Abstract
Neurological diseases resulting from proteins containing expanded polyglutamine (polyQ) are characteristically associated with insoluble neuronal inclusions, usually intranuclear, and neuronal death. We describe here oligomeric and polymeric aggregates formed in cells by expanded polyQ. These aggregates are not dissociated by concentrated formic acid, an extremely effective solvent for otherwise insoluble proteins. Perinuclear inclusions formed in cultured cells by expanded polyQ can be completely dissolved in concentrated formic acid, but a soluble protein oligomer containing the expanded polyQ and released by the formic acid is not dissociated to monomer. In Huntington's disease, a formic acid-resistant oligomer is present in cerebral cortex, but not in cerebellum. Cortical nuclei contain a polymeric aggregate of expanded polyQ that is insoluble in formic acid, does not enter polyacrylamide gels, but is retained on filters. This finding shows that the process of polymerization is more advanced in the cerebral cortex than in cultured cells. The resistance of oligomer and polymer to formic acid suggests the participation of covalent bonds in their stabilization.
Kennedy disease (1), Huntington's disease (2), and a number of other neuronal diseases (3) result from a protein containing an expanded series of glutamine repeats. Such a protein, different in each disease, confers a dominant gain of function whose most striking feature is the formation of insoluble aggregates, largely confined to inclusions in neuronal nuclei. These inclusions contain a variety of proteins, in addition to the mutant protein bearing the expanded polyglutamine (polyQ).
Two general mechanisms of stabilization of such protein aggregates have been proposed.
(i) Stabilization by noncovalent bonds: It was proposed by Perutz and coworkers (4) that aggregates are stabilized by hydrogen bonds between main-chain and side-chain amides of expanded polyQ aligned in the form of a β-sheet (polar zipper). Experiments performed in numerous laboratories have given some support to this interpretation, although it does not easily explain the association of expanded polyQ with proteins not bearing polyQ.
(ii) Stabilization by covalent bonds: Studies of involucrin, an epidermal protein that becomes incorporated into a polymeric structure by the action of transglutaminase, revealed that the protein is rich in glutamine (5) and frequently contains glutamine repeats (6, 7). This finding led to the proposal that any protein containing glutamine repeats could be a very good glutamine donor in a transglutaminase-catalyzed reaction with any protein whose lysine residues can participate as amine donors (8).
Our studies of glutamine repeats in synthetic peptides made by solid-phase synthesis showed that increasing the number of consecutive glutamine residues led to increased reactivity of each residue in a transglutaminase-catalyzed reaction; ultimately as many as 80% of the total glutamine residues participated in the reaction (9). Experiments by others showed that the addition of polyQ to the protein GST greatly increased its reactivity as a transglutaminase substrate (10). Increasing the length of synthetic polyQ increased the reactivity per Q residue in transglutaminase-catalyzed coupling to spermine (11). Our experiments on full-length huntingtin containing glutamine repeats of different length showed that, as in the peptides, the activity of huntingtin as a substrate for transglutaminase increased sharply with length of the polyQ (12).
Numerous other reports have provided supporting evidence linking diseases of polyQ to the action of transglutaminase (13–24). The role of transglutaminase has also been challenged (25). What is lacking to prove the role of transglutaminase in these diseases is a direct demonstration of covalently bonded polymers in the inclusions and ultimately the identification of the cross-linking isodipeptide in the polymers. We describe here the presence, in the inclusions of cultured cells making expanded polyQ and in the cortical neurons in Huntington's disease, of oligomers and polymer resistant to dissociation by concentrated formic acid.
The use of concentrated formic acid as a solvent for proteins insoluble in solutions of SDS appears to date from the work of Mokrasch (26). The ability of concentrated formic acid to dissolve aggregates containing polyQ was first demonstrated by Hazeki and coworkers (27). They showed that Cos cells transiently transfected with a construct expressing huntingtin exon 1, encoding expanded polyglutamine, accumulated aggregates of large molecular weight, which could be dissolved by a brief treatment with 100% formic acid. After removal of the formic acid by drying in vacuo, there remained, in addition to the monomeric huntingtin fragment, an expanded polyQ-containing protein of size thought to correspond to a dimer (27).
Methods
Construction of Cell Line PC12-Q205, Which Conditionally Produces Expanded PolyQ.
The plasmid pSG5hAR-M4 (28) contains a synthetically expanded polyQ flanked by the residues adjacent to the polyQ-containing region of the androgen receptor. This construct was kindly provided by H. Nakajima (Osaka Medical College, Osaka). The plasmid was altered to produce Met-(c-Myc)-56AA-205gln-17AA-(6xHis), and then ligated to an upstream ecdysone-inducible promoter derived from pIND (Invitrogen), whose neomycin-resistance gene had been substituted by a blastocidin-resistance gene. This final plasmid was designated as pIBQ205G.
PC12 (ATCC CRL-1721), derived from a pheochromocytoma of the rat adrenal gland, was transfected with plasmid pVgRXR (Invitrogen) and selected for growth in medium containing Zeocin. From the culture, a single colony was isolated twice and regrown. This line expressed the mammalian retinoid x-receptor mRNA as highly as the EcR 293 cells of Invitrogen. The zeocin-resistant PC12 clone was then transfected with pIBQ205G and selected for growth in the medium with blasticidin and Zeocin. After two consecutive colony isolations, a single cell was isolated under direct vision and expanded to obtain the line PC12-Q205, whose expression of Q205 was induced with ponasterone A.
Purification of Inclusions from Cultured Cells.
PC12-Q205 cells were cultivated for 6 days in the presence of 10 μM ponasterone A. The cells were then collected, washed, and suspended in isotonic Hepes buffer. The suspension was centrifuged and the pellet was suspended with the aid of sonication in Tris⋅HCl buffer. DNase I, RNase (Roche Applied Science), and 0.2% Triton X-100 were added and incubated for 1 h at 37°C. A solution of 6.6 M guanidine hydrochloride was added to the suspension with stirring. The suspension was cleared by centrifugation at 800 × g, and the supernatant was then centrifuged at 72,000 × g. The pellet was washed once in 5 M guanidine and once in PBS, suspended in a solution of 1.4 M NaCl containing 2% SDS and 5% 2-mercaptoethanol (2-ME), and boiled for 5 min. The inclusions, which were resistant to all of these treatments, were collected by centrifugation for 30 min at 100,000 × g, washed, and resuspended in water.
Analysis of Expanded PolyQ in the Brain of Patients with Huntington's Disease.
Frozen brain samples from patients with juvenile Huntington's disease were obtained from the Harvard Brain Tissue Resource Center, which is supported in part by Public Health Service Grant MN/NS 31862. Patients were a 16-year-old, grade 3 female, 3123; a 14-year-old, grade 3 female, 3815; an 18-year-old, grade 4 female, 3177; a 22-year-old, grade 4 female, 3482; and a 28-year-old, grade 3 male 3535. The human lymphoblastoid cell line SALDAV 080010, containing normal and expanded huntingtin (Q17/Q79), was provided by the Banque d'ADN et de Cellules de l'Unité 289 de Institut National de la Santé et de la Recherche Médicale (Hôpital de la Pitié-Salpêtrière, Paris).
Western blots.
For analysis of huntingtin, extracts of cortex (50 μg of protein) and cerebellum (80 μg of protein) were analyzed by the standard Western blotting method (29) using 4% acrylamide, unless specified otherwise. Huntingtin was stained by an anti-N-terminal antibody (30), diluted 1:1,500, and by the 1C2 antibody specific for expanded polyQ (31), diluted 1:2,500.
Isolation of nuclei.
Briefly, the gray matter of cortex or cerebellum was homogenized in isotonic sucrose buffer containing Brij. The homogenate was filtered through a 70-μm nylon mesh. The nuclei were sedimented by centrifugation at 850 × g for 10 min at 4°C, and the supernatant was collected as a source of cytoplasmic proteins. The crude nuclear pellet was resuspended and sedimented again under the same conditions. The nuclei in the resulting pellet were further purified by centrifugation through discontinuous gradients of either iodixanol (32) or sucrose (33), using a modification of a procedure described in ref. 34.
For scoring of inclusions, the crude homogenate was stained with the anti-N-terminal antibody and Hoechst 33258. The proportion of nuclei containing inclusions was determined by immunofluorescence microscopy.
Detection of polymer in nuclei isolated from cerebral cortex of Huntington's disease by filter retention assay.
After formic acid treatment of isolated nuclei, undissolved polymer was deposited on cellulose acetate, by using the well-known filter retention assay (35).
Quantitation of Oligomer in Cultured Cells and Oligomer and Polymer in Cerebral Cortex of Huntington's Disease by IR Fluorescence.
Nitrocellulose membranes for Western blots and cellulose acetate filters with retained polymer were incubated with 1C2 antibody, then with Alexa Fluor 680-labeled goat anti-mouse IgG and finally washed in Tris-buffered saline containing 0.05% Tween 20. This near IR fluorophore was excited at 680 nm and the emission at 702 nm was quantitated in channel 700 of the LI-COR Infrared Imaging System (Odyssey, Lincoln, NE). Signal intensity was proportional to the amount of expanded polyQ.
A complete description of the methods is available in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
Solubility in Concentrated Formic Acid of Peptides Containing PolyQ.
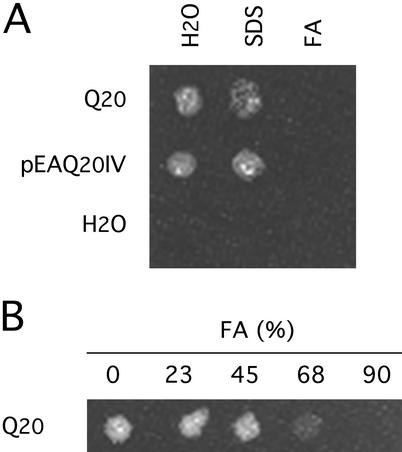
Peptides containing more than six consecutive Q residues made by solid-phase synthesis become increasingly insoluble in water. This is demonstrated for Q20 and p(pyro)EA Q20 IV (36) (Fig. 1). Each peptide was added in amounts sufficient to make a 2-mM solution in 100 μl of water or of a solution containing SDS and incubated for 5 min at 100°C. In both cases, most of the peptide remained insoluble. The addition of formic acid to 90%, instead of SDS, dissolved the peptides completely. The solvent effect of formic acid was appreciable at concentrations of >45% and complete at 90% (Fig. 1).
Figure 1.
Solubility of Q20 peptides in formic acid. Q20 and pEA Q20 IV were added to water and a solution of 2% SDS in an amount sufficient to make a 2-mM solution. The suspensions were centrifuged at 14,000 rpm for 5 min in an Eppendorf centrifuge. Any sediment was resuspended in water, and an aliquot was allowed to evaporate on a glass plate. The residue on the plate was photographed against a black background with overhead illumination. (A) Q20 and pEA Q20 IV were largely insoluble in water or in a solution of SDS. When 90% formic acid (FA) was substituted as solvent, both peptides dissolved, leaving no sediment. (B) Solubility of Q20 in different concentrations of formic acid (FA).
It may be concluded that if the insolubility of peptides containing polyQ is caused by the formation of hydrogen-bonded β-sheet, as proposed by Perutz et al. (4), or of secondary bonds of some other kind (37), these bonds are dissociated in 90% formic acid. The resistance of any polyQ-containing aggregate to 90% formic acid must be ascribed to stronger bonds. For example, epidermal corneocyte envelopes are stabilized by isopeptide cross-links of Nɛ(γ-glutamyl) lysine (38) introduced by transglutaminase (39). We prepared cornified envelopes from the tail and the ear of an adult mouse, by boiling the tissues in a solution containing 2% SDS and 2% 2-ME (40). The envelopes were then sedimented and incubated in 94% formic acid for 2 h at 37°C (106 envelopes per 200 μl). No appreciable difference either in the number or the morphology of the envelopes was detected after incubation in the formic acid. In what follows, we will designate as oligomer an aggregate of polyQ that is soluble but not converted to monomer by concentrated formic acid and resolved as a band of higher molecular weight in gel electrophoresis. We designate as polymer a larger aggregate that is insoluble in formic acid and unable to pass through a cellulose acetate filter of pore size 0.2 μm.
Dynamics of Inclusion Formation in Stably Transfected PC12-Q205 Cells.
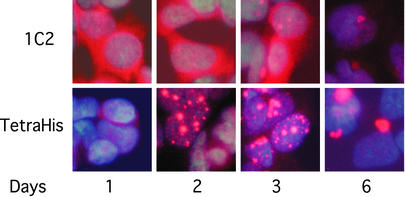
Synthesis of expanded polyQ by PC12-Q205 cells was initiated with ponasterone A. At different times, samples of the cells were stained with antibody to tetra-His, present in the C-terminal tag, and with antibody 1C2 (Fig. 2).
Figure 2.
Appearance and number of inclusions after induction of PC12-Q205 cells. Expression of the construct encoding Q205, flanked by 67 N-terminal and 17 C-terminal residues, followed by 6xHis, was induced from an ecdysone-inducible promotor, using ponasterone A. The 1C2 antibody, which is known to stain inclusions poorly, showed mainly diffuse cytoplasmic staining, declining with time, whereas staining by anti-tetra-His showed mainly inclusions, beginning as early as day 1, first increasing in number with time but later only in size, the number then being reduced.
Within 1 day, 1C2 revealed abundant cytoplasmic staining of expanded polyQ. In succeeding days, the intensity of cytoplasmic staining was reduced, but few stained inclusions were revealed. Intact inclusions are known to be poorly stained by 1C2 (41).
Staining by anti-tetra-His showed a different pattern, in which staining of inclusions predominated. These could be seen as early as day 1, but the inclusions steadily increased in size and number until day 3. The number decreased by day 6, when the inclusions were considerably larger and generally reduced to one per cell. The inclusions were associated mainly with nuclei, but were likely perinuclear rather than intranuclear.
Purification of Inclusions.
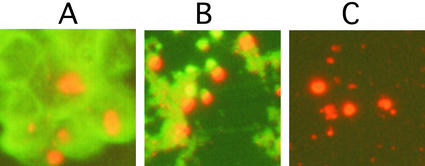
Inclusions were purified on the basis of their insolubility and identified by staining with antibody to c-Myc. We found that a commercial anti-rabbit IgG-FITC (Roche Applied Science) can stain cellular proteins nonspecifically, and we used it to monitor the purity of the inclusions. The sonicated cell suspension contained inclusions associated with abundant contaminating protein (Fig. 3A). Extraction with guanidinium chloride removed most contaminating protein (Fig. 3B) and subsequent extraction with SDS/2-ME removed nearly all of it (Fig. 3C).
Figure 3.
Purification of inclusions. Inclusions harvested 6 days after induction of PC12-Q205 cells were purified. The inclusions were stained with anti-c-Myc coupled indirectly to Cy3 (red) and nonspecific proteins were stained with anti-rabbit IgG-FITC (green). (A) No purification. (B) After extraction with guanidinium chloride. (C) After extraction with SDS/2-ME. (Magnifications: ×300.)
Presence in the Inclusions of Insoluble Monomer and Oligomer Containing Expanded PolyQ.
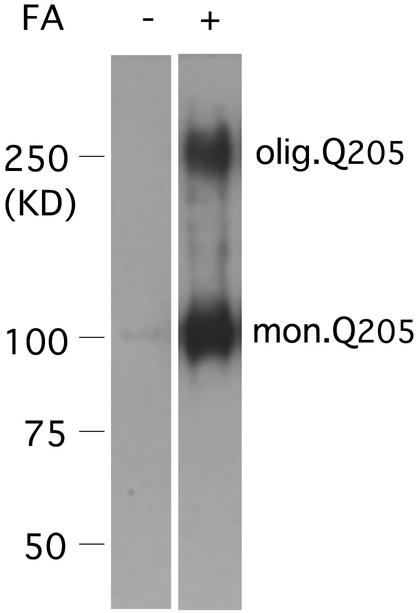
Purified inclusions were submitted to electrophoresis. There was no detectable signal after staining with 1C2 antibody (Fig. 4). This finding is not surprising because the inclusions had been insoluble in SDS/2-ME during their purification and could therefore not be expected to enter a polyacrylamide gel. When the inclusions were first treated with 90% formic acid for 1 h at 37°C, the formic acid removed under vacuum, and the residue neutralized, electrophoresis of the proteins showed an intense band of monomer and a smaller discrete band with mobility equaling 21% that of the monomer. Longer incubations in the presence of formic acid resulted in no further change. The effect of formic acid pretreatment may be explained by its ability to dissociate noncovalently stabilized aggregates otherwise unable to enter the gel, as well as by its ability to enhance immunostaining (42). After filtration of formic acid-treated extract through cellulose acetate filters, no retained aggregate could be detected.
Figure 4.
Synthesis by PC12-Q205 cells of an oligomer containing expanded polyQ. Purified inclusions, with or without formic acid (FA) treatment, were boiled in SDS/2-ME and submitted to electrophoresis through 7.5% polyacrylamide. After transfer to nitrocellulose, the proteins were stained with the antibody 1C2. In the absence of formic acid pretreatment, neither monomer nor oligomer was detected. After pretreatment with 90% formic acid for 1 h at 37°C, electrophoresis revealed a strong band of monomer corresponding to an apparent molecular mass of ≈100 kDa; the true molecular mass should be 36,340, but expanded polyQ reduces mobility in gel electrophoresis. In addition to the monomer, an oligomer with a considerably higher molecular mass is clearly stained by the 1C2 antibody. This film is overexposed to demonstrate the oligomer, but estimation of staining intensity by IR fluorescence showed that the oligomer was present in a range from 0.5% to 3% of monomer. The ratio of the mobility of the oligomer to that of the monomer was 0.21.
In contrast to the insolubility of the oligomer and monomer of the inclusions in the absence of formic acid pretreatment, both monomer and oligomer could readily be detected in the cytoplasm without formic acid pretreatment.
To see whether these results depended on the cell line used, we carried out transient transfections of 293 cells (human embryonic kidney) and examined the distribution of expanded polyQ by electrophoresis with and without formic acid pretreatment. Only after formic acid treatment were monomer and oligomer bands strongly stained. The ratio of the mobility of the oligomer to that of the monomer was 15%, a value similar to that obtained from PC12 cells. We concluded that formation of oligomer did not depend on properties exclusive to PC12 cells. The oligomer did not result from polyubiquitination of the monomer, because an antiubiquitin antibody (Dako), at a 1:1,000 dilution, failed to stain any proteins in the region of the oligomer, whereas it easily detected free ubiquitin and ubiquitinated proteins of red blood cells and lymphoblasts.
As in the case of PC12-Q205 cells, water-soluble monomer and oligomer could be detected in the cytosol.
Absence of Reassociation of Monomer After Removal of Formic Acid.
To be certain that oligomer dissociated by formic acid does not reform after removal of the formic acid, the dried residue after formic acid treatment of inclusions was redissolved in buffer containing SDS with or without neutralization and allowed to stand for various periods. The sample that still retained some acidity was then neutralized and both samples were subjected to electrophoresis. During storage of the formic acid-treated material at 4°C for up to 72 h under either condition, there was no detectable increase in the amount of oligomer. It was also found that when the migration was slowed during electrophoresis by lowering the applied voltage, there was similarly no tendency to reaggregation. These results showed that the dissociation of oligomer in formic acid is indeed stable (27). It is not clear why this should be. In addition to disaggregation of proteins, treatment by concentrated formic acid is known to result in formylation of serine residues (43, 44). There are five serine residues and three threonine residues in the flanking regions of the Q205. We have found that increasing the incubation time in formic acid resulted in more acidity after drying and dissolving in SDS solution. Perhaps this finding suggests that some kind of formylation had occurred during incubation.
The Partition of Expanded PolyQ Between the Cytosolic and Particulate Cell Compartments.
There have been numerous previous studies of the inclusions resulting from proteins with expanded polyQ (37, 45–47). None of these studies dealt with the role of oligomers containing expanded polyQ. We have studied the kinetics of the transfer of monomer and oligomer from the cytosol into the particulate compartment, which contains the inclusions induced in the cell line PC12-Q205. At intervals, duplicate cultures were trypsinized. The cells were washed in PBS containing a mixture of protease inhibitors (Complete, Roche Applied Science) and 100 mM EDTA and were sonicated for 2–3 s twice in 10 mM Tris⋅HCl buffer, pH 7.5, containing 1 mM EDTA, the same protease inhibitors and 1% Triton X-100. A cytosolic fraction and a particulate fraction containing the inclusions were then separated by centrifugation at 100,000 × g. Each fraction was dissolved in 90% formic acid and taken to dryness in vacuo. The proteins were then incubated in buffered SDS, neutralized, heated to 100°C for 5 min, and subjected to electrophoresis in SDS. After transfer to nitrocellulose and staining with the 1C2 antibody, the amounts of expanded polyQ in the monomer and oligomer bands were quantitated by near IR fluorescence.
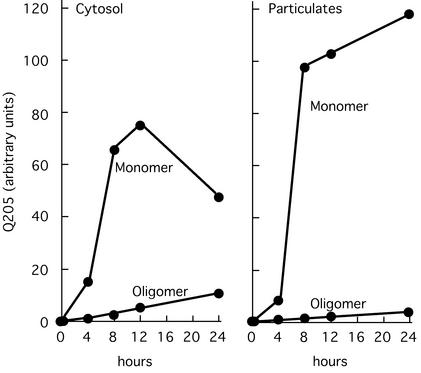
At 4 h after induction, monomer and oligomer both could be detected in the cytosol and the particulates (Fig. 5). In the cytosol, the amount of monomer reached its maximum at 12 h and then declined sharply (and continued to do so in longer term experiments), whereas monomer continued to increase in the particulates. The amount of oligomer in the youngest inclusions (4–8 h) equaled 30% of the oligomer in the entire cytosol. This pattern shows that oligomer participates in inclusion formation from the very beginning of the process; with time, both oligomer and monomer were removed from the cytoplasm into the inclusions, where the amount of monomer ultimately became 50 times larger than the amount of oligomer.
Figure 5.
Kinetics of appearance of monomer and oligomer in cytosol and particulates of induced PC12-Q205 cells. One day after inoculation, PC12-Q205 cells were induced by 10 μM ponasterone A and harvested by trypsinization after 0, 4, 8, 12, and 24 h of induction. Sonicated extracts were fractionated into soluble and particulate fractions (containing the inclusions) and both fractions were analyzed for Q205 by Western blotting. The amount of Q205 determined by IR fluorescence is shown per 5 μg of cellular protein. Monomer and oligomer were readily detected in both cytosol and particulates within 4 h of induction. The amount of monomer always exceeded the amount of oligomer, but the particulates account for an appreciable part of the total oligomer at the earliest time points (see text).
Soluble Oligomer Containing Expanded Huntingtin in the Cerebral Cortex of Patients with Juvenile Huntington's Disease.
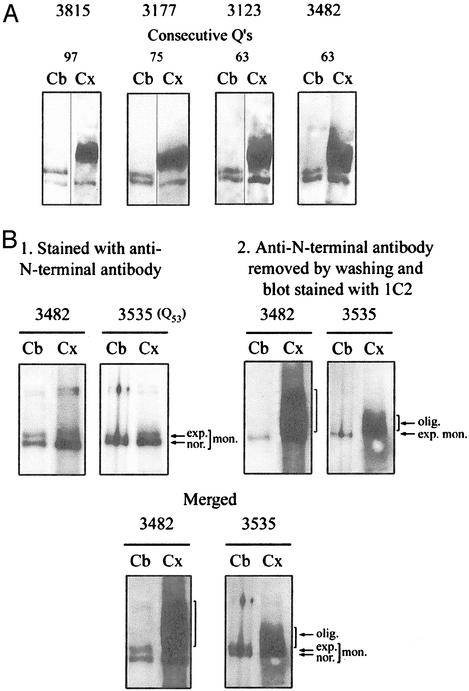
It has been reported (12, 31, 48) that in the cortex of patients with Huntington's disease, expanded huntingtin is replaced by a broad band or smear containing huntingtin and extending upward from the expected position of expanded huntingtin. We prepared extracts of cortex and cerebellum from four patients with juvenile Huntington's disease, obtained from the Harvard Brain Tissue Resource Center. We then dried them and incubated the residue in 96% formic acid for 1 h at 37°C. The free formic acid was evaporated, and the samples were dissolved in gel loading buffer and neutralized with Tris base. The proteins were resolved by PAGE and immunoblotting, using sequential staining by the anti-N-terminal antibody and the 1C2 antibody (Fig. 6A).
Figure 6.
Oligomeric state of soluble expanded huntingtin in cerebral cortex in Huntington's disease. (A) Crude extracts of cortex and cerebellum were treated with 96% formic acid. Western blots prepared after electrophoresis were stained consecutively with anti-N-terminal antibody (30) and 1C2. All specimens of cortex produced a broad oligomeric band of expanded huntingtin absent from the cerebellum. In contrast, all of the cerebellar specimens, but none of the cortical specimens, produced a clear band of monomeric expanded huntingtin. (B) Blots prepared after electrophoresis of formic acid-treated crude brain extracts were photographed after staining with the N-terminal antibody and again after staining with the 1C2 antibody. The expanded monomer, but not the oligomer, is clearly stained by the N-terminal antibody. The broad band of oligomer in the merged figure clearly extends above the corresponding monomer.
In the cerebellar extract, the normal and expanded monomeric huntingtins were detected as expected. But the cortical extract gave a broad band of formic acid-resistant oligomeric protein extending over a molecular mass range of ≈100 kDa and with mean mobility of 0.90 ± SD 0.03 that of the expanded huntingtin of the cerebellum (Fig. 6A). In the absence of formic acid treatment, the appearance of this band was unchanged. The mean mobility of the oligomeric form was not reduced as much with respect to monomeric huntingtin as was the mobility of the oligomer of cultured cells with respect to its monomer.
Sequential staining with the anti-N-terminal and 1C2 antibodies of blots prepared after electrophoresis of crude cortical extracts of cases 3482 and 3535 showed that a small amount of expanded monomer could be detected below the broad band of oligomer (Fig. 6B). We had previously shown by Southern analysis that the expanded cortical allele of case 3482 contained 63 CAGs, or only one more CAG than the corresponding allele of the cerebellum (49). This finding establishes that the broad band of oligomer cannot be explained as the result of cortical mosaicism. The oligomer was not appreciably stained by antibody to the N-terminal sequence (Fig. 6B) or by mAb 2166, directed against sequence C-terminal of the polyQ, or by an antiubiquitin antibody.
Quantitative analysis of the oligomer was carried out by IR fluorescence. The amounts of expanded polyQ in the form of oligomer in cortices 3482 and 3177 were 9.8- and 4.5-fold, respectively, the amount of expanded monomeric huntingtin of the cerebellum. After normalization for unequal content of normal huntingtin monomer in the lanes of cortex and cerebellum (a factor of 1.74 for 3482 and 1.4 for 3177), the values for the ratio of oligomer to monomer were 5.6 and 3.2. (see Table 2, which is published as supporting information on the PNAS web site).
To determine the subcellular location of the polymer, we isolated cortical nuclei by centrifugation through density gradients of sucrose or iodixanol. In the absence of formic acid pretreatment, the oligomer containing expanded polyQ could be detected only in the cortical cytoplasm. But if the isolated nuclei were first treated with formic acid, subsequent electrophoresis revealed a band corresponding to oligomer, another broad band of lower molecular weight, also stained by 1C2 antibody and presumably consisting of fragmented huntingtin, and a prominent band located at the top of the gel. These components are most likely located in the nuclear inclusions (see Fig. 8, which is published as supporting information on the PNAS web site).
Polymer Insoluble in Formic Acid Recovered from the Nuclei of Cerebral Cortex.
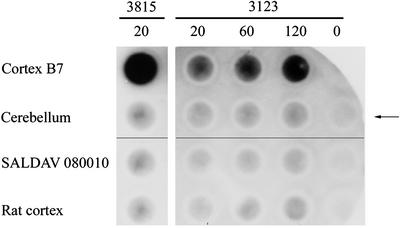
To examine and quantitate the component located at the top of the gel, we isolated nuclei from the cortex of patients 3815 and 3123 by density gradient centrifugation, incubated the nuclei in 96% formic acid, and submitted the samples to the filter retention assay (35, 50). In cortex of patient 3815, which contained inclusions in 20% of the nuclei, the presence of nuclear polymer insoluble in 96% formic acid and of size permitting its retention on the filter was revealed by a dark spot stained with the 1C2 antibody (Fig. 7). In contrast, nuclei prepared from the cerebellum of the same patient contained no inclusions and virtually no stainable polymer. Quantitative assay by IR fluorescence gave a value for cortex >100 times than that of cerebellum (Table 1). The lymphoblastoid cell line SALDAV 080010 and rat cortex gave low values.
Figure 7.
Polymeric state of expanded huntingtin in cerebral cortex in Huntington's disease. Purified nuclei of cortex and cerebellum of brain 3815 and of controls were extracted with 96% formic acid. The suspension was diluted with a 10-fold excess of Tris/SDS/2-ME, boiled for 5 min, and immediately passed through cellulose acetate filters. The retained material was visualized by chemiluminescence by using antibody 1C2 and the ECL plus kit. Control SALDAV 080010 was a lymphoblast line possessing a huntingtin with 79 glutamine residues. Rat cortex, whose huntingtin possesses Q8 (51), should not be stainable by 1C2. Cortex 3815: 20,000 nuclei gave a strong signal. The corresponding cerebellum, SALDAV 080010, and rat cortex gave weak signals. Cortex 3123: 120,000 nuclei produced a weaker signal than 20,000 nuclei of cortex 3815. Arrow indicates control without nuclei. Because chemiluminescence does not provide a quantitative measure of the amount of expanded polyQ, quantitation was performed by using IR fluorescence (see Table 1).
Table 1.
Formic acid-insoluble polymer of expanded polyQ in cortical nuclei of Huntington's disease: Quantitation of the polymer retained on cellulose acetate filters
| Source of polyQ | IR fluorescence, units
|
||
|---|---|---|---|
| Brain 3815, 20,000 nuclei, without background correction | Brain 3123, 120,000 nuclei
|
||
| Before background subtraction | After background subtraction | ||
| Cortex | 59.8 | 31.0; 40.9 | 25.2; 35.9 |
| Cerebellum | 0.5 | 8.8; 13.0 | 3.0; 8.0 |
| SALDAV 080010 | 0.6 | 5.2; 5.2 | 0; 0.2 |
| Rat brain | 2.7 | 5.8; 5.0 | —; — |
For brain 3123, two independent determinations were done.
Cortex 3123 was found to contain many fewer inclusions than cortex 3815. Counts of inclusions in 1,068 nuclei of cortex 3123 revealed their presence in only 1.96% of nuclei. Chemiluminescent staining was consistent with this difference (Fig. 7). Quantitation by IR fluorescence showed that the value for polymer of cortex 3123 was about one-tenth that of cortex 3815. Accordingly, 120,000 nuclei were used for its accurate measurement, and the sensitivity of the instrument had to be increased. In two independent experiments under these conditions, cortex 3123 gave highly significant values for retained polymer but the control values were appreciable for SALDAV 080010 and rat brain, whose huntingtin contains only Q8 (51) (Table 1). Because the value for IR fluorescence of rat brain did not increase with the number of nuclei applied, this value could be subtracted as background correction. When this was done, values for cortex were further elevated with respect to those of cerebellum. The cerebellar values, although quite low, may be real, whereas those for SALDAV 080010 were reduced to background. We may conclude that the nuclear inclusions in the cortex of juvenile cases of Huntington's disease contain a formic acid-insoluble polymer.
Concluding Remarks
Using 90–96% formic acid to dissociate aggregates stabilized by noncovalent bonds, we have demonstrated the existence of three types of aggregate-containing expanded polyQ: (i) an oligomer released by formic acid from inclusions produced by cultured cells synthesizing Q205; (ii) a water-soluble oligomer present in the cytoplasm of cerebral cortex affected by juvenile Huntington's disease but not in the cytoplasm of the corresponding cerebellum; this oligomer is also present in an insoluble form in nuclei (presumably their inclusions), from which it is released by formic acid; and (iii) a formic acid-insoluble polymer of too large a size to pass through a 0.2-μm filter, present in the nuclei of the same cortices, but not in the corresponding cerebellum or in lymphoblasts or in cultured cells synthesizing Q205. The amount of this polymer correlates with the number of nuclear inclusions. The oligomer of the nuclei is likely to be a precursor of this polymer.
Supplementary Material
Acknowledgments
This work was aided by a consortial grant from the National Institute of Neurological Disorders and Stroke (5 RO1 NS38566-03) (to H.G. and P.D.).
Abbreviations
- polyQ
polyglutamine
- 2-ME
2-mercaptoethanol
References
- 1.La Spada A R, Wilson E M, Lubahn D B, Harding A E, Fischbeck K H. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Gusella J F, MacDonald M E. Nat Rev Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 4.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert R L, Green H. Cell. 1986;46:583–589. doi: 10.1016/0092-8674(86)90884-6. [DOI] [PubMed] [Google Scholar]
- 6.Delhomme B, Djian P. Gene. 2000;252:195–207. doi: 10.1016/s0378-1119(00)00237-7. [DOI] [PubMed] [Google Scholar]
- 7.Djian P, Phillips M, Easley K, Huang E, Simon M, Rice R H, Green H. Mol Biol Evol. 1993;10:1136–1149. doi: 10.1093/oxfordjournals.molbev.a040069. [DOI] [PubMed] [Google Scholar]
- 8.Green H. Cell. 1993;74:955–956. doi: 10.1016/0092-8674(93)90718-6. [DOI] [PubMed] [Google Scholar]
- 9.Kahlem P, Terré C, Green H, Djian P. Proc Natl Acad Sci USA. 1996;93:14580–14585. doi: 10.1073/pnas.93.25.14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper A J, Sheu K R, Burke J R, Onodera O, Strittmatter W J, Roses A D, Blass J P. J Neurochem. 1997;69:431–434. doi: 10.1046/j.1471-4159.1997.69010431.x. [DOI] [PubMed] [Google Scholar]
- 11.Gentile V, Sepe C, Calvani M, Melone M A B, Cotrufo R, Cooper A J, Blass J P, Peluso G. Arch Biochem Biophys. 1998;352:314–321. doi: 10.1006/abbi.1998.0592. [DOI] [PubMed] [Google Scholar]
- 12.Kahlem P, Green H, Djian P. Mol Cell. 1998;1:595–601. doi: 10.1016/s1097-2765(00)80059-3. [DOI] [PubMed] [Google Scholar]
- 13.Chun W, Lesort M, Tucholski J, Faber P W, MacDonald M E, Ross C A, Johnson G V. Neurobiol Dis. 2001;8:391–404. doi: 10.1006/nbdi.2001.0390. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 15.Lesort M, Attanavanich K, Zhang J, Johnson G V. J Biol Chem. 1998;273:11991–11994. doi: 10.1074/jbc.273.20.11991. [DOI] [PubMed] [Google Scholar]
- 16.de Cristofaro T, Affaitati A, Cariello L, Avvedimento E V, Varrone S. Biochem Biophys Res Commun. 1999;260:150–158. doi: 10.1006/bbrc.1999.0851. [DOI] [PubMed] [Google Scholar]
- 17.Karpuj M V, Garren H, Slunt H, Price D L, Gusella J, Becher M W, Steinman L. Proc Natl Acad Sci USA. 1999;96:7388–7393. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeitner T M, Bogdanov M B, Matson W R, Daikhin Y, Yudkoff M, Folk J E, Steinman L, Browne S E, Beal M F, Blass J P, Cooper A J. J Neurochem. 2001;79:1109–1112. doi: 10.1046/j.1471-4159.2001.00673.x. [DOI] [PubMed] [Google Scholar]
- 19.Cariello L, de Cristofaro T, Zanetti L, Cuomo T, Di Maio L, Campanella G, Rinaldi S, Zanetti P, Di Lauro R, Varrone S. Hum Genet. 1996;98:633–635. doi: 10.1007/s004390050273. [DOI] [PubMed] [Google Scholar]
- 20.Cooper A J, Wang J, Pasternack R, Fuchsbauer H L, Sheu R K, Blass J P. Dev Neurosci. 2000;22:404–417. doi: 10.1159/000017470. [DOI] [PubMed] [Google Scholar]
- 21.Mastroberardino P G, Iannicola C, Nardacci R, Bernassola F, de Laurenzi V, Melino G, Moreno S, Pavone F, Oliverio S, Fesus L, Piacentini M. Cell Death Differ. 2002;9:873–880. doi: 10.1038/sj.cdd.4401093. [DOI] [PubMed] [Google Scholar]
- 22.Lesort M, Tucholski J, Miller M L, Johnson G V. Prog Neurobiol. 2000;61:439–463. doi: 10.1016/s0301-0082(99)00052-0. [DOI] [PubMed] [Google Scholar]
- 23.Karpuj M V, Becher M W, Springer J E, Chabas D, Youssef S, Pedotti R, Mitchell D, Steinman L. Nat Med. 2002;8:143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 24.Dedeoglu A, Kubilus J K, Jeitner T M, Matson S A, Bogdanov M, Kowall N W, Matson W R, Cooper A J, Ratan R R, Beal M F, et al. J Neurosci. 2002;22:8942–8950. doi: 10.1523/JNEUROSCI.22-20-08942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun W, Lesort M, Tucholski J, Ross C A, Johnson G V. J Cell Biol. 2001;153:25–34. doi: 10.1083/jcb.153.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mokrasch L C. Biochim Biophys Acta. 1965;107:608–610. doi: 10.1016/0304-4165(65)90207-2. [DOI] [PubMed] [Google Scholar]
- 27.Hazeki N, Tukamoto T, Goto J, Kanazawa I. Biochem Biophys Res Commun. 2000;277:386–393. doi: 10.1006/bbrc.2000.3682. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima H, Kimura F, Nakagawa T, Furutama D, Shinoda K, Shimizu A, Ohsawa N. J Neurol Sci. 1996;142:12–16. doi: 10.1016/0022-510x(96)00142-6. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Hoffner G, Kahlem P, Djian P. J Cell Sci. 2002;115:941–948. doi: 10.1242/jcs.115.5.941. [DOI] [PubMed] [Google Scholar]
- 31.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 32.Kegel K B, Meloni A R, Yi Y, Kim Y J, Doyle E, Cuiffo B G, Sapp E, Wang Y, Qin Z H, Chen J D, et al. J Biol Chem. 2002;277:7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 33.Valenzuela S M, Martin D K, Por S B, Robbins J M, Warton K, Bootcov M R, Schofield P R, Campbell T J, Breit S N. J Biol Chem. 1997;272:12575–12582. doi: 10.1074/jbc.272.19.12575. [DOI] [PubMed] [Google Scholar]
- 34.Vakakis N, Hearn M T, Veitch B, Austin L. J Neurochem. 1991;57:307–317. doi: 10.1111/j.1471-4159.1991.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 35.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 36.Parameswaran K N, Velasco P T, Wilson J, Lorand L. Proc Natl Acad Sci USA. 1990;87:8472–8475. doi: 10.1073/pnas.87.21.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Berthelier V, Yang W, Wetzel R. J Mol Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 38.Rice R H, Green H. Cell. 1977;11:417–422. doi: 10.1016/0092-8674(77)90059-9. [DOI] [PubMed] [Google Scholar]
- 39.Rice R H, Green H. Cell. 1979;18:681–694. doi: 10.1016/0092-8674(79)90123-5. [DOI] [PubMed] [Google Scholar]
- 40.Sun T T, Green H. Cell. 1976;9:511–521. doi: 10.1016/0092-8674(76)90033-7. [DOI] [PubMed] [Google Scholar]
- 41.Sieradzan K A, Mechan A O, Jones L, Wanker E E, Nukina N, Mann D M. Exp Neurol. 1999;156:92–99. doi: 10.1006/exnr.1998.7005. [DOI] [PubMed] [Google Scholar]
- 42.Kitamoto T, Ogomori K, Tateishi J, Prusiner S B. Lab Invest. 1987;57:230–236. [PubMed] [Google Scholar]
- 43.Orlando R, Kenny P T, Zagorski M G. Biochem Biophys Res Commun. 1992;184:686–691. doi: 10.1016/0006-291x(92)90644-z. [DOI] [PubMed] [Google Scholar]
- 44.Klunk W E, Pettegrew J W. J Neurochem. 1990;54:2050–2056. doi: 10.1111/j.1471-4159.1990.tb04910.x. [DOI] [PubMed] [Google Scholar]
- 45.Hazeki N, Nakamura K, Goto J, Kanazawa I. Biochem Biophys Res Commun. 1999;256:361–366. doi: 10.1006/bbrc.1999.0337. [DOI] [PubMed] [Google Scholar]
- 46.Kazantsev A, Walker H A, Slepko N, Bear J E, Preisinger E, Steffan J S, Zhu Y-Z, Gertler F B, Housman D E, Marsh J L, Thompson L M. Nat Genet. 2002;30:367–376. doi: 10.1038/ng864. [DOI] [PubMed] [Google Scholar]
- 47.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schilling G, Sharp A H, Loev S J, Wagster M V, Li S H, Stine O C, Ross C A. Hum Mol Genet. 1995;4:1365–1371. doi: 10.1093/hmg/4.8.1365. [DOI] [PubMed] [Google Scholar]
- 49.Kahlem P, Djian P. Neurosci Lett. 2000;286:203–207. doi: 10.1016/s0304-3940(00)01029-6. [DOI] [PubMed] [Google Scholar]
- 50.Wanker E E, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 51.Holzmann C, Maueler W, Petersohn D, Schmidt T, Thiel G, Epplen J T, Riess O. Biochem J. 1998;336:227–234. doi: 10.1042/bj3360227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.