Abstract
The germinal center (GC) reaction is crucial for T cell-dependent immune responses and is targeted by B cell lymphomagenesis. Here we analyzed the transcriptional changes that occur in B cells during GC transit (naïve B cells → centroblasts → centrocytes → memory B cells) by gene expression profiling. Naïve B cells, characterized by the expression of cell cycle-inhibitory and antiapoptotic genes, become centroblasts by inducing an atypical proliferation program lacking c-Myc expression, switching to a proapoptotic program, and down-regulating cytokine, chemokine, and adhesion receptors. The transition from GC to memory cells is characterized by a return to a phenotype similar to that of naïve cells except for an apoptotic program primed for both death and survival and for changes in the expression of cell surface receptors including IL-2 receptor β. These results provide insights into the dynamics of the GC reaction and represent the basis for the analysis of B cell malignancies.
The germinal center (GC) reaction of antigen-activated B lymphocytes is the hallmark of antibody-mediated immune responses to T cell-dependent antigens (1). Individuals with genetic defects impairing GC formation suffer from immunodeficiency (2), and transgenic mice lacking factors that are required for GC formation do not show affinity maturation of the antibody response or humoral memory (summarized in ref. 3). GC B cells are also thought to be involved in the pathogenesis of most types of human B cell malignancies, including diffuse large cell lymphoma, follicular lymphoma, and Burkitt lymphoma (4, 5).
The GC reaction starts when naïve B cells (IgM+IgD+) are activated by antigen receptor stimulation and receive costimulatory signals from immune helper cells (6–8). These events induce the B cell to transform into a centroblast (CB) that proliferates within the histologically defined “dark zone” of the GC (1, 9, 10); CBs express the Ki67 nuclear antigen and can be identified by the expression of the CD77 cell surface marker (11). It is generally thought that CBs revise their antigen receptors through somatic hypermutation of IgV region genes, a process that introduces mainly single nucleotide substitutions into the IgV gene to generate antibodies with a higher or lower affinity to the respective antigen (7, 12). CBs then develop into noncycling centrocytes (CCs), which compose the “light zone” of the GC (9) and are distinguished from CBs by their lack of expression of the CD77 and Ki67 markers (11). In the CC stage, newly generated antibody mutants are selected based on their ability to bind their cognate antigen with the help of follicular dendritic cells and T cells. A large fraction of GC B cells undergoes apoptosis as they have acquired deleterious somatic mutations in their IgV regions that abolish antigen binding, whereas CCs expressing high-affinity antibody mutants eventually differentiate into plasma cells or memory B cells. A fraction of CCs also switches from the expression of IgM and IgD to that of other Ig classes by somatic DNA recombination to generate antibodies with different effector functions. The high-affinity memory B cells released from the GC are long-lived and have acquired the potential to rapidly differentiate into Ig-secreting cells during secondary immune responses (13).
Current knowledge of the physiology of the GC reaction is based on: (i) genetic approaches that have identified molecules required for GC development; (ii) the characterization of GC subpopulations based on the expression of immunophenotypic markers; and (iii) in vitro experiments that attempt to recapitulate the regulatory aspects of in vivo GC development. Although these studies have provided fundamental information on the physiology of GCs, they are based on the analysis of individual or small numbers of genes, proteins, or signaling pathways and cannot fully address the complex dynamics of the GC reaction. To obtain a comprehensive view of GC function and generate a data set for comparing normal versus malignant B cells, we have tracked the expression of ≈12,000 genes during the GC reaction.
Methods
Magnetic Cell Separation and Flow Cytometry.
Tonsils were obtained from routine tonsillectomies performed at the Babies and Children's Hospital of Columbia-Presbyterian Medical Center. Informed consent was obtained from the patients and/or exempt from informed consent being residual material after diagnosis and fully anonymized. Tissue collection was approved by the institutional ethical committee. The specimens were kept on ice immediately after surgical removal. After mincing, tonsillar mononuclear cells (MCs) were isolated by Ficoll-Isopaque density centrifugation. The four B cell subpopulations were isolated by magnetic cell separation by using the MidiMACS system (Milteny Biotec, Auburn, CA); for details see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. Briefly, naïve B cells were isolated by depletion of GC B cells (CD10, CD27), memory B cells (CD27), plasma cells/blasts (CD27), and T cells (CD3), followed by a positive enrichment of IgD-positive cells. CB were isolated in a single step by magnetically labeling CD77-positive cells. To enrich for CC, tonsillar MCs were first depleted for CB (CD77), naïve, memory, and plasma cells/blasts (CD39), and T cells (CD3). In a second step, CC were enriched by CD10. Memory B cells were purified by depletion of GC B cells (CD10, CD38), plasma cells/blasts (CD38), and T cells (CD3), followed by a positive enrichment of CD27-positive cells. The purity of the isolated fractions was determined on a FACSCalibur (Becton Dickinson) and by Ig V region gene analysis (see Table 1, which is published as supporting information on the PNAS web site). In addition, 89% of CBs and 82% of CCs isolated from the same tonsils expressed the GC-specific Bcl6 protein (14), but were negative for TdT, except for 1% of CC as noted (15) (data not shown). To exclude the possibility that some of the genes defining the profiles actually correspond to message from contaminating T cells, we purified T cells from the tonsillar MCs of two individuals by magnetic cell separation (Milteny Biotec); only a few genes might have potentially been derived from T cells. The occurrence of macrophages/monocytes in any of the fractions is unlikely because message for CD14 or CD33 was not detectable in all 20 hybridizations.
Immunohistological Analysis.
Stainings were performed on formalin-fixed, paraffin-embedded tonsillar tissue sections. Antibodies used were: rabbit anti-N-terminal c-Myc (clone N-262) and rabbit anti-IL-2 receptor β (IL-2Rβ) (clone C-20) (both Santa Cruz Biotechnology), and mouse anti-Pax-5 (PharMingen). A full characterization of the specificity of antibodies and tissue distribution of c-Myc will be reported elsewhere.
Gene Expression Profiling.
Microarray hybridization using Affymetrix (Santa Clara, CA) U95A GeneChip arrays was performed as described (16). For data analysis, we used the GENES@WORK software platform (17), which is a pattern discovery-based gene expression analysis tool, as described (16). GENES@WORK is available through www.research.ibm.com/FunGen. The primary data are available through ICG.cpmc.columbia.edu.
Results
Isolation and Characterization of B Cell Subpopulations.
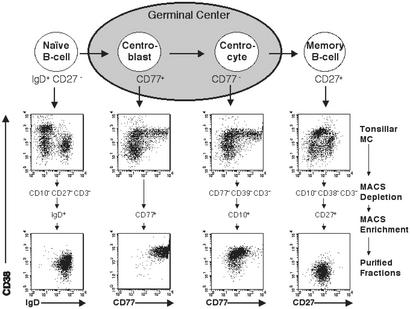
Based on recent characterizations of human B cell subsets (11, 18–20), we devised protocols enabling the purification of naïve B cells, GC CBs and CCs, and memory B cells from human tonsillar MCs by magnetic cell separation (Fig. 1), according to the following marker expression: naïve B cells were IgD+, CD38low, CD27−, CD10−, CD3−, and CD14−; CBs were CD77+ and CD38high; CCs were CD10+, CD38high, CD77−, CD39−, and CD3−; and memory B cells were CD27+, CD38low, CD10−, CD3−, and CD14−. These four B cell subpopulations were purified from the tonsils of five individuals, and their phenotypes were analyzed by flow cytometric analysis (Fig. 1). Further confirmation of their identity was obtained by: (i) nucleotide sequence analysis of Ig heavy-chain variable region (IgV) genes, which showed that naïve B cells expressed unmutated IgV genes (35/38 transcripts), whereas the majority of those derived from the CB (32/39), CC (29/41), and memory (36/42) fractions were somatically mutated as expected (11, 18) (see Table 1), and (ii) the observation that the vast majority of both CBs and CCs expressed Bcl6 protein (14) (see Methods). Together, these results indicate that these B cell populations represent well-defined phenotypic subsets as described (11, 15, 18–20).
Figure 1.
Isolation of tonsillar B cell subpopulations by magnetic cell separation. Tonsillar naïve B cells (IgD+, CD27−, CD38low), GC CB (CD38high, CD77+), GC CC (CD38high, CD77−), and memory B cells (CD27+, CD38low), were isolated by magnetic cell separation (see Methods). Flow cytometric analyses of representative isolations, tonsillar MCs before separation, and purified fractions are shown stained for CD38 and IgD, CD77, and CD27. Separation steps are summarized on the right.
Gene Expression Profile Analysis.
Total RNA samples from the purified naïve, CB, CC, and memory B cells were converted into labeled cRNAs and hybridized to Affymetrix U95A arrays representative of ≈12,000 genes (16). To determine whether the four B cell subsets could be identified based on their gene expression profiles, the 20 data sets corresponding to five samples each of the naïve, CB, CC, and memory B cells were analyzed by using the unsupervised mode of the pattern discovery-based GENES@WORK software (17). The first round of pattern discovery identified a pattern that clearly separated the 10 GC samples (CB and CC) from the 10 non-GC (naïve and memory) samples (Fig. 5a, which is published as supporting information on the PNAS web site). Subsequent unsupervised pattern discovery analyses of each of these two groups identified patterns that separated the naïve and memory B cell samples, and the CB and CC samples, the latter pattern composed of a smaller number of genes and thus of less statistically significance (17). The unsupervised pattern discovery and the phenotypes of the isolated cell populations were further validated by noting the correct expression patterns of genes known to be differentially expressed among naïve, GC, and memory B cells (Fig. 5a). Hierarchical clustering yielded results consistent with the unsupervised pattern discovery analyses (Fig. 5b). Taken together, these findings indicate that each of the purified B cell subpopulations displays a distinct gene expression profile that is consistent among individuals.
Naïve B Cell to CB Transition.
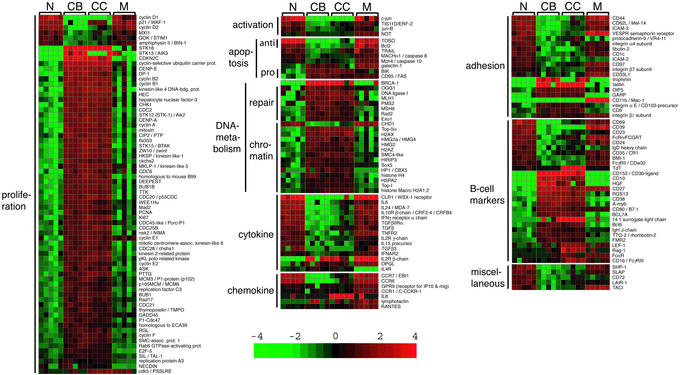
To identify changes in gene expression that occur in the transition from a naïve B cell to a GC CB, we compared these two subpopulations by supervised pattern discovery analysis using GENES@WORK, which allows the identification of differentially expressed genes between cell types defined a priori according to a given criterion (cell phenotype in this case) (see ref. 16). The naïve B cell → CB transition involves changes in the expression of 457 genes (Fig. 6, which is published as supporting information on the PNAS web site), which are organized into putative functional categories in Fig. 2 (and Fig. 7, which is published as supporting information on the PNAS web site).
Figure 2.
Supervised analysis of changes in gene expression during the GC transit of B cell subpopulations. The genes identified in the four individual transitions (Fig. 6) by supervised pattern discovery using GENES@WORK were merged to visualize their expression changes during GC transit. Color changes within a row indicate expression levels relative to the average of the sample population. Values are quantified by the scale bar that visualizes the difference in the zge score (expression difference/standard deviation) relative to the mean (0). Genes are ranked according to their zg score (mean expression difference of the respective gene between phenotype and control group/standard deviation). Shown are only those gene segments that differ 3-fold or more in their zg score. The gene expression changes among the subsets are shown according to functional or operational categories; for additional categories see Fig. 7.
The transition of a naïve to a GC B cell is associated with a dramatic change in the expression of genes associated with cell proliferation. Fifty-seven of 63 identifiable proliferation-associated genes were up-regulated in the GC B cells, including: (i) genes involved in DNA replication [e.g., PCNA (7-fold)]; (ii) cyclins A (7-fold), B1 and B2 (6- and 10-fold), E1 and E2 [9-fold over background (o.b.) and 7-fold] and F (3-fold) and various CDC genes, including CDC2 and CDC28 (8- and 3.5-fold, respectively); (iii) genes controlling the G1/S and G2/M transitions [e.g., DP-1 (3-fold), GADD45 (4-fold)]; and (iv) mitotic checkpoint kinases [e.g., BUB1 (5.5-fold), Mad3L (6-fold)], and mitotic genes that encode components of the centrosome, spindle, and kinetochore. Consistent with the change from a quiescent to a highly proliferative phenotype, the negative cell cycle regulator p21/WAF-1 was completely down-regulated in the CB fraction (30-fold). Also the tumor/growth suppressors BIN-1 and GOK/STIM1, not previously known to be expressed in B cells, were down-regulated.
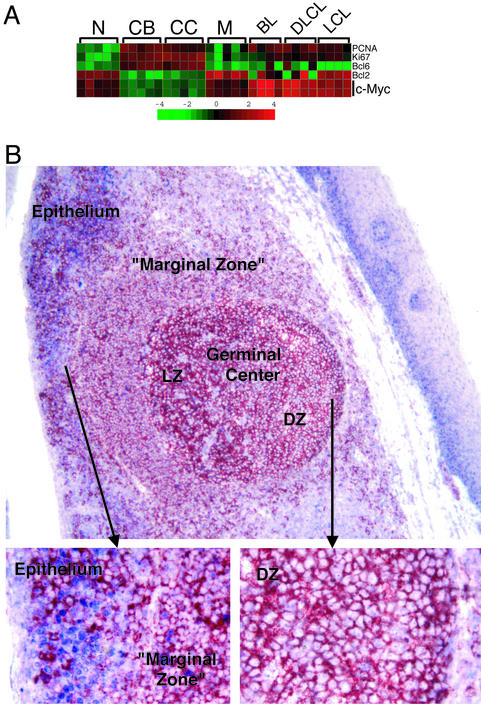
Surprisingly, the c-Myc protooncogene, which encodes a transcription factor thought to be ubiquitously expressed in proliferating cells, was not expressed in the CB. Whereas B cell lines and Burkitt lymphoma tumor biopsies contained high amounts of c-Myc mRNA as expected, the corresponding levels in CB were even lower than those in resting naïve or memory cells (Fig. 3A), suggesting that c-Myc expression may be actively suppressed in the CB. This result was confirmed by immunohistochemical analysis of human tonsil sections that showed that GC B cells, identified by CD20 staining, lack detectable levels of c-Myc protein in the nucleus (Fig. 3B); in contrast, nuclear c-Myc was readily detected in proliferating epithelial cells of the tonsils. These results were confirmed with three antibodies of nonoverlapping specificities (not shown). Expression of other Myc family members, such as N-Myc or L-Myc, did not appear to compensate for the absence of c-Myc in GC B cells, because mRNA for N-Myc and L-Myc were not detectable by microarray analysis in any of the four B cell populations, consistent with previous reports (21, 22).
Figure 3.
Analysis of c-Myc expression in normal and malignant B cells. (A) c-Myc mRNA expression in normal B cells and Burkitt lymphoma (BL), diffuse large B cell lymphoma (DLCL), and Epstein–Barr virus-transformed lymphoblastoid cell lines (LCL). c-Myc was represented by two probe sets on the U96A chip. Bcl2 and Bcl6, as well as the proliferation-associated genes PCNA and Ki67, are shown along with c-Myc as markers for the identity of the purified cell populations. The matrix is as in Fig. 2. (B) Immunohistochemical analysis of a tonsillar section for c-Myc (blue) and the B cell marker CD20 (red). Tonsillar epithelium, “marginal zone,” and GC light zone (LZ) and dark zone (DZ) are indicated (magnification ×10). (Insets) A higher magnification (×40) of the epithelium and the GC DZ, respectively. Note the expression of c-Myc in the nuclei of epithelial cells and the lack of expression in DZ CBs.
Genes associated with DNA repair were mostly up-regulated in the naïve B cell → CB transition, including the mismatch repair genes PMS2 (5-fold), MLH1 (6-fold), and MSH6 (3-fold), the base excision repair gene OGG1 (4-fold), and genes involved in aspects of replication, repair, and homologous recombination, namely BRCA-1 (26-fold o.b.), Rad2 (3-fold), Exo1 (5-fold), and DNA ligase 1 (7.5-fold).
The naïve B cell → CB transition is associated with significant and specific changes in the expression of genes controlling apoptosis. Genes known to exert proapoptotic functions, such as BIK (8.5-fold) and FAS/CD95 (17-fold o.b.), were up-regulated in the CB, whereas antiapoptotic genes were strongly down-regulated, including Bcl2 (6.5-fold) and TOSO (13.5-fold), the latter previously known to block CD95/FAS-mediated apoptosis in T cells (23).
A number of cytokines, chemokines, and their receptors were down-regulated in CB, including the chemokine receptors CCR6 (32-fold) (24) and CCR7 (27-fold), the class I cytokine receptor WSX-1/CLR1/TCCR (32-fold o.b.), a novel receptor expressed on CD4+ T cells that is required for Th1-type immune responses in mice (25), and the class II cytokine receptor CRF2–4 (9-fold), the β-subunit of the IL-10 receptor complex.
Eight genes associated with cell adhesion were down-regulated in the naïve B cell → CB transition, including CD44 (11-fold), CD62L (9-fold), and the semaphorin receptor VESPR (7-fold). A number of adhesion-related genes up-regulated in the CB were so far not known to be associated with B lymphocyte physiology (Fig. 2). These genes include trophinin and tastin (both 19-fold o.b.) that are involved in embryo implantation (26), as well as OIP5 and GARP (8- and 10-fold, respectively), two molecules whose structure suggest functions in cellular adhesion.
CB to CC Transition.
The CB → CC transition involved changes in the expression of only 19 genes (Fig. 6). Some of the genes up-regulated in CC, including TdT (9-fold o.b.), RAG-1 (12.5-fold o.b.), and 14.1 surrogate light chain (5-fold) (Fig. 6; see also Fig. 2), are known to be expressed in B cells undergoing Ig gene rearrangements (immature or “transitional” B cells), and their expression was previously reported in purified CC fractions (15). In addition, our results show that CC express the mRNA for lymphocyte enhancing factor-1 (9.5-fold), which is required for the transition of the pro-B cell to the pre-B cell stage (27). These findings are explained by the recent observation that immature B cells with phenotypic characteristic of CCs are present in peripheral lymphoid organs and copurify with CCs (see Discussion). The remaining 15 genes distinguishing CBs from CCs were expressed at levels similar to those in naïve or memory B cells, with few exceptions including the CC-specific expression of the chemokine IL-8 (160-fold o.b.) (see Fig. 2). The small number of differences between CBs and CCs compared with the naïve B cell → CB transition (19 vs. 457 genes) indicates that the CD77+ and CD77− GC B cell subsets are very similar in their gene expression profiles (see Discussion).
CC to Memory B Cell Transition.
Somatically mutated, antigen-selected B cells leave the GC as plasma cells or memory B cells. The CC → memory B cell transition involved 267 genes (Fig. 6). As evident from Fig. 2, most genes associated with activation, proliferation, and DNA metabolism regained expression levels comparable to those found in the naïve B cells, implying that most post-GC memory cells return to quiescence. The antiapoptotic genes Bcl2 (8.5-fold) and TOSO (4.5-fold) were also up-regulated in the CC → memory B cell transition, returning to levels of expression similar to those in naïve cells.
The chemokine receptor CCR6 (7-fold) as well as the cytokine receptors WSX-1/CLR1/TCCR (21.5-fold o.b.) and CRF2–4/IL-10R β chain (4.5-fold) also regained expression levels comparable to those found in naïve cells. Among the adhesion-associated molecules, the CD11b/Mac-1/complement receptor type 3 gene was found to be strongly up-regulated in the CC → memory B cell transition (12.5-fold o.b.); CD11b was recently shown to be expressed on a subpopulation of human peripheral blood memory B cells (28).
Naïve B Cell to Memory B Cell Transition.
To identify directly the phenotypic differences between pre-GC and post-GC B cells, we compared the gene expression profiles of naïve and memory B cells. Supervised pattern discovery revealed only 62 differentially expressed genes (Fig. 6). Overall, there was no change in the expression of antiproliferative and pro-proliferative genes or of genes involved in DNA metabolism. Conversely, the apoptosis-inducing genes that were up-regulated in the naïve B cell → CB transition (i.e., CD95/FAS and BIK) remain expressed at similar levels in the memory B cells (Fig. 2). However, the antiapoptotic genes Bcl2 and TOSO are similarly expressed in both naïve and memory cells. Thus, the memory B cell, in contrast to the naïve B cell, contains mRNA from both antiapoptotic and proapoptotic genes.
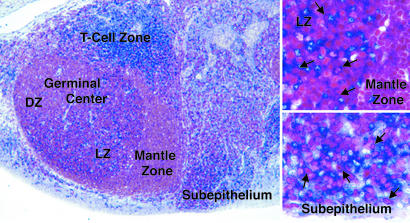
Four genes involved in chemotaxis and responsiveness to cytokines were up-regulated in the memory cells, including the osteoprotegerin ligand (OPGL) (9-fold o.b.) and IL-2Rβ (95-fold o.b.). mRNA for IL-2Rβ was found to be up-regulated in the GC fractions and, to a greater extent, in the memory B cells (Fig. 2). Immunohistochemical analysis of tonsillar sections showed that individual B cells, as defined by the expression of the B cell marker Pax-5, expressed IL-2Rβ within both the GC light zone and resting B cells of the subepithelium, the site of memory B cells in tonsils (29) (Fig. 4). Together, these observations suggest that IL-2Rβ is up-regulated on some GC B cells in the light zone and that these eventually may differentiate into memory cells.
Figure 4.
Immunohistochemical analysis of tonsillar tissue sections for IL-2Rβ. GCs, GC dark zone (DZ) and light zone (LZ), mantle zone, T cell zone, and tonsillar subepithelium are indicated (magnification ×10). Pax-5 (red), a B cell marker, and IL-2Rβ (blue). (Insets) A higher magnification (×40) of the GC LZ and the subepithelium, respectively. Arrows indicate Pax-5/IL-2Rβ double-positive cells.
Several cell adhesion molecules were differentially expressed in the two cell types. Naïve cells expressed mRNA for protocadherin-9/VRA-11 (19-fold), a cell adhesion molecule identified in the nervous system, whereas memory B cells specifically contained message for CD11b/Mac-1 (12.5-fold o.b.). As evident from Figs. 2 and 7, the majority of the genes that discriminate naïve and memory B cells were already up-regulated in GC cells and remain expressed in memory B cells.
Discussion
In this study, we have attempted a characterization of the GC reaction by gene expression profiling, an approach that is ideally suited for addressing the complexity of biological processes in which multiple pathways are sequentially activated. This approach was previously used to characterize the signature of GC B cells by comparing them to quiescent and in vitro-activated B cells (30, 31); although naïve and memory B cell samples were included in that study, analysis of the different stages of GC development was not performed, conceivably because only one sample for each subpopulation was available. The results presented here provide a detailed analysis of each of the known stages of GC B cell development in vivo.
The Naïve B Cell.
Substantial evidence indicates that naïve B cells are resting and lack mRNAs that encode proliferation-promoting genes (29). Nonetheless, our results indicate that naïve B cells express proliferation-inhibitory genes (e.g., p21/WAF-1) as well as tumor or growth suppressor genes (BIN1, GOK1) whose expression has not been described in B cells. In addition, naïve cells express mRNA for the various immediate early response genes (c-jun, ERF-2) and cyclins (D1 and D2) that are required for G1- to S-phase transition. These findings suggest that naïve B cells are actively maintained in a quiescent state by the expression of growth-inhibitory genes, but are poised for rapid induction of the GC response by expression of activation genes that are required to initiate cell proliferation. The results herein also indicate that naïve cells express a complex set of cytokine and chemokine receptors, as well as various adhesion molecules, almost all of which are down-regulated in GC B cells. Thus, the naïve B cell seems to be equipped to respond to signals that induce its activation and determine its homing to specific peripheral lymphoid organs.
Phenotype of GC CBs.
The gene expression profiles described here substantiate earlier reports on the proliferative status of CB (1, 9, 10). Nonetheless, our results show that the proliferative activity of CB has unique properties that set it apart from other cell types. Most notable is the lack of expression of the c-Myc protooncogene (Fig. 3), a result that is in contrast with previous reports showing c-Myc RNA and protein in GC B cells (32, 33). This discrepancy may be caused by: (i) the use of RT-PCR (32, 33), a technique too sensitive to minimal cell contamination to be used for the analysis of purified B cell subpopulations (see Supporting Text) and (ii) cross-reactivity of the anti-c-Myc antibody used with other molecules, as suggested by the fact that c-Myc was localized to the “nuclear membrane” (33) as opposed to the more typical diffuse nuclear distribution shown here. The lack of c-Myc expression in CBs is particularly surprising in view of the fact that c-Myc is thought to be required for cell proliferation and cell growth (34) and therefore would be expected to be essential for CBs, which are relatively large cells with one of the fastest proliferative rates among eukaryotic cells (35). On speculative terms, this finding can be explained by the recent observation that c-Myc can induce genomic instability and recombination (36), an activity that may not be desirable in GC B cells that have to undergo physiologic DNA double-strand breaks to execute somatic hypermutation and Ig switch recombination. Notably, the lack of c-Myc expression in GC B cells has implications for understanding the pathogenesis of Burkitt lymphoma because it suggests that chromosomal translocations may lead to ectopic, rather than deregulated, expression of c-Myc as currently postulated (37).
The changes in gene expression profiles that characterize the naïve to GC transition indicate a radical change from an antiapoptotic program to a markedly proapoptotic one. This proapoptotic phenotype is the result of both the induction of proapoptotic genes as well as by the down-regulation of antiapoptotic genes. Among the latter, our results identify TOSO (23), a member of the Ig superfamily, as a player in the regulation/execution of B cell apoptosis in the GC. These results provide a mechanistic basis for the model, previously proposed based only on in vitro data, that GC B cells are primed to undergo programmed cell death if not rescued (38).
A previously unrecognized aspect of the phenotype of GC B cells is the expression of a specific profile of adhesion receptor genes including several that had not been previously associated with B lymphocyte physiology (see Results). This finding implies that a distinct adhesion molecule repertoire is presented on the cell surface of GC B cells, possibly to facilitate specific interactions with accessory cells as well as trafficking within the GC microenvironment. Some of these adhesion molecules may characterize topographically distinct GC subpopulations and influence the spread of GC-derived tumors.
CBs Versus CCs.
The two major GC B cell subpopulations, CBs and CCs, were previously identified based on various independent criteria, including their topographical distribution within the GC (CBs in the dark zone and CCs in the light zone), proliferative status (active for CBs and quiescent for CCs), and differential expression of the CD77 and CD44 markers (1, 9–11, 39). Of these phenotypic traits, the differential CD77 expression was the only one readily exploitable for cell purification and was therefore used in this study to isolate CBs and CCs. It was thus surprising that CB and CC populations isolated by virtue of differential CD77 expression and validated by typical GC markers (e.g., CD10, CD38high, and mutated IgV genes) showed a largely common gene expression profile differing in the expression of only 19 genes. Considering that supervised gene selection methods can identify differentially expressed genes only if they are uniformly expressed within each of the two cell types under study, a possible explanation for this result is that either the CB and/or the CC fractions may represent heterogeneous cell populations. This hypothesis is supported by: (i) the existence of CC subpopulations characterized by the differential expression of transcription factors such as Bcl6, IRF-4/MUM-1 (40), Blimp-1 (41), and IL-2Rβ (Fig. 4), and (ii) the observation that the CC fraction contains mRNA species characteristic of immature B cell precursors (RAG-1, TdT, surrogate light chain, and lymphocyte enhancing factor-1) (ref. 15 and Fig. 2), suggesting the presence of immature B cells in peripheral lymphoid organs where they are localized outside the GC microenvironment (42–44). Taken together, our results and these observations indicate that the CD77 marker does not identify the two histologically defined populations of GC B cells. Whereas CD77+ GC cells appear homogeneous, CD77− GC cells comprise subpopulations that need further characterization.
Memory B Cells.
Previous studies showed that memory B cells differ from naïve cells in several aspects, including their size, Ig mRNA levels, response to in vitro activation stimuli, expression of the tumor necrosis factor receptor family member CD27 (18, 20, 29, 45, 46), and expression of CD80 and CD11b on a subset of cells (28). Our results confirm these few known differences, but indicate that the overall gene expression profiles of naïve and memory B cells are remarkably similar (Fig. 6). This finding suggests that the GC reaction does not produce drastic changes in the cell phenotype, but rather represents a selection process largely based on the ability of B cells to hypermutate IgV genes to increase their affinity for the antigen.
Our results reveal that a subset of memory B cells express the IL-2Rβ chain, which is part of the IL-2R complex and is also involved in the response to IL-15 (47). Because this molecule is also expressed in a subset of cells localized in the GC light zone, it is possible that IL-2Rβ-expression identifies late CCs that are committed to the memory lineage or differentiated memory B cells that are about to leave the GC. The identification of IL-2Rβ as a marker of a subset of memory B cells further suggests that the memory B cell pool includes distinct subsets with distinct roles in secondary immune responses (18, 28, 48–50).
Implications for the Study of GC-Derived B Cell Malignancies.
Finally, the gene expression profiles of the normal B cell subsets established here are especially valuable for the study of transformed B cells because most types of B cell malignancies are derived from cells that have passed through the GC (4, 5). Several studies aimed at identifying prognostic subgroups of B cell lymphoma have compared these tumors with GC B cells and in vitro-activated B cells (30, 51–53). However, these two populations are not adequate to represent the multiple stages of GC development that occur in vivo. For instance, the profiles from isolated B cell subpopulations have allowed us to propose a memory B cell origin for B cell chronic lymphocytic leukemia (16). The profiles of four distinct B cell subpopulations shown here should give a more precise reference for studies on the cellular origin of the various B cell malignancies, the identification of genes involved in oncogenic transformation, and gene products of potential therapeutic relevance.
Supplementary Material
Acknowledgments
We thank Ashlyn Celestine for help with the immunohistological stainings and Richard Baer for comments on the manuscript. U.K. is a recipient of a fellowship granted by the Human Frontiers Science Program. G.C. is a recipient of an Esther Aboodi Associate Professor Fellowship.
Abbreviations
- GC
germinal center
- CB
centroblast
- CC
centrocyte
- MC
mononuclear cell
- o.b.
over background
- IL-2Rβ
IL-2 receptor β
Note Added in Proof.
During the review process, a paper was published by Feldhahn et al. (54), who analyzed genomewide gene expression profiles of human B cell subpopulations by using sequential analysis of gene expression (SAGE).
References
- 1.MacLennan I C. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Notarangelo L D, Hayward A R. Clin Exp Immunol. 2000;120:399–405. doi: 10.1046/j.1365-2249.2000.01142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosco-Vilbois M H, Bonnefoy J Y, Chvatchko Y. Immunol Rev. 1997;156:127–136. doi: 10.1111/j.1600-065x.1997.tb00964.x. [DOI] [PubMed] [Google Scholar]
- 4.Küppers R, Klein U, Hansmann M L, Rajewsky K. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson F, Sahota S, Zhu D, Ottensmeier C, Chapman C, Oscier D, Hamblin T. Immunol Rev. 1998;162:247–259. doi: 10.1111/j.1600-065x.1998.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 6.Berek C, Ziegner M. Immunol Today. 1993;14:400–404. doi: 10.1016/0167-5699(93)90143-9. [DOI] [PubMed] [Google Scholar]
- 7.Rajewsky K. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 8.Kelsoe G. Immunol Today. 1995;16:324–326. doi: 10.1016/0167-5699(95)80146-4. [DOI] [PubMed] [Google Scholar]
- 9.Kroese F G, Timens W, Nieuwenhuis P. Curr Top Pathol. 1990;84:103–148. doi: 10.1007/978-3-642-75519-4_5. [DOI] [PubMed] [Google Scholar]
- 10.Stein H, Gerdes J, Mason D Y. Clin Haematol. 1982;11:531–559. [PubMed] [Google Scholar]
- 11.Pascual V, Liu Y J, Magalski A, de Bouteiller O, Banchereau J, Capra J D. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner S D, Neuberger M S. Annu Rev Immunol. 1996;14:441–457. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- 13.McHeyzer-Williams M G, Ahmed R. Curr Opin Immunol. 1999;11:172–179. doi: 10.1016/s0952-7915(99)80029-6. [DOI] [PubMed] [Google Scholar]
- 14.Cattoretti G, Chang C C, Cechova K, Zhang J, Ye B H, Falini B, Louie D C, Offit K, Chaganti R S, Dalla-Favera R. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 15.Meffre E, Papavasiliou F, Cohen P, de Bouteiller O, Bell D, Karasuyama H, Schiff C, Banchereau J, Liu Y J, Nussenzweig M C. J Exp Med. 1998;188:765–772. doi: 10.1084/jem.188.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein U, Tu Y, Stolovitzky G A, Mattioli M, Cattoretti G, Husson H, Freedman A, Inghirami G, Cro L, Baldini L, et al. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Califano A, Stolovitzky G, Tu Y. Proc Int Conf Intell Syst Mol Biol. 2000;8:75–85. [PubMed] [Google Scholar]
- 18.Klein U, Rajewsky K, Küppers R. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tangye S G, Liu Y J, Aversa G, Phillips J H, de Vries J E. J Exp Med. 1998;188:1691–1703. doi: 10.1084/jem.188.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agematsu K, Hokibara S, Nagumo H, Komiyama A. Immunol Today. 2000;21:204–206. doi: 10.1016/s0167-5699(00)01605-4. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman K A, Yancopoulos G D, Collum R G, Smith R K, Kohl N E, Denis K A, Nau M M, Witte O N, Toran-Allerand D, Gee C E, et al. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 22.Smith R K, Zimmerman K, Yancopoulos G D, Ma A, Alt F W. Mol Cell Biol. 1992;12:1578–1584. doi: 10.1128/mcb.12.4.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hitoshi Y, Lorens J, Kitada S I, Fisher J, LaBarge M, Ring H Z, Francke U, Reed J C, Kinoshita S, Nolan G P. Immunity. 1998;8:461–471. doi: 10.1016/s1074-7613(00)80551-8. [DOI] [PubMed] [Google Scholar]
- 24.Krzysiek R, Lefevre E A, Bernard J, Foussat A, Galanaud P, Louache F, Richard Y. Blood. 2000;96:2338–2345. [PubMed] [Google Scholar]
- 25.Chen Q, Ghilardi N, Wang H, Baker T, Xie M H, Gurney A, Grewal I S, de Sauvage F J. Nature. 2000;407:916–920. doi: 10.1038/35038103. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M N, Sato T, Nakayama J, Klier G, Mikami M, Aoki D, Nozawa S. Genes Dev. 1995;9:1199–1210. doi: 10.1101/gad.9.10.1199. [DOI] [PubMed] [Google Scholar]
- 27.Reya T, O'Riordan M, Okamura R, Devaney E, Willert K, Nusse R, Grosschedl R. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 28.Bar-Or A, Oliveira E M, Anderson D E, Krieger J I, Duddy M, O'Connor K C, Hafler D A. J Immunol. 2001;167:5669–5677. doi: 10.4049/jimmunol.167.10.5669. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y-J, Barthelemy C, de Bouteiller O, Arpin C, Durand I, Banchereau J. Immunity. 1995;2:239–248. doi: 10.1016/1074-7613(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 30.Shaffer A L, Rosenwald A, Hurt E M, Giltnane J M, Lam L T, Pickeral O K, Staudt L M. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- 31.Ma C, Staudt L M. Philos Trans R Soc London B. 2001;356:83–89. doi: 10.1098/rstb.2000.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Valdez H, Guret C, de Bouteiller O, Fugier I, Banchereau J, Liu Y J. J Exp Med. 1996;183:971–977. doi: 10.1084/jem.183.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cutrona G, Dono M, Pastorino S, Ulivi M, Burgio V L, Zupo S, Roncella S, Ferrarini M. Eur J Immunol. 1997;27:234–238. doi: 10.1002/eji.1830270135. [DOI] [PubMed] [Google Scholar]
- 34.Johnston L A, Prober D A, Edgar B A, Eisenman R N, Gallant P. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y-J, Zhang J, Lane P J, Chan E Y, MacLennan I C. Eur J Immunol. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 36.Vafa O, Wade M, Kern S, Beeche M, Pandita T K, Hampton G M, Wahl G M. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 37.Boxer L M, Dang C V. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y-J, Joshua D E, Williams G T, Smith C A, Gordon J, MacLennan I C. Nature. 1989;342:929–931. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
- 39.Feuillard J, Taylor D, Casamayor-Palleja M, Johnson G D, MacLennan I C. Int Immunol. 1995;7:121–130. doi: 10.1093/intimm/7.1.121. [DOI] [PubMed] [Google Scholar]
- 40.Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, Pacini R, Alunni C, Natali-Tanci L, Ugolini B, et al. Blood. 2000;95:2084–2092. [PubMed] [Google Scholar]
- 41.Calame K L. Nat Immunol. 2001;2:1103–1108. doi: 10.1038/ni1201-1103. [DOI] [PubMed] [Google Scholar]
- 42.Yu W, Nagaoka H, Jankovic M, Misulovin Z, Suh H, Rolink A, Melchers F, Meffre E, Nussenzweig M C. Nature. 1999;400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 43.Monroe R J, Seidl K J, Gaertner F, Han S, Chen F, Sekiguchi J, Wang J, Ferrini R, Davidson L, Kelsoe G, Alt F W. Immunity. 1999;11:201–212. doi: 10.1016/s1074-7613(00)80095-3. [DOI] [PubMed] [Google Scholar]
- 44.Cattoretti G, Parravicini C, Bonati A, Buscaglia M, Zuliani G, Plebani A, Delia D, Rilke F. Eur J Immunol. 1989;19:493–500. doi: 10.1002/eji.1830190313. [DOI] [PubMed] [Google Scholar]
- 45.Arpin C, Banchereau J, Liu Y J. J Exp Med. 1997;186:931–940. doi: 10.1084/jem.186.6.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurer D, Fischer G F, Fae I, Majdic O, Stuhlmeier K, Von Jeney N, Holter W, Knapp W. J Immunol. 1992;148:3700–3705. [PubMed] [Google Scholar]
- 47.Nelson B H, Willerford D M. Adv Immunol. 1998;70:1–81. doi: 10.1016/s0065-2776(08)60386-7. [DOI] [PubMed] [Google Scholar]
- 48.McHeyzer-Williams L J, Cool M, McHeyzer-Williams M G. J Exp Med. 2000;191:1149–1166. doi: 10.1084/jem.191.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dono M, Zupo S, Leanza N, Melioli G, Fogli M, Melagrana A, Chiorazzi N, Ferrarini M. J Immunol. 2000;164:5596–5604. doi: 10.4049/jimmunol.164.11.5596. [DOI] [PubMed] [Google Scholar]
- 50.Weller S, Faili A, Garcia C, Braun M C, Le Deist F F, de Saint Basile G G, Hermine O, Fischer A, Reynaud C, Weill J. Proc Natl Acad Sci USA. 2001;98:1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 52.Rosenwald A, Alizadeh A A, Widhopf G, Simon R, Davis R E, Yu X, Yang L, Pickeral O K, Rassenti L Z, Powell J, et al. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shipp M A, Ross K N, Tamayo P, Weng A P, Kutok J L, Aguiar R C, Gaasenbeek M, Angelo M, Reich M, Pinkus G S, et al. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 54.Feldhahn N, Schwering I, Lee S, Wartenberg M, Klein F, Wang H, Zhou G, Rowley J D, Hescheler J, Kronke M, et al. J Exp Med. 2002;196:1291–1305. doi: 10.1084/jem.20020881. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.