Summary
Transcriptional elongation and termination by RNA polymerase (RNAP) are controlled by interactions among the nascent RNA, DNA, and RNAP that comprise the ternary transcription elongation complex (TEC). To probe the effects of co-transcriptionally-folded RNA hairpins on elongation as well as the stability of the TEC, we developed a single-molecule assay to monitor RNA elongation by Escherichia coli RNAP molecules while applying controlled loads to the nascent RNA that favor forward translocation. Remarkably, forces up to 30 pN, twice those required to disrupt RNA secondary structure, did not significantly affect enzyme processivity, transcription elongation rates, pause frequencies, or pause lifetimes. These results indicate that ubiquitous transcriptional pausing is not a consequence of the formation of hairpins in the nascent RNA. The ability of the TEC to sustain large loads on the transcript reflects a tight binding of RNA within the TEC and has important implications for models of transcriptional termination.
Introduction
Transcription constitutes the first step in gene expression and is highly regulated. Regulation of the elongation phase of transcription is mediated, in part, by interactions in the TEC involving the RNA (Artsimovitch and Landick, 2000). Elongation is frequently interrupted by pauses of varying durations, at least some of which play regulatory roles. Pausing can act as a governor to slow rates of polymerization, helping to synchronize transcription and translation in prokaryotes (Landick et al., 1985), bind cofactors to modify transcription (Artsimovitch and Landick, 2002; Bailey et al., 1997; Marr and Roberts, 2000), and facilitate the cotranscriptional folding of transcripts (Pan and Sosnick, 2006). Two classes of defined pauses with regulatory functions have been identified. Hairpin-stabilized pauses occur when self-complementary RNA structures form at the exit channel and help inhibit nucleotide addition (Artsimovitch and Landick, 2000; Toulokhonov et al., 2001; Toulokhonov and Landick, 2003). Backtracking pauses occur when the enzyme encounters a weak RNA:DNA hybrid, biasing the enzyme to move upstream on the DNA template and extrude the nascent RNA into the nucleotide entry channel (Komissarova and Kashlev, 1997; Reeder and Hawley, 1996).
Frequent, short-lifetime (<20 s) pauses have been identified in single-molecule experiments (Adelman et al., 2002; Neuman et al., 2003). These “ubiquitous” pauses have been found to be sequence-dependent (Herbert et al., 2006) and independent of RNAP backtracking (Neuman et al., 2003; Shaevitz et al., 2003). Although hairpins are predicted to form frequently in mRNA (Rivas and Eddy, 2000) and are known to stabilize some pauses (Chan and Landick, 1993), the extent to which they contribute to ubiquitous pausing is unknown.
RNA polymerase must maintain a tight association with the growing RNA while extending it for thousands of nucleotides, yet readily release the transcript upon recognition of specific termination sequences or the binding of termination factors (von Hippel, 1998). RNA is bound to the TEC via an 8–9 nt RNA:DNA hybrid formed in the active-site cleft (Korzheva et al., 1998). Crosslinking and structural studies suggest that RNA is further stabilized by protein contacts (Gnatt et al., 2001; Korzheva et al., 2000). The extensive contacts between DNA and RNAP enable the DNA to sustain large external loads (Neuman et al., 2003; Wang et al., 1998). The nascent RNA chain might support less force than the DNA, in principle, given that the enzyme makes significantly fewer stabilizing contacts to the RNA – roughly one-third of the number made with DNA (Gnatt et al., 2001). However, the stability of the TEC against mechanical disruption of the RNA has not previously been probed.
TEC stability is dramatically modulated during termination, ultimately leading to release of the bound transcript. In prokaryotes, termination follows the formation of a stable RNA hairpin followed by a uridine- (U-) rich tract, and consequently a weak RNA:DNA hybrid (Yarnell and Roberts, 1999), or from the action of Rho protein, which translocates along the nascent RNA until it reaches the polymerase, whereupon it induces transcript dissociation (Richardson, 2002). In either case, transcript release is conjectured to be caused by forces exerted on the RNA, produced either by hairpin folding or by Rho displacement, leading to forward translocation of the enzyme in the absence of continued RNA synthesis (Park and Roberts, 2006; Yarnell and Roberts, 1999). Alternatively, termination may be produced by an allosteric mechanism, where the hairpin or Rho factor binds to polymerase and destabilizes the TEC (Toulokhonov et al., 2001). By applying an external force to RNA, one can probe differences between direct mechanical and indirect allosteric effects.
To gauge the strength of RNA interactions with the TEC and ascertain the effects of RNA secondary structure on transcription, we developed a new variant of the single-molecule transcription assay for E. coli RNAP. In our experimental geometry, force is applied by an optical trap directly to the elongating RNA chain, as opposed to the DNA template. Employing this assay, we found that RNAP continues to transcribe despite comparatively high loads applied to the RNA, indicating that force alone (up to 30 pN) did not induce termination. Force-extension curves of transcripts demonstrate that RNA secondary structure was completely disrupted for forces higher than 18 pN, in broad agreement with single-molecule measurements performed on RNA and DNA hairpins. Transcriptional pauses were characterized over a range of forces applied to the RNA, and compared directly to pauses in an assay where assisting load was applied instead to template DNA. Pause lifetime and probability distributions measured in these two assays are statistically indistinguishable, suggesting that RNA structure exerts comparatively little influence on ubiquitous transcriptional pausing.
Results
RNA-pulling Assay Shows the TEC Can Sustain High Loads
Force was applied between the bead-attached polymerase molecule and the nascent RNA, mediated by a DNA molecule hybridized to the RNA that served as a ‘handle’ (Figure 1A). We also employed a second assay (Neuman et al., 2003) where force was applied between the polymerase and the upstream end of the DNA template (Figure 1B). In both cases, a stalled TEC, comprised of biotin-tagged RNAP (Tolic-Norrelykke et al., 2004), template DNA, and a 29-nt–long nascent RNA transcript, was bound to an avidin-coated bead. For the RNA-pulling assay, the 5′ end of the RNA was hybridized to a complementary overhang on a double-stranded DNA (dsDNA) handle molecule that was bound at its distal end to the coverglass through a digoxigenin-antibody linkage. The DNA template remained free at both ends. For the DNA-pulling assay, the template DNA was attached directly to the coverglass surface at its upstream terminus: in this case, the growing RNA chain remained free at its 5′ end.
Figure 1. Cartoon of experimental assays and data records (not to scale).

(A) The RNA-pulling assay. RNAP (green) transcribing a DNA template (blue) is attached to a bead via an avidin-biotin linkage (yellow/black). Nascent RNA (red) from the polymerase is hybridized to a 3-kb-long DNA handle (green) via a 25-base overhang; the distal end of the handle is attached via a digoxigenin-antibody linkage (black/purple) to the coverglass of a flow cell mounted on a 3D piezo stage. The optical trap (pink) holds the bead at a fixed offset from the trap center, producing a constant restoring force. The stage is moved by feedback to compensate for any elongation of the tether.
(B) The DNA-pulling assay for assisting load. The upstream end of the DNA template is attached to the coverglass surface via a digoxigenin-antibody linkage; the nascent RNA remains unbound and free to form secondary structure.
(C) 9 representative transcription records (of N = 202) from the RNA-pulling assay (red), showing elongation of the nascent RNA (in nt) vs. time. Note transcriptional pauses.
(D) 6 representative transcription records (of N = 87) from the DNA-pulling assay (blue), showing progress along the DNA (in bp) vs. time. Note transcriptional pauses.
For these experiments, transcription was restarted in stalled TECs by the addition of saturating levels of ribonucleotides to surface-tethered complexes (1 mM NTPs), after which a bead was optically trapped and its position monitored during elongation. Data were acquired using a feedback arrangement that maintained constant force on the RNAP by displacing the stage to compensate for transcriptional motion. Records obtained with the RNA-pulling assay were qualitatively similar those from the DNA-pulling assay (compare Figures 1C, 1D), with periods of uniform elongation rate interspersed with pauses. Lengthy transcription records were obtained for loads on the RNA as high as 30 pN; beyond this, the frequency of tether rupture increased dramatically, likely reflecting a disruption of the antibody linkage. A monoclonal variant of this linkage is known to break quickly at forces near 24 pN (Neuert et al., 2006). In contrast, the 25 bp RNA:DNA hybrid between the nascent chain and the handle is expected to support shearing forces in excess of ~40 pN (Lang et al., 2004), while the biotin-avidin linkage supports forces in excess of ~160 pN (Florin et al., 1994).
When sufficiently high tension was applied, progressive lengthening of the RNA was observed (F = 22 pN; Figure 2A). However, when the force was abruptly reduced, the apparent transcript length decreased dramatically and remained nearly invariant (F = 7 pN; Figure 2A). Once force was restored, this length returned to a level commensurate with the value extrapolated from the period prior to force reduction, indicating that RNA elongation had continued throughout the interval of low force at the same rate. From this result, we conclude that the precipitous drop in extension is caused by the formation of extensive secondary (and possibly tertiary) structure in the RNA, causing the growing chain to develop a highly compact shape at low loads.
Figure 2. Secondary structure in the nascent RNA is suppressed at high force.
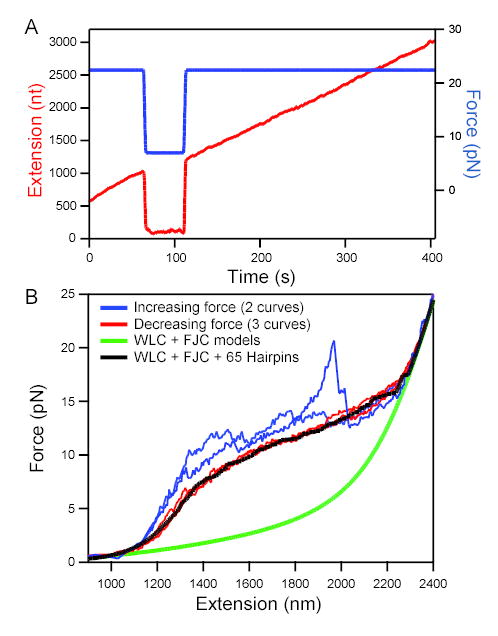
(A) RNA extension (red; left axis) vs. time at high and low loads. Elongation took place at an initial force of 22 pN (blue; right axis). When force was reduced to 7 pN at 75 s, apparent elongation ceased. When high force was restored at 110 s, extension returned to the position extrapolated from earlier elongation, showing that transcription had continued during the low-force period with no significant change in extension as secondary structure formed.
(B) Force-extension curves (FECs) for a tether that had stalled prematurely (taken at ~120 nm/s). Stretching curves (increasing force; 2 examples shown) displayed distinct features with each pull (blue traces). In contrast, relaxation curves (decreasing force; 3 examples shown) displayed very reproducible behavior (red traces). Model FECs: A worm-like chain (WLC, representing dsDNA) plus a freely-jointed chain (FJC, fit to 2,200 nt ssRNA) fits the data only in low and high force limits (green line). The additional effect of forming 65 random hairpins (loop 4 nt; Gaussian distribution of stem lengths centered at 10.5 bp, std. dev. 3 bp) reproduces the plateau seen in the experimental data (black line) (see Supplemental Material).
Secondary Structure is Removed Above F = 18 pN
To quantitate RNA elongation, we needed to establish the force sufficient to disrupt most secondary structure. Force-extension curves (FECs) for RNA transcribed by a complex that had stalled prematurely were measured. Curves obtained using monotonically increasing force showed a series of jagged sawtooth features that were not reproducible in successive pulls (Figure 2B). Such features, reaching up to 20 pN, correspond to the opening of secondary structures that form in the absence of tension (Onoa et al., 2003). By contrast, curves obtained with monotonically decreasing force were consistently reproducible and displayed a broad force plateau for intermediate loads, reminiscent of the plateaus for ssDNA (Dessinges et al., 2002), the Tetrahymena ribozyme (Onoa et al., 2003) and E. coli 16S rRNA structures (Harlepp et al., 2003).
To model the force plateau and determine the force at which RNA secondary structure is disrupted, we computed theoretical FECs in the absence or presence of numerous hairpins formed at random in a load-dependent manner, based on the elastic properties of the single- and double-stranded nucleic acids comprising the RNA-dsDNA tether (Figure 2B; see also Supplementary Material). This simple model reproduces the basic features of the plateau, consistent with the hypothesis that it arises from the formation of multiple, short RNA hairpins as load is relieved. The propensity to form secondary structure sets the minimal load necessary to recover the full transcriptional extension of the RNA; based on our data, forces beyond 18 pN generate records free of secondary structure. This value is consistent with results from a recent study of DNA hairpins under load (Woodside et al., 2006).
Backtracking Is Inhibited in the RNA-pulling Assay
Pulling the 5′ end of the RNA away from the polymerase tends to promote forward (transcriptionally downstream) motion of the polymerase along DNA. Thus, the effect of applying force to the RNA should be similar to that of applying an assisting load that pulls the upstream end of the DNA away from the polymerase, as in the experimental geometry of (Neuman et al., 2003). Assisting forces on the DNA are known to suppress transcriptional backtracking (Shaevitz et al., 2003). For this reason, we anticipated that loads applied to RNA would also inhibit pauses associated with backtracking. To test this, we measured elongation in the RNA-pulling assay in the presence of inosine triphosphate (ITP), a GTP-analog that induces long-duration, backtracking pauses associated with misincorporation. Addition of 200 μM ITP to the reaction buffer did not result in a statistically significant increase in the long pause density, in contrast to previous data for a hindering-load assay (Table 1), indicating that pauses in the RNA-pulling assay are not due to backtracking.
Table 1.
Effect of ITP on single-molecule assays with loads applied to RNA or DNA
| Assay | Force (pN) direction | [ITP] (μM) | Pause density (kb−1) (N) |
|---|---|---|---|
| RNA-pulling | 28 | 0 | 0.12 ± 0.03 (46) |
| RNA-pulling | 28 | 200 | 0.20 ± 0.10 (4) |
| DNA-pulling | 28 assisting | 0 | 0.20 ± 0.10 (24) |
| DNA-pulling* | 8 assisting | 0 | <0.03 (1) |
| DNA-pulling* | 8 assisting | 200 | 0.15 ± 0.10 (2) |
| DNA-pulling* | 8 hindering | 0 | 0.95 ± 0.21 (56) |
| DNA-pulling* | 8 hindering | 200 | 1.46 ± 0.29 (26) |
data from (Shaevitz et al., 2003).
Comparison of RNA-pulling and DNA-pulling Pause Data
The role played by RNA structure in ubiquitous pausing was probed by collecting transcriptional elongation records while applying F = 26–30 pN to the RNA, sufficient to disrupt secondary structure. The pause lifetime distribution was determined by a pause-finding algorithm that compares the dwell time in a record at a given position with a velocity-dependent threshold, scoring a pause whenever the threshold is exceeded (Adelman et al., 2002); this distribution was normalized by the total transcription time to supply the pause frequency (Figure 3). We compared pause distributions from the RNA-pulling assay with an assisting-load DNA-pulling assay. RNA is considerably more compliant than dsDNA of comparable length for the loads explored here, giving rise to larger thermal fluctuations in extension at a given load. To compare pauses scored in the two different types of assay, we numerically imposed a comparable level of noise on the DNA-pulling records prior to analysis (see Experimental Procedures). In the presence of this additional noise, pauses >4 s duration were reliably detected. A comparison of pause lifetime duration distributions obtained from RNA- and DNA-pulling assays agreed statistically (Figure 3, see Experimental Procedures). Pause lifetime distributions for both assays were fit by double-exponential functions, and were consistent with previous measurements (Neuman et al., 2003).
Figure 3. Normalized pause lifetime distributions for RNA-pulling (red) and DNA-pulling (blue) assays at 26–30 pN load.

Lifetime distributions were scaled by total number of seconds transcribed (RNA: t = 14,608 s, N = 132; DNA: tt = 12,042 s, N = 122) to supply the overall frequency (pauses/s). Bin widths were ≥1 sec and scaled to ensure ≥6 counts per bin; statistical errors were computed from √N. Inset: Semi-logarithmic plot for DNA (filled triangles) and RNA (open squares). Fits to DNA data: Double exponential τ = 0.9 ± 0.4 and 5.3 ± 1.6 s (χν2 = 0.81; ν= 3; p(χν2) = 0.48; 4 parameters), single exponential (not shown) τ = 2.7 ± 0.3 (χν2 = 3.1; ν = 5; p(χν2) = 0.009; 2 parameters). Fits to RNA data: Double exponential τ = 0.6 ± 0.2 and 3.8 ± 0.2 s (χν2 = 0.99; ν = 3; p(χν2) = 0.39; 4 parameters); single exponential (not shown) τ = 2.4 ± 0.3 (χν2 = 3.31; ν = 5; p(χν2) = 0.003; 2 parameters).
Elongation Rates and Pauses Are Independent of Force
In models where pausing represents an off-pathway state, distinct from the normal elongation pathway, entry into the pause state is in kinetic competition with elongation (Herbert et al., 2006; Neuman et al., 2003). To ensure that loads applied to the RNA did not affect elongation kinetics, we measured the transcriptional velocity between pauses as a function of force, using previously established methods (Neuman et al., 2003). Velocity was approximately invariant over the accessible range (Figure 4A), with a global mean rate of 8.5 ± 2.5 nt/s, comparable to the rate of the DNA-pulling assay performed here (10.3 ± 3.5 bp/s) and to values from other single-molecule studies (Abbondanzieri et al., 2005; Adelman et al., 2002; Neuman et al., 2003).
Figure 4. Force dependence of velocity, pause characteristics, and processivity.
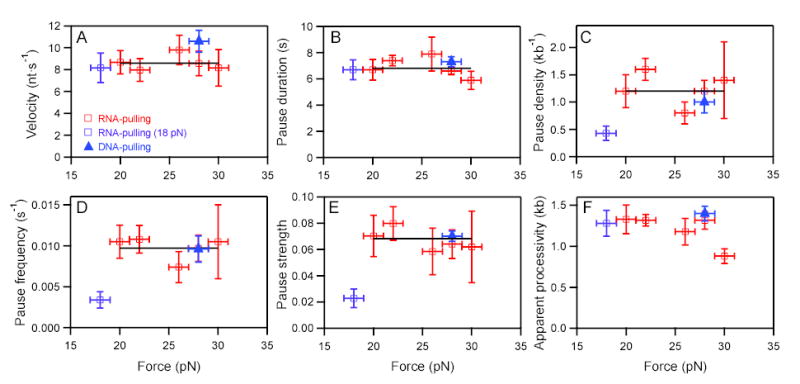
RNA-pulling data (open squares), DNA-pulling data (filled triangles). Fits to weighted means (black horizontal lines) rather than to lines (nonzero slope) were justified by F tests. The low force point at 18 pN was excluded from fits because it was deemed too close to the opening force for the most stable hairpins. Estimated errors represent std. errs. in (A, B) and bootstrap errors in (C-F).
(A) Mean velocity vs. force; avg. = 8.6 ± 0.7 nt/s.
(B) Mean pause duration vs. force; avg. = 6.9 ± 0.2 s.
(C) Pause density vs. force; avg. = 1.2 ± 0.1 kb−1.
(D) Pause frequency vs. force; avg. = 9.7 ± 0.9 ×10−3 s−1.
(E) Pause strength (pause duration multiplied by frequency) vs. force; avg. = 68 ± 7 ×10−3.
(F) Apparent processivity (distance to tether rupture) vs. force.
We also measured pause frequency (pauses/s), pause density (pauses/kb transcribed), pause duration, and pause strength (defined here as pause duration multiplied by frequency) as functions of load applied to the RNA (Figure 4B-E): these quantities were all fairly constant over the accessible force range. Pause characteristics for the RNA-pulling assay were similar to those found in the DNA-pulling assay, as well as generally consistent with previous measurements where assisting and hindering load forces were applied to template DNA (see Supplemental Material) (Neuman et al., 2003).
Apparent Processivity Is Independent of Force on RNA
One current model for transcriptional termination invokes a mechanical displacement of the RNA 3′ end from the enzyme active site (hypertranslocation), driven, for example, by terminator-hairpin folding or by Rho-driven translocation (Park and Roberts, 2006; Richardson, 2002; Yarnell and Roberts, 1999). This model predicts that the application of significant force to the nascent RNA would induce RNAP to release the transcript prematurely. We tested this possibility by determining the apparent enzyme processivity, i.e., the average elongation of the RNA transcript prior to rupture, for forces of 18 to 30 pN. Processivity was found to be largely independent of force (Figure 4F). However, for forces beyond 30 pN, processivity decreased, likely due to premature breakage of the surface linkage. The apparent processivity agreed, within error, with the equivalent measure for the DNA-pulling assay, indicating that forces ≤30 pN applied to the RNA did not substantially reduce enzyme processivity. Moreover, U-rich tracts encountered in the transcript (up to 6 nt within an 8-nt window) were not strongly correlated with rupture position (see Supplementary Results).
Discussion
Ubiquitous Pausing Unaffected by Force on Nascent RNA
Heretofore, single-molecule studies of transcriptional pausing have exerted forces only between the enzyme and the template DNA (Adelman et al., 2004; Forde et al., 2002; Neuman et al., 2003; Shaevitz et al., 2003). The most recent studies show that the brief, ubiquitous pauses which occur roughly once per 100 bases are not a consequence of enzyme backtracking. However, that work could not exclude the possibility that ubiquitous pauses might be associated with RNA hairpin formation. Indeed, hairpin formation would seem to be a plausible explanation for ubiquitous pausing, given the high frequency for the predicted formation of hairpins in co-transcriptionally folded RNA, the well-characterized regulatory pauses produced by hairpins known to exist in biosynthetic operons [so-called ‘Class I’ pauses, (Artsimovitch and Landick, 2000)], and the long-recognized role of hairpins in termination (Lee and Yanofsky, 1977).
Here, we studied transcription by single RNAP molecules subjected to forces exerted between the enzyme and the nascent RNA chain sufficient to disrupt all secondary structure (18–30 pN; Figure 2B). Interestingly, RNAP continued to exhibit ubiquitous pausing under these circumstances. Because forces in the RNA-pulling assay have the same directional sign as loads applied to the upstream end of the template DNA in a DNA-pulling assay, they tend to supply assisting loads that suppress enzyme backtracking. That suppression was confirmed by the addition of the nucleotide analog ITP, which induces long-duration pauses at low or hindering loads on DNA (Shaevitz et al., 2003), but which failed to generate additional pausing here.
Comparisons among transcriptional records obtained when loads are applied to DNA and to RNA are challenging, due to the increased level of thermal noise encountered in the latter, arising from the comparatively high elastic compliance of RNA, which limits the time resolution for pauses to a few seconds. Nevertheless, parallel studies of the two pulling assays obtained at equivalent loads and analyzed with equivalent noise levels gave remarkably similar results, with similar pause lifetimes and densities (Figure 2, Table 1).
If hairpins were responsible for ubiquitous pausing at low forces, it is still formally possible that the removal of hairpins at high force is somehow compensated by an effect of external load, which might substitute for the role of hairpin formation by pulling directly on the RNA. In this interpretation, the effect of external load (or hairpin folding) is to transiently displace the 3′ end of RNA from the active site, producing a hypertranslocated state that renders polymerase incompetent for further elongation (Artsimovitch and Landick, 2000). However, any such compensation of load for hairpin folding would need to be fairly exact, because pause rates scarcely differ between DNA-pulling assays (where no load is applied to RNA) and RNA-pulling assays. Moreover, there is no effect of load on pause frequency in DNA-pulling assays (Neuman et al., 2003). One would expect the degree of hypertranslocation-induced pausing to be closely correlated with the amount of load placed on the RNA, which is contrary to observation (Figure 4), at least over the range of force that could be usefully explored here. Nevertheless, it remains possible that load on the RNA induces hypertranslocation pausing, but that this phenomenon saturates for forces above 20 pN, beyond which pause characteristics are independent of force: we consider this unlikely.
The available single-molecule data suggest that neither RNA hairpins, backtracking, nor hypertranslocation are responsible for ubiquitous pauses. Potential mechanisms for ubiquitous pausing have been proposed; these include fraying of the RNA 3′ end away from the active site, conformational rearrangements of the so-called “bridge helix” and “trigger loop,” and hyperextension of the RNA:DNA hybrid (Bar-Nahum et al., 2005; Gnatt et al., 2001; Herbert et al., 2006; Landick, 2004; Neuman et al., 2003). The absence of hairpin, backtrack, and hypertranslocation effects is consistent with the idea that ubiquitous pausing represents an elemental pause state from which subsequent rearrangements (e. g., hairpin formation or backtracking) create long-lived pauses (Artsimovitch and Landick, 2000; Erie, 2002; Toulokhonov and Landick; 2003; Herbert et al., 2006).
Given their frequency in nascent RNA, why don’t the majority of hairpins stabilize pauses? Disruption of the his pause hairpin does not completely abolish the associated pause (Chan and Landick, 1993; Toulokhonov and Landick, 2003), and the effect of the hairpin appears to require a precisely spaced interaction with the β flap of RNAP near the RNA exit channel (Toulokhonov et al., 2001). Thus, hairpin pausing would require both a sequence signal to trigger entry into the elemental pause state and the subsequent formation of an appropriately spaced hairpin (Artsimovitch and Landick, 2000; Erie, 2002; Herbert et al., 2006; Toulokhonov and Landick, 2003). In this scenario, any RNA hairpins that do not coincide with an elemental pause signal will not affect pausing. Furthermore, hairpins that form near an elemental pause signal but fail to interact allosterically with the enzyme will not stabilize the pause.
A Sliding Clamp May Help Maintain Association of RNA with the TEC
Even when forces up to 30 pN are applied to the nascent RNA, RNAP is able to maintain a functional TEC, with negligible changes in the kinetics, pause characteristics, or processivity. These loads are comparable to the maximum forces that have been applied to template DNA [30–35 pN; (Neuman et al., 2003)]. They are also larger than the forces sufficient to shear a 10-bp-long DNA duplex [~20 pN at low loading rates; (Strunz et al., 1999)]. The DNA:RNA heteroduplex inside RNAP is just 8–9 bp long; pulling the RNA away from RNAP should exert a shearing force on this hybrid. Clearly, significant protein-nucleic acid interactions must contribute to keeping the RNA stably bound.
To explain this stability, we favor a sliding clamp model where protein-nucleic acid contacts prevent TEC dissociation (Korzheva et al., 2000). Crosslinking studies show the bacterial enzyme to be in close proximity to the RNA at both the front and rear ends of the RNA:DNA heteroduplex (Korzheva et al., 1998). Studies of eukaryotic RNA polymerase II suggest analogous stabilizing contacts with phosphates along the RNA:DNA hybrid, and spatial confinement inside the protein channel may provide additional stabilization (Gnatt et al., 2001; Kireeva et al., 2000). Pulling the RNA strand from a confined protein channel would be inhibited by steric clashes between bases as the two strands of the hybrid attempted to slide past one another. Slippage of the entire hybrid would be discouraged by further contacts between protein and DNA, and because reducing the hybrid length is energetically unfavorable.
In the context of the sliding clamp, our results offer a natural explanation for the failure of most hairpins to induce dissociation except at intrinsic terminators (Korzheva et al., 2000). As the terminator hairpin starts to fold, it exerts a force on the RNA; we estimate the maximal force from such a hairpin to be ~20 pN (see Supplemental Material). For the majority of transcript sequences, such forces are insufficient to lead to RNA release, as shown by our data. At terminator sites, however, the RNA in the hybrid consists of a U-rich tract that forms a weaker hybrid, and offers minimal steric hindrance when sliding past complimentary adenosine bases in DNA, leading to release at lower forces. Mechanisms that destabilize the RNA:DNA hybrid could further facilitate RNA release, including hairpin-stem invasion (Korzheva et al., 2000), allostery (Toulokhonov et al., 2001), or forward translocation (Santangelo and Roberts, 2004; Yarnell and Roberts, 1999). Future RNA-pulling experiments using defined terminator sequences may clarify the role of force in intrinsic termination.
Rho protein has been proposed to induce termination by pulling the RNA from the polymerase, hydrolyzing 1 ATP/nt translocated (Richardson, 2002). In a cell, Rho ATPase can expend ~80 pN·nm per ATP, which corresponds to an upper-bound force of ~130 pN exerted over a distance of 0.6 nm (the separation of bases in ssRNA). The actual force developed is likely to be less than this amount, but nevertheless must exceed a lower bound of 30 pN, set by these measurements, to cause release of RNA through a force-dependent mechanism in the absence of a weak hybrid.
Co-transcriptional Folding Studied with the RNA-pulling Assay
In closing, we note that the RNA-pulling assay geometry developed for this work is able to apply a range of controlled forces to the nascent RNA during transcription. The current study focused on the use of force in the high range (F ≥ 20 pN), sufficient to disrupt secondary structure in transcripts during elongation. However, the data of Figure 2 indicate that it is also feasible to carry out transcription under essentially unloaded conditions (F < 7 pN), where the growing RNA chain folds co-transcriptionally. The complex shapes of stretching curves performed after transcript growth reveal that substantial structure is formed (Figure 2B). By comparing features observed to unfold during the initial pull with those that later reform and open during subsequent pulls, it should be possible to characterize co-transcriptional folding elements in a variety of RNA structures of interest.
Experimental Procedures
Experimental Assays
Preparation of samples and calculation of tether length for the DNA-pulling assay were performed as described (Neuman et al., 2003). The 4,954 bp template DNA for the RNA-pulling assay was created by PCR from plasmid pRL732, which contains the rpoB gene following the T7A1 promoter (Neuman et al., 2003). The promoter site was located at positions 1093–1144, leading to a maximum possible RNA transcript of 3,811 nt. The DNA handle was created by autosticky PCR (Lang et al., 2004) of the M13mp18 plasmid using two primers: one had a 5′ digoxigenin linker molecule and the other primer was 50 nt long with a 24 nt hybridization region, followed by an abasic site and a 25 nt overhang. The PCR product was a 3,057 bp DNA with a 25 nt single stranded overhang which was complementary to the initial 25 bases of the nascent RNA generated from the pRL732 template.
Buffers, beads, reagents, and flow cells were prepared as in (Neuman et al., 2003); these were common to both assays unless noted. Stalled transcription complexes (8 μL at 2–4 nM, see Supplemental Experimental Procedures) were mixed with approximately equimolar quantity of DNA handle and a 3-fold molar excess of 500 nm avidin DN-coated beads for 1 hr. Flow cells were incubated with 20 μg/ml antidigoxigenin polyclonal antibody (Roche Molecular Biochemicals) dissolved in 100 mM phosphate buffer (pH 8.0) and were washed after 1 hr with 200 μl transcription buffer (50 mM HEPES [pH 8.0], 130 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 20 μg/ml heparin, 5 mg/ml BSA). Microscope flow cells were then perfused with 12 μL of diluted complex-handle-bead mixture and incubated for 1 hr, followed by a final wash with 200 μL transcription buffer. TECs were pre-screened for single tethers (see Supplemental Experimental Procedures).
Data Collection
Data were collected using the apparatus described in (Neuman et al., 2003). A position-sensitive diode (Pacific Silicon Sensors) was used to detect displacements in two dimensions in the specimen plane (Neuman et al., 2005). We estimate the uncertainty in force due to calibration errors and bead size variations at ~5–10%. Position data were filtered at 1 kHz by an 8-pole lowpass Bessel filter, acquired at 2 kHz using custom software (written in LabVIEW), and analyzed using Igor. Data were boxcar-averaged over a 20-pt window to supply a 100 Hz signal that controlled stage feedback for the force clamp. Tether lengths were calculated as described in Supplemental Experimental Procedures.
Pause Detection and Data Analysis
Initial and terminal pauses were excluded from analysis. Long pauses (>25 s) were scored by eye to circumvent problems associated with long-term drift. Short pauses (4–25 s) were determined by an automated algorithm (Adelman et al., 2002). In brief, the dwell time was computed at every position in a record and smoothed with a 10 s boxcar filter. Peaks in the dwell time distribution correspond to pauses; pause durations were determined by integrating these peaks. For RNA-pulling assays, pauses >4 s could be detected reliably; this limit was confirmed by simulations, as follows. Artificial records were generated with from a model pathway consisting of a single elongation step acting in kinetic competition with transitions to two pause states (this model generates a constant elongation rate interspersed with pauses drawn from a double-exponential distribution). Transcription elongation rates, pause entry/exit rates, and branching ratios were adjusted to match the experimental data of (Neuman et al., 2003). The noise power spectrum was determined from a stalled RNA-pulling tether and the corresponding spatial distribution was added to the simulated records. Analysis by the automated algorithm confirmed that 94% of pauses were detected with a 3% false-positive rate above 4 s.
Noise levels in RNA- and DNA-pulling records were evaluated by plotting the SD of position computed in 10 s windows against the average template position. Noise in RNA records increased faster with elongation, and was twice that of the equivalent DNA records by ~1,300 nt. Noise affects pause detection efficiency. To facilitate comparisons of pausing with RNA records, the magnitude of noise in a DNA record was artificially increased by first subtracting a smoothed (10 s filter) version of the record, then rescaling the residual according to [new noise = residual × (1 + (# bases)/1300 nt)], and adding it back to the smoothed record.
To compare pause lifetime distributions (Figure 3), we applied the Kuiper variant of the Kolmogorov-Smirnov (KS) test, which measures the separation of the normalized cumulative distribution functions for the two unbinned datasets (Press, 2002). The latter test has the advantage that there is no loss of information associated with binning. The KS test returned p = 0.60, indicating a less than 40% chance that the two experimental distributions were inconsistent with the same parent distribution. For a simulation of two datasets drawn from an identical parent distribution and containing the same number of points as the experimental data (NDNA = 132 and NRNA = 122), the KS test returned p = 0.53 ± 0.30.
Supplemental Data
Supplemental Data include text, experimental procedures, three figures, and Supplemental References and can be found with this article online at www.cell.com.
Supplementary Material
Acknowledgments
The authors thank the Block lab for discussions, especially M. Woodside for suggestions on DNA handles and discussions of RNA structure, A. La Porta for help with data analysis, J. Shaevitz for help with instrumentation, K. Herbert, W. Greenleaf, and E. Abbondanzieri for discussions of transcription, and M. Valentine, P. Fordyce, G. Liou and R. Neuman for critical reading of the text. MHL was supported by a Molecular Biophysics Training Grant through the NIH. This work was supported by an NIGMS grant to SMB.
References
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci U S A. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Yuzenkova J, La Porta A, Zenkin N, Lee J, Lis JT, Borukhov S, Wang MD, Severinov K. Molecular mechanism of transcription inhibition by Peptide antibiotic microcin j25. Mol Cell. 2004;14:753–762. doi: 10.1016/j.molcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- Dessinges MN, Maier B, Zhang Y, Peliti M, Bensimon D, Croquette V. Stretching single stranded DNA, a model polyelectrolyte. Phys Rev Lett. 2002;89:248102. doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- Erie DA. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim Biophys Acta. 2002;1577:224–239. doi: 10.1016/s0167-4781(02)00454-2. [DOI] [PubMed] [Google Scholar]
- Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- Forde NR, Izhaky D, Woodcock GR, Wuite GJ, Bustamante C. Using mechanical force to probe the mechanism of pausing and arrest during continuous elongation by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2002;99:11682–11687. doi: 10.1073/pnas.142417799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Harlepp S, Marchal T, Robert J, Leger JF, Xayaphoummine A, Isambert H, Chatenay D. Probing complex RNA structures by mechanical force. Eur Phys J E Soft Matter. 2003;12:605–615. doi: 10.1140/epje/e2004-00033-4. [DOI] [PubMed] [Google Scholar]
- Herbert, K. M., La Porta, A., Wong, B. J., Neuman, K. C., Landick, R., and Block, S. M. (2006). Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell, in press. [DOI] [PMC free article] [PubMed]
- Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3' end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Nudler E, Nikiforov V, Goldfarb A. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harb Symp Quant Biol. 1998;63:337–345. doi: 10.1101/sqb.1998.63.337. [DOI] [PubMed] [Google Scholar]
- Landick R. Active-site dynamics in RNA polymerases. Cell. 2004;116:351–353. doi: 10.1016/s0092-8674(04)00121-7. [DOI] [PubMed] [Google Scholar]
- Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci U S A. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F, Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977;74:4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- Neuert G, Albrecht C, Pamir E, Gaub HE. Dynamic force spectroscopy of the digoxigenin-antibody complex. FEBS Lett. 2006;580:505–509. doi: 10.1016/j.febslet.2005.12.052. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Block SM. Measurement of the effective focal shift in an optical trap. Optics Letters. 2005;30:1318–1320. doi: 10.1364/ol.30.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Jr, Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci U S A. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press, W. H. (2002). Numerical recipes in C : the art of scientific computing, 2nd edn (Cambridge [Cambridgeshire] ; New York, Cambridge University Press).
- Reeder TC, Hawley DK. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- Rivas E, Eddy SR. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics. 2000;16:583–605. doi: 10.1093/bioinformatics/16.7.583. [DOI] [PubMed] [Google Scholar]
- Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz T, Oroszlan K, Schafer R, Guntherodt HJ. Dynamic force spectroscopy of single DNA molecules. Proc Natl Acad Sci U S A. 1999;96:11277–11282. doi: 10.1073/pnas.96.20.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic-Norrelykke SF, Engh AM, Landick R, Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J Biol Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- von Hippel PH. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci U S A. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


