of new virulence genes in the pathogen (6, 7). The high level of diversity and the number of nonconservative substitutions in the RPS2 gene are consistent with such selection. It must be stressed, however, that to understand the exact nature of the evolutionary dynamics of the RPS2 locus more knowledge about wild A. thaliana populations and the polymorphisms they harbor is required. Moreover, the A. thaliana–P. syringae interaction is a model system for studying plant–pathogen interactions (13, 22), and this interaction has not been observed in nature. Thus, there is currently no knowledge about whether this pathogen is a selective force in the natural populations where evolution of this locus has occurred.
The level of diversity found at the RPS2 locus (as well as in other plant resistance genes) will provide a rich opportunity for the study of the evolution of plant–pathogen interactions. In addition, sequence data for different RPS2 alleles offer a unique chance to study both evolutionary and genetic aspects of a known functional gene. Studies in which evolutionary methods to evaluate diversity are combined with knowledge of gene structure and function have great potential for elucidating the molecular mechanisms of disease resistance. By determining the specific amino acid substitutions that alter plant–pathogen interactions, one can make inferences about how the RPS2 gene governs recognition of the pathogen and triggers the defense response in the host. These functional studies, in turn, can help define evolutionary mechanisms that lead to the evolution of plant disease resistance.
Acknowledgments
We thank Juan Carlos Lopez for help with sequencing, Vasantha Aaron, Rodolfo Zentella, David Brooks, and Tara Robertson for help with genetic analysis of UIE132 and Ws-0, Doug Hayworth for helpful discussion, and members of Schaal lab for comments on manuscript. This research was supported by National Institutes of Health Grant GM52536 (to B.N.K).
ABBREVIATIONS
- avr, avirulence gene
π, average number of pairwise nucleotide differences
- θ
average number of segregating sites
Footnotes
Mauricio, R., Stahl, E., Bergelson, J. & Kreitman, M., Ninth International Conference on Arabidopsis Research, June 24–28, 1998, Madison, WI, abstr. 57.
References
- 1.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 2.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 3.Dangl J L. Curr Top Microbiol Immunol. 1994;192:99–118. doi: 10.1007/978-3-642-78624-2_5. [DOI] [PubMed] [Google Scholar]
- 4.Leach J E, White F F. Annu Rev Phytopathol. 1997;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 5.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson J N, Burdon J J. Nature (London) 1992;360:121–125. [Google Scholar]
- 7.Frank S A. Trends Genet. 1992;8:213–219. doi: 10.1016/0168-9525(92)90236-w. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel B N, Bent A F, Dahlbeck D, Innes R W, Staskawicz B J. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu G L, Katagiri F, Ausubel F M. Mol Plant–Microbe Interact. 1993;6:434–443. doi: 10.1094/mpmi-6-434. [DOI] [PubMed] [Google Scholar]
- 10.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 11.Mindrinos M, Katagiri F, Yu G, Ausubel F M. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 12.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Stattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 13. Glazebrook J, Rogers E E, Ausubel F M. Annu Rev Genet. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- 14.Saghai-Maroof M A, Soliman K M, Jorgensen R A, Allard R W. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelstein B, Gillespie D. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whalen M, Innes R, Bent A, Staskawicz B. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strand A E, Leebens-Mack J, Milligan B G. Mol Ecol. 1997;6:113–118. doi: 10.1046/j.1365-294x.1997.00153.x. [DOI] [PubMed] [Google Scholar]
- 18.Franklin F C H, Lawrence M J, Franklin-Tong V E. In Rev Cytol. 1995;158:1–63. [Google Scholar]
- 19.Bergelson J, Stahl E, Dudek S, Kreitman M. Genetics. 1998;148:1311–1323. doi: 10.1093/genetics/148.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajima F. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel B N. Trends Genet. 1996;12:63–69. doi: 10.1016/0168-9525(96)81402-8. [DOI] [PubMed] [Google Scholar]