Abstract
The transition from transcription initiation to elongation involves phosphorylation of the large subunit (Rpb1) of RNA polymerase II on the repetitive carboxyl-terminal domain. The elongating hyperphosphorylated Rpb1 is subject to ubiquitination, particularly in response to UV radiation and DNA-damaging agents. By using computer modeling, we identified regions of Rpb1 and the adjacent subunit 6 of RNA polymerase II (Rpb6) that share sequence and structural similarity with the domain of hypoxia-inducible transcription factor 1α (HIF-1α) that binds von Hippel–Lindau tumor suppressor protein (pVHL). pVHL confers substrate specificity to the E3 ligase complex, which ubiquitinates HIF-α and targets it for proteasomal degradation. In agreement with the computational model, we show biochemical evidence that pVHL specifically binds the hyperphosphorylated Rpb1 in a proline-hydroxylation-dependent manner, targeting it for ubiquitination. This interaction is regulated by UV radiation.
The von Hippel–Lindau tumor suppressor protein (pVHL)-associated complex, which contains elongin B, elongin C, cullin-2, and Rbx-1 (1–3) is a primary ubiquitin ligase for ubiquitination of the α subunits of the hypoxia-inducible transcription factors (HIFs) (4–6). During normoxia, translated HIF-αs are hydroxylated on conserved proline residues located within L(XY)LAP motifs by the O2, Fe(II), and 2-oxoglutarate-regulated Egl-9 family of prolyl hydroxylases (7, 8), resulting in their ubiquitination and degradation. During hypoxia, proline hydroxylation is inhibited; HIF-αs are not ubiquitinated, and they accumulate and regulate transcription of the HIF-responsive genes (4–6, 9–12). Loss of pVHL function in VHL disease leads to the accumulation of HIF-αs during normoxic conditions, causing constitutive induction of HIF-responsive genes, including angiogenic vascular endothelial growth factor (VEGF) (13, 14). This functioning, in turn, contributes to the formation of highly vascular tumors such as hemangioblastomas, angiomas, and renal clear cell carcinomas (RCCs) (15).
von Hippel–Lindau disease is also associated with pheochromocytomas, nonmalignant tumors of adrenal medulla chromaffin cells, which synthesize and release large quantities of catecholamines and produce cardiovascular pathologies (16, 17). The molecular mechanism of the augmented catecholamine production is unknown. Recently, we presented evidence that pVHL regulates expression of the rate-limiting enzyme in catecholamine biosynthesis, tyrosine hydroxylase (TH), and in pheochromocytoma-derived (PC12) cells (18, 19). Low levels of pVHL, resulting from expression of VHL antisense RNA, correlate with more efficient transcription of the full-length TH transcripts (19). In contrast, high levels of overexpressed pVHL block transcript elongation between exons 6 and 8 of the TH gene (18). The presence of the elongation arrest site within this region of the TH gene has been confirmed by using in vitro transcriptional analysis (20).
Processive elongation of the initiated transcripts involves reversible hyperphosphorylation of tandemly repeated heptapeptides on the carboxyl-terminal domain (CTD) of subunit 1 of RNA polymerase II (Rpb1) within the RNA polymerase II complex (21). This elongation-competent, hyperphosphorylated Rpb1 is ubiquitinated in a transcription-dependent manner (22, 23). In particular, ubiquitination of the hyperphosphorylated Rpb1 is induced by UV radiation and DNA damage (24–26), suggesting that Rpb1 ubiquitination may play a role in the transcription-coupled repair (27). In yeast, ubiquitination is mediated by a HECT-class Rsp5 ubiquitin ligase (28); however, the nature of the E3 ligase in mammalian cells is unknown. We hypothesized that the hyperphosphorylated Rpb1 may be a substrate for pVHL-associated E3 ubiquitin-ligase activity.
Here, we identify a region of the Rpb1/Rpb6 subunits of RNA polymerase II that shares sequence and structural similarity with the pVHL binding domain of HIF-1α, and show that the pVHL-associated complex interacts specifically with the hyperphosphorylated Rpb1, leading to its ubiquitination.
Materials and Methods
Cell Cultures and Reagents.
PC12 cell clones (18, 19) and 786-O RCC cells were described (1), and were used at the cell density of 1.5–2.5 × 105 per cm2. UV irradiations (15 J/m2) were performed in a UV Crosslinker (FB-UVXL-1000, Fisher Biotech, Pittsburgh). N-Cbz-l-Leu-l-Leu-l-norvalinal (CbzLLn; 10 μM) was added 30 min before UV irradiation. This medium was removed immediately before the irradiation and the same medium was returned after irradiation.
Iron chloride, cobalt chloride, zinc chloride, desferrioxamine, 2,2′-dipyridyl, ascorbic acid, 2-oxoglutarate, and CbzLLn were purchased from Sigma. Reagents used in ubiquitination reaction were purchased from Boston Biochem (Boston) or Affiniti Research Products (Hamhead, Exeter, Devon, U.K.). The following antibodies were used as follows: H14 (Research Diagnostics, Flanders, NJ); C21 and anti-cullin-2 (Santa Cruz Biotechnology); Ig32 anti-pVHL (PharMingen); 12CA5 anti-hemagglutinin (HA) (Roche Molecular Biochemicals); anti-Rbx1 (Zymed); anti-elongin C (Signal Transduction, Lexington, KY); anti-elongin B polyclonal (custom made by Alpha Diagnostic, San Antonio, TX); anti-HIF-2α (Novus Biologicals, Littleton, CO); anti-ubiquitin (StressGen Biotechnologies, Victoria, Canada); and mouse anti-rabbit IgG (clone RG-96, Sigma). Anti-mouse secondary antibody-bound agarose was from Sigma. Synthetic biotinylated peptides were made by Alpha Diagnostic.
In Vitro pVHL-Peptide Binding Reaction.
Ten micrograms of biotinylated peptide was incubated with streptavidin-coated Dynabeads (M-280, Dynal, Great Neck, NY) in a buffer (25 μl) containing 20 mM Tris at pH 8, 100 mM NaCl, 0.5% Nonidet P-40, and 1 mM EDTA for 1 h at room temperature. Washed beads were incubated with WT [pRC-cytomegalovirus (CMV) expression vector; Invitrogen] or mutated pVHL (pCI- neo-CMV expression vector; Promega), translated in vitro by using [35S]methionine and TNT reticulocyte lysate (Promega). Binding reaction products were washed extensively in the same buffer and analyzed for bound [35S]pVHL by using SDS/PAGE. For the peptide hydroxylation step, immobilized peptide was first incubated in the hypotonically prepared cellular extract from PC12 cells as described below in the presence of 100 μM each of FeCl2, ascorbic acid, and 2-oxoglutarate for 1–2 h at 30 or 37°C.
Preparation of Extracts.
Intact nuclei were isolated as described (18, 19). The nuclei were resuspended in a half-pellet volume of low-salt buffer (20 mM Hepes, pH 7.9/20 mM NaCl/1 mM EDTA/20% glycerol), to which a half-pellet volume of high-salt buffer (20 mM Hepes, pH 7.9/1 M NaCl/1 mM EDTA/20% glycerol) was added. Proteins were extracted for 30 min at 4°C, followed by digestion of genomic DNA and RNA with DNase and micrococcal nuclease (15 and 88 units, respectively, per 100 μl of nuclear pellet volume) for 60 min, to release DNA-bound RNA polymerase II complexes. The digestion produced DNA fragments of <600 bp, as estimated by ethidium bromide staining on agarose gel. Extracts were centrifuged twice at 21,000 × g for 30 min at 4°C and dialyzed.
Total cellular extracts were prepared by using hypotonic lysis (20 mM Tris, pH 7.5/5 mM KCl/1.5 mM MgCl2/1 mM DTT and standard protease inhibitors) at 4°C and were homogenized by using 40 strokes of a tight-pestle Dounce homogenizer. The lysates were digested with DNase and micrococcal nuclease as described above, and centrifuged twice at 21,000 × g.
Denatured lysates were obtained by boiling pellets in 3 vol of SDS lysis buffer (1% SDS/50 mM Tris, pH 7.5/0.5 mM EDTA/1 mM DTT) for 10 min. The lysates were then diluted with immunoprecipitation buffer (see below) and centrifuged at 21,000 × g for 30 min.
Immunoprecipitations.
For all immunoprecipitation reactions, the agarose beads were precoated with BSA, and the primary antibodies were preconjugated with the secondary antibodies. Reactions were performed in buffer containing 50 mM Hepes at pH 7.8, 150 mM NaCl, 5 mM MgCl2, 20% (vol/vol) glycerol, and 0.1% Triton X-100 (immunoprecipitation buffer) and washed in the same buffer containing in addition 0.5% Triton, 0.5% Igepal, and 0.5% sodium deoxycholate, or a buffer containing 0.5% Igepal and NaCl from 150 to 900 mM. Immunoprecipitated proteins were eluted by boiling in SDS sample buffer, resolved by electrophoresis in SDS/4–22% polyacrylamide gradient gels, and detected by immunoblotting.
For hydroxylation of endogenous Rpb1, 150 μg of total protein extract was incubated in the presence of prolyl hydroxylase cofactors or inhibitors for 15–30 min at 30°C. The extracts were then processed for the immunoprecipitation reaction with anti-HA antibody. For elutions of Rpb1 from pVHL-associated complexes, proteins coimmunoprecipitated with anti-HA antibody from PC12VHL(WT) nuclear extracts were incubated in 40 mM Tris buffer in the presence of the respective peptides for 1.5 h. The eluates were analyzed by SDS/PAGE. To dephosphorylate Rpb1, extracts were incubated with 25 units of alkaline phosphatase (Roche Molecular Biochemicals) with or without 10 mM NaF for 30 or 60 min at room temperature. Dephosphorylated extracts were used for immunoprecipitations with anti-HA antibodies.
In Vitro Ubiquitination.
Four-hundred micrograms of total cellular extract was immunoprecipitated with anti-HA or H14 antibody, followed by four washes with high detergent immunoprecipitation buffer and two washes in buffer containing 50 mM Tris at pH 8 and 3 mM DTT. The immunoprecipitated complexes were resuspended in a final reaction volume of 50 μl containing ATP-regenerating buffer, 5 μg⋅μl−1 ubiquitin, 100 ng⋅μl−1 ubiquitin aldehyde, CbzLLn, and 16 μl of purifed rabbit reticulocyte fraction II, incubated for 2 h at 37°C, washed in 50 mM Tris, pH 8.0/3 mM DTT buffer, and analyzed by SDS/PAGE.
Computational Analysis.
The PROSITE server (29) was used to identify proteins containing proline hydroxylation motifs. HIF-1α secondary structures were predicted by using the Psi-PRED (30) server. The sequence of the human Rpb1 subunit (GenBank accession no. NP_000928) was first aligned optimally with the yeast Rpb1 structure (PDB ID code 1I50, chain A, 60% identical with mostly conservative substitutions), and then used to build the optimal structurally biased sequence alignment with the sequences of the human HIF-α factor (GenBank accession no. BAB70608). HIF-1α residues 530–577 were aligned with the yeast Rpb1 (structure 1I50, chain A) by using the loopp program and structurally biased sequence alignment (refs. 31–33; loopp is available at www.tc.cornell.edu/CBIO/loopp). Residues 571–679 of HIF-1α were aligned with the structure of the human Rpb6 subunit (structure 1qkl, chain A) by using the 3D-PSSM server (34).
Results
Similarity of Rpb1/Rpb6 Subunits and HIF-1α Oxygen-Dependent Degradation Domain (ODDD).
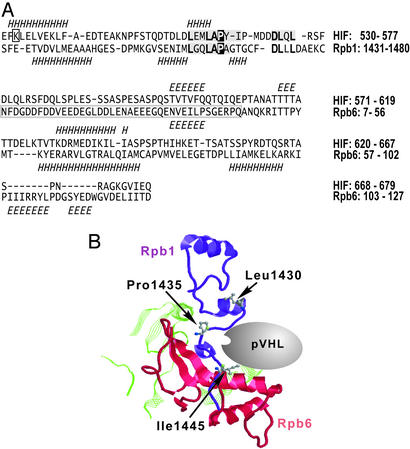
The human, murine, and yeast Rpb1 subunits contain an analogous, L(XY)LAP, motif located amino-terminal to the binding site for Rpb6 and to the beginning of the unstructured CTD (Fig. 1; ref. 35). Comparing the sequence of the HIF-1α ODDD with representative libraries of protein structures identified a region with similarities to a fragment of Rpb1 and the adjacent Rpb6 subunit. The ≈50-aa Rpb1 counterpart is 30% identical and contains the L(XY)LAP motif, including P1465 as a counterpart of the HIF-1α P564 residue. The HIF-1α secondary structures, as predicted by the psi-pred method (30), are also consistent with those of Rpb1 and Rpb6. The plausible pVHL binding pocket between Rpb1 and Rpb6 with the critical pVHL-binding motif (Fig. 1B) is located on the surface of the RNA polymerase II complex (see the legend to Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). The estimated statistical significance of individual alignments of the HIF-1α sequence into the Rpb1 and Rpb6 structures is low (Fig. 7). However, two weak matches into adjacent structural domains of the RNA polymerase II complex make the overall prediction stronger than suggested by the individual estimates of significance. This prediction is further strengthened by the recently published partial structure of the ODDD peptide and pVHL complex (36, 37), which suggests that ODDD exists in an extended conformation and reveals that the adjacent DLQL motif stabilizes the pVHL binding. A closely related motif (DLLL) is found in the predicted Rpb1 counterpart of ODDD.
Figure 1.
Computational prediction of the pVHL-binding pocket in the RNA polymerase II complex. (A) Sequence-to-structure alignments of the HIF-1α ODDD fragment into the carboxyl-terminal fragments of human Rpb1 and Rpb6 subunits. The Rpb1 and Rpb6 secondary structures are indicated below, and the predicted HIF-1α secondary structures are shown above their sequences. H, α (and other)-helices; E, extended β-strands. HIF-1α motifs that make contact with the pVHL complex (including the Pro-564 residue) are shaded and, if conserved in Rpb1, bold. The critical HIF-1α residues (L559, L562, P564, and D571) are conserved in the Rpb1 structure. The K532 residue, ubiquitinated on HIF-1α, is boxed. The human and yeast Rpb6 structures (PDB ID codes 1QKL and 1I50, chain F, respectively) are different by an additional β-strand occurring only on the human Rpb6 structure (boxed fragment). (B) The predicted pVHL-binding pocket (Rpb1, purple; Rpb6, red; other fragments in contact with the binding pocket are green). The critical proline residue and the flanking amino acids are indicated by using ball and stick models of their side chains. The numbering of residues is according to the yeast Rpb1 structure with the yeast Leu-1430, Pro-1435, and Ile-1445 residues corresponding to Leu-1460, Pro-1465, and Leu-1475 of the human Rpb1, respectively.
pVHL Binds Hyperphosphorylated Rpb1 in a Proline Hydroxylation-Dependent Manner.
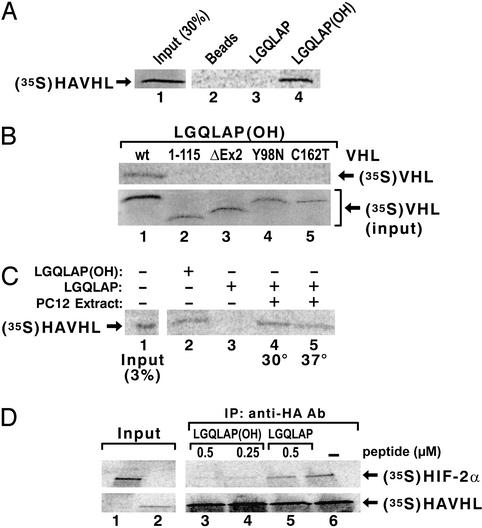
The immobilized Rpb1 peptide (amino acids 1440–1475) containing the hydroxylated proline binds to WT [35S]pVHL (Fig. 2A), but not to pVHL with deletions of exon 3 or exon 2 or with point mutations within the β (C162T) or α (Y98N) domain (Fig. 2B). The nonhydroxylated peptide does not bind pVHL, but it acquires pVHL-binding properties after incubation with PC12 cell extracts in the presence of Fe(II), ascorbic acid, and 2-oxoglutarate (Fig. 2C). The hydroxylated, but not the nonhydroxylated, peptide competes with full-length [35S]HIF-2α for [35S]pVHL binding (Fig. 2D). [35S]pVHL and Rpb6 do not interact under these conditions.
Figure 2.
pVHL binds to the Rpb1 synthetic 36-aa peptide with hydroxylated P1465. (A) Binding of [35S]pVHL to the Rpb1 peptide hydroxylated (lane 4), or nonhydroxylated (lane 3), on P1465. (B) Binding of the in vitro-translated mutated forms of pVHL to the hydroxylated peptide. To ensure that the amounts of the labeled mutant proteins used in the peptide-binding reactions were the same as for the WT pVHL, the amounts of the lysate with radioactively labeled mutant proteins used in the binding reactions were normalized accordingly by using the PhosphorImager quantification. (C) Hydroxylation of the Rpb1 peptide in extract from PC12 cells. HAVHL, HA-tagged pVHL. (D) Competition experiment of [35S]HIF-2α–[35S]pVHL binding by hydroxylated (lanes 3 and 4) or nonhydroxylated (lane 5) peptide.
Coimmunoprecipitation experiments using anti-pVHL antibodies in nuclear extracts from PC12 cells overexpressing HA-tagged human pVHL, or in control vector-transfected PC12 cells (18), reveal that both anti-VHL and anti-HA antibodies are able to coimmunoprecipitate hyperphosphorylated Rpb1 as detected by the H14 antibody, which is specific for phosphoserine-5 within the CTD repeats (refs. 38–40; Fig. 3A). In contrast, pVHL fails to coimmunoprecipitate the nonphosphorylated Rpb1 as detected by the C21 antibody (Fig. 3 A and B), which is specific for the nonphosphorylated peptide sequence from the CTD. The pVHL–Rpb1 complex is stable in high salt (up to 900 mM NaCl washes), consistent with other pVHL-binding proteins (1–3) (Fig. 3B). Dephosphorylation of extracts with alkaline phosphatase greatly attenuates binding of pVHL to the hyperphosphorylated Rpb1, and does not induce binding of the hypophosphorylated Rpb1 (Fig. 3C). pVHL–Rpb1 complex is also formed in extracts derived from RCC cells, either expressing endogenous truncated and nonfunctional pVHL or stably transfected with HA-tagged pVHL (ref. 1; Fig. 3D).
Figure 3.
pVHL specifically interacts with the hyperphosphorylated Rpb1 in nuclear extracts from PC12 and RCC cells. Coimmunoprecipitations (IP) using monoclonal antibodies against HA or pVHL or mouse anti-rabbit IgG (RG-96) in nuclear extracts from PC12 cells overexpressing human HA-tagged pVHL [PC12 VHL (WT)] (A and B), or control PC12 cells stably transfected with an empty vector (A). (C) Dephosphorylation of Rpb1 in PC12 cellular extracts for the indicated times by treating the extracts with alkaline phosphatase (AP) in the absence (lanes 3, 4, 7, and 8) or presence (lanes 6 and 9) of NaF. Treated extracts were subjected to immunoprecipitations using anti-HA antibodies. (D) Anti-HA immunoprecipitations in nuclear extracts from RCC 786-O cells lacking pVHL function (lanes 1 and 2) or from cells stably transfected with HA-pVHL (1) (lanes 3 and 4). PC12 and indicated RCC cells were pretreated with 10 μM CbzLLn for 6 h to increase accumulation of the hyperphosphorylated Rpb1. The immunoprecipitates were washed with high-detergent immunoprecipitation buffer (A, C, and D), or immunoprecipitation buffers containing up to 900 mM NaCl and 0.5% Igepal (B). Blots were probed with the indicated antibodies, human (h)pVHL and rat (r)pVHL, respectively.
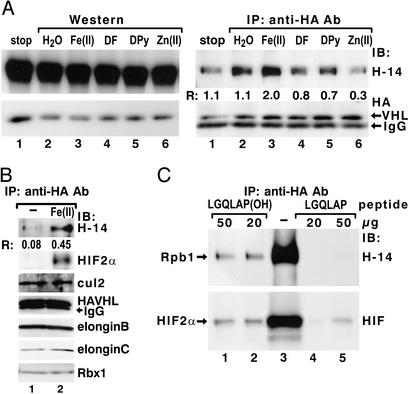
Incubation of cellular lysates in the presence of Fe(II), 2-oxoglutarate, and ascorbic acid substantially increases the formation of the pVHL–Rpb1 complex as determined by coimmunoprecipitation reactions (Fig. 4 A and B), whereas this complex is inhibited in the presence of iron chelators, desferrioxamine, and 2,2′-dipyridyl (41), or by the addition of ZnCl2, a potent divalent inhibitor of collagen prolyl hydroxylases (ref. 42; Fig. 4A Right). These treatments do not affect the total amount of hyperphosphorylated Rpb1 in the extracts (Fig. 4A Left). Pretreatment of lysates with cofactors of prolyl hydroxylases augments both the association of pVHL with Rpb1 and the formation of the pVHL–HIF-2α complex (Fig. 4B). However, these treatments do not affect the formation of the pVHL–elongin BC, cullin-2, and Rbx-1 complex (Fig. 4B), and they do not induce binding of pVHL to the hypophosphorylated Rpb1 (data not shown). The synthetic 36-aa P1465-hydroxylated Rpb1 peptide elutes Rpb1 and HIF-2α from the anti-HA immunoprecipitated complex, whereas the nonhydroxylated peptide is only marginally effective (Fig. 4C). These data indicate that, similar to HIF-α, hyperphosphorylated Rpb1 binds pVHL in a proline hydroxylation-dependent manner.
Figure 4.
pVHL binds Rpb1 in a proline hydroxylation-dependent manner. (A) Preincubation of PC12 cellular extracts with FeCl2, ascorbic acid, and 2-oxoglutarate (100 μM each) (lane 3), or with 100 μM each of iron chelators: desferrioxamine (DF, lane 4) and 2,2′-dipyridyl (DPy, lane 5), or ZnCl2 (lane 6), followed by Western blot analysis (Left) or coimmunoprecipitations (IP) with anti-HA antibodies (Right). IB, immunoblotting antibody. (B) Coimmunoprecipitation of the components of pVHL-associated complex by using anti-HA antibody in cellular lysates (lane 1) or lysates treated under hydroxylating conditions with Fe(II), ascorbate, and 2-oxoglutarate, as in A. Immunoblots were probed with the indicated antibodies. (C) Elution of hyperphosphorylated Rpb1 and HIF-2α with a hydroxylated 36-aa Rpb1 peptide. R describes the ratio of the signal detected with H14 antibody to the signal detected with anti-HA antibody, as quantified by using optical density measurements.
pVHL Regulates Ubiquitination and Accumulation of Hyperphosphorylated Rpb1.
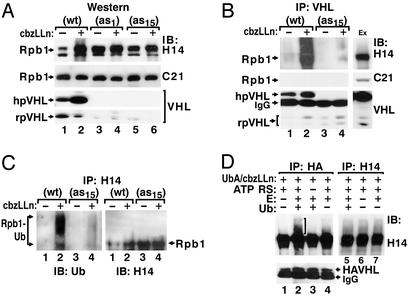
The amount of hyperphosphorylated, but not of hypophosphorylated, Rpb1 in PC12 cells correlates inversely with the levels of pVHL (Fig. 5A). Cells overexpressing pVHL exhibit low levels of constitutively accumulated hyperphosphorylated Rpb1, whereas cells expressing reduced levels of pVHL (19) exhibit high levels of Rpb1 detected with H14 (Fig. 5 A and C). After treatment with the proteasomal inhibitor CbzLLn, cells expressing high levels of pVHL accumulate the more slowly migrating forms of Rpb1, whereas cells expressing low levels of pVHL do not. In contrast, steady-state levels of the hypophosphorylated form of Rpb1 are not affected by CbzLLn treatment, and are independent of pVHL levels (Fig. 5A). Formation of the pVHL-Rpb1 complex is proportional to the concentration of pVHL and increases in cells treated with CbzLLn (Fig. 5B). Consistent with these data, in vivo ubiquitination of the hyperphosphorylated Rpb1 correlates with the levels of pVHL in a CbzLLn-dependent manner (Fig. 5C).
Figure 5.
Accumulation and ubiquitination of hyperphosphorylated Rpb1 in cells with different levels of pVHL. (A) Western blot analysis of hyperphosphorylated (H14) and hypophosphorylated (C21) Rpb1 in nuclear extracts from PC12VHL(WT) or two different clones of PC12VHL antisense (as) cells. (B) Coimmunoprecipitations using anti-pVHL antibody from nuclear extracts of WT and antisense cells. Ex, extract. (C) Immunoprecipitation of ubiquitinated forms of hyperphosphorylated Rpb1 from denatured cellular lysates by using H14 antibody. (D) In vitro ubiquitination reactions on protein complexes coimmunoprecipitated by using anti-HA (lanes 1–4) or H14 (lanes 5–7) antibodies from cellular extracts from PC12VHL(WT) cells. H14 antibody does not coimmunoprecipitate pVHL. UbA, ubiquitin aldehyde; E, purified enzymatic fraction II from reticulocyte lysate; ATP RS, ATP-regenerating solution. The bracket marks ubiquitinated forms of Rpb1.
To further investigate whether the pVHL complex directly ubiquitinates hyperphosphorylated Rpb1, protein complexes were coimmunoprecipitated with either anti-HA or H14 antibody, washed stringently, and subjected to in vitro ubiquitination (Fig. 5D). The more slowly migrating forms of Rpb1 are detected only with the full ubiquitination-reaction-containing complexes coimmunoprecipitated with anti-HA, but not with H14 (Fig. 5D, lanes 2 and 5). The ubiquitinated forms of Rpb1 are not detected if ATP-regenerating buffer or ubiquitin is omitted (Fig. 5D, lanes 3 and 4). These data show that pVHL targets hyperphosphorylated Rpb1 for ubiquitination.
UV Irradiation Induces pVHL–Rpb1 Interaction.
In cells overexpressing pVHL, UV irradiation induces an early and transient increase in the accumulation of hyperphosphorylated Rpb1 detected with H14, which decreases during 8 h of recovery (Fig. 6A). In contrast, in cells with reduced levels of pVHL, hyperphosphorylated Rpb1 does not increase after UV irradiation, but declines with a delay beginning after 6 h of recovery (Fig. 6A). The disappearance of Rpb1 in pVHL-overexpressing cells is prevented by proteasomal inhibitors, indicating that the loss of Rpb1 results from proteasomal degradation (Fig. 6A). Levels of the hypophosphorylated Rpb1 are not affected by UV exposure and do not depend on the concentration of pVHL. UV irradiation increases the amount of Rpb1 coimmunoprecipitated with pVHL, in the presence and absence of proteasomal inhibitors (Fig. 6B). The UV stimulus clearly increases ubiquitination of hyperphosphorylated Rpb1 in cells overexpressing pVHL, but fails to induce its ubiquitination in cells expressing low levels of pVHL (Fig. 6C). These data indicate that pVHL contributes to the processing of the RNA polymerase complex in response to UV stress.
Figure 6.
Rpb1–pVHL interactions in response to the UV treatment. (A) Western blot analysis of Rpb1 and pVHL in nuclear extracts from cells in the absence (Upper) or presence (Lower) of CbzLLn. (B) Coimmunoprecipitation of hyperphosphorylated Rpb1 with anti-HA antibody in nuclear extracts from cells treated with UV for the indicated times. Blots were probed with indicated antibodies. (C) Ubiquitination of the hyperphosphorylated Rpb1 in response to the UV treatment in PC12VHL(WT) and antisense cells. Two hours after UV irradiations denatured cellular lysates were immunoprecipitated with H14 antibodies and the immunoblots were probed with anti-ubiquitin antibody.
Discussion
Our findings extend the role of the pVHL complex. They show that, in addition to its role as an E3 ubiquitin ligase, which regulates the accumulation of HIF-α protein (9–12), and thereby expression of hypoxia-inducible genes, the pVHL complex can function as an E3 ligase, which targets the hyperphosphorylated Rpb1 for ubiquitination and degradation. Importantly, binding of pVHL to the full-size Rpb1 requires hydroxylation of proline-1465 within Rpb1 and phosphorylation of the CTD. To date, the proteins that pVHL targets for ubiquitination include, in addition to HIF-αs, a subfamily of deubiquitinating enzymes (43, 44) and activated atypical protein kinase C (45).
The exact role that ubiquitination of hyperphosphorylated Rpb1 by pVHL plays in the function of the RNA polymerase complex remains to be determined. Because ubiquitination of Rpb1 occurs in a transcription-dependent manner (22, 23), and because our earlier observations indicate that pVHL levels affect in vivo elongation of TH transcripts (18, 19), we anticipate that ubiquitination of Rpb1 by pVHL complex is likely to regulate efficient transcript elongation through elongation-pause and -arrest sites of specific genes. In particular, it may be involved in the regulation of TH transcript elongation (18–20). Such potential role of the pVHL–Rpb1 interaction is supported by the fact that pVHL binds in the pocket between Rpb1 and Rpb6, and that Rpb6 promotes elongation through arrest sites by binding to the elongation factor TFIIS (46). The pVHL-binding site is located on the surface of the elongating RNA polymerase II complex, and thus is accessible for pVHL binding during transcription. Interaction of pVHL with the RNA polymerase II complex may also locally titrate elongins B and C from elongation factor SIII (elongin ABC), thereby providing another mechanism by which pVHL could inhibit transcription elongation, as proposed based on in vitro studies (47).
We also anticipate that the pVHL–Rpb1 interaction has a more universal role and may regulate genes other than TH. The pVHL–Rpb1 interaction is regulated by UV stress, thus pVHL may play a role in the regulation of transcription complexes (transcription-coupled repair) under conditions of DNA damage, such as UV irradiation. In this respect, pVHL-negative cells undergo apoptosis in response to UV treatment, whereas the pVHL-positive cells do not (48). Our data also suggest a molecular mechanism by which the loss of pVHL function in von Hippel–Lindau disease may result in tumorigenesis.
Our results demonstrate that antisense cells having decreased levels of pVHL accumulate hyper- but not hypophosphorylated Rpb1, resulting in decreased ubiquitination of Rpb1. The most consistent explanation for this finding is that the antisense cells have decreased pVHL-associated E3 ligase activity toward the hyperphosphorylated Rpb1. However, it cannot be excluded that pVHL may affect the activity and/or expression of some kinases or phosphatases involved in the phosphorylation of CTD. In view of the role of CTD phosphorylation in pVHL binding, this last possibility might be an attractive regulatory mechanism increasing the pVHL–Rpb1 interaction under conditions of reduced amounts of pVHL.
These data provide biochemical evidence that Rpb1 can be modified by proline hydroxylation. It is unclear whether proline hydroxylation requires CTD phosphorylation. Two major groups of proline hydroxylases have been identified to date: the endoplasmic reticulum collagen proline hydroxylases (42) and the Egl-9-like group of proline hydroxylases involved in O2-dependent regulation of HIF-α (7, 8). Both groups hydroxylate prolines in an O2-, Fe(II)-, and 2-oxoglutarate-dependent manner; howeve, hydroxylases involved in collagen maturation are less sensitive to O2 levels and are functional even under hypoxic conditions (42). In contrast, the HIF prolyl hydroxylases appear to be strictly O2-sensitive, and their activities are inhibited by a decrease in pO2 (7). At this time, it has not been possible to measure the O2-sensitivity of prolyl hydroxylation of Rpb1 and pVHL binding because hyperphosphorylated Rpb1 disappears rapidly from the hypoxic extracts by an as yet unknown mechanism (M.F.C.-K., unpublished results).
Computational models and experimental observations demonstrate significant structural similarity between the ODDD of HIF-1α and Rpb1/Rpb6. However, the HIF-1α pVHL-binding peptide needs to be rotated along a single bond in the central “bulge” to bring it into a good agreement with its predicted yeast Rpb1 counterpart. These different conformations of the pVHL-binding domains, as well as variations in the structure between human and yeast Rpb6, suggest the existence of some differences in the pVHL-binding mechanism between HIF-1α and Rpb1/Rpb6.
In summary, our work shows that the pVHL complex binds the hyperphosphorylated large subunit of the RNA polymerase II complex, in a proline hydroxylation- and CTD phosphorylation-dependent manner, targeting it for ubiquitination. These results indicate that pVHL plays a role in the regulation of gene expression and cellular function.
Supplementary Material
Acknowledgments
We thank Glenn Doerman for preparing the figures. The HA-VHL expression construct and RCC clones stably expressing HA-VHL were a gift from W. G. Kaelin, Jr. This work was supported by the following grants to M.F.C.-K.: National Institutes of Health Grants HL58687 and HL66312, American Cancer Society Research Scholar Grant GMC-101430, and a von Hippel–Lindau Family Alliance Research Grant. J.M. acknowledges support from the Children's Hospital Research Foundation Trustee Grant.
Abbreviations
- pVHL
von Hippel–Lindau protein
- TH
tyrosine hydroxylase
- PC12
phoechromocytoma cell line
- CTD
carboxyl-terminal domain
- Rpb1 or Rpb6
subunit 1 or 6 of RNA polymerase II
- HA
hemagglutinin
- CbzLLn
N-Cbz-l-Leu-l-Leu-l-norvalinal
- HIF
hypoxia-inducible factor
- RCC
renal cell carcinoma
- ODDD
oxygen-dependent degradation domain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G., Jr Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 2.Pause A, Lee S, Worrell R A, Chen D Y, Burgess W H, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 4.Cockman M E, Masson N, Mole D R, Jaakola P, Chang G-W, Clifford S C, Maher E R, Pugh C W, Ratcliffe P J, Maxwell P H. J Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 5.Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M F, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohh M, Park C W, Ivan M, Hoffman M A, Kim T Y, Huang L E, Pavletich N, Chau V, Kaelin W G., Jr Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 7.Epstein A C R, Gleadle J M, McNeill L A, Hewiston K S, O'Rourke J, Mole D R, Mukherji M, Metzen E, Wilson M I, Dhanda A, et al. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 8.Taylor S. Gene. 2001;275:125–132. doi: 10.1016/s0378-1119(01)00633-3. [DOI] [PubMed] [Google Scholar]
- 9.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara J M, Lane W S, Kaelin W G., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 10.Jaakkola P, Mole D R, Tian Y M, Wilson M I, Gielbert J, Gaskell S J, Kriegsheim A V, Hebestreit H F, Mukherji M, Schofield C J, et al. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 11.Yu F, White S B, Zhao Q, Lee F S. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruick R K, McKnight S L. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 13.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Jr, Goldberg M A. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wizigmann-Voos S, Breier G, Risau W, Plate K H. Cancer Res. 1995;55:1358–1364. [PubMed] [Google Scholar]
- 15.Yang H, Kaelin W G., Jr Cell Growth Differ. 2001;12:447–455. [PubMed] [Google Scholar]
- 16.Walther M M, Reiter R, Keiser H R, Choyke P L, Venzon D, Hurley K, Gnarra J R, Reynolds J C, Glenn G M, Zbar B, Linehan W M. J Urol. 1999;162:659–665. doi: 10.1097/00005392-199909010-00004. [DOI] [PubMed] [Google Scholar]
- 17.Eisenhofer G, Walther M M, Huynh T-T, Li S-T, Bornstein S R, Vortmeyer A, Mannelli M, Goldstein D S, Linehan W M, Lenders J W M, Pacak K. J Cell Endocrinol Metab. 2001;86:1999–2008. doi: 10.1210/jcem.86.5.7496. [DOI] [PubMed] [Google Scholar]
- 18.Kroll S L, Paulding W R, Schnell P O, Barton M C, Conaway J W, Conaway R C, Czyzyk-Krzeska M F. J Biol Chem. 1999;274:30109–30114. doi: 10.1074/jbc.274.42.30109. [DOI] [PubMed] [Google Scholar]
- 19.Bauer A L, Paulding W R, Striet J B, Schnell P O, Czyzyk-Krzeska M F. Cancer Res. 2002;62:1682–1687. [PubMed] [Google Scholar]
- 20.Hawryluk P, Luse D. M.S. thesis. Cleveland: Case Western Reserve Univ.; 2002. [Google Scholar]
- 21.Dahmus M. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 22.Mitsui A, Sharp P A. Proc Natl Acad Sci USA. 1999;96:6054–6059. doi: 10.1073/pnas.96.11.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K B, Wang D, Lippard S J, Sharp P A. Proc Natl Acad Sci USA. 2002;99:4239–4244. doi: 10.1073/pnas.072068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratner J N, Balasubramanian B, Corden J, Warren S L, Bergman D B. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 25.Bregman D B, Halaban R, Van Gool A J, Henning K A, Friedberg E C, Warren S L. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rockx D A, Mason R, van Hoffen A, Barton M C, Citterio E, Bregman D B, van Zeeland A A, Vrieling H, Mullenders L H F. Proc Natl Acad Sci USA. 2000;97:10503–10508. doi: 10.1073/pnas.180169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svejstrup J Q. Nat Rev. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 28.Beaudenon S L, Huacani M R, Wang G, McDonnell D P, Huibregtse J M. Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist C J, Hofmann K, Bairoch A. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones D T. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 31.Meller J, Elber R. Proteins Struct Funct Genet. 2001;45:241–261. doi: 10.1002/prot.1145. [DOI] [PubMed] [Google Scholar]
- 32.Meller J, Elber R. Learning, Observing, and Outputting Protein Patterns (loopp), a Program for Protein Recognition and Design of Folding Potentials. Ithaca, NY: Cornell Univ.; 2000. , Version 2.0. [Google Scholar]
- 33.Frary A, Nesbitt T C, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert K B, Tanksley S D. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 34.Kelley L A, MacCallum R M, Sternberg M J E. J Mol Biol. 2000;299:501–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 35.Cramer P, Bushnell D A, Kornberg R D. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 36.Min J H, Yang H, Ivan M, Gertler F, Kaelin W G, Jr, Pavletich N P. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 37.Hon W C, Wilson M I, Harlos K, Claridge T D, Schofield C J, Pugh C W, Maxwell P H, Ratcliffe P J, Stuart D I, Jones E Y. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 38.Kim E, Du L, Bregman D B, Warren S L. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bregman D B, Du L, Van der Zee S, Warren S L. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 41.Franklin T J, Morris W P, Edwards P N, Large M S, Stephenson R. Biochem J. 2001;353:333–338. doi: 10.1042/0264-6021:3530333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kivirikko K I, Pihlajaniemi T. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Na X, Wang D, Schoen S R, Messing E M, Wu G. J Biol Chem. 2002;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Wang D, Na X, Schoen S R, Messing E M, Wu G. Biochem Biophys Res Commun. 2002;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 45.Okuda H, Saitoh K, Hirai S, Iwai K, Takaki Y, Baba M, Minato N, Ohmo S, Shuin T. J Biol Chem. 2001;276:43611–43617. doi: 10.1074/jbc.M107880200. [DOI] [PubMed] [Google Scholar]
- 46.Ishiguro A, Nogi Y, Hisatake K, Muramatsu M, Ishihama A. Mol Cell Biol. 2000;20:1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 48.Schoenfeld A R, Parris T, Eisenberger A, Davidowitz E J, De Leon M, Talasazan F, Devarajan P, Burk R D. Oncogene. 2000;19:5851–5857. doi: 10.1038/sj.onc.1203985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.